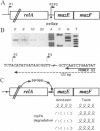
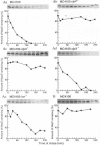
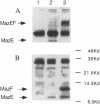
Abstract
"Addiction modules" consist of two genes. In most of them the product of one is long lived and toxic while the product of the second is short lived and antagonizes the toxic effect; so far, they have been described mainly in a number of prokaryotic extrachromosomal elements responsible for the postsegregational killing effect. Here we show that the chromosomal genes mazE and mazF, located in the Escherichia coli rel operon, have all of the properties required for an addiction module. Furthermore, the expression of mazEF is regulated by the cellular level of guanosine [corrected] 3',5'-bispyrophosphate, the product of the RelA protein under amino acid starvation. These properties suggest that the mazEF system may be responsible for programmed cell death in E. coli and thus may have a role in the physiology of starvation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv M., Giladi H., Schreiber G., Oppenheim A. B., Glaser G. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol Microbiol. 1994 Dec;14(5):1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- Bejarano I., Klemes Y., Schoulaker-Schwarz R., Engelberg-Kulka H. Energy-dependent degradation of lambda O protein in Escherichia coli. J Bacteriol. 1993 Dec;175(23):7720–7723. doi: 10.1128/jb.175.23.7720-7723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar I., Miller C., Engelberg-Kulka H. Frameshifting in the expression of the Escherichia coli trpR gene. Mol Microbiol. 1992 Oct;6(19):2777–2784. doi: 10.1111/j.1365-2958.1992.tb01457.x. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Bech F. W., Jørgensen S. T., Løbner-Olesen A., Rasmussen P. B., Atlung T., Boe L., Karlstrom O., Molin S., von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986 Aug;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Nielsen A., Thorsted P., Wagner E. G. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J Mol Biol. 1992 Aug 5;226(3):637–649. doi: 10.1016/0022-2836(92)90621-p. [DOI] [PubMed] [Google Scholar]
- Geuskens V., Mhammedi-Alaoui A., Desmet L., Toussaint A. Virulence in bacteriophage Mu: a case of trans-dominant proteolysis by the Escherichia coli Clp serine protease. EMBO J. 1992 Dec;11(13):5121–5127. doi: 10.1002/j.1460-2075.1992.tb05619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E. I., Panzer H. A. The F factor of Escherichia coli carries a locus of stable plasmid inheritance stm, similar to the parB locus of plasmid RI. Mol Gen Genet. 1988 Oct;214(2):353–357. doi: 10.1007/BF00337735. [DOI] [PubMed] [Google Scholar]
- Gottesman M. E., Adhya S., Das A. Transcription antitermination by bacteriophage lambda N gene product. J Mol Biol. 1980 Jun 15;140(1):57–75. doi: 10.1016/0022-2836(80)90356-3. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Clark W. P., de Crecy-Lagard V., Maurizi M. R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993 Oct 25;268(30):22618–22626. [PubMed] [Google Scholar]
- Gottesman S., Maurizi M. R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992 Dec;56(4):592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Minimizing proteolysis in Escherichia coli: genetic solutions. Methods Enzymol. 1990;185:119–129. doi: 10.1016/0076-6879(90)85013-e. [DOI] [PubMed] [Google Scholar]
- Jaffé A., Ogura T., Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985 Sep;163(3):841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Gottesman S., Pumphrey J., Rudikoff S., Clark W. P., Maurizi M. R. The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988 Oct 15;263(29):15226–15236. [PubMed] [Google Scholar]
- Kroh H. E., Simon L. D. The ClpP component of Clp protease is the sigma 32-dependent heat shock protein F21.5. J Bacteriol. 1990 Oct;172(10):6026–6034. doi: 10.1128/jb.172.10.6026-6034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnherr H., Maguin E., Jafri S., Yarmolinsky M. B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol. 1993 Oct 5;233(3):414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- Lehnherr H., Yarmolinsky M. B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y., Miyakawa K., Nishimura Y., Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993 Nov;175(21):6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi M. R., Clark W. P., Kim S. H., Gottesman S. Clp P represents a unique family of serine proteases. J Biol Chem. 1990 Jul 25;265(21):12546–12552. [PubMed] [Google Scholar]
- Metzger S., Dror I. B., Aizenman E., Schreiber G., Toone M., Friesen J. D., Cashel M., Glaser G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988 Oct 25;263(30):15699–15704. [PubMed] [Google Scholar]
- Metzger S., Schreiber G., Aizenman E., Cashel M., Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989 Dec 15;264(35):21146–21152. [PubMed] [Google Scholar]
- Roberts R. C., Ström A. R., Helinski D. R. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J Mol Biol. 1994 Mar 18;237(1):35–51. doi: 10.1006/jmbi.1994.1207. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarría M. J., de Torrontegui G., Giménez-Gallego G., Díaz-Orejas R. Structural and functional comparison between the stability systems ParD of plasmid R1 and Ccd of plasmid F. Mol Gen Genet. 1991 Mar;225(3):355–362. doi: 10.1007/BF00261674. [DOI] [PubMed] [Google Scholar]
- Schoulaker-Schwarz R., Dekel-Gorodetsky L., Engelberg-Kulka H. An additional function for bacteriophage lambda rex: the rexB product prevents degradation of the lambda O protein. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4996–5000. doi: 10.1073/pnas.88.11.4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G., Metzger S., Aizenman E., Roza S., Cashel M., Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991 Feb 25;266(6):3760–3767. [PubMed] [Google Scholar]
- Schreiber G., Ron E. Z., Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr Microbiol. 1995 Jan;30(1):27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- Svitil A. L., Cashel M., Zyskind J. W. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993 Feb 5;268(4):2307–2311. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisted T., Nielsen A. K., Gerdes K. Mechanism of post-segregational killing: translation of Hok, SrnB and Pnd mRNAs of plasmids R1, F and R483 is activated by 3'-end processing. EMBO J. 1994 Apr 15;13(8):1950–1959. doi: 10.1002/j.1460-2075.1994.tb06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S., Nishimura Y., Ohtsubo E. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol. 1992 Jul;174(13):4205–4211. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S., Ohtsubo H., Ohtsubo E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988 Apr;170(4):1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L., Bernard P., Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol. 1994 Mar;11(6):1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Wojtkowiak D., Georgopoulos C., Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993 Oct 25;268(30):22609–22617. [PubMed] [Google Scholar]
- Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991 Mar 25;266(9):5980–5990. [PubMed] [Google Scholar]
- Yarmolinsky M. B. Programmed cell death in bacterial populations. Science. 1995 Feb 10;267(5199):836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]