Significance
Kinesins are a superfamily of ATP-dependent motors that are important for a wide variety of microtubule-based functions in eukaryotic cells. Kinesins have evolved to allow variable tuning of their motor properties, but the link between molecular variation and motor function is largely unknown. To understand this link, we have studied an essential mitotic kinesin, kinesin-5, which is the target of anticancer drugs. We used cryo-electron microscopy to visualize directly sequential conformational changes of structural elements during the motor ATPase cycle. We have identified the contribution of kinesin-5–specific variations to motor function indicating that kinesins indeed are precisely tuned according to cellular function. This insight will be important in designing kinesin-specific inhibitors in different disease contexts.
Keywords: molecular motors, macromolecular assemblies, mitosis, cancer
Abstract
Kinesins are responsible for a wide variety of microtubule-based, ATP-dependent functions. Their motor domain drives these activities, but the molecular adaptations that specify these diverse and essential cellular activities are poorly understood. It has been assumed that the first identified kinesin—the transport motor kinesin-1—is the mechanistic paradigm for the entire superfamily, but accumulating evidence suggests otherwise. To address the deficits in our understanding of the molecular basis of functional divergence within the kinesin superfamily, we studied kinesin-5s, which are essential mitotic motors whose inhibition blocks cell division. Using cryo-electron microscopy and determination of structure at subnanometer resolution, we have visualized conformations of microtubule-bound human kinesin-5 motor domain at successive steps in its ATPase cycle. After ATP hydrolysis, nucleotide-dependent conformational changes in the active site are allosterically propagated into rotations of the motor domain and uncurling of the drug-binding loop L5. In addition, the mechanical neck-linker element that is crucial for motor stepping undergoes discrete, ordered displacements. We also observed large reorientations of the motor N terminus that indicate its importance for kinesin-5 function through control of neck-linker conformation. A kinesin-5 mutant lacking this N terminus is enzymatically active, and ATP-dependent neck-linker movement and motility are defective, although not ablated. All these aspects of kinesin-5 mechanochemistry are distinct from kinesin-1. Our findings directly demonstrate the regulatory role of the kinesin-5 N terminus in collaboration with the motor’s structured neck-linker and highlight the multiple adaptations within kinesin motor domains that tune their mechanochemistries according to distinct functional requirements.
Nucleotide triphosphates are the fuel that powers the cell’s machinery. Conversion of this fuel into mechanical work, i.e., mechanochemistry, depends on individual machines and the functional context in which they have evolved. Indeed, elucidation of the mechanochemistry of a particular machine provides critical insight into both its functions and modes of regulation. Kinesins are a superfamily of motors that use ATP to undertake microtubule (MT)-based work. Kinesins operate throughout the cell cycle in many contexts and can generate force toward the MT plus or minus end and also depolymerize MTs (1). The kinesin mechanochemical engine—the motor domain (MD)—is highly conserved, and conformational changes in the active site during the motor’s ATPase cycle are transmitted to other parts of the MD to generate force (2). Most of our current knowledge about kinesin mechanochemistry comes from studies of the superfamily founding member, the transport motor kinesin-1 (K1) (2). However, accumulating evidence suggests that small modifications within kinesin MDs have profound effects on their cellular function. The molecular basis of such adaptations is largely unknown.
Our interests focus on kinesin-5s (K5s), which cross-link MTs and are essential for the formation and maintenance of bipolar mitotic spindles in most eukaryotes (3). Like K1s, K5s have an N-terminal MD followed by a neck-linker (NL) that connects the MD to the rest of the motor. However, K1s are dimeric, whereas K5 tetramerization is mediated by the coiled-coil that follows the K5 NL and gives rise to a dumbbell-shaped molecule with pairs of MDs at either end (3, 4). However, it is not simply the K5 tetrameric structure, with each MD pair taking ATP-driven steps toward the MT plus-ends (5), that is required for its mitotic functions: K5 MDs have specific mechanochemical properties that are matched to their spindle functions (6). Typically, K5s are slow motors thought to work in teams when cross-linking and sliding MTs (5). The MT-stimulated ATPase turnover of monomeric K5 is slower than that of K1 (5–9/s vs. ∼50/s) (7–9), with phosphate release being the rate-limiting step (6–8/s) (10). K5 dimer stepping is concomitantly slower than that of K1 (∼100 nm/s vs. ∼650 nm/s unloaded velocity) and has a different response to external load (11, 12).
A complete molecular explanation for these divergent properties remains elusive. Although it is well established that ATP binding induces NL docking along the MD toward the MT plus end in plus-end kinesins, different physical properties of the NL and variations in nucleotide-dependent conformational changes affect modes of force generation (3). Recently, the collaborative role of the K1 MD N terminus and, in particular, its proximal portion, called the “cover strand,” has emerged. The cover strand forms a short β-sheet, the cover-neck bundle (CNB) with the proximal section of the NL to assist docking and thus force production (13, 14). The K5 N terminus is longer than the N terminus of K1s (19 vs. 9 residues), but we recently described K5 CNB formation on ATP (AMPPNP) binding (15), and a role for the K5 CNB in force generation was inferred indirectly using a K1/K5 chimera (16).
Fundamental questions remain concerning the fate of the CNB throughout the kinesin ATPase cycle and more generally about how information is conveyed from the MD catalytic center to the site of force generation at the CNB. Deciphering the K5-specific molecular mechanism also is of great interest because vertebrate K5-specific inhibitors bind to the highly variable loop L5 that regulates the K5 ATPase cycle and NL movement (17–19). These inhibitors allosterically block ATPase activity and directional movement (20, 21), arrest mitosis (22), and currently are being investigated in cancer clinical trials (3). Using cryo-electron microscopy (cryo-EM), we have addressed deficiencies in our understanding of kinesin mechanochemistry by directly visualizing MT-bound K5 in its ATP hydrolysis transition and ADP-bound states. Combined with our previous characterization of the ATP-binding step of MT-bound K5 (15), we provide a comprehensive picture of the molecular mechanism of K5 throughout its ATPase cycle and highlight critical molecular adaptations in comparison with other kinesins, particularly K1s. Unexpectedly, we demonstrate a role for the N terminus beyond force generation in controlling the NL conformation of K5s throughout their ATPase cycle. We also identify the role of the short helix α0 proximal to the active site in conveying conformational changes to the N terminus. Using biophysical methods, we directly demonstrate the kinetic role of the N terminus in assisting NL movement and hence in supporting efficient K5 activity. We also highlight intrinsic differences in the NL of K5, as compared with K1, that contribute to the very different mechanochemistries of these functionally divergent motors.
Results and Discussion
To obtain a comprehensive description of the conformational changes of the MT-bound human K5 MD during its ATPase cycle, we used cryo-EM and image processing to calculate 3D reconstructions of the ATP hydrolysis transition state (with ADP aluminum fluoride, ADP.AlFx) and the ADP-bound state at 9.2-Å and 10-Å resolution, respectively (Table S1 and Fig. S1). In interpreting these structure, we (i) identified and tracked critical elements in the K5 MD by localization in 3D reconstructions and difference maps of a covalently bound undecagold label at each of three engineered cysteines at the N terminus (A9C), L5 (T126C), and NL (V365C) (Figs. 1A and 2A, Fig. S2, and Table S1) and (ii) generated pseudoatomic models for each nucleotide state validated by quantitative docking and consistency with the positions of the gold densities (Figs. 1B and 2B and Table S2). In each case, the asymmetric unit of these reconstructions, a K5 MD–αβ-tubulin dimer, is the basis of our description. This global analysis of conformational changes combines our current studies with previous work characterizing the ATP-binding step (15).
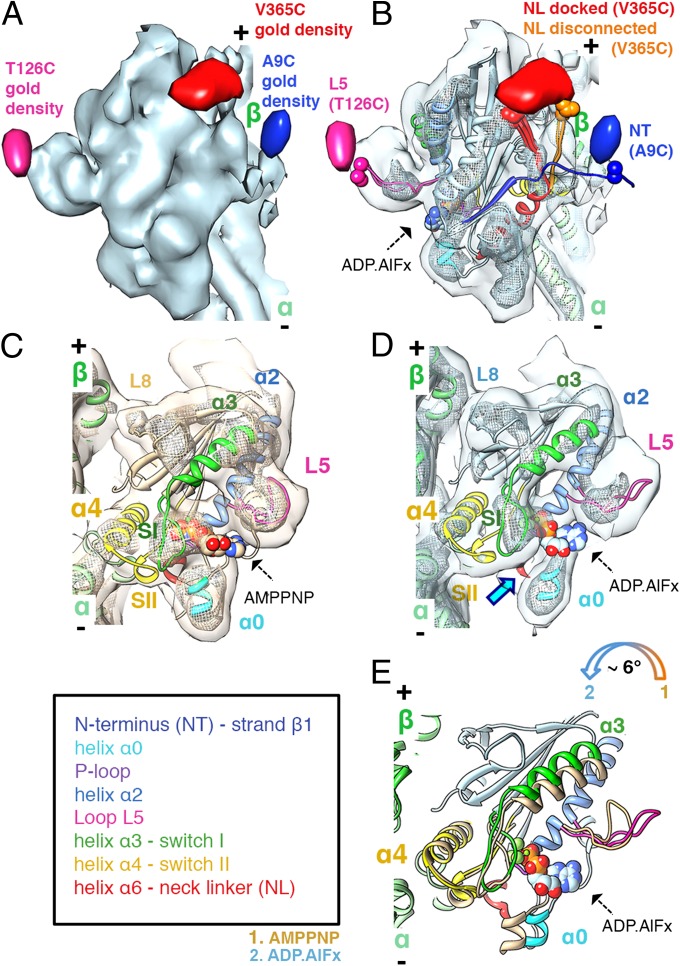
Fig. 1.
The ATP hydrolysis transition state: MD tilting, L5 uncurling, and CNB destabilization. (A) MT-bound K5 MD in the ADP.AlFx state (light blue surface: 1.1σ contour) with gold difference maps superimposed (blue: 2.3σ; pink: 2.2σ; red: 2.2σ contour). In all figures, the MT plus end is towards the top. (B) Pseudo-atomic models of K5 MD (color-coded as in the key) bound to an αβ-tubulin dimer (green ribbon) docked into the reconstruction (contour as in A; mesh: 3.3σ contour, ∼1/3 of the volume). Residues to which gold clusters were covalently attached and the nucleotide analogue are shown in space-filling representation. NL conformers clustered into docked (red) and disconnected (orange) conformations are indicated (SI Materials and Methods). (C and D) View towards the nucleotide-binding pocket of the AMPPNP (15) (C: coral surface: 1σ contour, mesh: 3σ contour, to depict the same molecular volumes as in B) and ADP.AlFx (D: contoured as in B). The blue arrow indicates the absence of contact between helix α0 and switch I in the ADP.AlFx state, which is present in the AMPPNP state. (E) Comparison of K5 MD models of ADP.AlFx (colored as in the key) and AMPPNP states [coral (15)], superimposed on helices α4. ATP hydrolysis triggers a tilt of the MD towards the MT lattice (colored arrow). L5 uncurls from its arch-like conformation upon ATP hydrolysis (comparison based on superimposed helices α2). Displacements of helix α3/switch I and helix α0 are caused by MD tilting.
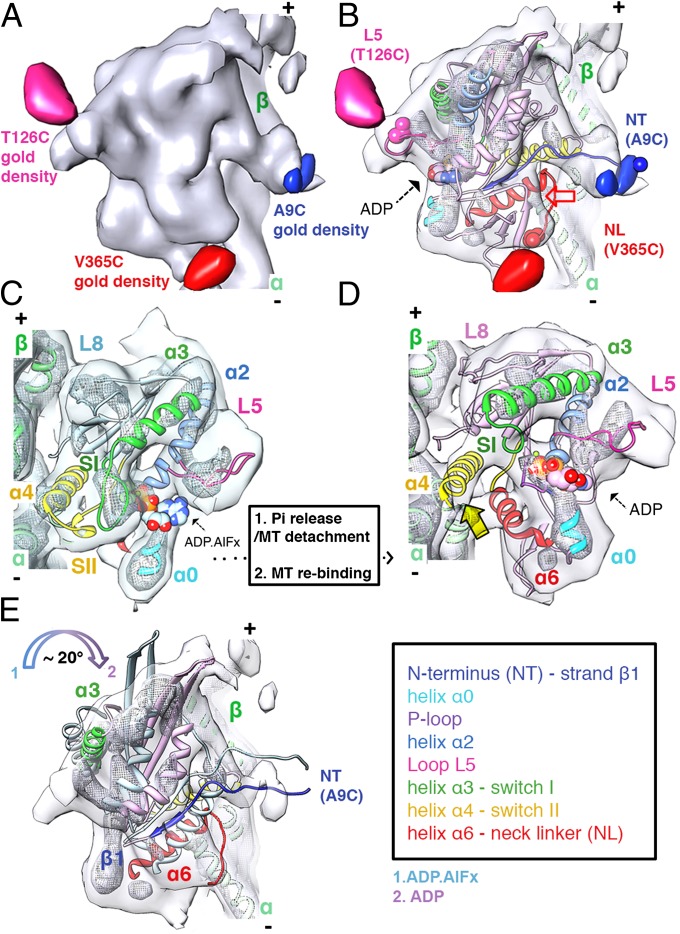
Fig. 2.
The ADP MT weak-binding state. (A) ADP state (1.5σ contour, the same volume as in Fig. 1A) with gold density difference maps superimposed (blue: 1.5σ; pink: 2.5σ; red: 1.9σ contours). (B) Pseudo-atomic model of K5 MD bound to an αβ-tubulin dimer (surface contoured as in A; mesh 3σ contour 1/3 of the surface volume). The red arrow indicates the absence of EM density accommodating the N-terminal part of the NL and the C-terminal part of helix α6 to which it is attached. (C and D) Comparison of K5 MD pseudoatomic models of the ADP.AlFx (C) and ADP (D) states; surfaces and meshes are contoured as in Fig. 1C (C, ADP.AlFx) and Fig. 1B (D, ADP). The yellow arrow indicates the lack of density for switch II that therefore is absent in our ADP model. (E) The ADP MT weakly bound state is rotated clockwise with respect to the MT axis compared with the ADP.AlFx state (color-coded arrows). The surface is contoured at 2σ; for clarity, only some of the secondary structural elements are shown.
The ATP Hydrolysis Transition State.
The triangular K5 MD bound to ADP.AlFx mimicking the ATPase hydrolysis transition state points toward the MT plus end (Fig. 1). The main MT-binding contacts are formed by helix α4 and loop L8 (Fig. 1 C–E), as is consistent with a tight binding state and previously shown for K1 (23, 24). The curved density protruding above the active site is L5 (Fig. 1 A, B, and D; gold density on T126C is shown in pink), whereas the active site itself is occupied and compact (Fig. 1D). The N terminus extends perpendicularly to the MD long axis, confirmed by the A9C-labeled gold density (Fig. 1 A and B), and has weaker EM density than the rest of the MD, likely reflecting its flexibility and disorder in this nucleotide state. Its unstructured conformation has never been captured in crystal structures, so we generated a model (Asn6–Gly16) that gives the best fit to the EM density and satisfies the gold density constraint (SI Materials and Methods).
Although the NL is directed toward the MT plus end, it adopts two discrete conformations. The first, in which the NL is docked along the MD, is essentially the same as that seen in AMPPNP (15, 25); the second is slightly disconnected from the MD (Fig. 1B). In support of these discrete NL conformations, the elongated density corresponding to the gold label on V365C stretches between the MD and the disconnected density (Fig. 1 A and B). The relative occupancy of the disconnected NL conformation was estimated at 0.6 (SI Materials and Methods). Using biophysical methods, we previously concluded that ATP binding produced NL docking and that ATP hydrolysis leads to further redistribution of NL orientations (15); our ADP.AlFx reconstruction is consistent with these data. Modeling of the NL conformation generated several conformational clusters, the top-ranked of which corresponds to the NL-docked conformation and the second to the disconnected conformation (Fig. 1B, Fig. S3, and SI Materials and Methods). Based on these NL models, we estimated that the NL can shift ∼13 Å at this early stage of ATP hydrolysis (Fig. 1B).
L5 Uncurling and Initiation of NL Release as ATP Hydrolysis Begins.
To determine conformational changes that occur as ATP hydrolysis proceeds, we compared the ADP.AlFx K5 conformation with our previous AMPPNP-bound structure (Fig. 1 C–E). At the resolution of our reconstructions, any differences between these structures in the nucleotide-binding loops are subtle (Fig. 1 C–E and Movie S1). Even so, these small changes are transmitted from the active site and are amplified in the rest of the MD. First, in agreement with previous FRET experiments (8), a slight tilt of the MD (∼6°) toward the MT lattice is seen in the ADP.AlFx structure (Fig. 1E and Fig. S4). This tilt of the MD occurs around the static helix α4 contact with the MT, and small changes also are observed in secondary structures adjacent to the active site, including helices α3 and α0 (Fig. 1 C–E). In the AMPPNP reconstruction, density connects helix α0 and the switch I loop, a connection that is broken in the ADP.AlFx reconstruction (Fig. 1D). In the MD primary structure, helix α0 is preceded by the N terminus via strand β1 (26) and offers a direct route for structural communication from the nucleotide-binding site to the motor N terminus. Allosteric structural communication from the active site also causes larger conformational changes in the elements that regulate the motor (i.e., L5) and control force generation (i.e., the NL and the N terminus), which together form the CNB (15). As ATP hydrolysis is initiated, L5 moves away from the MD surface and uncurls from its arch-like structure (Fig. 1 C–E and Movie S1). At the CNB, the K5 N terminus moves ∼20 Å from its plus end–pointing location upon ATP hydrolysis, as is consistent with previous FRET data (Fig. 1B and Fig. S5) (15); this movement coincides with loosening between the NL and the body of the MD, thereby disrupting the CNB. Nevertheless, the remaining N terminus–NL interaction is sufficient to limit major NL reorientation (Fig. 1B).
The ADP State: A View of the Weakly MT-Bound Motor Conformation.
In the K5 ADP state (Fig. 2), the major MT contact points are still helix α4 and loop L8 (Fig. 2 C and D). Although helix α4 is not completely modeled in the available ADP-bound K5 MD crystal structures because of the absence of MTs (26), the length of helix α4 remains constant throughout the K5 MT-bound ATPase cycle, including the ADP state, as previously shown for K1 (24). We thus modeled a full-length helix α4 (residues 279–304) in our docked K5 MD coordinates (Fig. 2C). In the active site, however, there is no obvious density corresponding to the preceding switch II loop, suggesting that it is flexible and is not involved in coordinating the bound ADP (Fig. 2C). Additionally, helix α3 is elongated, causing retraction of the switch I loop (Fig. 2C). This five-turn conformation of helix α3 also is seen in ADP-bound K5 MD crystal structures but not in any other MT-bound K5 state. Together, these two features of the ADP reconstruction—the disorder of switch II and the shortening of switch I—contribute to an open conformation of the active site. Above the active site, the flattened density corresponding to L5 (gold density on T126C, shown in pink) protrudes from the MD, its tip unconnected to helix α3 (Fig. 2 B and C).
The K5 N terminus is perpendicular to the MD long axis in the ADP reconstruction (Fig. 2 A and B). This conformation appears to be stable, as judged by its strong, long density that is still visible at higher density thresholds (Fig. 2 B and D). Intriguingly, the conformation of the 19-amino acid K5 N terminus has not been seen in any crystal structures, many of which contain ADP (26), likely because of its flexibility in solution. The stabilization of the K5-ADP N terminus on MT association probably is caused by stabilization of helix α4, appropriate conformations of L5 and helix α0, and also electrostatic attraction between the negatively charged MT surface and the basic N terminus (theoretical pI = 9.52). In contrast the NL, which is directed toward the MT minus end, and the C terminus of helix α6, to which it is connected, are not well defined (Fig. 2B), suggesting some flexibility. However, NL-attached gold density is readily visualized, suggesting that NL flexibility is only partial.
Flexibility of switch II, shortening of switch I, and flexibility at the C terminus of helix α6 (Fig. 2 B and C) could loosen the helix α4–MT interaction and thus provide a structural explanation for low MT affinity in all kinesins in the ADP state (7, 23). However, it is not immediately obvious whether our ADP reconstruction captures the motor just before MT detachment [and immediately after Pi release, the rate-limiting step of the K5 monomer ATPase cycle (7)] or just after it reassociates with the MT. Although the loosening of the MD–MT contacts described above could be a feature of the posthydrolysis MD conformation that leads to MT release, the large conformational differences between the K5 MD in the ADP and ADP.AlFx reconstructions help explain the relationship of these two structures. First, the MD in the ADP state is rotated clockwise by ∼20° (Fig. 2D and Fig. S4); second, this rotation accompanies a major reorientation of the NL from a plus end- to a minus end-directed position. However, release of the NL away from its plus end-directed position after power stroke conformation must occur in solution: Were such a conformational change to occur before MT release, the plus end-directed force generation associated with ATP-induced NL docking would be reversed in a futile conformational cycle. This reasoning supports the conclusion that our ADP model represents the transient complex formed by the K5 ADP MD as it binds MTs from solution, before ADP release, as discussed below (8). This idea is consistent with the conformation of the ADP-bound K5 MD crystal structure (26), i.e., the conformation of the MD before MT reattachment, in which the NL also points toward the MT minus end.
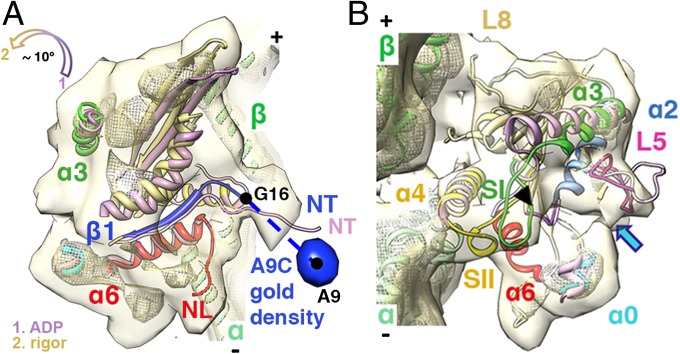
Conformational Changes on ADP Release: ADP to Rigor Transition.
Comparison between our previous rigor reconstruction and our ADP structure allows us to describe conformational changes that accompany the relatively rapid MT-stimulated ADP release step (∼40–75/s) (7, 8). Although the shape of the K5 MD is similar in the ADP and rigor states (Figs. 2B and 3A), the MD is tilted toward the MT lattice by ∼10° after ADP release (Fig. 3A, Fig. S4, and Movie S2). Several other conformational changes occur when ADP is released to reestablish tight MT binding. At the active site, helix α3 uncoils to lengthen switch I, and this lengthening in turn helps stabilize the structured conformation of switch II (Fig. 3B). L5 collapses into the empty active site and, as previously described (15), interacts with this more extended conformation of switch I (Fig. 3B) and also contacts density associated with helix α0 (Fig. 3B). The interaction with helix α0 also provides evidence of transmission of structural information about the absence of bound nucleotide to the N terminus and NL. The overall position of the K5 N terminus is not substantially affected when ADP is released (Fig. 3A). However, although both the N terminus and NL have the same orientation before and after ADP release, the apparent flexibility of each is reversed: In the ADP state, the N terminus is more stable than the NL (Fig. 2B), whereas in rigor, the NL is more stable than the N terminus, as shown by their corresponding strong and weak electron microscopy densities (Fig. 3A). The stabilization of the NL in rigor is related to the reestablishment of complete density corresponding to helix α6. The K5 ADP–rigor transition shows no evidence of the large-scale twisting of the central β-sheet that is observed in the evolutionarily related region of myosin (27). This comparison does not exclude smaller changes in the kinesin β-sheet (e.g., ref. 28) which are beyond the resolution of our current reconstructions.
Fig. 3.
Conformational changes associated with ADP release. (A and B) K5 MD pseudo-atomic models of the ADP (light purple) and rigor states are superimposed on helices α4 and docked into the rigor reconstruction (15); surface: 0.94σ contour; mesh: 3σ contour. (A) The position of the gold density (blue surface, 1.4σ contour) attached to the N terminus via A9C at ∼23 Å or approximately seven amino acids away from the first residue in our rigor model (G16) confirms our previous assignment of the N terminus in the rigor reconstruction (15). The color-coded arrow indicates the direction of the MD tilt around the helix α4 and towards the MT lattice upon ADP release. (B) Upon ADP release, L5 moves towards the empty nucleotide-binding pocket, based on superimposition of helices α2. The black arrowhead and the blue arrow indicate a connection between L5 and switch I (SI) and between L5 and helix α0, respectively. Helix α3/switch I, helix α4, and helix α0 of the ADP model also are shown for comparison.
The K5 N Terminus Is Required for Efficient NL Docking and Motility.
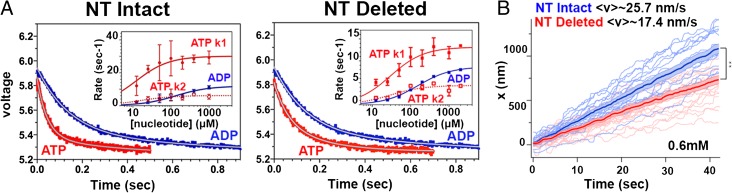
To test the idea that the K5 N terminus plays a coordinating role in the motor’s mechanochemistry, we monitored both the kinetics of ATP-induced NL docking and NL reorientation on ATP hydrolysis using K5 MD constructs labeled with tetramethyl rhodamine maleimide (TMR) and changes in TMR fluorescence (15). The first construct, NT-Intact, described previously (15), is a monomeric K5 MD construct that contains two reactive cysteines at positions 256 in the MD beta core (V256C) and 365 in the NL (V365C),. The second construct, NT-Deleted, is similar except that the first 17 N-terminal residues have been deleted. Their MT-activated steady-state ATPase activity shows that the NT-Deleted construct is characterized by a kcat that is 53% lower (4.9 ± 0.5/s versus 3.2 ± 0.4/s) and a K0.5,MT that is more than fivefold larger (0.81 ± 0.26 µM versus 4.1 ± 1.1 µM) than in NT-Intact (Fig. S6). Mixing TMR-labeled NT-Intact with ATP produces a biphasic decrease in TMR fluorescence; this measurement implies that NL movement occurs in two phases: NL docking when ATP binds followed by NL reorientation on hydrolysis (15). In contrast, mixing with ADP produces a monophasic decrease in fluorescence (Fig. 4A). As expected, the amplitude of the change in fluorescence is the same in both cases, because ATP hydrolysis and Pi release ultimately generate K5-ADP. The rate constants for both phases of the ATP transient vary hyperbolically with [ATP], defining maximum rate constants of 30.1 ± 2.0/s and 6.1 ± 0.6/s (Fig. 4A, Left). The corresponding rate constant for the ADP transient also varies hyperbolically with [ADP], with an extrapolated rate constant of 11.9 ± 1.1/s (Fig. 4A, Left). Analysis of the NT-Deleted construct produced qualitatively similar results (Fig. 4A, Right): ATP binding also produced a biphasic decrease in fluorescence, and the rate constants for both phases also varied with [ATP], defining maximum extrapolated rates of 13.1 ± 0.9 and 4.5 ± 0.4/s, respectively. However, although the rate constant for the slower phase with the NT-Deleted construct is minimally different, that for the faster phase is nearly 130% lower than for NT-Intact. In the case of the ADP transient (Fig. 4A, Right), the rate constant also varies hyperbolically with [ADP], defining a maximum rate of 8.5 ± 0.3/s. These data demonstrate that the K5 N terminus assists ATP-dependent NL movement, so that its deletion perturbs NL structural transitions and presumably force production.
Fig. 4.
Effect of deletion of the K5 N terminus on kinetic parameters and motility. (A) The jagged curves represent the fluorescence transient observed by mixing TMR-labelled NT-Intact MT (Left) and TMR-labelled NT-Deleted:MT (Right) complex with 800 μM ATP or ADP, and the smooth curves are fit to a double (red) or single (blue) exponential rate equation. (Insets) Plots of rate constant versus [ATP] or [ADP]. (B) Individual MT gliding traces for NT-Intact (blue) and NT-Deleted (red) constructs. Each set of traces (n = 17 and 22, respectively) was averaged together pointwise to extract average behavior [bold traces: blue, 25.7 nm/s; red, 17.4 nm/s; error bars (light-colored zones) indicate SEM; **P < 0.001]. Individual frames were 85 ms apart (SI Materials and Methods).
To assess motor functionality directly, we compared the activity of the NT-Intact and NT-Deleted constructs in an in vitro MT gliding assay (Fig. 4B and Movies S3 and S4). Our data using automated MT tracking (SI Materials and Methods and Fig. S7) show that the tracks for the NT-Deleted construct are qualitatively similar to those for the NT-Intact construct, but their average speed is slower (25.7 nm/s for NT-Intact compared with 17.4 nm/s for NT-Deleted, P < 0.001) (Fig. 4B), as is consistent with rates measured for this construct in the steady-state ATPase assay (Fig. S6). Thus, the K5 N terminus is required to assist NL docking and reorientation kinetically and hence to ensure efficient force generation and motility.
Molecular Adaptations of Kinesin MDs: Model of Coordination Between NL and N Terminus for K5 Force Generation and Motility.
The comprehensive visualization of the mechanochemical cycle of the MT-bound human K5 described here reveals remarkable plasticity within the MD and highlights important molecular adaptations of kinesin motors. In particular, our data reveal the extent of conformational coordination between the NL and N terminus that contributes to K5 function (Fig. 5). The NL and N terminus form the CNB in the MT-bound ATP state (13–16), and our current work now shows the surprisingly transient nature of the CNB during the K5 ATPase cycle. As ATP hydrolysis is initiated, subtle, coupled conformational changes of the NL and N terminus occur: The CNB partially melts, thereby destabilizing the plus end-directed conformation of the NL (Fig. 5, step A). Our data show that throughout the ATPase cycle, NL movement can be controlled not only by the large-scale seesaw motion of the entire MD (15, 29) but also by more subtle destabilization of the CNB. The N terminus is structurally linked to the active site by helix α0, suggesting the allosteric route by which the nucleotide state of the K5 MD is communicated to the N terminus. Hence, our ADP.AlFx reconstruction supported by our biophysical measurements provides the first direct evidence that the N terminus plays a key role in K5 mechanochemistry, both by assisting NL docking for force generation (Fig. 5, step D) and by inducing its gradual undocking as the ATPase cycle proceeds.
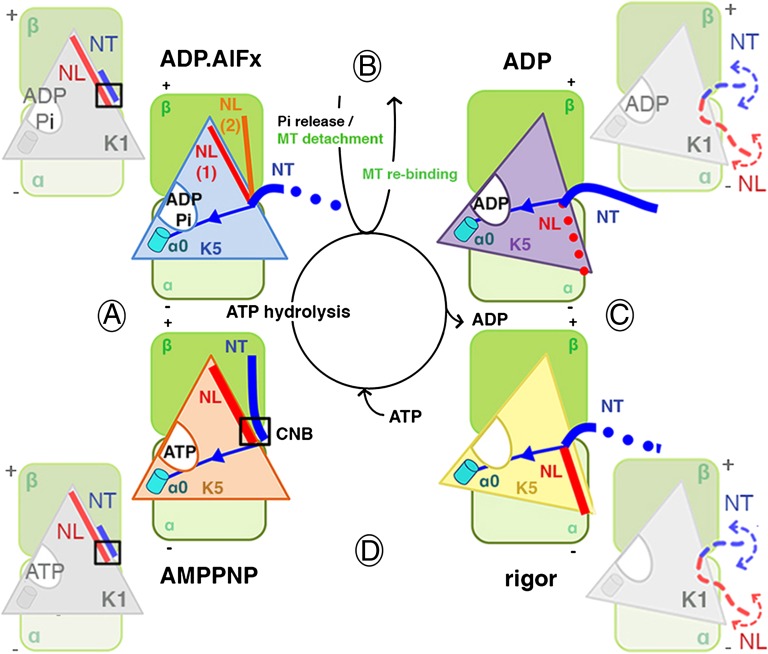
Fig. 5.
Molecular adaptations of kinesin motors: N terminus/NL conformational coupling during the mechanochemical cycle. The central schematic illustrates the MT-based ATPase cycle: ATP hydrolysis (step A), Pi release/MT detachment and MT re-binding (step B), ADP release (step C), and ATP binding (step D). In the corners of the figure, the major nucleotide-dependent conformational changes in the K5 N terminus and NL are depicted. The large seesawing motion of the MD at steps B and D is shown by rotation of the K5 MD on the αβ-tubulin dimer. The proposed route of structural communication between helix α0 proximal to the active site via strand β1 is shown. Stability of the N terminus/NL is depicted as a solid line; flexibility is indicated by a dotted line. CNB formation is indicated by a black square. The outer cycle depicts the conformational changes in the K1 MD (24, 30). In contrast to K5, however, both the K1 N terminus and NL are highly flexible in both the ADP and rigor states. The exact route of structural communication between the K1 helix α0 and N terminus has not been described.
NL reorientation toward the MT minus end is completed in solution following phosphate release and MT detachment (Fig. 5, step B). After NL reorientation and MT rebinding, the K5 N terminus appears quite stable and could block reversion of the NL toward the MT plus end. This stabilization of the K5 N terminus also could be part of the trigger that stimulates ADP release. MT-stimulated ADP release results in tilting of the MD toward the MT to form a high-affinity nucleotide-free complex (Fig. 5, step C). In this rigor state, the NL conformation is visualized more clearly than in the ADP state but remains directed toward the MT minus end. On the other hand, the K5 N terminus is flexible and thus is less well defined in our rigor reconstruction. However, our biophysical analyses of the K5 mutant lacking the N terminus demonstrate that on ATP binding and the accompanying seesawing of the MD (Fig. 5, step D), the N terminus is important for the efficient NL docking that powers K5 motility. In this state, the CNB is reformed, with both the NL and N terminus pointing toward the MT plus end. In summary, the interdependence of NL and N terminus movement and flexibility seen in our reconstructions emphasizes the important regulatory role played by the long K5-specific N terminus in force generation: It assists NL docking when ATP binds (Fig. 5, step D), triggers NL undocking on ATP hydrolysis (Fig. 5, step A), and physically limits search space of the flexible NL in the ADP state, ensuring its minus end-directed conformation (Fig. 5, step B). These movements appear to be controlled, at least in part, by the relay of information from the active site by helix α0. Thus, the NL undergoes a large, ordered movement back and forth along the K5 MD during one ATPase cycle orchestrated by lower-amplitude movement of the N terminus (Fig. 5, compare steps A and D).
Conclusion
Previous subnanometer-resolution cryo-EM reconstructions of the MT-bound K1 MD led to the seesaw model of plus end-directed kinesin mechanochemistry (24), and K1s were the first family in which the importance of CNB formation for force generation was characterized (13, 14). The seesaw-related conformational changes we observe in the K5 MD at the ADP.AlFx-to-ADP transition and at the rigor-to-AMPPNP transition are likely to be shared by all plus end-directed kinesins (29). However, the other conformational changes we have visualized—rolling of the K5 MD, unfurling of L5, and the coordinated motions of the NL and N terminus—are K5 specific. Although the flexibility of the K5 NL varies in different nucleotide states, we have visualized its orientation directly throughout the motor’s ATPase cycle. In contrast, the shorter K1 NL is more flexible and thus is not visualized in rigor and ADP states (24, 30), implying intrinsic differences in the mechanical properties of K1 and K5 NLs (Fig. 5). This idea is supported further by our data showing that, even when K5 CNB formation is not possible in our NT-deleted mutant, NL docking and motility are observed, albeit less efficiently than with the NT-intact MD (Fig. 4). Globally, K1 and K5 mechanochemistries share conserved nucleotide-binding motifs, large seesaw movements, and CNB formation. Recent studies also suggest that K1 and K5 have similar processivities (31). However, many other aspects of their enzymologies and molecular mechanisms are very different: The K1 N terminus is required for force generation, whereas our data suggest that the K5 N terminus plays a regulatory role at multiple steps throughout the motor’s ATPase cycle. Disruption of the CNB as soon as ATP hydrolysis occurs might destabilize K5 dimeric motors on the MT track, possibly explaining their sensitivity to load compared with K1 (11). The K5 N terminus could interact with the C terminus of the full-length protein in the context of the K5 tetramer to control stepping (32) and also could be subject to cell cycle-dependent regulation (Fig. S8). Additional differences elsewhere in the MDs, for example in L5, are allosterically amplified to produce motors with very different mechanical outputs. Drug binding to the K5 L5 likely indirectly blocks movement elsewhere in the MD and hence the motor’s ATPase and directional motility.
Molecular adaptations within the kinesin MD such as those highlighted in our study indicate that the properties of an individual motor cannot be extrapolated directly from those of the so-called K1 paradigm. Such adaptations are likely to operate across the whole superfamily to support functional divergence. Future work will be directed toward elucidating the molecular contribution of these divergent regions of kinesins in diverse functional settings.
Materials and Methods
Samples were prepared for cryo-EM and kinetics/biophysical experiments as described previously (8, 15, 17). Full details of our methods for 3D reconstructions (24, 33) and for atomic model building are provided in SI Materials and Methods. The kinetics of NL movement was measured as previously described (15) using TMR-labeled MD complexes. The gliding assay (34) was done in a flow cell in which K5 constructs were attached via penta-His antibodies. Motion of the fluorescent, rhodamine-labeled MTs (Cytoskeleton Inc.) was monitored using an inverted microscope (TE2000; Nikon). Additional details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Charles Sindelar (Yale University) for reconstruction algorithms; Maya Topf, Elena Orlova, and other members of the Birkbeck electron microscopy group for helpful discussions; and Dan Safer for undecagold. This work was funded by Biotechnology and Biological Sciences Research Council Grant BB/H005137/1 and National Institutes of Health Grants GM102875-07 (to S.S.R.) and GM64624 (to S.P.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The cryo-EM reconstructions reported in this paper have been deposited in the Electron Microscopy Data Bank [accession nos. 2533 (ADPAlFx native), 2534 (ADPAlFx, gold L5), 2535 (ADPAlFx, gold NL), 2536 (ADPAlFx, gold Nter), 2537 (ADP native), 2538 (ADP, gold L5), 2539 (ADP, gold NL), 2540 (ADP, gold Nter), 2541 (AMPPNP, gold Nter), and 2542 (rigor, gold Nter)]. The docked coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [accession nos. 4ck6 (ADPAlFx, docked NL), 4ck7 (ADPAlFx, disconnected NL), and 4ck5 (ADP)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319848111/-/DCSupplemental.
References
- 1.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10(10):682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 2.Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288(5463):88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 3.Goulet A, Moores C. New insights into the mechanism of force generation by kinesin-5 molecular motors. Int Rev Cell Mol Biol. 2013;304:419–466. doi: 10.1016/B978-0-12-407696-9.00008-7. [DOI] [PubMed] [Google Scholar]
- 4.Acar S, et al. The bipolar assembly domain of the mitotic motor kinesin-5. Nat Commun. 2013;4:1343. doi: 10.1038/ncomms2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapitein LC, et al. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 6.Cahu J, Surrey T. Motile microtubule crosslinkers require distinct dynamic properties for correct functioning during spindle organization in Xenopus egg extract. J Cell Sci. 2009;122(Pt 9):1295–1300. doi: 10.1242/jcs.044248. [DOI] [PubMed] [Google Scholar]
- 7.Cochran JC, et al. Mechanistic analysis of the mitotic kinesin Eg5. J Biol Chem. 2004;279(37):38861–38870. doi: 10.1074/jbc.M404203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenfeld SS, Xing J, Jefferson GM, King PH. Docking and rolling, a model of how the mitotic motor Eg5 works. J Biol Chem. 2005;280(42):35684–35695. doi: 10.1074/jbc.M506561200. [DOI] [PubMed] [Google Scholar]
- 9.Cross RA. The kinetic mechanism of kinesin. Trends Biochem Sci. 2004;29(6):301–309. doi: 10.1016/j.tibs.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Cochran JC, Krzysiak TC, Gilbert SP. Pathway of ATP hydrolysis by monomeric kinesin Eg5. Biochemistry. 2006;45(40):12334–12344. doi: 10.1021/bi0608562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8(5):470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Block SM, Asbury CL, Shaevitz JW, Lang MJ. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc Natl Acad Sci USA. 2003;100(5):2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang W, Lang MJ, Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16(1):62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Khalil AS, et al. Kinesin’s cover-neck bundle folds forward to generate force. Proc Natl Acad Sci USA. 2008;105(49):19247–19252. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulet A, et al. The structural basis of force generation by the mitotic motor kinesin-5. J Biol Chem. 2012;287(53):44654–44666. doi: 10.1074/jbc.M112.404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hesse WR, et al. Modular aspects of kinesin force generation machinery. Biophys J. 2013;104(9):1969–1978. doi: 10.1016/j.bpj.2013.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behnke-Parks WM, et al. Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J Biol Chem. 2011;286(7):5242–5253. doi: 10.1074/jbc.M110.192930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waitzman JS, et al. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys J. 2011;101(11):2760–2769. doi: 10.1016/j.bpj.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson AG, Naber N, Cooke R, Pate E, Rice SE. The conserved L5 loop establishes the pre-powerstroke conformation of the Kinesin-5 motor, eg5. Biophys J. 2010;98(11):2619–2627. doi: 10.1016/j.bpj.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cochran JC, Gatial JE, 3rd, Kapoor TM, Gilbert SP. Monastrol inhibition of the mitotic kinesin Eg5. J Biol Chem. 2005;280(13):12658–12667. doi: 10.1074/jbc.M413140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwok BH, et al. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol. 2006;2(9):480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- 22.Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286(5441):971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 23.Rosenfeld SS, Rener B, Correia JJ, Mayo MS, Cheung HC. Equilibrium studies of kinesin-nucleotide intermediates. J Biol Chem. 1996;271(16):9473–9482. doi: 10.1074/jbc.271.16.9473. [DOI] [PubMed] [Google Scholar]
- 24.Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci USA. 2010;107(9):4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parke CL, Wojcik EJ, Kim S, Worthylake DK. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J Biol Chem. 2010;285(8):5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner J, et al. Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J Biol Chem. 2001;276(27):25496–25502. doi: 10.1074/jbc.M100395200. [DOI] [PubMed] [Google Scholar]
- 27.Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126(Pt 1):9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim ED, et al. Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core. J Biol Chem. 2010;285(24):18650–18661. doi: 10.1074/jbc.M109.092072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sindelar CV. A seesaw model for intermolecular gating in the kinesin motor protein. Biophys Rev. 2011;3(2):85–100. doi: 10.1007/s12551-011-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402(6763):778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 31.Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci USA. 2011;108(39):16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21(2):154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sindelar CV, Downing KH. The beginning of kinesin’s force-generating cycle visualized at 9-A resolution. J Cell Biol. 2007;177(3):377–385. doi: 10.1083/jcb.200612090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leduc C, Ruhnow F, Howard J, Diez S. Detection of fractional steps in cargo movement by the collective operation of kinesin-1 motors. Proc Natl Acad Sci USA. 2007;104(26):10847–10852. doi: 10.1073/pnas.0701864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.