Significance
Riboswitches are short RNA sequences for ligand-dependent modulation of gene expression in cis. This study demonstrates that an artificial riboswitch, a ligand-dependent self-cleaving ribozyme (aptazyme), can knockdown expression of an adeno- (DNA) virus early and a measles (RNA) virus structural gene, impacting biological outcomes, i.e. inhibiting viral genome replication and infectivity, respectively. It is the first report of riboswitches for replication control of human-pathogenic viruses and of their function in fully cytoplasmic (virus) systems. For future applications, aptazymes can be customized in other viruses facilitating analyses of viral gene functions or as a safety switch in oncolytic viruses. Because of their small size and RNA-intrinsic activity, we propose aptazymes as an alternative for inducible promoters in eukaryotic gene expression control.
Abstract
Aptazymes are small, ligand-dependent self-cleaving ribozymes that function independently of transcription factors and can be customized for induction by various small molecules. Here, we introduce these artificial riboswitches for regulation of DNA and RNA viruses. We hypothesize that they represent universally applicable tools for studying viral gene functions and for applications as a safety switch for oncolytic and live vaccine viruses. Our study shows that the insertion of artificial aptazymes into the adenoviral immediate early gene E1A enables small-molecule–triggered, dose-dependent inhibition of gene expression. Aptazyme-mediated shutdown of E1A expression translates into inhibition of adenoviral genome replication, infectious particle production, and cytotoxicity/oncolysis. These results provide proof of concept for the aptazyme approach for effective control of biological outcomes in eukaryotic systems, specifically in virus infections. Importantly, we also demonstrate aptazyme-dependent regulation of measles virus fusion protein expression, translating into potent reduction of progeny infectivity and virus spread. This not only establishes functionality of aptazymes in fully cytoplasmic genetic systems, but also implicates general feasibility of this strategy for application in viruses with either DNA or RNA genomes. Our study implies that gene regulation by artificial riboswitches may be an appealing alternative to Tet- and other protein-dependent gene regulation systems, based on their small size, RNA-intrinsic mode of action, and flexibility of the inducing molecule. Future applications range from gene analysis in basic research to medicine, for example as a safety switch for new generations of efficiency-enhanced oncolytic viruses.
Universally applicable, cis-acting and simple genetic switches implementing small-molecule–mediated control of viral gene expression would represent powerful tools for studying virus genetics in cell culture and in the host as well as for developing safe oncolytic viruses and virus vaccines. For analyses of viral gene functions, the insertion of such small-molecule–triggered switches should enable conditional knockdown of viral genes in vitro and in vivo with superior efficiency, timing, and systemic activity as well as less off-target effects compared with gene knockdown by RNA interference (RNAi). By now, the required genetic engineering methods for insertion of such cis-acting switches into virus genes have become available for most viruses. In the context of therapeutic applications, replication-competent viruses used for tumor destruction would benefit from genetic safety switches. Such oncolytic viruses originate from increasing numbers of virus families and results of current late-stage efficacy trials fuel the rising clinical interest in the field (1). Although safety in patients has been convincingly shown for current viruses, successful translation into clinical routine will depend on oncolytic viruses with increased potency and/or applications of viruses at higher titers. Therefore, safety will be of increasing concern. However, antiviral drugs enabling virus control in case of emerging side effects during treatment are available for very few viruses only. Thus, a drug-inducible safety switch is a valuable tool to facilitate therapeutic applications of virus strains for which pathogenicity is of concern (refs. 2, 3, and references therein) and of new generations of oncolytic viruses that were engineered for enhanced potency by different strategies for which side effects in patients cannot be predicted (1, 4). Furthermore, transient immunosuppression is being investigated as a means to block antiviral immunity thereby enhancing oncolysis (5). However, this strategy bears an increased risk of uncontrolled virus replication, which makes a drug-triggered genetic safety switch desirable as a clinical countermeasure. Likewise, such genetic safety switches could also be used for new generations of live virus vaccines with increased safety.
Ideally, ligand-inducible gene switches should be of short sequence, universally applicable in both DNA and RNA viruses, remain functional at high copy numbers after viral genome replication, and possess a simple mode of action independent of complex protein–nucleic acid interactions. Here, we propose artificial riboswitches known as aptazymes as powerful genetic switches. Aptazymes consist of a self-cleaving ribozyme sequence linked to a ligand-binding aptamer domain (6, 7). Hence, they enable conditional cleavage of RNA, including mRNAs, and fulfill all of the above-mentioned criteria. Moreover, they can be engineered for ON- or OFF-switch activity by modification of the aptamer–ribozyme linking sequences (8–10). Compared with well-established inducible promoters, as used for example in the Tet system (11), aptazymes are much smaller (approximately 100 bp) and do not require the expression of recombinant, potentially immunogenic transcription factors. Furthermore, they can be customized for responsiveness to different small-molecule ligands by exchange of the aptamer domain (12, 13). Importantly, due to their RNA-intrinsic mode of action they should be applicable in both DNA and RNA viruses. Moreover, inducible promoters, but not aptazymes, were reported to lose regulation at high copy numbers and during DNA virus replication (14–17), likely resulting from imbalances of stoichiometry of the required protein–DNA interactions. Aptamers alone were reported to enable virus control by ligand-dependent modulation of regulatory elements, as shown for a tomato virus (18). However, this strategy has to be optimized individually for the selected virus genome context and will likely not be applicable for most viruses. In contrast, aptazymes can be universally applied by insertion into the untranslated region (UTR) of the viral target gene, enabling direct regulation of mRNA stability and translation.
Aptazymes have been extensively developed and studied in vitro, in bacteria, and in yeast (6, 12, 13, 19–22). Their application in mammalian systems, however, has been challenging. This is mostly because aptazymes selected in vitro, in bacteria, or in yeast showed loss or strong reduction of regulation when applied in mammalian cells (20, 22–24). Previous reports on aptazyme applications in mammalian cells were reporter gene studies or studies on regulation of therapeutic gene expression (17, 23, 25, 26).
In this study, we investigated the utility of aptazymes as tools for conditional shutdown of viral gene expression in the biological context of virus replication control. Specifically, we analyzed aptazyme regulation of the immediate early gene E1A of adenoviruses and of the gene encoding the fusion glycoprotein F of measles viruses (MVs) and investigated how regulation of these genes translated into control of viral replication, infectivity, spread, and cytotoxicity. We chose adenovirus and measles virus because (i) this allowed us to explore aptazyme activity in the context of DNA viruses replicating in the nucleus and RNA viruses replicating in the cytoplasm, respectively; (ii) both viruses are being developed as oncolytic agents in preclinical and clinical studies (1); and (iii) antivirals effective in patients are not available for either virus.
Results
Aptazyme Insertion into the Adenoviral Immediate Early Gene E1A Enables Ligand-Dependent Shutdown of E1A Expression and Viral DNA Replication.
Our first aim was to establish aptazyme regulation of a DNA virus early gene enabling ligand-inducible shutdown of virus replication. We used human adenovirus serotype 5, a popular oncolytic agent, as model DNA virus (Fig. 1A) and inserted the OFF-switch aptazyme P1-F5 (23) into the viral E1A gene. The adenoviral immediate early E1A proteins play a key role in initiating virus infection as they induce transcription of further early viral genes and modify several host cell functions, both of which are required for viral DNA replication (27). P1-F5 possessed the best fold-induction rate at the time this study was initiated, which was between three- and sixfold in reporter assays (17, 23). We hypothesized that insertion of this aptazyme into the E1A gene would not affect virus replication in the absence of the P1-F5–activating ligand theophylline (theo). Upon its addition, the ligand should trigger activation of the aptazyme’s self-cleaving activity, resulting in degradation of E1A mRNA, inhibition of E1A protein expression, and ultimately inhibition of virus genome replication (Fig. 1B and Fig. S1A). We inserted the aptazyme (Az viruses) into the 5′- and/or 3′-UTR of the mutant E1A gene E1AΔ24 (Fig. 2A), commonly used in oncolytic adenoviruses because it attenuates viral replication in normal quiescent, but not tumor cells (28, 29). As reference, matching viruses were generated containing the constitutively active ribozyme (Rz viruses) instead of the aptazyme. A control virus contained unmodified E1A UTRs.
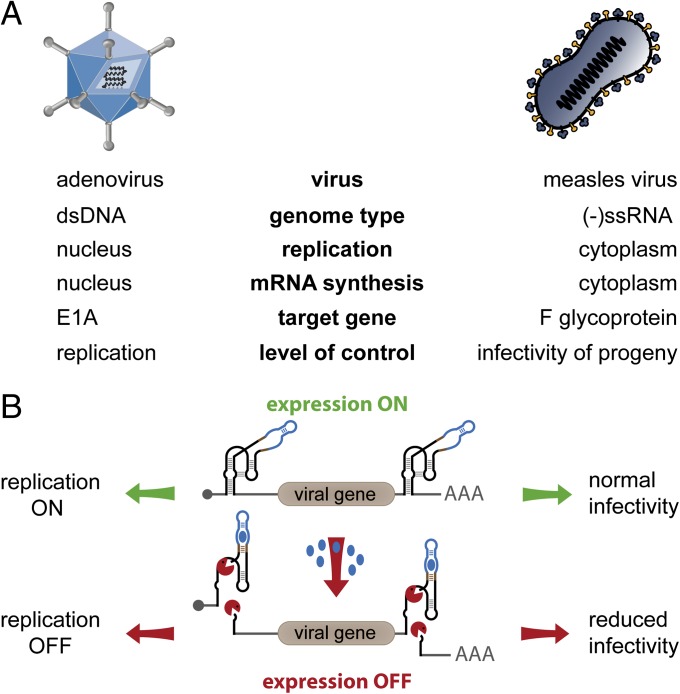
Fig. 1.
Schematic outline of the strategy for aptazyme-mediated shutdown of viral genes and functions. (A) Key features of adenovirus and measles virus infection cycle. Target genes for aptazyme control and their influence on viral infection are indicated. (B) Shutdown of viral gene expression is achieved using a cis-acting synthetic RNA switch, specifically an OFF-switch aptazyme. This ligand-dependent self-cleaving ribozyme is inserted into the UTRs of the viral transcription unit. During infection, the viral genes are transcribed in the absence (expression ON), but not in the presence (expression OFF) of the small-molecule ligand (blue dots). This allows replication control of adenoviruses through regulation of the E1A gene and control of progeny infectivity of measles viruses by regulation of the F gene.
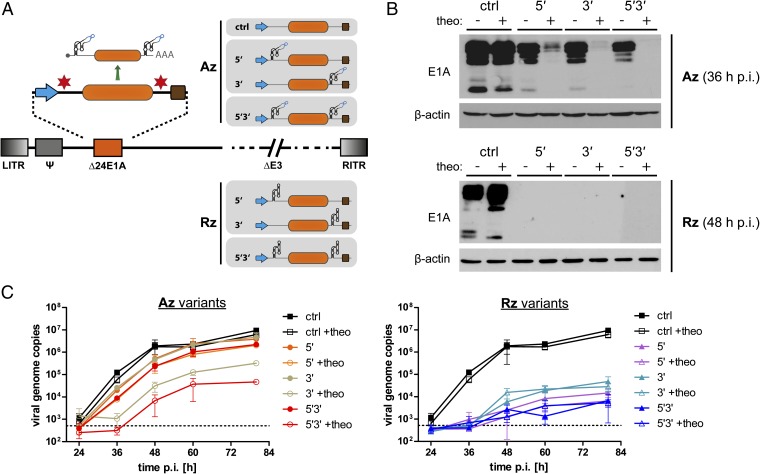
Fig. 2.
Aptazyme-mediated control of adenoviral E1A gene expression and DNA replication. (A) Schematic outline of oncolytic adenoviruses containing a regulatable E1A transcription unit. Insertion sites for the P1-F5 aptazyme (Az) or the parental ribozyme (Rz) were in the 5′- and/or 3′-UTR (nomenclature: 5′/3′/5′3′) of the E1A gene (orange box) driven by the adenoviral E1A promoter (blue arrow). Brown box, polyadenylation signal. ctrl, no aptazyme insertion. LITR/RITR, left/right inverted terminal repeat; Ψ, packaging signal; Δ24E1A, E1A gene with a 24-bp deletion for tumor selectivity; ΔE3, E3 region deleted (not required for virus replication); other viral genes and elements are not shown (dashed lines). (B and C) SK-MEL-28 cells were infected with the oncolytic adenovirus variants using 10 TCID50/cell (B) or 1 TCID50/cell (C). Cells were cultured in the absence (−) or presence (+) of 3 mM theophylline and harvested at indicated time points postinfection (p.i.). (B) For analysis of E1A expression, total lysates (20 μg) were assayed by Western blotting for E1A protein expression; human β-actin was detected as loading control. Multiple bands for E1A represent multiple splice variants. Time points of cell harvest were chosen according to replication kinetics of adenoviruses in these cells. Results for the Rz viruses are shown at 48 h p.i. to demonstrate stringent inhibition of gene expression by the ribozyme. (C) Viral genome copy numbers were determined by qPCR and are presented relative to cellular DNA content as determined for each sample individually. Symbols show mean values, error bars SD of three samples (n = 3). Dashed line, background signal of mock-infected cells.
All viruses could be produced to normal titers in E1-complementing cells, showed similar infectivity in these cells, and were used for infection studies in the melanoma cell line SK-MEL-28. Protein expression analysis showed that ribozyme insertion abrogates E1A expression independently of both insertion site and theo addition (Fig. 2B, Lower). Insertion of the aptazyme per se resulted in somewhat reduced E1A expression, but importantly strong inhibition of E1A expression was achieved by theo addition (Fig. 2B, Upper). Regulation by theo was best for the 5′3′ construct, for which we also showed dose-dependent regulation (Fig. S2A), and the least pronounced with the 5′ construct. To explore how the aptazyme-mediated block of E1A expression translates into inhibition of viral DNA replication, we quantified viral genome copies during a single viral replication cycle (Fig. 2C). For the Rz viruses, we observed a strong reduction of genome copy numbers compared with the control virus, which was 200- to 2,000-fold at 80 h postinfection (p.i.). For the Az viruses, we observed a minimal delay in DNA replication in absence of theo. Addition of theo resulted in minor, intermediate, and strong reduction of viral genome copy numbers for the Az 5′, 3′, and 5′3′ viruses, respectively, which was dose dependent (as shown for the Az 5′3′ virus; Fig. S2B).
These results are in accord with the observed E1A protein expression profiles, validating virus replication as biological readout for aptazyme-mediated gene regulation. Theo-mediated regulation of viral replication of the Az 5′3′ virus at 80 h p.i. was approximately 200-fold. Overall, these results demonstrate that aptazyme insertion into both 5′- and 3′-UTRs of the viral E1A gene is suitable for conditional shutdown of viral gene expression, facilitating indirect control of viral DNA replication. In fact, potent inhibition of adenoviral DNA replication was achieved even using an aptazyme that possesses modest induction rates like P1-F5.
Aptazyme Shutdown of Adenoviral E1A Expression Inhibits Infectious Virus Particle Production and Oncolysis.
For applications as safety switch in the context of oncolytic viruses or virus vaccines, it is essential that aptazyme regulation of viral genes translates into inhibition of infectious viral particle production and spread-dependent cytotoxicity. Indeed, infectious progeny production of the Az 5′3′ virus in SK-MEL-28 cells was completely blocked in a dose-dependent manner by theo at 48 h p.i. (Fig. 3A and Fig. S2C). At 80 h p.i., progeny viruses were produced in the presence of theo, but remained at approximately 60-fold lower titers than in the absence of theo. Titers of the Az 5′3′ virus in the absence of theo did not reach the titers of the control virus, indicating that aptazyme insertion per se resulted in some degree of attenuation, as expected from the E1A expression and virus genome replication data (Fig. 2 B and C). Note that the positioning of the aptazyme in the E1A UTRs was not optimized and it remains to be investigated whether insertion site optimization improves adenovirus replication in the absence of the ligand. Infectious particle production of the control virus was not affected by theo and was completely inhibited for the Rz 5′3′ virus (Fig. 3A).
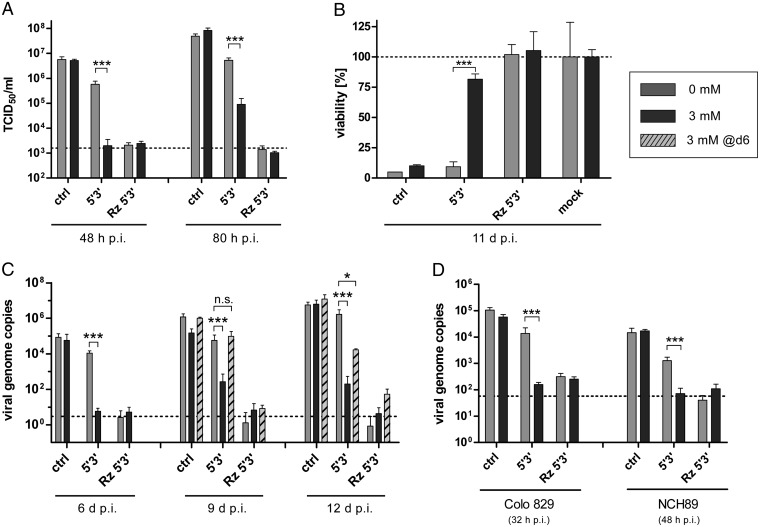
Fig. 3.
Aptazyme-mediated control of adenoviral replication, particle production, and oncolysis. For analysis of aptazyme-regulated production of infectious viral progenies (A), viral cytotoxicity (B), or viral genome replication (C and D), SK-MEL-28 (A–C, melanoma), Colo 829 (D, melanoma), or NCH89 (D, glioblastoma) cells were infected using the indicated oncolytic adenovirus variants (see Fig. 2A). Infected cells were cultured in the absence (gray bar) or presence of 3 mM (black bar) theophylline added either after infection or 6 d p.i. (dashed bar). Cells were harvested at indicated time points p.i. Significance for theophylline-dependent regulation for individual oncolytic adenovirus variants is indicated with *P < 0.05, ***P < 0.001. Columns show mean values, error bars SD of three samples (n = 3). (A) Cells were infected at 1 TCID50/cell. Total infectious progeny particles produced were determined by titration in HEK293 cells. Dashed line, input virus as measured for infected cells harvested 1 h p.i. (B) Cells were infected at 50 TCID50/cell, allowing determination of viral cytotoxicity by staining of surviving cells 11 d p.i. with crystal violet. Crystal violet intensity was quantified and normalized to mock-infected cells (mock, set as 100% for theophylline-treated and untreated groups, dashed line). (C and D) Viral genome copy numbers were determined at indicated time points p.i. (C, 0.01 TCID50/cell; D, 1 TCID50/cell) by qPCR and are presented relative to cellular DNA content as determined for each sample individually. Dashed line, background signal of mock-infected cells. Infections were performed at low titers (C) to enable several virus replication cycles without eradication of cell monolayers. Time points of cell harvest in D differed between cell types because of differences in adenovirus replication kinetics. Adenovirus replication is faster in Colo 829 cells than in NCH89 or SK-MEL-28 cells as determined in a pilot time-course experiment.
Next, we investigated regulation of oncolytic activity of adenoviruses by the aptazyme. In a spread-dependent cytotoxicity assay allowing for several rounds of virus replication (Fig. 3B), we observed almost complete cell killing by the control virus without any insertion, but no loss in cell viability after infection with the Rz 5′3′ virus, both independently of theo. Importantly, for the Az 5′3′ virus, we observed cytotoxicity similar to the control virus in the absence of theo, but high cell viability in the presence of theo, establishing a switch from 10 to 80% viability. In conclusion, aptazyme insertion into the E1A gene resulted in effective ligand-dependent control of infectious virus particle production and oncolytic activity of adenoviruses.
Flexible Shutdown of Adenoviral Infection by Aptazyme Regulation of E1A Expression.
In the previous assays, theo was added to infected cell cultures immediately after infection. We next investigated whether established adenovirus infection can be switched off by using the aptazyme, resembling the application as a safety switch (Fig. 3C). By quantification of viral genome copy numbers as readout for virus replication, we confirmed potent aptazyme-dependent theo-induced inhibition at 6, 9, and 12 d p.i. when theo was added immediately after infection. When adding theo at 6 d p.i., i.e., during full adenovirus replication, viral genome copies were not affected 3 d later, but were eventually strongly reduced (100-fold, 12 d p.i.). These results demonstrate that aptazyme-dependent regulation of E1A expression can mediate shutoff of established adenovirus infection, although with a delay of 3–6 d. This delay could be attributed to the nature of adenoviral replication, which proceeds independently of E1A once early steps of infection are established, whereas subsequent infection of progeny viruses is blocked at the immediate early stage of the replication cycle.
Aptazyme Control of Adenoviral Replication in Different Cell Lines.
Our results demonstrate potent control of adenovirus replication by aptazyme regulation of E1A expression in SK-MEL-28 cells using the P1-F5 aptazyme. When testing a panel of other tumor cell lines, we observed similar replication control in some cell lines. In Colo 829 melanoma and NCH89 glioblastoma cells, aptazyme-mediated inhibition of viral genome replication after theo addition was similar to that using the ribozyme virus, demonstrating that our results are not restricted to SK-MEL-28 melanoma cells (Fig. 3D). However, regulation of virus genome amplification was not observed in A549 and Capan-1 cells (Fig. S3A). Importantly, this was not due to failure of the aptazyme to regulate E1A protein expression, because we could demonstrate theo-dependent inhibition of E1A expression for the Az 5′3′ virus in both cell lines (Fig. S3B). We hypothesized that this cell-line–dependent lack of replication control despite functional regulation of E1A expression must be due to a favorable cellular milieu for adenovirus replication and could be overcome in two ways: By improving aptazymes for increased self-cleaving activity in presence of the ligand (this can only be addressed in the future once such aptazymes become available, see Discussion) or by reducing the baseline level of E1A expression. Toward this end, we could show for Capan-1 cells that replacing the E1A promoter with weaker human promoters resulted in reduced E1A baseline expression (Fig. S3C) and retained aptazyme-mediated regulation of E1A expression (Fig. S3D), thereby enabling aptazyme-mediated inhibition of adenovirus replication as analyzed by viral genome amplification (Fig. S3E) and infectious virus progeny production (Fig. S3F). Overall, these results clearly demonstrate the general applicability of aptazymes as safety switches for blocking adenovirus replication, although improved switches are required to establish the full potential of this approach.
Aptazyme Insertion into the Measles Virus Fusion Gene Results in Ligand-Dependent Shutdown of Gene Expression, Viral Infectivity, and Viral Spread.
We next investigated whether applications of aptazymes for shutdown of viral gene expression can be extended to RNA viruses. Importantly, aptazyme activity has so far not been demonstrated in cytosolic RNA systems or in the context of RNA genomes. We used measles virus based on the Edmonston B vaccine strain (MV) as model virus (Fig. 1A), because viruses derived from this strain are being clinically developed as oncolytic agents (1). We inserted the OFF-switch aptazyme P1-F5 into the viral fusion (F) gene. The F protein mediates fusion of the viral and host cell membranes after virus attachment, resulting in viral entry into the cell. Furthermore, F proteins of infected cells trigger cell fusion with neighboring cells resulting in syncytia formation. We hypothesized that ligand-induced activation of the aptazyme results in degradation of the F mRNA and thus in loss of F protein expression, fusion activity, virus infectivity, and ultimately spread (Fig. 1B and Fig. S1B). Note that in this setting, replication of the input viruses should not be affected, but rather infectivity of the progeny virus generation lacking F proteins. Regulating F protein expression has the technical advantage of allowing for validation of aptazyme-mediated regulation in transient transfection studies using a fusion assay as readout.
To address our hypothesis, we inserted the P1-F5 aptazyme, the constitutively active ribozyme, or inactive mutants thereof (point mutation in the active center) into both 5′- and 3′-UTRs of the F gene, initially within an expression plasmid (Fig. 4A and Fig. S4A). A control plasmid contained only the cloning sites. We could demonstrate ligand-dependent inhibition of syncytia formation for the 5′3′ aptazyme construct in a fusion assay (Fig. S4B). Here, F expression plasmids are transfected together with plasmids encoding the MV attachment protein H and GFP, yielding fluorescent syncytia when all proteins are expressed (30). Results similar to the 5′3′ aptazyme construct were obtained with F expression plasmids containing only one aptazyme in either the 5′- or the 3′-UTR (Fig. S4B). The ribozyme construct did not yield syncytia irrespective of the ligand, whereas the ribozyme and aptazyme mutants and the empty control constructs readily produced syncytia. These results indicate that conditional expression of F by aptazyme regulation is functional and translates into control of syncytia formation.
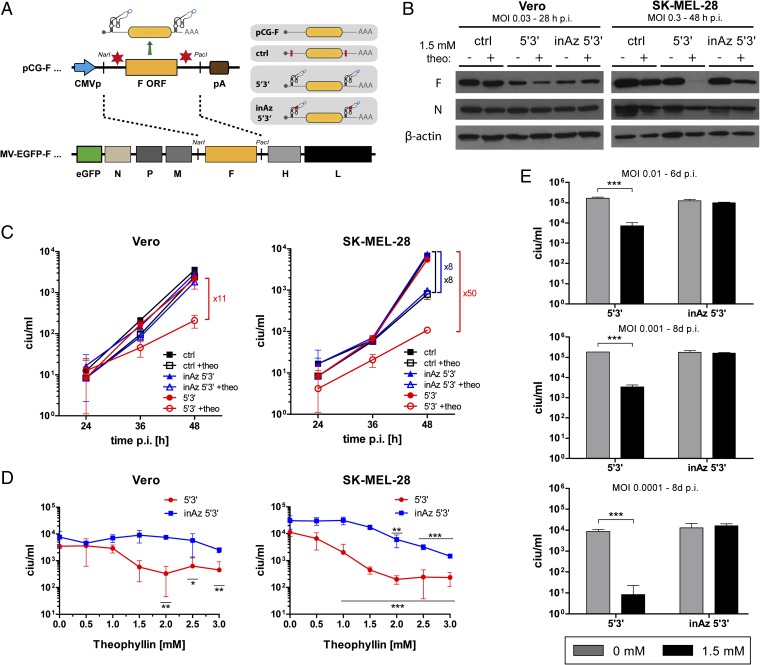
Fig. 4.
Aptazyme-mediated control of measles virus infectivity and spread. (A) Schematic outline of F expression plasmids (pCG-F, pCG-F ctrl/5′3′/inAz5′3′) and measles virus (MV) genomes (MV-EGFP-F, MV-EGFP-F ctrl/5′3′/inAz5′3′) containing a regulatable F transcription unit. Insertion sites for the P1-F5 aptazyme (Az) or the inactive mutant thereof (inAz) were in the 5′- and 3′-UTRs of the F gene (yellow box). Blue arrow, CMV promoter; brown box, polyadenylation signal; ctrl, modified pCG-F plasmid with introduced restriction sites for Az/Rz insertion (red bar); red dot in inAz 5′3′, inactivating point mutation. For all MV variants, EGFP is inserted upstream of the N ORF in an additional transcription unit. N, P, M, H, and L represent MV genes. (B–E) Vero (B–D) and SK-MEL-28 (B–E) cells were infected with indicated MV variants at 0.03 ciu/cell or 0.3 ciu/cell (B, SK-MEL-28) and cultured in the absence or presence of 1.5 mM (B and E) or 3 mM (C) or various (D) theophylline concentrations. Cells were harvested at indicated time points. Columns/symbols show mean values, error bars SD of three samples (n = 3). (B) For analysis of MV F protein expression, total lysates (40 μg) were assayed by Western blotting for MV F expression; MV N and β-actin were detected as internal controls. Time points of cell harvest differed between cell types because of a more rapid cell lysis by measles virus infection in Vero cells. (C) Produced infectious progeny particles in cells and supernatant were determined by titration on Vero cells. (D) Concentration-dependent production of infectious progeny particles. Cells were harvested 48 h p.i. and subjected to titration of total infectious progenies on Vero cells. (E) For assaying long-term control of MV infection, infections at indicated low multiplicities of infection (MOI) were performed allowing several rounds of virus replication and titers were determined at the indicated time points. (D and E) Significance for theophylline-dependent regulation for individual MV variants is indicated with *P < 0.05, **P < 0.01, ***P < 0.001.
We then produced viruses with F genes containing aptazymes (Azs) or inactive mutant aptazymes (inAzs) in both 5′- and 3′-UTRs (Fig. 4A). The control virus contained the cloning sites in the UTRs. These viruses were generated at similarly high titers, whereas a virus with 5′3′ ribozyme insertions could not be rescued. The latter could also not be established when the F protein was complemented by F plasmid cotransfection, indicating that the nascent, unpackaged MV (+)RNA antigenome must be efficiently cleaved by the ribozyme after transfection of MV production plasmids (31). Infection studies in Vero and SK-MEL-28 cells showed theo-dependent shutdown of F protein expression for the Az 5′3′ virus (Fig. 4B). In Vero cells, aptazyme insertion per se resulted in somewhat lower F protein expression. In SK-MEL-28 cells, addition of theo alone had nonspecific effects on virus replication as deduced from reduction of both N and F proteins. Nevertheless, the specific aptazyme-mediated control of F protein expression after infection with the Az 5′3′ virus was clearly visible. A comparison of F and N protein expression for the Az 5′3′ virus also indicates that aptazyme-mediated regulation is at the mRNA level rather than by degradation of the (+)RNA antigenome during virus infection, as the latter would cause reduced secondary transcription from the replicated (−)RNA genome and, consequently, similar reduction of F and N protein levels. As newly synthesized (+)RNA antigenomic RNA molecules are cotranscriptionally packaged, the MV nucleocapsid structure prevents proper folding of the aptazyme sequence as required for self-cleavage activity.
Next, we analyzed how control of F expression translates into shutdown of MV infectivity using infectious particle production in a single-step growth curve as readout based on the fact that the infectivity of the progeny particles essentially depends on F protein (Fig. 4C). At 48 h after infection of Vero cells, we observed a prominent, 11-fold reduction of infectious particle production after addition of theo specifically for the Az 5′3′ virus. In SK-MEL-28 cells, we observed an 8-fold reduction of infectious particle production for the control and inAz 5′3′ virus after addition of theo, which was in line with the slight reduction of F and N protein expression (Fig. 4B). However, 48 h p.i. reduction of infectious particle production by theo was 50-fold for the Az 5′3′ virus, confirming ligand-dependent regulation via the aptazyme. Regulation was not observed for viruses containing single aptazymes in either the 5′- or the 3′-UTR of the F gene (Fig. S4C). Syncytia formation was not affected by aptazyme regulation (Fig. S4D), indicating that low amounts of newly generated F protein are sufficient to induce syncytia in a single-step growth experiment.
Considering the aptazyme-independent effects of 3 mM theo, we performed a dose–response experiment showing that 1.5 mM theo facilitated effective aptazyme-dependent regulation of MV infectivity, but lower nonspecific virus attenuation (Fig. 4D). Consequently, theo resulted in superior regulation of infectious particle production at 1.5 mM compared with 3 mM. To better demonstrate how aptazyme shutdown of F expression leads to a block of MV infection and spread, we performed a multistep growth curve after low titer virus inoculation of SK-MEL-28 cells. In this setting, measurement of the ultimately produced infectious particles depends on several rounds of infection for which functional F protein is required. Indeed, we observed a powerful inhibition of spread-dependent infectious MV production by theo for the Az 5′3′ virus, but not for the inAz 5′3′ virus (Fig. 4E), thus establishing long-term control of MV infection. Here, aptazyme-mediated shutdown of MV replication by theo was more potent at lower virus titers. In conclusion, these results demonstrate that aptazymes facilitate conditional knockdown of RNA virus genes translating into biological readouts, here ligand-induced shutdown of MV spread.
Discussion
Our study shows that a model aptazyme enables small-molecule–triggered shutdown of DNA virus immediate early gene and RNA virus glycoprotein expression, resulting in inhibition of viral replication and infectivity, respectively, and consequently of viral spread and cytotoxicity. Conditional expression of the adenoviral gene E1A, the master regulator of adenoviral replication, was achieved with increasing efficiency by inserting the aptazyme into the 5′-UTR, 3′-UTR, or both UTRs. These data very well match our previously reported results for regulation of reporter genes, pointing at an universal applicability of aptazymes for gene regulation (17). Of note, insertion positions for the aptazyme within the UTRs were not optimized, revealing that interference of neighboring sequences with proper folding and activity of the aptazyme is not a major concern. A key question we addressed in this study was how small-molecule–triggered, aptazyme-mediated regulation of viral gene expression as an input signal affects viral functions as biological output. Whereas single aptazyme insertion moderately affected viral replication, 5′3′ double insertion translated into a dramatic, dose-dependent inhibition of adenoviral genome replication, infectious particle production, and cytotoxicity in SK-MEL-28 cells. Corroborated by the maximal replication inhibition achieved with the constitutively active ribozyme, our results thus provide a proof of concept that aptazyme-mediated regulation of viral gene expression can effectively affect viral functions. Importantly, we extended the applicability of aptazyme-mediated regulation to RNA viruses and fully cytoplasmic gene expression systems: aptazyme-mediated shutdown of F glycoprotein expression in Vero and SK-MEL-28 cells translated into reduced infectivity of progeny viruses and a dramatic attenuation of virus spread in a multistep infection assay using SK-MEL-28 cells.
Considering these results, we suggest aptazymes as novel tools for exploring virus gene functions in experimental infections and as a safety switch for new generations of efficiency-enhanced oncolytic viruses. Careful analysis of our results, however, reveals several current limitations regarding the utilization of the P1-F5 aptazyme, which was the best available aptazyme at the time this study was initiated: (i) In line with our previous results (17), aptazyme-mediated shutdown of E1A was less effective than shutdown by the parental ribozyme, which executed maximal repression due to constitutive self-cleavage activity. This provides proof of concept for the aptazyme approach as such, but implies that the P1-F5 aptazyme represents a suboptimal switch, clearly leaving a window of opportunity for improvement. (ii) In two of four further tested cell lines, A549 and Capan-1 cells, aptazyme-mediated reduction of E1A expression did not translate into shutdown of viral genome replication, although the ribozyme control virus was capable of replication shutdown in all tested cell lines. This input–output disconnect likely results from a lower E1A input necessary to drive adenoviral replication or from higher E1A baseline expression in these cells. (iii) Similarly, shutdown of glycoprotein F expression did not translate into inhibition of syncytia formation during primary measles virus infection, indicating that F protein at the observed reduced concentrations is sufficient to induce syncytia, while not supporting infectivity of progeny viruses. However, results from an RNAi study demonstrate that effective down-regulation of F protein expression translates into a lack of syncytia formation (32). (iv) Baseline expression of regulated genes, i.e., in absence of the ligand, can be affected by aptazyme insertion to varying degrees. (v) Aptazyme activation by applying ligands to established infections allowed for replication control, but in a delayed fashion.
Essentially, all these issues can be addressed by optimizing the aptazyme for higher induction rates and thus improved gene shutdown potency. Clearly, the fact that the constitutively active ribozyme caused potent inhibition of virus replication in all cases indicates that the aptazyme approach should be universally functional with optimized aptazymes. Artificial aptazymes have been mostly developed in vitro and in bacteria, where aptazymes with high induction rates were generated by high-throughput selection (12, 13, 20, 33). However, only a few aptazymes have been reported to be functional in mammalian cells. The aptazyme P1-F5 used here was obtained by manual selection from only 192 aptazyme constructs with random communication sequences. Thus, it is very likely that high-throughput selection of highly diverse libraries will yield new generations of high-fidelity aptazymes. Optimized aptazymes should also be responsive to lower concentrations of theo, thus avoiding nonspecific effects on cell and virus behavior, as observed in this study. A key advantage of aptazymes is that they can be customized to respond to alternative ligands. To this end, aptamers with high affinity for a diversity of small-molecule ligands, e.g., for antibiotics like tetracycline, kanamycin, neomycin, and tobramycin, have been reported (34). Moreover, aptamers for further ligands of interest, ideally pharmaceuticals with known safety, bioavailability, and cell permeability profile, can be developed by affinity selection of RNA libraries (35). These aptamers can then be exploited for the development of novel aptazymes. In this line, Smolke and coworkers have reported the customization of hammerhead ribozyme-based aptazymes for regulation of reporter genes and therapeutic genes by exchange of the theo aptamer domain with a tetracyclin aptamer (21, 25). Yokobayashi and coworkers have recently reported a guanine-inducible aptazyme featuring a 29.5-fold induction rate (36). Comprehensive efforts of directed evolution of aptazymes in mammalian cells using high throughput techniques will eventually lead to the identification of optimized aptazymes, ultimately allowing for more effective regulation of biological outputs.
Several other aspects of our regulation system may be further improved in the context of basic research or therapeutic applications. First, the exact positions in the UTRs for the aptazyme insertion(s) should be systematically optimized to minimize reductions in baseline gene expression after aptazyme insertion possibly resulting from adverse effects on mRNA translation or stability independent of ligand-induced aptazyme regulation. Second, adjustment of baseline viral gene expression levels can improve aptazyme control of biological outcomes, as conceptionally shown here for adenoviral E1A in control of viral infection in Capan-1 cells using promoter-modified viruses. Alternatively, adenoviral genes other than E1A that more directly link gene expression input to virus replication output can be regulated. In this context, the DNA polymerase or IVa2 genes could be of interest, as their knockdown RNAi proved more effective than that of E1A (37, 38). Similarly, aptazyme regulation of the measles virus P or L polymerase genes or of the nucleocapsid N gene may facilitate conditional virus replication, as indicated by RNAi studies (39, 40). Finally, the pace of shutting down established virus infections can most certainly be accelerated considerably by combined control of several viral genes blocking viral infections simultaneously at different stages of the virus replication cycle.
For exploring virus gene functions in experimental infections, aptazymes offer key advantages compared with protein-dependent inducible promoters (as discussed in the Introduction) and RNAi (41). First, ligand-induced regulation of gene expression by aptazymes functions in cis and is thus mechanistically likely to be more effective than RNAi regulation, although systematic comparisons will have to be performed to prove this assumption. Second, aptazyme-mediated gene shutdown is induced by small-molecule ligands that are less intrusive than nucleic acid transfection and should exert fewer nonspecific off-target effects than siRNAs. Furthermore, small molecules as triggers allow for more precise timing of virus gene shutdown. Most importantly, in vivo applications of RNAi are hampered by delivery problems, whereas small molecules triggering aptazyme-mediated shutdown of viral genes should be systemically available and thus facilitate in vivo studies investigating the role of genes in virus tropism, spread, pathogenicity, or immune escape.
The aptazyme regulation system presented in this study may find therapeutic application in conjunction with oncolytic viruses. Although clinical trials have generally demonstrated the safety of present oncolytic viruses, they have also revealed that viruses with increased potency are required to establish therapeutic efficacy in patients (1, 4). For such viruses, it is paramount to ensure safety. Considering the shortage of antiviral drugs, aptazymes might establish a necessary safety mechanism. Such safety switches are of interest for clinical development of certain candidate oncolytic viruses like Coxsackievirus A21 and vesicular stomatitis virus, that have shown potent antitumor activity, but also fatal toxicity in animal models (refs. 2, 3, and references therein). Note that in these cases promoter targeting is not possible due to their RNA genomes. Moreover, extensive research efforts focus on enhancing the efficiency of established oncolytic viruses. Strategies toward this end include enhancement of virus cell binding, improvement of virus spread, attenuation of IFN sensitivity, and insertion of therapeutic genes (1, 4). Because it is often difficult to predict potential side effects of such modifications, e.g., resulting from infection of previously not susceptible cells by tropism-modified viruses or from the induction of overshooting immune responses, clinical implementation of efficiency-enhanced oncolytic viruses can benefit from or even require a safety switch. Lately, various cell types with intrinsic tumor tropism are being exploited as carrier cells for oncolytic virus delivery. Once infected, they protect the virus from the host’s immune system during transport before releasing progeny particles at the tumor site (42). In this context, transient aptazyme-mediated inhibition of virus replication in carrier cells could improve their migration to the tumor and avoid premature release of progeny viruses, also adding to overall efficacy and safety of oncolytic viruses.
In summary, we establish artificial riboswitches as a novel tool for drug-regulated expression of viral genes and resulting viral functions. Regulation of adenoviral E1A and measles virus glycoprotein F enabled control of viral replication and spread. Importantly, aptazyme-regulated gene expression was functional in the context of both DNA and RNA viruses and thus was shown to be fully active in cytoplasmatic systems. Hence, as an appealing alternative to protein- and RNAi-dependent gene regulation systems, we propose aptazymes as universally applicable tools to study viral gene functions and to allow safety of new generations of efficiency-enhanced oncolytic viruses.
Materials and Methods
Mammalian Cell Culture Maintenance.
Human cell lines HEK293 (embryonic kidney), SK-MEL-28 (malignant melanoma), Capan-1 (pancreatic cancer), A549 (lung adenocarcinoma), NCH89 (glioblastoma), and monkey cell line Vero (African green monkey kidney) were cultivated in Dulbecco’s Modified Eagle’s Medium (Invitrogen) supplemented with 10% (vol/vol) heat-inactivated FBS (PAA), 100 IU/mL penicillin, and 100 µg/mL streptomycin (both from Invitrogen). The human cell line Colo 829 (malignant melanoma) was cultivated in RPMI 1640 medium supplemented as described above. Cells were grown at 37 °C in a humidified atmosphere of 5% (vol/vol) CO2.
Cloning and Production of Adenoviruses.
Nomenclature was used as follows: Rz, hammerhead ribozyme N107 (43); Az, theophylline-dependent aptazyme P1-F5 (23). The term “in” describes a mutated Az or Rz sequence harboring the A14G point mutation within the catalytic core of the Rz domain, which was reported to inactivate Rz self-cleavage (43). All cloning steps were conducted by applying standard molecular biology techniques. Oligonucleotides were synthesized by Eurofins MWG Operon. Cloned plasmids were purified by standard anion exchange columns (Qiagen) and verified by DNA sequencing (GATC Biotech). Restriction enzymes were obtained from Fermentas, DNA polymerase was obtained from BioCat.
For cloning of OAd (oncolytic adenovirus)-Δ24E1A variants (Az 5′/3′/5′3′, Rz 5′/3′/5′3′), pCDNA 3.1(+) (Invitrogen) and pS-Δ24E1A (pShuttle containing the adenoviral E1A gene with a 24-bp deletion and the E1B transcription unit) (44) were used as cloning vectors. For a schematic outline of the OAd-Δ24E1A variants, see Fig. 2A. First, the E1A transcription unit was cloned into pCDNA 3.1(+) via SspI and KpnI. Then, Rz was inserted into pCDNA-E1A by PCR into the 5′ or 3′ using primer pairs encoding the Rz sequence (5′-E1A_N107_fw, CGAAACGCGCTTCGGTGCGTCCTGGATTCCACTGCTATCCACTTCTCCTCCGAGCCGCTCC and 5′-E1A_N107_rv, TCCTATTTGGGACTCGTCAGCTGGATGTACCTGCACCTCAGAACTCTACTCGCTGGCACTCAAG or 3′-E1A_N107_fw, CGAAACGCGCTTCGGTGCGTCCTGGATTCCACTGCTATCCACGTGTGGTTAACGCCTTTGTTTGC and 3′-E1A_N107_rv, TCCTATTTGGGACTCGTCAGCTGGATGTACCTGCACCTCAGACGCAATCACAGGTTTACACC). Then, Az (P1-F5) was introduced by PCR using primer pairs encoding the P1-F5 sequence (P1-F5_fw, AAGGCCCTTGGCAGGGTTCCTGGATTCCACTGCTATCCAC and P1-F5_rev, TCGGCTGGTATGGCTTTCGTCCTATTTGGGACTCGTCAG). For multimerization, pCDNA-E1A Az 5′/Rz 5′ was combined with pCDNA-E1A Az 3′/Rz 3′ via XbaI. Then, cloned pCDNA variants were subcloned into pS-Δ24E1A via the unique restriction sites SspI and KpnI.
For cloning of OAd-Δ24E1A promoter variants, wild-type E1A promoter was replaced from position 343–524 (nucleotide positions refer to NCBI Sequence AC_000008.1) with the SV40 or TKs promoter insulated by a synthetic polyA signal.
Subsequently, adenoviral particle variants were generated by homologous recombination in BJ5183 bacteria as described before (45) using the cloned pS-Δ24E1A or pS-Δ24E1A-promoter variants and pAdEasy-1 (Promega).
Virus particle production, verification, and quantification were performed as previously described (17). Quality of virus preparation was comparable as shown by similar ratios of physical virus particles to infectious virus particles: ctrl, 11; Az 5′, 4; Az 3′, 5; Az 5′3′, 6; Rz 5′, 10; Rz 3′, 11; Rz 5′3′, 13; SV40 ctrl, 15; SV40 5′3′, 14; TKs ctrl, 18; TKs 5′3′, 18; virus particle yield was in the same range for all OAd variants.
Cloning and Production of MVs.
Nomenclature was used as described above. For cloning of MV-EGFP-F variants (ctrl, 5′3′, inAz 5′3′, Rz 5′3′, and inRz 5′3′), pCG-F and pcpNSe ld-EGFP (46) based on the Edmonston B vaccine lineage strain) were used as cloning vectors. For a schematic outline of the MV-EGFP-F variants, see Fig. 4A. First, restriction sites were inserted into the UTRs of the F gene according to the rule of six by site-directed mutagenesis using primer pairs encoding the new restriction sites and spacer (pCG-F_HindIII_rv, GAAAGCTTTTCTGGGATTCCCGGTGTC and pCG-F_BsrGI_fw, GATCTGTACATCAAGACTCATCCAATGTCCATC or pCG-F_NheI_rv, GCTAGCGAGTTGTAGAGGATCAGAGCG and pCG-F_AflII_fw, GATCGACTTAAGTTGAAACACAAATGTCCCACAAGTC). Upon combining both generated plasmids, the resulting pCG-F HBNA plasmid (ctrl) was used as cloning vector for subsequent insertion of Rz or Az. Therefore, Rz or Az were amplified from psiCHECK-2_Rz or psiCHECK-2_Az (23) using various primer pairs for 5′ (Rz-HindIII_fw, GATCAAGCTTCTGAGGTGCAGGTACATCCAGC plus Rz-C_BsrGI_rv, GATCTGTACAGGTGGATAGCAGTGGAATCCAGG or Az-CC_BsrGI _rv, GATCTGTACAGGGTGGATAGCAGTGGAATCCAGG) and 3′ insertion (Rz_NheI_fw, GATCGCTAGCCTGAGGTGCAGGTACATCCAG plus Rz-C_AflII_rv, GATCCTTAAGGGTGGATAGCAGTGGAATCCAGG or Az-CC_AflII_rv, GATCCTTAAGGGGTGGATAGCAGTGGAATCCAGG). The resulting pCG-F variants (pCG-F Az/Rz 5′/3′/5′3') were then used for subcloning into pcpNSe ld-EGFP. In brief, pCG-F Az/Rz 5′/3′/5′3′ were digested with NarI and PacI and the F transcription unit was isolated and exchanged in pcpNSe ld-EGFP via NarI and PacI. Generated pcpNSe ld-EGFP-F Az/Rz 5′/3′/5′3′ plasmids were sequence verified. Viral particles were produced according to a modified rescue system (47). Vero cells were transfected with the respective pcpNSe-based genomic plasmid and pCG-N, pCG-P, pCG-L, and pcDI-DsRed using FuGENEHD (Roche) reagent. The recombinant MV particles were amplified in Vero cells. Viral stocks from passage 3 were used for experiments, for verification of viral genomes by RT-PCR and PCR, and for sequencing of the F transcription unit. Virus titers were determined by titration on Vero cells [cell infectious units (ciu)/mL] as previously described (48). Virus particle yield was in the same range for all MV-EGFP-F variants.
Infections.
All transient infections were performed in growth medium containing 2% (vol/vol) FBS. Respective cell lines were seeded in the appropriate multiwell plates to 70% confluence. The next day, infections were carried out using indicated tissue culture infectious dose 50 (TCID50) per cell or multiplicity of infection (MOI). Adenoviral infections were performed using 1 TCID50/cell with the following exceptions: For immunoblot analysis, SK-MEL-28 cells were infected using 10 TCID50/cell, because of their lower susceptibility to adenovirus infection and lower sensitivity of this readout. Infectious titers were optimized for long-term experiments (0.01 TCID50/cell, Fig. 3C) and cytotoxicity assays (50 TCID50/cell, Fig. 3B) to allow viral replication over 12 d without eradication of monolayer cultures and detection of cytotoxicity at 11 d postinfection, respectively. Measles virus infections were performed at 0.03 ciu/cell with the following exception: For immunoblot analysis, SK-MEL-28 cells were infected at 0.3 ciu/cell because of their lower susceptibility to measles virus infection and lower sensitivity of this readout. Lower titers were used for long-term experiments (0.01–0.0001 ciu/cell, Fig. 4E) to allow viral replication over 6 or 8 d without eradication of monolayer cultures. Infection growth medium containing 10% (vol/vol) FBS was added with or without theophylline 2 h p.i. When indicated, theophylline was added at later time points. For long-term experiments, 80% of medium was changed every 2 d. Infected cells were harvested or stained at indicated time points followed by sample preparation for subsequent analysis.
Immunoblot.
For analysis of adenoviral E1A protein expression, infected cells were lysed in 100 µL radioimmunoprecipitation assay (RIPA) lysis buffer. Indicated amounts of total lysates were analyzed by immunoblot. E1A expression was determined by probing the membrane with a monoclonal antibody specific for adenoviral E1A protein (sc-25, clone M73; Santa Cruz Biotechnology).
For analysis of MV F protein expression, infected cells were lysed in 100 µL RIPA lysis buffer. Total lysates (40 µg) were analyzed by immunoblot. F expression was determined by probing the membrane with an anti-F antibody. As internal infection control, N expression was determined by probing the membrane with an anti-N antibody (32) (kind gifts from R. Cattaneo, Mayo Clinic, Rochester, MN).
Generally, equal loading was confirmed by using a mouse monoclonal antibody specific for human β-actin (C4; MP Biomedical). Antibody binding was visualized using chemiluminescence (Pierce ECL, Thermo Fisher Scientific).
Quantitative PCR.
For analysis of adenoviral genome copy numbers, DNA was purified using the QIAamp Blood Mini kit (Qiagen) following the manufacturer’s instructions. Viral genome copy numbers were determined by quantitative real-time PCR as previously described (17) using the 7300 Real Time PCR system (Applied Biosystems). Data were analyzed using the 7300 System SDS software (Applied Biosystems).
Infectious Progeny Production.
To analyze the production of progeny viruses upon adenoviral infection, burst assays were performed. Infected cells were harvested with supernatant and after three freeze/thaw cycles, infectious viral particles were quantified by titration for infectious progenies in HEK293 cells. As internal control, infected cells were harvested 1 h p.i. and processed accordingly.
For analysis of infectious progeny production upon MV infection, burst assays were performed with one freeze/thaw cycle similar to the one for adenoviral infections described above. Here, harvested cells were titrated on Vero cells for produced infectious particles.
Cytotoxicity Assay.
Virus spread-dependent cytotoxicity was determined when cell lysis was visible at the lowest virus dilution using crystal violet staining as previously described (17).
Fusion Assay.
To confirm functionality of the cloned pCG-F variants, fusion assays were performed. SK-MEL-28 cells were cotransfected with respective pCG-F variants, pCG-H and pEGFP-N1 (Clontech). As negative control, cells were transfected without pCG-F. Transfected cells were subsequently cultured in the absence or presence of 3 mM theophylline. Phase contrast and fluorescence photos were taken 50 h posttransfection and merged (BIOREVO BZ-9000E; Keyence).
Statistical Analysis.
All statistical analyses were performed of logarithmized data using GraphPad Prism 5.00 software (GraphPad Software). Differences between indicated groups were analyzed using two-way ANOVA analysis of variance with Bonferroni posttest. Values of *P < 0.05, **P < 0.01, ***P < 0.001 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Helmholtz Association (Helmholtz University Group Grant VH-NG-212) (to D.M.N.), the Peter und Traudl Engelhorn Stiftung (PhD fellowship) (to P.K.), the German Cancer Aid (Max Eder Research program, Grant 108307) (to G.U.), and the VolkswagenStiftung (Lichtenberg Professorship) (to J.S.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318563111/-/DCSupplemental.
References
- 1.Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat Biotechnol. 2012;30(7):658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14(11):1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 3.Kelly EJ, Nace R, Barber GN, Russell SJ. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J Virol. 2010;84(3):1550–1562. doi: 10.1128/JVI.01788-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattaneo R, Miest T, Shashkova EV, Barry MA. Reprogrammed viruses as cancer therapeutics: Targeted, armed and shielded. Nat Rev Microbiol. 2008;6(7):529–540. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiocca EA. The host response to cancer virotherapy. Curr Opin Mol Ther. 2008;10(1):38–45. [PubMed] [Google Scholar]
- 6.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4(6):453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 7.Wieland M, Hartig JS. Artificial riboswitches: Synthetic mRNA-based regulators of gene expression. ChemBioChem. 2008;9(12):1873–1878. doi: 10.1002/cbic.200800154. [DOI] [PubMed] [Google Scholar]
- 8.Vinkenborg JL, Karnowski N, Famulok M. Aptamers for allosteric regulation. Nat Chem Biol. 2011;7(8):519–527. doi: 10.1038/nchembio.609. [DOI] [PubMed] [Google Scholar]
- 9.Chang AL, Wolf JJ, Smolke CD. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotechnol. 2012;23(5):679–688. doi: 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link KH, Breaker RR. Engineering ligand-responsive gene-control elements: Lessons learned from natural riboswitches. Gene Ther. 2009;16(10):1189–1201. doi: 10.1038/gt.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toniatti C, Bujard H, Cortese R, Ciliberto G. Gene therapy progress and prospects: Transcription regulatory systems. Gene Ther. 2004;11(8):649–657. doi: 10.1038/sj.gt.3302251. [DOI] [PubMed] [Google Scholar]
- 12.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96(7):3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew Chem Int Ed Engl. 2009;48(15):2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 14.Rohmer S, Mainka A, Knippertz I, Hesse A, Nettelbeck DM. Insulated hsp70B′ promoter: Stringent heat-inducible activity in replication-deficient, but not replication-competent adenoviruses. J Gene Med. 2008;10(4):340–354. doi: 10.1002/jgm.1157. [DOI] [PubMed] [Google Scholar]
- 15.Fecker LF, et al. Efficient and selective tumor cell lysis and induction of apoptosis in melanoma cells by a conditional replication-competent CD95L adenovirus. Exp Dermatol. 2010;19(8):e56–e66. doi: 10.1111/j.1600-0625.2009.00977.x. [DOI] [PubMed] [Google Scholar]
- 16.Fecker LF, et al. Efficient melanoma cell killing and reduced melanoma growth in mice by a selective replicating adenovirus armed with tumor necrosis factor-related apoptosis-inducing ligand. Hum Gene Ther. 2011;22(4):405–417. doi: 10.1089/hum.2010.108. [DOI] [PubMed] [Google Scholar]
- 17.Ketzer P, Haas SF, Engelhardt S, Hartig JS, Nettelbeck DM. Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res. 2012;40(21):e167. doi: 10.1093/nar/gks734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, White KA. Riboswitching on RNA virus replication. Proc Natl Acad Sci USA. 2007;104(25):10406–10411. doi: 10.1073/pnas.0704178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl. 2008;47(14):2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 20.Link KH, et al. Engineering high-speed allosteric hammerhead ribozymes. Biol Chem. 2007;388(8):779–786. doi: 10.1515/BC.2007.105. [DOI] [PubMed] [Google Scholar]
- 21.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA. 2007;104(36):14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittmann A, Suess B. Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol Biosyst. 2011;7(8):2419–2427. doi: 10.1039/c1mb05070b. [DOI] [PubMed] [Google Scholar]
- 23.Ausländer S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst. 2010;6(5):807–814. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 24.Wieland M, Ausländer D, Fussenegger M. Engineering of ribozyme-based riboswitches for mammalian cells. Methods. 2012;56(3):351–357. doi: 10.1016/j.ymeth.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci USA. 2010;107(19):8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura Y, Kumar D, Yokobayashi Y. Synthetic mammalian riboswitches based on guanine aptazyme. Chem Commun (Camb) 2012;48(57):7215–7217. doi: 10.1039/c2cc33140c. [DOI] [PubMed] [Google Scholar]
- 27.Berk A. Adenoviridea: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 2355–2394. [Google Scholar]
- 28.Heise C, et al. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med. 2000;6(10):1134–1139. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- 29.Fueyo J, et al. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene. 2000;19(1):2–12. doi: 10.1038/sj.onc.1203251. [DOI] [PubMed] [Google Scholar]
- 30.Springfeld C, et al. Oncolytic efficacy and enhanced safety of measles virus activated by tumor-secreted matrix metalloproteinases. Cancer Res. 2006;66(15):7694–7700. doi: 10.1158/0008-5472.CAN-06-0538. [DOI] [PubMed] [Google Scholar]
- 31.Radecke F, et al. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14(23):5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leber MF, et al. MicroRNA-sensitive oncolytic measles viruses for cancer-specific vector tropism. Mol Ther. 2011;19(6):1097–1106. doi: 10.1038/mt.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu H, Furukawa K, Breaker RR. Engineered allosteric ribozymes that sense the bacterial second messenger cyclic diguanosyl 5′-monophosphate. Anal Chem. 2012;84(11):4935–4941. doi: 10.1021/ac300415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012;2012:748913. doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107(9):3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 36.Nomura Y, Zhou L, Miu A, Yokobayashi Y. Controlling mammalian gene expression by allosteric Hepatitis Delta Virus Ribozymes. ACS Synth Biol. 2013;2(12):684–689. doi: 10.1021/sb400037a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckstein A, et al. Inhibition of adenovirus infections by siRNA-mediated silencing of early and late adenoviral gene functions. Antiviral Res. 2010;88(1):86–94. doi: 10.1016/j.antiviral.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Kneidinger D, Ibrišimović M, Lion T, Klein R. Inhibition of adenovirus multiplication by short interfering RNAs directly or indirectly targeting the viral DNA replication machinery. Antiviral Res. 2012;94(3):195–207. doi: 10.1016/j.antiviral.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otaki M, Sada K, Kadoya H, Nagano-Fujii M, Hotta H. Inhibition of measles virus and subacute sclerosing panencephalitis virus by RNA interference. Antiviral Res. 2006;70(3):105–111. doi: 10.1016/j.antiviral.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuter T, Weissbrich B, Schneider-Schaulies S, Schneider-Schaulies J. RNA interference with measles virus N, P, and L mRNAs efficiently prevents and with matrix protein mRNA enhances viral transcription. J Virol. 2006;80(12):5951–5957. doi: 10.1128/JVI.02453-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: Strike and counterstrike. Nat Biotechnol. 2007;25(12):1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willmon C, et al. Cell carriers for oncolytic viruses: Fed Ex for cancer therapy. Mol Ther. 2009;17(10):1667–1676. doi: 10.1038/mt.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen L, et al. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431(7007):471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki K, Alemany R, Yamamoto M, Curiel DT. The presence of the adenovirus E3 region improves the oncolytic potency of conditionally replicative adenoviruses. Clin Cancer Res. 2002;8(11):3348–3359. [PubMed] [Google Scholar]
- 45.He TC, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95(5):2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devaux P, Hodge G, McChesney MB, Cattaneo R. Attenuation of V- or C-defective measles viruses: Infection control by the inflammatory and interferon responses of rhesus monkeys. J Virol. 2008;82(11):5359–5367. doi: 10.1128/JVI.00169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin A, Staeheli P, Schneider U. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J Virol. 2006;80(12):5708–5715. doi: 10.1128/JVI.02389-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaufmann JK, et al. Chemovirotherapy of malignant melanoma with a targeted and armed oncolytic measles virus. J Invest Dermatol. 2013;133(4):1034–1042. doi: 10.1038/jid.2012.459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.