Significance
We demonstrate that chronic cocaine, including cocaine self-administration, induces poly(ADP-ribose) polymerase-1 (PARP-1) in the nucleus accumbens (NAc). Using a combination of viral-mediated gene transfer and pharmacological tools, we show that upregulation of PARP-1 in NAc dramatically enhances behavioral responses to cocaine, whereas downregulation of PARP-1 has the opposite effect. We used chromatin immunoprecipitation sequencing to map genome-wide binding of PARP-1 in NAc. The data demonstrate upregulation of PARP-1 binding across the genome after cocaine administration and identify numerous target genes for PARP-1. Among these is sidekick-1 (SDK1), previously implicated in regulating synaptic connections during development. We confirm SDK1 induction in NAc after chronic cocaine and demonstrate its ability to promote both cocaine’s behavioral effects and induction of dendritic plasticity in NAc.
Keywords: histone PARylation, drug addiction, medium spiny neurons
Abstract
Many of the long-term effects of cocaine on the brain’s reward circuitry have been shown to be mediated by alterations in gene expression. Several chromatin modifications, including histone acetylation and methylation, have been implicated in this regulation, but the effect of other histone modifications remains poorly understood. Poly(ADP-ribose) polymerase-1 (PARP-1), a ubiquitous and abundant nuclear protein, catalyzes the synthesis of a negatively charged polymer called poly(ADP-ribose) or PAR on histones and other substrate proteins and forms transcriptional regulatory complexes with several other chromatin proteins. Here, we identify an essential role for PARP-1 in cocaine-induced molecular, neural, and behavioral plasticity. Repeated cocaine administration, including self-administration, increased global levels of PARP-1 and its mark PAR in mouse nucleus accumbens (NAc), a key brain reward region. Using PARP-1 inhibitors and viral-mediated gene transfer, we established that PARP-1 induction in NAc mediates enhanced behavioral responses to cocaine, including increased self-administration of the drug. Using chromatin immunoprecipitation sequencing, we demonstrated a global, genome-wide enrichment of PARP-1 in NAc of cocaine-exposed mice and identified several PARP-1 target genes that could contribute to the lasting effects of cocaine. Specifically, we identified sidekick-1—important for synaptic connections during development—as a critical PARP-1 target gene involved in cocaine’s behavioral effects as well as in its ability to induce dendritic spines on NAc neurons. These findings establish the involvement of PARP-1 and PARylation in the long-term actions of cocaine.
Chronic cocaine exposure is marked by persistent changes in gene expression and altered neuronal morphology within the rodent nucleus accumbens (NAc), a central component of the brain’s reward circuitry (1, 2). Important aspects of cocaine’s effects on gene transcription appear to be mediated via chromatin remodeling (2–11). Thus, cocaine has been shown to regulate chromatin structure and gene expression in NAc through changes in histone acetylation, phosphorylation, and methylation. However, a role for enzymes controlling poly(ADP-ribosyl)ation have not to date been investigated.
Poly(ADP-ribose) polymerase-1 (PARP-1), the founding member of the PARP family, is a a ubiquitous and abundant nuclear protein found in most cell types that binds to DNA, histones, and other proteins (12). PARP-1 catalyzes the NAD+-dependent synthesis of a polymer, called poly(ADP-ribose) or PAR, on target proteins (13). Although historically studied in the context of DNA damage detection and repair, PARP-1 has more recently been shown to play at least two important roles in the regulation of gene transcription: as a histone modifier and as a component of enhancer/promoter regulatory complexes (14). Histones are PARylated by PARP-1 at specific lysine residues, whereas ADP ribosyl hydrolases and poly(ADP-ribose) glycohydrolases (PARGs) degrade PAR. The PAR modification is read by zinc-finger motifs or macrodomains, which then regulate chromatin structure and gene transcription. Accordingly, histone PARylation is considered an additional component of the histone code (15).
Previously, it has been reported that PARP-1 and its PARylation activity are critical in neuronal viability as well as in learning and memory (16, 17). PARylation induces a fast and transient decondensation of chromatin structure and enables the transcription needed to form long-term memory in Aplysia and mammals (18, 19). It has been shown recently that PARylation is increased in the hippocampus after memory consolidation and long-term potentiation (LTP), whereas inhibition of PARylation blocked memory and LTP induction (17). Based on these findings, and on the role of other histone modifications in cocaine action, we sought to determine if PARP-1 and PAR are required for the maladaptive learning associated with addiction.
Results
Regulation of PARP-1 Activity in NAc by Chronic Cocaine Administration.
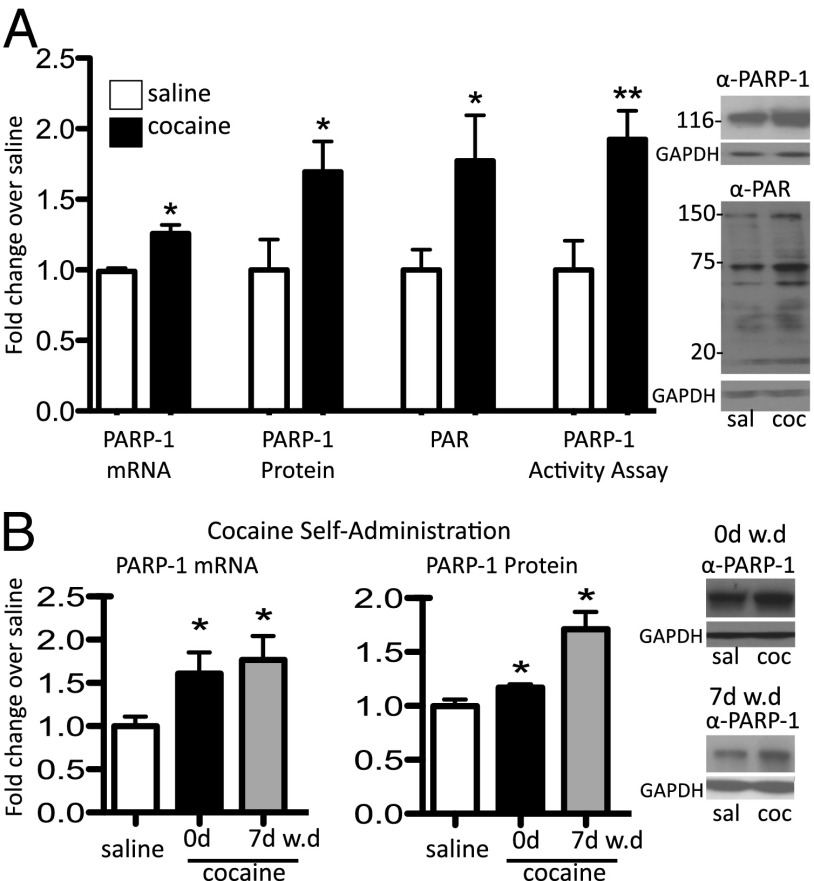
We administered cocaine to mice for 7 d and analyzed the animals 30 min after the last dose. We found a significant increase in PARP-1 mRNA and protein levels in NAc (Fig. 1A). No other PARP isoform displayed significant regulation in response to chronic cocaine. Consistent with increased PARP-1 expression, PARP-1 activity and its PAR mark were also induced (Fig. 1A). We next measured PARP-1 levels in rats trained to self-administer cocaine. We found increased levels of PARP-1 mRNA and protein in NAc 24 h after the last cocaine exposure (Fig. 1B). Even greater effects were seen in animals withdrawn from cocaine for 7 d and then re-exposed to the drug and examined 24 h later.
Fig. 1.
PARP-1 expression and activity are up-regulated in NAc by cocaine. (A) PARP-1 mRNA, protein, and activity, as well as PAR levels, were analyzed in mouse NAc 30 min after repeated (7 d) cocaine (20 mg/kg i.p.) administration. (B) PARP-1 mRNA and protein levels were measured in rat NAc 24 h after 10 d of cocaine self-administration or after 7 d of withdrawal with animals given a challenge dose of cocaine 24 h before analysis. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Data are presented as mean ± SEM (n = 7–11). *P < 0.05, **P < 0.01.
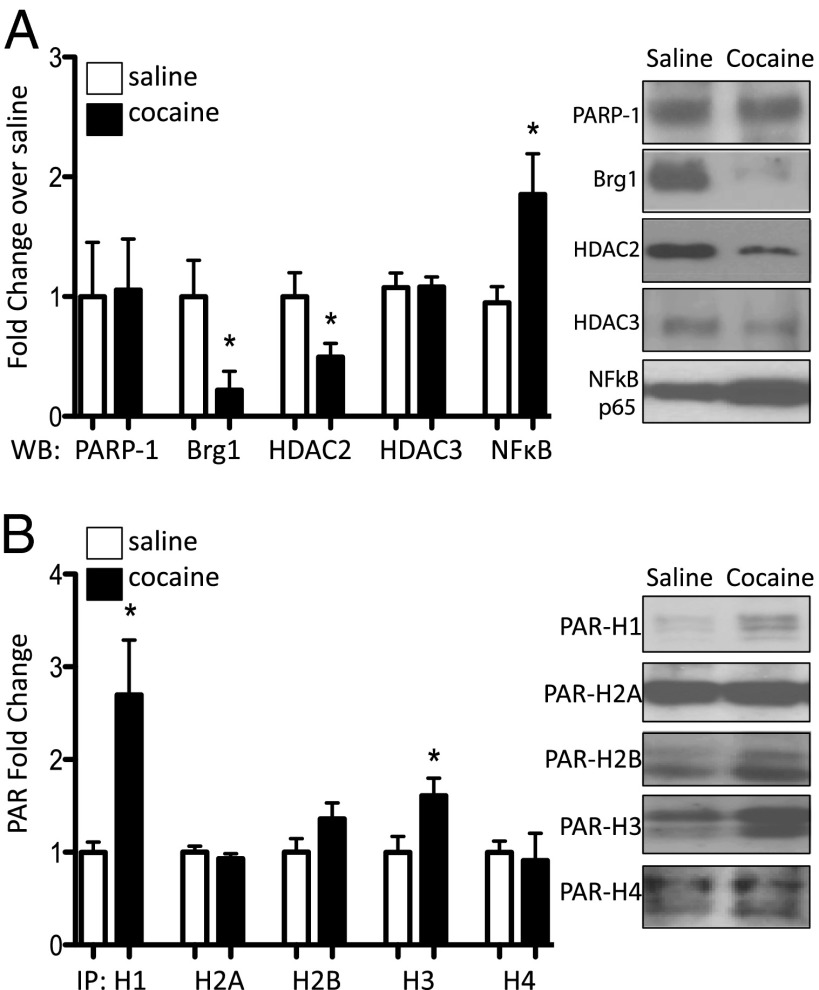
PARP-1 has been implicated in both activation and repression of gene transcription by virtue of which regulatory proteins are complexed with it (20–23). We thus investigated if the composition of complexes containing PARP-1 in NAc is changed by chronic cocaine. Consistent with previous studies in other tissues (20), we found that PARP-1 is associated with Brg1 and HDAC2, a presumed repressive complex, in NAc under baseline conditions and that levels of such complexes are reduced dramatically after chronic cocaine (Fig. 2A). Work in HeLa cells has shown that PARP-1 interactions with NF-κB subunits are required for NF-κB–dependent activation of target genes (23). We found that chronic cocaine increases PARP-1 association with NF-κB (Fig. 2A), a transcription factor induced in NAc under these conditions (24). These findings suggest that chronic cocaine enhances the incorporation of PARP-1 into activating complexes.
Fig. 2.
Chronic cocaine induces changes in PARP-1 complexes, consistent with a permissive transcriptional environment. (A) Immunoprecipitation (IP) of PARP-1 from NAc extracts of control and chronic cocaine-treated mice followed by Western blots for Brg1, HDAC2, HDAC3, or NF-κB p65 subunit. All data were normalized to PARP-1 and expressed as mean ± SEM (n = 5–7 independent samples/group with each sample representing tissue of three animals). (B) Levels of PAR present on histones H1, H2A, H2B, H3, and H4 after chronic cocaine. Each histone subunit was IPed and Western blotted for PAR. Data are normalized to total amount of each histone subunit pulled down (images shown in Fig. S1) and expressed as mean ± SEM (n = 4–7 independent samples/group as above). *P < 0.05.
In addition to its role as a transcriptional coregulator, PARP-1 is able to modulate transcription by adding PAR to potentially all histone subunits, thus changing chromatin structure at target genes (13, 25). Addition of the highly negatively charged PAR to histones is thought to repel the DNA, leading to an opening of chromatin structure. It was therefore of interest to determine if chronic cocaine regulates the PARylation of histones in NAc. We found that cocaine induction of PARP-1 in mouse NAc is associated with increased PARylation of H1 and H3, with no changes seen at other histone subunits (Fig. 2B and Fig. S1). These observations further support the view that chronic cocaine induces a permissive chromatin structure, in this case through selective histone PARylation.
PARP-1 Activity in NAc Regulates the Behavioral Response to Cocaine.
We next investigated the consequences of altered levels of PARP-1 in NAc on the behavioral effects of cocaine. Overexpression of PARP-1 selectively in the NAc of adult mice, using viral-mediated gene transfer, increased levels of PARP-1 expression and of PAR (Fig. S2). Such overexpression dramatically increased locomotor responses to cocaine, cocaine-induced conditioned place preference (CPP)—which provides an indirect measure of drug reward—and self-administration of low doses of cocaine (Fig. 3 A–C). Although an increase in drug self-administration could reflect either an increase or a decrease in the rewarding value of a drug, our findings of increased CPP along with increased self-administration, and, in particular, increased self-administration at low but not high drug doses, provide strong support for the interpretation that PARP-1 induction in NAc increases rewarding responses to cocaine.
Fig. 3.
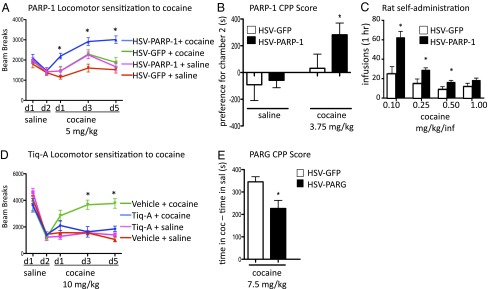
PARP-1 controls behavioral responses to cocaine. (A) Locomotor response to cocaine (5 mg/kg i.p.) in mice injected intra-NAc with HSV-GFP or HSV-PARP-1. (B) Conditioned place preference for cocaine in mice injected intra-NAc with HSV-GFP or HSV-PARP-1. (C) Number of self-administered infusions of cocaine by rats, trained to self-administer cocaine and then injected intra-NAc with HSV-GFP or HSV-PARP-1, and tested 2–5 d later for self-administering elevating intravenous cocaine doses. (D) Locomotor responses to cocaine in mice with intra-NAc infusion of TIQ-A or vehicle. (E) Conditioned place preference for cocaine in mice injected intra-NAc with HSV-GFP or HSV-PARG. Data are presented as mean ± SEM (n = 7–10). *P < 0.05.
To obtain the converse type of information, we blocked PARP-1 PARylation activity through the intra-NAc infusion of the PARP-1 inhibitor Tiq-A. This manipulation successfully decreased PARylation levels in NAc (17) (Fig. S3) and completely blocked locomotor responses to higher cocaine doses (Fig. 3D). In addition, we virally overexpressed PARG, which removes the PAR mark (25) in NAc. PARG overexpression decreased PAR levels in this brain region (Figs. S2 and S4) and attenuated the rewarding effects of cocaine in the CPP paradigm (Fig. 3E).
Genome-Wide Mapping of PARP-1 Binding in NAc.
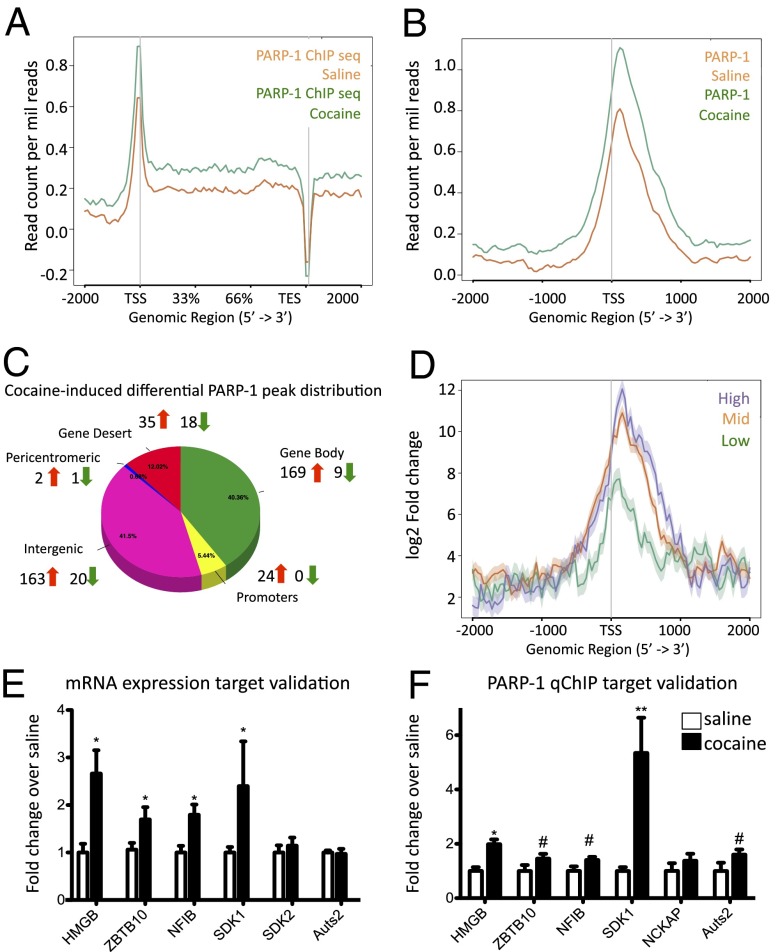
We turned to chromatin immunoprecipitation sequencing (ChIP-seq) to map the distribution of PARP-1 genome-wide and identify downstream molecular target genes in NAc of saline- and cocaine-treated mice. The pattern of PARP-1 binding at target genes is consistent with published data from cultured cells (26) with a peak at transcription start sites (TSSs) (Fig. 4A). We observed a striking increase in PARP-1 binding after chronic cocaine across the genome and specifically at the TSS (Fig. 4B). The overall distribution of PARP-1 peaks was similar between control and cocaine-treated animals with ∼40% of the peaks residing in gene bodies (Fig. 4C). The remaining 60% of identified peaks reside throughout intergenic regions (gene deserts, pericentromeres, repetitive sequences, etc.) and gene promoters. To verify our biochemical data (Fig. 2), and confirm commonly held assumptions primarily determined from peripheral tissues and cultured cells concerning the activating role of PARP-1 on gene expression (26), we used RNA sequencing (RNA-seq) data derived from NAc (27) with genes grouped by their levels of mRNA expression: low, medium, and high. We found that gene expression was positively correlated genome-wide with PARP-1 enrichment (Fig. 4D). We then used diffReps analysis (28) (see Materials and Methods for more details) to examine cocaine-induced differential regulation of PARP-1 enrichment throughout the genome. Cocaine treatment was associated with greater PARP-1 peak numbers compared with control conditions (see Table S1 for complete gene list), suggesting profound global alterations in PARP-1–DNA interactions after cocaine, consistent with our biochemical data (Fig. 1). Gene ontology analysis revealed that several interesting categories of genes, including several related to neuronal projections and cytoskeleton, showed induced PARP-1 binding in response to chronic cocaine (Fig. S5).
Fig. 4.
ChIP-seq identification of PARP-1 targets in NAc after chronic cocaine administration. (A) Next-generation sequencing plot (https://code.google.com/p/ngsplot/) of PARP-1 binding to genes genome-wide in NAc under saline and chronic cocaine conditions with a higher-magnification view of the TSS (B). (C) Cocaine-induced differential PARP-1 peak distribution. Arrows show the number of genes at which PARP-1 binding is significantly increased or decreased after cocaine. (D) Correlation of PARP-1 peaks with gene expression, the latter based on RNA-seq data. (E) Validation of mRNA expression of PARP-1 target genes after chronic cocaine (n = 8 per group). (F) Validation of ChIP-seq findings by qChIP for PARP-1 binding at key target genes after chronic cocaine (n = 5–7 per group). #P < 0.08, *P < 0.05, **P < 0.01.
Because our findings indicate that cocaine induces a permissive transcriptional environment in part via PARP-1 induction, we examined how a major mark of transcriptional activation, histone H3 lysine 4 trimethylation (H3K4me3), might overlap with PARP-1 enrichment. By use of ChIP-seq, we demonstrated enrichment of H3K4me3 at TSSs genome-wide, as would be expected (26), and identified 45 genes that displayed significant up-regulation of both PARP-1 and H3K4me3 binding in NAc after chronic cocaine (see Table S2 for complete gene list). We selected several of the most significantly regulated genes and validated their mRNA induction in NAc after chronic cocaine by quantitative PCR (qPCR) analysis of independent biological samples (Fig. 4E). We also validated increased PARP-1 binding seen by ChIP-seq data by performing quantitative ChIP (qChIP) on independent biological samples (Fig. 4F). All of these validated genes are previously unexplored in cocaine models, thus confirming that our ChIP-seq datasets reveal previously unknown mechanisms for the molecular basis of cocaine action.
Sidekick-1: A New Substrate of Cocaine Action.
One intriguing gene validated by qPCR and qChIP is sidekick-1 (SDK1), which is induced more than twofold in NAc by chronic cocaine. The distribution of PARP-1 binding across the entire SDK1 gene, and cocaine-induced increases in such binding at four distinct sites on the gene, are shown in Fig. S6. In contrast to SDK1, we observed no change in the levels of mRNA expression of the related sidekick isoform, SDK2, in response to chronic cocaine (Fig. 4E). Moreover, this lack of induction in response to chronic cocaine of SDK2 expression was associated with the lack of induction of both PARP-1 binding (measured by qChIP) and H3K4me3 binding (measured by ChIP-seq) to the SDK2 gene after chronic cocaine.
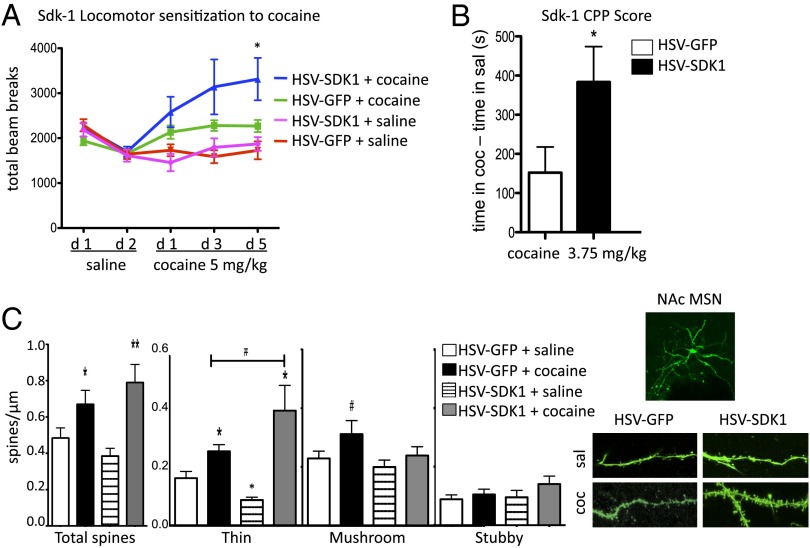
SDK1 is a cell adhesion molecule concentrated at synapses that directs lamina-specific synaptic connections in retina (29, 30). Our data represent previously unidentified SDK1 regulation in the brain outside of the retina and a previously unidentified demonstration of its regulation by PARP-1. To determine the behavioral relevance of SDK1 induction in NAc in cocaine action, we used viral-mediated gene transfer to overexpress SDK1 in this brain region. Mice overexpressing SDK1 in NAc showed increased locomotor responses to cocaine (Fig. 5A) as well as increased cocaine CPP (Fig. 5B).
Fig. 5.
SDK1 is a PARP-1 target that regulates the behavioral response to cocaine through altered spine dynamics. (A) Locomotor response to cocaine in mice injected intra-NAc with HSV-GFP or HSV-SDK1 at indicated cocaine dose (n = 7–10). (B) Conditioned place preference for cocaine in mice injected intra-NAc with HSV-GFP or HSV-SDK1 at indicated cocaine dose (n = 9–11). (C) Dendritic spine analysis of animals injected intra-NAc with HSV-GFP or HSV-SDK1 and treated chronically with saline or cocaine (20 mg/kg i.p.; n = 4–5 mice per group, n = 3–4 neurons per mouse). #P < 0.08, *P < 0.05, **P < 0.01.
Repeated administration of cocaine increases the density of dendritic spines on NAc medium spiny neurons (MSNs) (1, 31, 32), a process associated with functional changes at excitatory glutamatergic synapses in these neurons (33–35) and sensitized behavioral responses to the drug (1, 31, 32). Although PARP-1 has not previously been linked to cocaine-induced dendritic plasticity, its binding partner NF-κB has been so connected (24). We sought to determine if a change in spine number or morphology of NAc MSNs was influenced by SDK1 induction. As expected, chronic cocaine treatment increased spine density of NAc MSNs in both herpes simplex virus (HSV)-GFP and HSV-SDK1 groups compared with saline. However, in mice overexpressing SDK1, chronic cocaine produced a significantly larger increase in thin spine density (Fig. 5C), indicating that this may be one mechanism by which SDK1 induction in NAc promotes behavioral responses to cocaine.
Discussion
Results of the present study establish a key role for PARP-1 and associated changes in PARylation in NAc in mediating the behavioral and neural effects of chronic cocaine administration. We show that PARP-1 is induced in this brain region by chronic exposure to cocaine, including cocaine self-administration. PARP-1 protein is increased to a greater extent than PARP-1 mRNA. This might suggest additional mechanisms of cocaine regulation of PARP-1, for example, an increase in its translation or a decrease in its degradation. Cocaine induction of PARP-1 expression is associated with increased levels of its catalytic activity. PARP-1 induction by cocaine is associated with alterations in transcriptional complexes formed by this enzyme as well as with altered global levels of PARylation of certain histone subunits, modes of regulation consistent with a more permissive state of gene transcription in response to chronic cocaine administration.
We mapped PARP-1 binding in NAc genome-wide by use of ChIP-seq. Cocaine increases PARP-1 binding, compared with control conditions, as indicated by the increase in read counts after chronic cocaine. PARP-1 binding shows peaks around the TSSs of genes, coincident with H3K4me3 binding. We found significant overlap in genes showing elevated PARP-1 binding and H3K4me3 binding after chronic cocaine. This list of overlapping genes reveals cocaine regulation of several interesting gene families, including those involved in actin dynamics and chromatin remodeling. We validated several of these genes and confirmed that they are bona fide cocaine targets.
SDK1, a transmembrane protein of the Ig superfamily and a homophilic adhesion molecule, is one previously unidentified cocaine target gene identified in this screen. We validated its induction by cocaine in an independent cohort via qPCR. In contrast to SDK1, cocaine did not alter the expression of the other known sidekick isoform, SDK2. Through viral-mediated gene transfer, we then directly demonstrated a role for SDK1 in NAc in promoting the behavioral effects of cocaine. Further investigations revealed the ability of SDK1 to also promote cocaine-induced dendritic plasticity of NAc medium spiny neurons. These findings support a previously unappreciated role of SDK1 in mediating plasticity in the adult brain, specifically in controlling neural and behavioral adaptations to cocaine. We hypothesize that PARP-1, upon its induction by chronic cocaine, induces numerous cytoskeleton-related target genes in NAc including SDK1 and that increased expression of SDK1 promotes dendritic growth by increasing homophilic interactions between SDK1-containing synapses, similar to what has been reported in kidney podocytes (36).
Several chromatin modifications have been shown to be altered in NAc by cocaine, including histone acetylation, methylation, and phosphorylation (2–11). Here, we add PARylation as another module of the histone code that is regulated robustly in NAc in response to chronic cocaine exposure. PARylation of histones has been studied primarily following the introduction of DNA strand breaks, limiting the discussion of PARylation to genotoxic stress. However, results of the present study add to more recent evidence that PARylation has essential functions in the regulation of transcription and chromatin remodeling that go far beyond a limited role in genotoxic stress responses.
In summary, we demonstrate that induction of PARP-1 turns on a gene network in NAc that controls neural and behavioral responses to cocaine. Increased expression of PARP-1 after repeated cocaine administration promotes cocaine preference, in part through the transcriptional activation of genes, such as SDK1. SDK1 is known to be involved in synaptic formation, and we show that SDK1 regulates dendritic plasticity in the NAc. This study thereby establishes a link between PARP-1 transcriptional activity and spine dynamics in the brain. Together, these findings implicate PARP-1 and its numerous target genes as potential sites of pharmacological intervention in the treatment of cocaine addiction.
Materials and Methods
Animals.
For all experiments, 9- to 11-wk-old C57BL/6J male mice (The Jackson Laboratory) were group-housed (five per cage) in a colony room set at a constant temperature (23 °C) on a 12 h light/dark cycle (lights on from 7:00 AM to 7:00 PM) with ad libitum access to food and water. All protocols were approved by the Institutional Animal Care and Use Committee at Icahn School of Medicine at Mount Sinai and at State University of New York at Buffalo. Chronic cocaine treatment is defined as seven daily i.p. injections of cocaine at 20 mg/kg unless otherwise stated.
RNA Isolation, Reverse Transcription, and qPCR.
Bilateral 14-gauge NAc punches were obtained and frozen on dry ice. Samples were homogenized in TRIzol and processed according to manufacturer instructions (Invitrogen). RNA was purified with RNAeasy Micro columns (Qiagen) and reverse-transcribed using an iScript Kit (Bio-Rad). cDNA was quantified by qPCR using SYBR green. Each reaction was run in duplicate and analyzed following the standard ΔΔCt method using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a normalization control.
Western Blotting.
Frozen NAc punches were homogenized in 30 μL of homogenization buffer containing 320 mM sucrose, 5 mM Hepes buffer, 1% SDS, phosphatase inhibitor mixtures I and II (Sigma), and protease inhibitors (Roche) using an ultrasonic processor (Cole Parmer). Protein concentrations were determined using a DC protein assay (Bio-Rad), and 10–30 μg of protein was loaded onto 4–15% gradient Tris⋅HCl polyacrylamide gels for electrophoresis fractionation (Bio-Rad). Proteins were transferred to nitrocellulose membranes, which were incubated at 4 °C in 5% (wt/vol) bovine serum albumin blocking buffer and then overnight with primary antibodies. Thereafter, they were thoroughly washed with tris-buffered saline plus 0.1% Tween-20, incubated with secondary antibodies (1:50,000) for 1 h at room temperature, and washed again. Resulting blots were quantified by densitometry using National Institutes of Health ImageJ. Data were normalized to levels of GAPDH, which were not affected by cocaine treatment.
Viral-Mediated Gene Transfer.
Expression plasmids were subcloned into HSV vectors and packaged into high-titer viral particles as described previously (8). Mice were positioned in small-animal stereotaxic instruments under ketamine (100 mg/kg) and xylazine (10 mg/kg) anesthesia, and their cranial surfaces were exposed. Thirty-three–gauge syringe needles were bilaterally lowered into NAc to infuse 0.5 μL of virus at a 10° angle (anterior/posterior, +1.6 mm; medial/lateral, +1.5 mm; dorsal/ventral, −4.4 mm from Bregma). Infusions occurred at a rate of 0.1 μL/min. Animals receiving HSV injections were allowed to recover for at least 24 h after surgery.
PARP-1 Activity Assay.
A chemiluminescent PARP-1 Assay Kit was used according to manufacturer instructions (BPS Biosciences). Briefly, bilateral 14-gauge NAc punches were collected 30 min after the last dose of chronic cocaine and sonicated on ice at 30% amplitude in 10 vol of 1× PARP buffer supplied with a kit containing protease inhibitor mixture, 0.4 M NaCl, and 1% Triton X-100. Twenty micrograms of protein in three replicates per sample was used. Ten animals (20 NAc punches) were pooled per group.
Immunoprecipitations.
A coimmunoprecipitation kit (Roche) was used as follows. Bilateral 14-gauge NAc punches were lysed by douncing in 300 μL of provided lysis buffer and centrifuged, and the supernatant was transferred to a clean tube. Preclearance was accomplished by incubation with protein G-agarose for 3 h on a rotator at 4 °C. The beads were centrifuged, and the supernatant was transferred to fresh tubes to be incubated with 5 μL of anti–PARP-1 antibody (Cell Signaling) for 1 h before 50 μL of a homogeneous protein G–agarose suspension was added and incubated overnight at 4 °C on a rotator. The complexes were centrifuged and the supernatant was removed, the beads were washed two times with lysis buffer 1, two times with lysis buffer 2, and once with lysis buffer 3. Protein sample buffer was added, and the samples were boiled for 3 min. Complexes were then analyzed by Western blotting.
Conditioned Place Preference.
Place conditioning was performed as described previously (8) with slight modifications. Several visual and nonvisual (tactile) cues enabled the animals to distinguish two separate chambers of the apparatus. All conditioning and test sessions were performed under dim illumination. Baseline preference was monitored 1 d before surgery at 12:00 PM; the mice did not show a preference for either side of the chamber at baseline. Following surgery and recovery, mice were conditioned for 30 min over 2 d to the saline-paired side in the morning and the cocaine-paired side (3.75 or 7.5 mg/kg i.p. as indicated in figures and legends) in the afternoon. On the final day, mice were placed again in the central compartment at 12:00 PM and allowed to move freely between the two chambers for 20 min. CPP scores were calculated as time spent in the cocaine-paired chamber minus time spent in the saline-paired chamber.
Locomotor Sensitization.
After recovery from surgery, mice were habituated in the locomotor box for 2 d for 30 min and then monitored for locomotor activity after a saline injection using the Photobeam Activity System (San Diego Instruments). For the HSV–PARP-1 and HSV-SDK1 overexpression experiments, cocaine (5 mg/kg i.p.) was then administered daily in the locomotor box for 5 d, and locomotor activity was monitored for 30 min. For PARP-1 inhibition, we used Thieno[2,3-c]isoquinolin-5-one (Tiq-A) (Sigma-Aldrich), a PARP-1 antagonist. Five min before each behavioral test, 0.5 μL of a 250 nM solution of Tiq-A was microinjected into NAc over 60 s through each cannula, and the infusion cannulae were left for an additional 60 s to minimize backflow. Cocaine (10 mg/kg) was then administered, and mice were monitored for locomotor activity 30 min each day for 5 d.
Self-Administration.
Rats were trained to self-administer (SA) cocaine (1 mg/kg/infusion i.v.) for 2 h/day for 10 d. The NAc was then injected with either HSV-GFP (n = 7) or HSV–PARP-1 (n = 9) equally counterbalanced for SA performance. On day 2–5 of viral expression, rats were put through a cocaine dose–response using four escalating doses of cocaine (0.1, 0.25, 0.5, and 1 mg/kg/infusion i.v.). Each SA dose session was 1 h followed by a 5-min time-out before the lights and cues came back on to signal the beginning of the next session. All doses were given in descending order once a day for a total of 3 days.
qChIP.
Published methods were used (8, 37). Two 14-gauge NAc punches from each mouse were placed in 1% formaldehyde in 1× PBS to cross-link DNA with associated proteins. After 12 min on the rotator, 2 M glycine was added to stop the fixation for 5 min. The punches were then placed on ice and rinsed five times with ice-cold 1× PBS. Tissue from five animals was pooled at this point and homogenized in SDS lysis buffer (10% SDS, 1 M Tris⋅HCl, 0.5 M EDTA) with a desktop sonicator. ChIP dilution buffer (10% Triton X-100, 5 M NaCl, 1 M Tris⋅HCl, pH 8.1, 0.5 M EDTA, 10% SDS, and protease inhibitors) was added, and the chromatin underwent high-powered sonication with a Bioruptor at 4 °C for 25 cycles of 30 s on/30 s off on high power. The machine was allowed to cool for 25 min before another round of 15 cycles. Conjugated magnetic beads were used to immunoprecipitate PARP-1 with the ChIP-validated anti–PARP-1 antibody (rabbit polyclonal anti-PARP, ab6079) overnight in block solution (0.5% BSA). The immunoprecipitation (IP) reaction was collected with a magnetic rack and washed, and the input chromatin and the immunoprecipitated DNA were reverse–cross-linked at 65 °C overnight. The DNA was then purified with RNase, Proteinase K, and the Qiagen PCR purification kit.
ChIP, Library Preparation, and Sequencing.
Freshly dissected NAc punches were cross-linked with formaldehyde and prepared for PARP-1 or H3K4me3 ChIP as described above. Bilateral 14-gauge NAc punches were pooled from 10 mice for ChIP-seq as described (37). PARP-1 immunoprecipitated DNA and total (input) genomic DNA were prepared for ChIP-seq using an Illumina kit according to the manufacturer instructions. Each experimental condition was performed in triplicate. Briefly, each sample underwent end repair followed by addition of an A base to the 3′ end. Proprietary adapters were then ligated to the ends, followed by size selection on a 2% agarose gel. The range of excision was 250–350 bp. After DNA clean up, samples were amplified with 21 cycles of PCR. Amplification and size selection were confirmed with a BioAnalyzer. The resulting libraries were sequenced on an Illumina HiSEq. 2000 with a 100-bp read length.
ChIP-seq Data Analysis.
The 100-bp reads were first analyzed by the pipeline software of Illumina for quality filtering and then aligned to the reference genome. Additional preprocessing steps were performed as described previously (37). Differential analysis between cocaine and saline groups was performed with an in-house program called diffReps. For more details, please refer to http://code.google.com/p/diffreps/ (24). Briefly, diffReps uses a sliding-window strategy to identify all genomic regions, genome-wide, that display differential binding levels of a histone mark between two conditions in a manner independent of peak calling or gene regions. The negative binomial distribution is used to model the within-group variation and between-group differences for discrete counting data. Before testing, diffReps obtains global statistics about the ChIP-seq samples under study and filters out the regions with read counts below the mean plus 2 SDs. It then scores all of the windows with fixed sizes and picks up those with a P value <1e-4 as candidate differential windows. Any two overlapping or adjacent candidate windows are merged to make a differential site. Finally, multiple testing corrections are applied to all differential sites on the genome, and a false discovery rate <10% is used as a cutoff. In this study, diffReps was used with a window size of 200 bp and a moving size of 20 bp to obtain a high-resolution profiling of PARP-1 and H3K4me3 regulation by cocaine.
Spine Analysis.
For dendritic spine analysis following HSV-GFP or HSV-SDK1 injection, mice were treated with “saline” (five treatments of saline intraperitoneally) or “chronic cocaine” (five treatments of 20 mg/kg cocaine intraperitoneally) over the course of 3 d, as this injection protocol has previously been demonstrated to increase spine density on NAc neurons within the time frame of HSV transgene expression (8). Mice used for dendritic spine analysis were killed 30 min after the last treatment. Spine analysis was performed as described previously (8). Briefly, dendritic segments 50–150 μm away from the soma were randomly chosen from HSV-infected cells that express GFP. Images were acquired on a confocal LSM 780 (Carl Zeiss) for morphological analysis using NeuronStudio with the rayburst algorithm. NeuronStudio classifies spines as thin, mushroom, or stubby on the basis of the following values: (i) aspect ratio, (ii) head-to-neck ratio, and (iii) head diameter. Spines with a neck are classified as either thin or mushroom, and those without a significant neck are classified as stubby. Spines with a neck are labeled as thin or mushroom on the basis of head diameter.
Statistical Analysis.
One- or two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test or two-way Student t tests were used whenever two groups were being compared.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319703111/-/DCSupplemental.
References
- 1.Robinson TE, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 2004;47(Suppl 1):33–46. doi: 10.1016/j.neuropharm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 2.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12(11):623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar A, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Renthal W, et al. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28(29):7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stipanovich A, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453(7197):879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60(6):961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renthal W, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62(3):335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun H, et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci. 2012;32(48):17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogge GA, Wood MA. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology. 2013;38(1):94–110. doi: 10.1038/npp.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogge GA, Singh H, Dang R, Wood MA. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci. 2013;33(15):6623–6632. doi: 10.1523/JNEUROSCI.4472-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraus WL. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20(3):294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 14.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113(6):677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 15.Hottiger MO. ADP-ribosylation of histones by ARTD1: An additional module of the histone code? FEBS Lett. 2011;585(11):1595–1599. doi: 10.1016/j.febslet.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 16.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase-1 in the nervous system. Neurobiol Dis. 2000;7(4):225–239. doi: 10.1006/nbdi.2000.0324. [DOI] [PubMed] [Google Scholar]
- 17.Fontán-Lozano A, et al. Histone H1 poly[ADP]-ribosylation regulates the chromatin alterations required for learning consolidation. J Neurosci. 2010;30(40):13305–13313. doi: 10.1523/JNEUROSCI.3010-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen-Armon M, et al. Long-term memory requires polyADP-ribosylation. Science. 2004;304(5678):1820–1822. doi: 10.1126/science.1096775. [DOI] [PubMed] [Google Scholar]
- 19.Wang SH, et al. NGF promotes long-term memory formation by activating poly(ADP-ribose)polymerase-1. Neuropharmacology. 2012;63(6):1085–1092. doi: 10.1016/j.neuropharm.2012.06.050. [DOI] [PubMed] [Google Scholar]
- 20.Hang CT, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto T, Kakizawa T, Hashizume K. Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol Cell Biol. 1999;19(4):2644–2649. doi: 10.1128/mcb.19.4.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soldatenkov VA, et al. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem. 2002;277(1):665–670. doi: 10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- 23.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem. 2001;276(49):45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 24.Russo SJ, et al. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29(11):3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messner S, Hottiger MO. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011;21(9):534–542. doi: 10.1016/j.tcb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39(5):736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen L, et al. 2013. Epigenetic regulation of cocaine action in mouse nucleus accumbens. Soc Neurosci Abs, 2013 Neuroscience Meeting Planner (Society for Neuroscience, Washington, DC), Program no. 59.02. Available at www.sfn.org/∼/media/SfN/Documents/Annual%20Meeting/FinalProgram/FullAbstractPDFs/AbstractPDFs_Poster_Sat_PM.ashx. Accessed January 8, 2014.
- 28.Shen L, et al. diffReps: Detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8(6):e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamagata M, Weiner JA, Sanes JR. Sidekicks: Synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110(5):649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- 30.Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: Expression and role of contactins. J Neurosci. 2012;32(41):14402–14414. doi: 10.1523/JNEUROSCI.3193-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalivas PW. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci. 2009;10(8):561–572. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- 32.Russo SJ, et al. The addicted synapse: Mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010;33(6):267–276. doi: 10.1016/j.tins.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ungless MA, Whistler JL, Malenka RC, Bonci A. Single cocaine exposure in vivo induces long-term potentiation in dopamine neurons. Nature. 2001;411(6837):583–587. doi: 10.1038/35079077. [DOI] [PubMed] [Google Scholar]
- 34.Thomas MJ, Malenka RC. Synaptic plasticity in the mesolimbic dopamine system. Philos Trans R Soc Lond B Biol Sci. 2003;358(1432):815–819. doi: 10.1098/rstb.2002.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee BR, Dong Y. Cocaine-induced metaplasticity in the nucleus accumbens: Silent synapse and beyond. Neuropharmacology. 2011;61(7):1060–1069. doi: 10.1016/j.neuropharm.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufman L, et al. The homophilic adhesion molecule sidekick-1 contributes to augmented podocyte aggregation in HIV-associated nephropathy. FASEB J. 2007;21(7):1367–1375. doi: 10.1096/fj.06-7191com. [DOI] [PubMed] [Google Scholar]
- 37.Maze I, et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci USA. 2011;108(7):3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.