Significance
The CLRC complex is essential for heterochromatin formation in the yeast Schizosaccharomyces pombe. Its well-known role in placing methyl marks on histone H3 lysine 9 at heterochromatic loci is attributed to one of its components, cryptic loci regulator 4. However, it also contains an E3 ubiquitin ligase, a less understood activity of this complex. Here, we describe the organization of this seven-component complex and determine the crystal structure of delocalization of Swi6 1 (Dos1), a key subunit involved in targeting CLRC. We identify Dos2 as the central component of the complex and point of contact with Stc1, which bridges CLRC to the RNAi-induced transcriptional silencing complex, and show that heterochromatin formation is dependent on an exposed surface of Dos1. These results provide an unprecedented, high-resolution functional annotation of CLRC.
Keywords: epigenetics, transcriptional gene silencing
Abstract
Repressive histone H3 lysine 9 methylation (H3K9me) and its recognition by HP1 proteins are necessary for pericentromeric heterochromatin formation. In Schizosaccharomyces pombe, H3K9me deposition depends on the RNAi pathway. Cryptic loci regulator 4 (Clr4), the only known H3K9 methyltransferase in this organism, is a subunit of the Clr4 methyltransferase complex (CLRC), whose composition is reminiscent of a CRL4 type cullin-RING ubiquitin ligase (CRL) including its cullin Cul4, the RING-box protein Pip1, the DNA damage binding protein 1 homolog Rik1, and the DCAF-like protein delocalization of Swi6 1 (Dos1). Dos2 and Stc1 have been proposed to be part of the complex but do not bear similarity to canonical ubiquitin ligase components. CLRC is an active E3 ligase in vitro, and this activity is necessary for heterochromatin assembly in vivo. The similarity between CLRC and the CRLs suggests that the WD repeat protein Dos1 will act to mediate target recognition and substrate specificity for CLRC. Here, we present a pairwise interaction screen that confirms a CRL4-like subunit arrangement and further identifies Dos2 as a central component of the complex and recruiter of Stc1. We determined the crystal structure of the Dos1 WD repeat domain, revealing an eight-bladed β-propeller fold. Functional mapping of the putative target-binding surface of Dos1 identifies key residues required for heterochromatic silencing, consistent with Dos1’s role as the specificity factor for the E3 ubiquitin ligase.
The regulation of heterochromatin is essential for proper chromosome segregation, telomere maintenance, genomic stability, and cell fate determination (1). In the fission yeast Schizosaccharomyces pombe, there are three major heterochromatic regions: the centromeres, telomeres, and the cryptic (silent) mating-type locus. Centromeric heterochromatin formation and silencing is tightly regulated by the interplay of protein complexes of the transcription, RNA interference, and chromatin modification machineries (2–4). Cryptic loci regulator 4 (Clr4) is the only histone H3K9 methyltransferase in S. pombe and therefore absolutely required for heterochromatin regulation. Clr4 has been found to be part of a multiprotein complex termed “Clr4 methyltransferase complex” (CLRC) (5–10). Members of this complex are as essential for heterochromatin formation as is Clr4 itself (11). In addition to the methyltransferase Clr4, CLRC comprises the cullin scaffold protein Cul4, the RING finger protein Pip1, and WD-40 β-propeller proteins Rik1 and delocalization of Swi6 1 (Dos1)/Clr8/Raf1 as well as Dos2/Clr7/Raf2. Dos2 contains a replication foci targeting sequence domain and is distantly similar to human nuclear receptor activator 4, but otherwise shows no homology to known proteins. A small protein, Stc1, was shown to mediate the interaction between CLRC and the RNAi-induced transcriptional silencing (RITS) complex and is required for RNAi-dependent H3K9 methylation (12).
Rik1, Cul4, Pip1, and Dos1 show strong resemblance to subunits of Cullin-RING ubiquitin ligases (CRLs), the largest family of multisubunit E3 ubiquitin ligases (13). CLRC is most similar to the human CRL4 complex, which contains the cullin CUL4 that serves as a scaffold to bring the E2-ubiquitin–conjugating enzyme in proximity to its substrate (14). In CRL4, the RING finger subunit RBX1/2, bound to the C terminus of CUL4, recognizes the E2 enzyme, whereas an adaptor subunit, the DNA damage binding protein 1 (DDB1), bound to the CUL4 N terminus, recruits a variety of WD-40 substrate receptors, known as DCAFs (DDB1 CUL4 associated factors) that recognize specific substrates (15–18). The best characterized DCAF, DDB2, acts as a DNA damage sensor, binding pyrimidine dimers at UV lesions, and as part of the RBX1/CUL4/DDB1/DDB2 complex (CRL4DDB2), ubiquitylates histones and DNA repair proteins (19). The structure of CRL4DDB2 displays a U-shaped architecture, with the DCAF DDB2 recognizing damaged DNA through its β-propeller while bound to the adapter DDB1 (19). By analogy, it has been proposed that Rik1 assumes the function of DDB1 and that Dos1 is the DCAF involved in target recognition in the CLRC complex (20, 21). Biochemical and genetic data show that CLRC is an active ubiquitin ligase and that the ligase activity is required for heterochromatin formation (7, 8). However, functional bona fide targets of CLRC remain unknown.
Here, we present a pairwise interaction screen that reveals the CLRC subunit arrangement. We show that the similarities to CRL4 extend to the organization of the complex. Importantly, we find that the non-CRL4–like subunit Dos2 assumes a central position inside the complex and interacts strongly with Stc1. The placement of Dos1 in the complex suggests a role as the specificity factor for the ubiquitin ligase. We present the crystal structure of the Dos1 WD repeat domain—an eight-bladed β-propeller—and demonstrate through structure-guided mutagenesis that an exposed surface of Dos1, which does not contact any of the known CLRC components, is required for heterochromatic silencing.
Results
Subunit Interactions in the CLRC Complex.
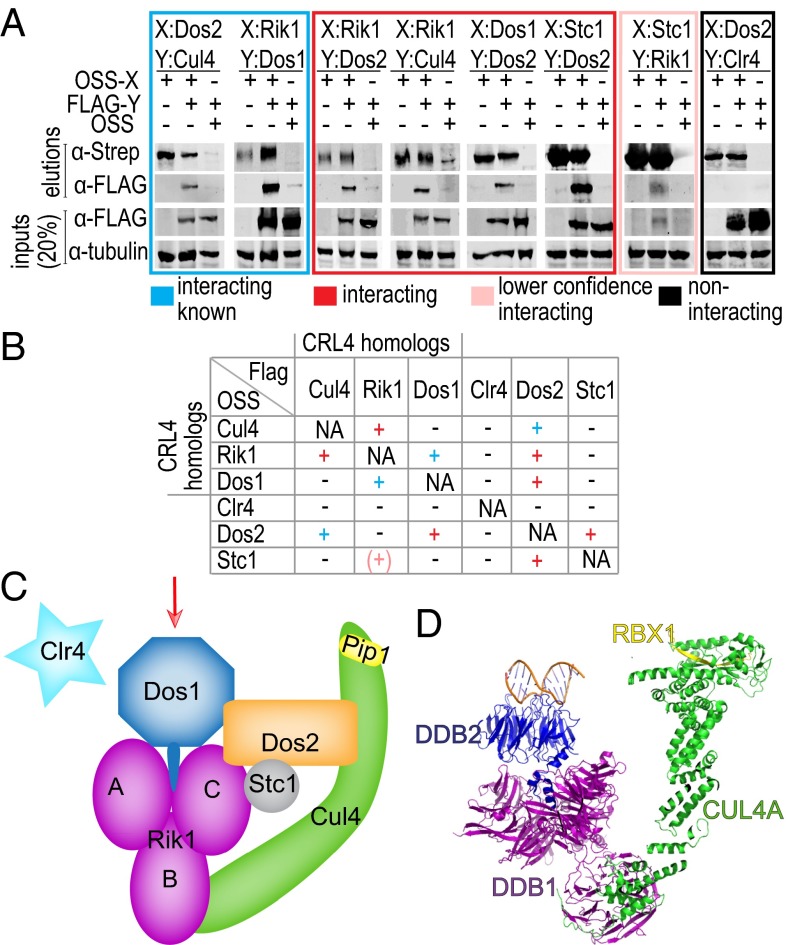
To understand the detailed architecture of CLRC, we designed a screen to test the pairwise interactions between components of the CLRC complex. Using the Multi-BAC system (22), we coexpressed one member of the pair as an N-terminal One STrEP Sumo (OSS) fusion and the other with a FLAG tag and vice versa. The interaction was then assayed by affinity purification via the One STrEP tag and detection of the two subunits by Western blot as shown in Fig. 1 A and B and SI Appendix, Fig. S1.
Fig. 1.
Subunit arrangement in the CLRC complex. (A) Testing of pairwise interactions between differently tagged CLRC subunits after coexpression and pull-down using the OneSTrEP SumoStar (OSS)-tagged protein as bait. Boxes group different classes of interaction results (new: red, lower confidence; new: pink, previously reported: blue, no interaction: black) (B) Matrix summarizing all pull-downs performed in this work color coded as in A. (C) Schematic diagram showing the subunit arrangement in the CLRC complex based on the results of the pairwise interaction screen. The arrow indicates the putative target recognition surface of Dos1 (see below). (D) The structure of the CLR4DDB2 complex (PDB ID code 4A0K). Equivalent proteins of the CRL4 and CLRC complexes are shown in similar colors.
Overall, our assay identifies several previously unknown interactions and confirms interactions that were reported previously (Fig. 1 A and B). The architecture expected by analogy to CRL4 is largely recapitulated. Unexpectedly, Dos2 turns out to be a hub of the complex by interacting with all CLRC subunits except Clr4. Rik1 is the second most connected node. Our results place Stc1, previously reported as the bridge between the RITS and CLRC complexes (12), as a peripherally associated member of CLRC interacting solidly with Dos2, and less reliably with Rik1 (see below). Surprisingly, we have not been able to detect any interactions between Clr4 and the other subunits of CLRC. This might be due to the effects of tags, requirements for posttranslational modifications, or weak, transient interactions, which escape detection by this method.
Based on the analogy to the human DDB1-CUL4A interaction in the CRL4 complex, the Rik1-Cul4 interaction observed here is consistent with its role as a scaffold of a functional E3 ligase complex. In CLRC, Rik1 also interacts with Dos1, confirming a previous interaction by a yeast two-hybrid assay (6). Overall, the Cul4-Rik1 and Rik1-Dos1 interactions are analogous to the interactions between human CUL4A, DDB1, and DDB2 in CRL4DDB2 (23) (Fig. 1). Dos2, which has no analogous subunit in CRL4DDB2, interacts with all other subunits of CLRC except Clr4. The Dos2-Cul4 interaction is consistent with a previous result from a yeast two-hybrid screen (5). This is the only previously reported direct interaction involving Dos2, although a requirement for Dos2 in nuclear localization of Dos1 has also been reported (6). Consistent with this, Dos2 also directly interacts with Dos1. Dos2 also interacted with Rik1, albeit less reliably because the interaction scored above background only in the OSS-Rik1/FLAG-Dos2 combination. Of particular interest is the direct interaction between Dos2 and Stc1 because the binding partner of Stc1 in the CLRC complex has been elusive thus far. OSS-Stc1 also interacted with FLAG-Rik1, but this interaction was not recapitulated when tags were reversed and was observed in only one of two independent experiments. Immunoprecipitation assays in various mutant backgrounds has identified Stc1 as a peripheral component of CLRC that depends on Rik1, Dos1, and Dos2 for association with the complex (12). Our results are consistent with these observations and pinpoint Dos2 as the main recruiter of Stc1 with a likely weaker contribution by Rik1. Recent results show that Stc1 binds RITS by interacting with Ago1 through its Zn fingers on one hand and that it binds CLRC through its acidic domain on the other hand (24). Our data suggest that this interaction very likely involves Dos2. Based on these observations we propose a model where CLRC and RITS cooperate through a Dos2–Stc1–Ago1 axis.
The pairwise interactions between the CLRC components that we observed are consistent with the overall architecture of the analogous human CRL4 (Cul4-DDB1-DDB2) complex whose structure is known, while adding a set of previously unobserved interactions between members of the complex, especially those with no known equivalent with CRL4 (Fig. 1 C and D). The ability to recapitulate interaction data reported previously increases our confidence that our method reveals true interactions between the components of the CLRC complex. Moreover, our data support the proposal that Dos1 is the structural and functional equivalent of the β-propeller protein DDB2, as suggested previously (20), and therefore the proposal that Dos1 acts as the DCAF for target recognition in CLRC appears likely.
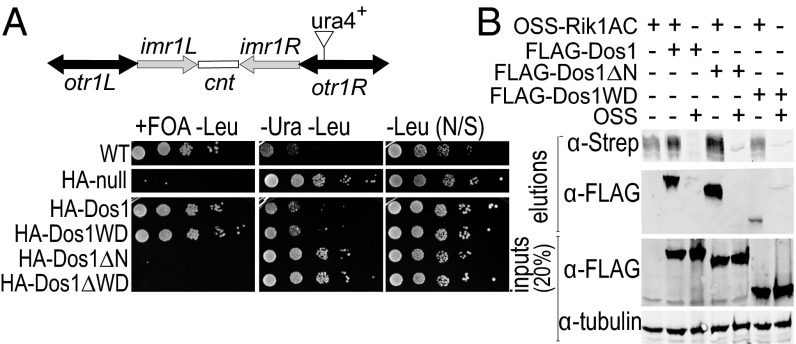
WD Repeat Domain of Dos1 Is Necessary but Not Sufficient to Rescue the dos1 Null Phenotype in S. pombe.
To understand the role of Dos1 in the CLRC complex and its domain architecture, we expressed and purified the full-length protein and subjected it to limited proteolysis. We identified two distinct fragments: one corresponding to the WD40 repeat domain [residues 213–638 (Dos1WD)] and the other corresponding to a longer fragment encompassing residues 66–638 (Dos1∆N) (SI Appendix, Fig. S2 A and B). Dos1 is essential for heterochromatin formation at S. pombe centromeres (5–9). To determine whether these fragments retain the in vivo silencing activity of the full-length protein, we tested whether they could rescue a dos1 null phenotype in S. pombe using ura4 reporter-silencing assays (25). When heterochromatin is formed, ura4 is silenced, allowing cells to grow in the presence of fluoroorotic acid (FOA). In contrast, when ura4 is expressed, it converts FOA to a toxic metabolite and prevents cell growth, indicating a loss of silencing. As shown in Fig. 2A, dos1 cells transfected with a plasmid expressing HA-tagged full-length Dos1 (HA-Dos1) complement the null mutant. By contrast, HA-tagged Dos1WD (HA-Dos1WD) or a Dos1 fragment lacking the WD repeat domain (HA-Dos1∆WD) expressed from a plasmid did not silence the ura4 reporter gene. This shows that the WD40 domain is essential, but not sufficient, for Dos1 function in centromeric heterochromatin formation in S. pombe. In contrast, Dos1∆N was able to rescue the dos1 null phenotype (Fig. 2A and SI Appendix, Fig S2C). Analysis of additional Dos1 truncations revealed that larger truncations of Dos1 did not restore the silencing defect (SI Appendix, Table S1). Thus, our results indicate that Dos1∆N contains the minimal fragment of Dos1 that can rescue the silencing defect in dos1 null cells.
Fig. 2.
The WD40 repeat domain of Dos1 is essential but not sufficient for heterochromatin formation at the S. pombe centromere. (B) Schematic diagram of S. pombe centromere 1. The position of the centromeric otr1R::ura4 reporter insertion used in this study is indicated. Comparative growth assay of the serially diluted dos1 null strain with the centromeric otr1R::ura4 reporter expressing the indicated Dos1 fragments from a plasmid. Strains were examined for growth on pombe glutamate media (PMG) lacking leucine and supplemented with 1 g/L 5-FOA (+FOA –Leu), PMG media lacking uracil and leucine (−Ura −Leu), and PMG media lacking leucine (–Leu). Cells were always grown on a PMG medium lacking leucine to select for Dos1 expressing plasmid. (C) OSS-Rik1AC was coexpressed with FLAG-Dos1 truncations and pulled down with Strep-Tactin beads to detect whether the interactions are still preserved in Dos1 truncations.
WD Repeat Domain of Dos1 Is Not Sufficient for Stable Interaction with Rik1.
Based on the analogy to the CRL4DDB2 complex, we argued that the loss of silencing for Dos1WD is likely a result of loss of interaction with Rik1. To test this, we performed pull-down assays with FLAG-tagged versions of full-length Dos1 and its two N-terminal truncations. Because full-length Rik1 is hard to obtain as a soluble protein, we expressed a form of Rik1 that contains the two WD repeat domains A and C that are predicted to interact with Dos1 based on the analogy to the CRL4DDB2 complex with an N-terminal OSS-tag (Rik1AC, Fig. 1D) (SI Appendix, Materials and Methods). Fig. 2B shows that although both full-length Dos1 and Dos1∆N interact with Rik1AC, the fragment of Dos1 that contains the WD repeat alone, Dos1WD, displays a much weakened interaction with Rik1AC. These results correspond very well with our observations from the in vivo rescue experiments and indicate that Dos1∆N is the minimum Dos1 fragment that interacts with Rik1 in a similar fashion to the interaction between DDB1 and DDB2 of the CRL4DDB2 complex. Thus, it is likely that the region between residues 66 and 213 of Dos1 harbors the helix–loop–helix motif that has been found in DDB1-associated DCAFs to provide the major DDB1-DCAF interaction interface (23, 26).
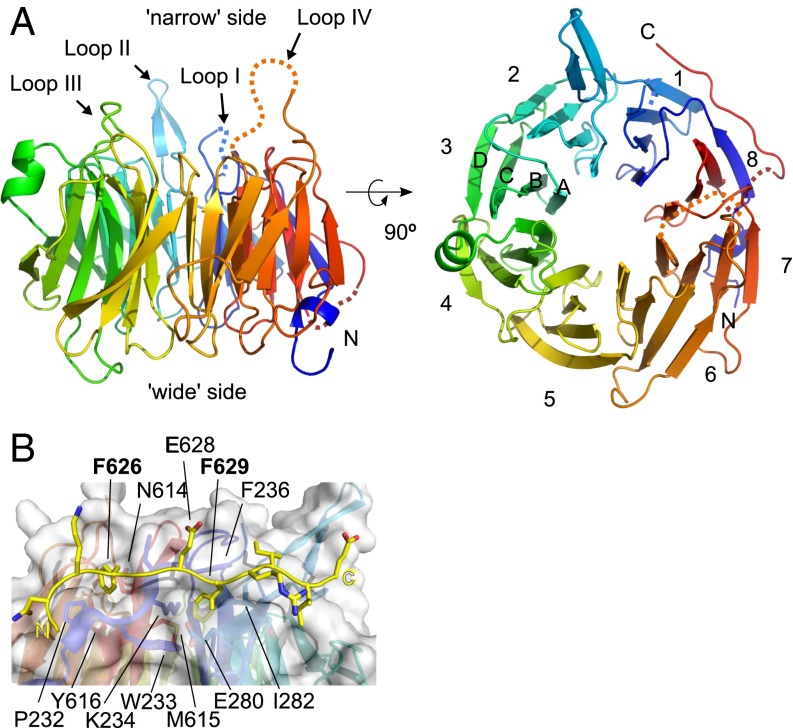
WD Repeat Domain of Dos1 Is an Eight-Bladed β-Propeller.
In our efforts to understand the function of Dos1 in CLRC, we determined the crystal structure of Dos1WD at 2.0 Å resolution (SI Appendix, Table S2). The structure of Dos1WD reveals an eight-bladed β-propeller (blades 1–8), with each blade consisting of four anti-parallel β strands (A–D) (Fig. 3A). The blades assemble to form a toroidally shaped structure with a solvent accessible central channel, where one side of the toroid is wider (“wide” side) than the other side (“narrow” side). In general, the surfaces of the wide and narrow sides contain loops that connect the β strands and are important for either protein–protein or protein–nucleic acid interactions (27). Similar to other β-propellers, the N and C termini of Dos1WD come together to form the last blade, known as the “velcro closure,” which has been shown to stabilize the β-propeller (27).
Fig. 3.
Dos1WD is an eight-bladed β-propeller. (A) Cartoon representation of the eight-bladed Dos1 β-propeller, viewed from the side (Left) and rotated 90° and viewed from the narrow side of the propeller (Right). Blades 1–8 are formed by four β strands (A–D) as labeled on blade 3. (B) The C-terminal tail of Dos1 latches blades 1 and 8. The side chain of the Phe626 and Phe629, which fit into hydrophobic pockets formed by the propeller and the two glutamates (Glu628 and Glu633), which are facing outward, are shown with carbon atoms in yellow and oxygen atoms in red. The view is from the narrow side of the propeller.
Unique to Dos1WD is an extended 20-residue C-terminal tail (residues 619–638). Residues 619–623 and 634–638 were not visible in the structure; however, the intervening segment, residues 624–633, forms an extended structure that “latches” blades 1 and 8 together on the side of the propeller by using Phe626 and Phe629 as anchors fit into hydrophobic pockets formed by the two blades (Fig. 3B). Specifically, Phe626 interacts with residues Pro232 and His235 from the D strand and Asn614 and Tyr616 from the C strand of blade 8. Phe629 interacts with Trp233 and Lys234 from the D strand and Met615 on the C strand of blade 8, along with Phe255 of the β strand of blade 1. The deletion of the C-terminal tail or alanine substitution of either Phe626 or Phe629 shows a moderate silencing defect in ura4 reporter assays (SI Appendix, Table S1). The fact that the C-terminal deletion mutant was also quite unstable (SI Appendix, Fig. S9) suggests a role for the latch in stabilizing Dos1.
On the wide side of the propeller, all of the loops that are connecting the β strands are ordered in the structure. By contrast, on the narrow side of the propeller, there are two loops that are disordered and two loops that are highly flexible, as indicated by their high B factors (SI Appendix, Fig. S3). Loop I connecting the B and C strands of blade 1 (residues 263–269) and loop IV connecting blade 6 to blade 7 (residues 536–546) are disordered. Loop II connects blade 1 and 2 (residues 289–299) and extends outward to contact another molecule in the crystal. Loop III (residues 414–422) resides on blade 4 between β strands B and C (Fig. 3 and SI Appendix, Fig. S3). Because the loops on the β-propellers are frequently involved in target recognition (27), these disordered and flexible loops on the narrow side of Dos1WD are expected to be important for Dos1 function.
Comparison of Dos1WD with DDB2 of CRL4 Identifies Putative Substrate and Rik1-Binding Surfaces.
Given the structural and functional analogy between CRL4 and CLRC, it is likely that Dos1 is the putative substrate specificity factor of the CLRC complex owing to its β-propeller domain. To identify the binding surfaces for Rik1 (analogous to DDB1 in CLR4DDB2) and the putative substrate on Dos1, we superimposed the structure of Dos1WD onto DDB2 in the CLR4DDB2 complex (19) (PDB ID code 4A0K) (SI Appendix, Fig. S4).
Based on this superposition, the Rik1-binding surface of Dos1WD is on the wide side of the toroid. In the CLR4DDB2 complex, DDB2 contacts DDB1 through a helix–loop–helix motif that is N-terminal to the β-propeller. The analogous region is not present in the Dos1WD construct that we used for crystallization. However, the N terminus emanates from the Rik1-facing side of the protein and, as we have shown above, a longer fragment of Dos1 is still able to interact with Rik1 (Fig. 2C). In addition, three mutants of Dos1 (R518A, R576A, T495I), which cannot form heterochromatin due to loss of Rik1 binding (20) map to the wide side of the toroid (SI Appendix, Fig. S5). Arg518 and Arg576 are immediately adjacent to one another in space, extending out into solvent on the predicted Rik1-binding surface (SI Appendix, Fig. S5). Superimposition of Dos1WD with DDB2 from the CRL4DDB2 complex reveals that these residues in DDB2 are precisely at the DDB1-DDB2 interface. In contrast, Thr495 is located near the central pore of the β-propeller on the A strand of blade 6 with its side chain interacting with residues Thr504 and Trp554. This hydrogen bond network is highly likely to be critical for the stability of the WD fold (SI Appendix, Fig. S5), and thus the T495I mutation might destabilize the local or overall fold of the domain.
Structural Similarities Suggest a Protein-Binding Surface on Dos1.
Dos1WD shows the strongest structural similarity to the human F-box protein Fbw7 and its homolog in budding yeast, Cdc4, which function in cell cycle regulation by guiding the ubiquitylation of cyclin E (28, 29). The putative substrate-binding surface of Dos1WD, the narrow side of the toroid, shows an electrostatic surface charge that is predominantly negative (Fig. 4). This indicates that Dos1WD is unlikely to bind negatively charged substrates such as nucleic acids or phosphopeptides, which are the targets of DDB2 or Fbw7 and Cdc4, respectively (Fig. 4). Another eight-bladed β-propeller protein is Sif2p, a structural component of the SET3C corepressor complex in Saccharomyces cerevisiae that has histone deacetylase activity (30, 31). Sif2p interacts with the N terminus of Sir4p and competes with Sir4p function at telomeres. Because S. pombe does not have a Sir4 homolog, we cannot draw direct parallels to Sif2p function for Dos1. Although seven-bladed, the β-propellers of EED and WDR5, which recognize histone tails, are structurally similar to Dos1 (32–36). These WD40 domains accommodate methyl-lysine or arginine residues in a pocket near the central channel using aromatic residues and main-chain carbonyls, which form a ring around the pore (32–36). Although the Dos1WD pore contains a ring of carbonyls, it lacks the numerous hydrophobic residues that EED and WDR5 display. Moreover, the Dos1WD pore is significantly larger than the pore in WDR5 (for example, 12 Å versus 7 Å in diameter), owing to the presence of the additional blade. Nevertheless, the electrostatic characteristics of the Dos1WD substrate-binding surface appear to be most suited for recognizing a basic peptide or a positively charged protein surface that would involve contacts to the numerous loops on the Dos1 surface.
Fig. 4.
Comparison of surface electrostatic potentials of different propellers. Surface electrostatic potentials of the Dos1WD putative target-binding site, DDB2 DNA-binding site (PDB ID code 4A0K), Fbw7 phospho-peptide–binding site (PDB ID code 3V7D), Sif2p (PDB ID code 1R5M), and WDR5 (PDB ID code 3UVN) were compared. Surface electrostatic potentials were calculated using the APBS plugin in Pymol and mapped onto the solvent-accessible surface of the proteins (contoured from −5 to +5 eV) (49).
Structure-Based Mutants on the Putative Target-Binding Surface Show Defects in Silencing.
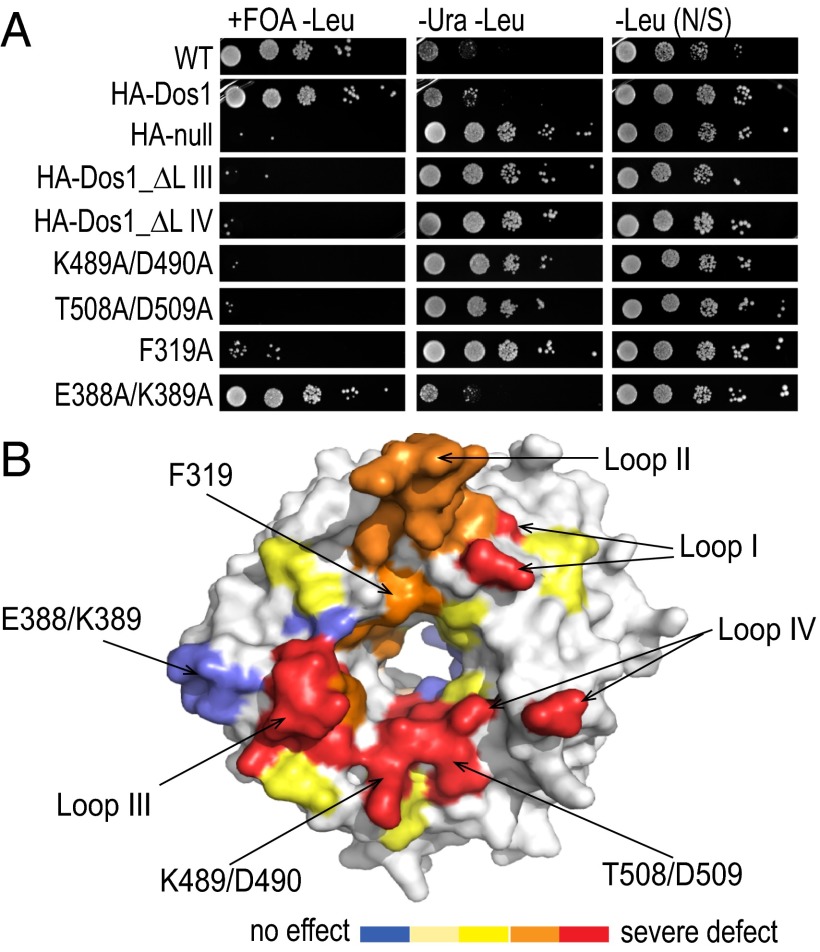
To understand whether the putative target-binding surface is important for Dos1 function, we designed structure-based mutations on the surface of the putative target binding site and tested these mutants’ effect on silencing in vivo using the ura4 reporter-silencing assay as described above. Specifically, the flexible loops I–IV were either deleted or connected with glycine linkers in a way that would not disrupt the overall fold of the protein. We also designed single or double alanine substitutions based on conservation and solvent accessibility on the surface, at the opening of the pore, and within the pore. As a control, we also tested the T495I, R518A, and R576A mutants on the Rik1-binding surface (20). To detect the less sensitive mutants, we monitored the growth of cells at both 30 °C and 37 °C. To confirm that the mutations on the putative target-binding surface that we introduced did not affect the overall fold of the protein or interactions with CLRC, we tested the interaction of four representative mutants with Rik1 and Dos2 by pull-down and showed that these mutants can still interact with Rik1 and Dos2 (SI Appendix, Fig. S6). In addition, the behavior of the OSS-Dos1 K489A/D490A mutant on a size exclusion column was indistinguishable from wild-type OSS-Dos1 (SI Appendix, Fig. S7), indicating that the stability and folding of Dos1 are not affected in this mutant.
The phenotypes were divided into five groups: strong defect at 30 °C (“strong”); mild defect at 30 °C and strong defect at 37 °C (“moderate”); strong defect at 37 °C only (“mild”); mild defect at 37 °C (“very mild”); and no effect on silencing (SI Appendix, Table S1 and Fig. S8). Deletion of loops I, III, or IV had the strongest defect in ura4+ silencing even though the proteins were produced (Fig. 5 and SI Appendix, Table S1 and Fig S9). Strikingly, the alanine substitutions with the strongest defects in silencing were adjacent to or within the opening of the central pore on the putative target-binding surface. Single or double alanine substitutions with the most severe silencing defects included the double substitutions Lys489/Asp490 and Ile491/Asn492 on the surface loop connecting blades 5 and 6 and Thr508/Asp509 on the adjacent loop of blade 6 that faces into the pore (Fig. 5). Surprisingly, substitution of His493, which is just at the opening of the pore in this region, had no effect. On the other hand, the single alanine substitution of Phe416, whose aromatic ring points into the pore, was almost as severe as deletion of the entire loop III (emanating from blade 4) on which it resides (Fig. 5 and SI Appendix, Figs. S8 and S10). These results indicate that a region extending from loop III to loop IV, including a patch in between at the pore entrance on blades 5 and 6, is critical for silencing. In all cases, we confirmed that protein expression was intact (SI Appendix, Fig. S9).
Fig. 5.
The putative target-binding surface of Dos1 is important for heterochromatic silencing. (A) Representative results from comparative growth assays of the serially diluted dos1 null strain with the centromeric otr1R::ura4 reporter expressing Dos1 mutants as indicated from a plasmid. Strains were examined for growth on a PMG medium lacking leucine and supplemented with 5-FOA (+FOA –Leu), PMG medium lacking uracil and leucine (−Ura −Leu), and PMG medium lacking leucine (–Leu). Cells were always grown on PMG medium lacking leucine to select for Dos1-expressing plasmid (SI Appendix, Table S1). (B) Surface representation of Dos1WD mapped mutations colored by the degree of the silencing defect (red: severe defect; orange: less severe defect; yellow: mild defect; light yellow: very mild defect in silencing; blue: no effect in ura4+ silencing).
A second patch of substitutions, which had moderate defects in silencing, is located on blades 1 and 2 at the opening or slightly inside the central pore and adjacent to loop II (Fig. 5 and SI Appendix, Table S1). This cluster includes Thr301, Asp304, and Phe319 on blade 2 and His344/K345 on the loop connecting blades 1 and 2 deeper in the pore. Mutation of adjacent residues Thr340/Asp341 had a much milder effect, and substitution of Lys359, which is adjacent to Asp304, had no effect. Other residues that had no effect on silencing include Glu397, Tyr450, and Gln553, deep within the pore, as well as residues on the Rik1 surface near the pore but away from the presumptive Rik1-binding site (His278, Asn559, and Glu599). Although highly conserved among fungi, residues Glu385 and Glu388/Lys389, located on a loop connecting blades 3 and 4 but facing out on the side of the propeller, had no effect on silencing (Fig. 5 and SI Appendix, Fig. S10). Mutations on blade 1, including Asn237/Glu238 and V244, had mild effects, but are probably affecting the stability of the velcro closure (SI Appendix, Fig. S10). The three mutations isolated previously that affect Rik1 binding—T495I, R518A, and R576A—showed mild to very mild effects by our assay (SI Appendix, Fig. S8). We conclude that substrate recognition by Dos1 would involve loops I, III, and IV and the residues proximal to these loops at the pore opening.
Discussion
The histone methyltransferase complex CLRC is essential for heterochromatin formation in S. pombe and is recruited to pericentromic heterochromatin, in part, by the RITS complex. Apart from its important role in depositing methyl marks on histone tails to enable binding of the HP1 homologs, Swi6 and Chp2, CLRC contains several components with less established roles in heterochromatin assembly. The architecture of the complex emerging from the interaction analysis described here, along with sequence similarities noted earlier, point to an interesting parallel to the human CRL4 complex that also includes the E3 ligase Cul4 as well as RBX1, DDB1, and DDB2. The structure of Dos1 that we presented here extends this similarity even further because it has a β-propeller structure, albeit an eight-bladed one rather than a seven-bladed one as for DDB2. It therefore seems likely that Dos1 acts as a DCAF in the CLRC complex. Our structure-based mutational analysis subsequently identified the surface of Dos1 that would interact with Cul4 targets on the opposite side from the Rik1 interaction surface.
In human and mouse CRL4DDB2, DDB2 acts as a DCAF to recognize specific DNA targets, and Cul4 then ubiquitylates nearby histones to open up nucleosomes for repair (37–39). However, in other cases, the target of the DCAF is also the substrate for Cul4. In humans, Cdt2 acts as the DCAF for another Cul4-DDB1 complex, CRL4Cdt2, and interacts with the H4K20 methyltransferase PR-SET7/SET8, which is ubiquitylated and subsequently degraded (40–42). In S. pombe, the DCAF Cdt2 in the CRL4Cdt2 complex interacts with Epe1, a JmjC family protein that is enriched at heterochromatic boundaries and is regulated by ubiquitylation of CRL4Cdt2, leading to its degradation (43). Thus, Dos1 might interact with an entity that brings CLRC to the vicinity of the Cul4 E3 ligase substrate or would bring CLRC to the Cul4 substrate itself.
Another possible candidate for a DCAF function is Dos2. TAP-Dos2 purification identified a complex of Rik1, Cdc20, and Mms19 without detectable Dos1 (44) whereas an identical purification strategy shows ubiquitin ligase activity on H2B (8). However, it is not clear whether Dos1 is present in this latter purification.
H2B remains a possible target of Dos1 because it is a substrate for a Rik1-TAP–purified CLRC E3 ligase activity in vitro (8). Consistent with this is the negative electrostatic character of the target-binding surface (Fig. 4). An interesting parallel is the requirement for H2B ubiquitylation by RNF20-BRE1 for H3K4 methylation by Set1 and H3K79 methylation by Dot1 (reviewed in ref. 45). H3K4 methylation levels are elevated in cul4 mutants, indicating that Cul4 may have a role in regulating H3K4 methylation (8, 9). Indeed, the H3K4 demethylase, Lid2, has been shown to interact with CLRC (21).
Establishment of pericentromeric heterochromatin requires recruitment of the RNAi machinery to an actively transcribing locus. However, in the cotranscriptional gene-silencing scenario, RNAi is then thought to maintain heterochromatin through replication by inhibiting transcription elongation. This results in subsequent release of RNA pol-II (46) to allow replication fork progression. An interesting possibility is a role for targeting of the E3 ligase of CLRC to RNA pol-II in the displacement step, and PolII-associated factors are found in a complex with Rik1 (44). Another Dos1 target candidate is Nup189, which has been found to interact with Dos1 in a yeast two-hybrid screen (5). Finally, Dos1 has also been reported to interact with Ams2, a CENP-A–loading factor (47). Proper CENP-A loading depends on Dos1, Dos2, and the DNA polymerase ε-subunit Cdc20. Whether the interaction between Ams2 and Dos1 is direct is not clear, however.
Given its pervasive role in heterochromatic silencing at telomeres and at the cryptic mating-type locus as well as at the centromere, it is likely that the Dos1–Rik1–Cul4 complex has numerous targets involved in chromatin organization and transcription and potentially in replication, recombination, and repair. Our results indicate a central role for Dos1 in mediating this targeting. On the other hand, it is also clear that both Rik1 and Cul4 participate in additional complexes that could potentially compete for interaction with Dos1, thus altering target specificity. Here we present the refined architecture of CLRC provided by interaction assays and by the structure of Dos1, as well as its functional annotation. This will provide the basis for further elucidation of the structure and function of CLRC and its targets in the establishment and maintenance of heterochromatin domains.
Materials and Methods
Pull-Down Assays.
For pull-down assays, protein pairs were expressed in insect cells using the MultiBAC expression system (48). OSS-tagged proteins were used as bait and pulled down using streptactin beads. Details of the experiments can be found in SI Appendix, Materials and Methods.
Protein Expression and Purification.
Codon-optimized S. pombe Dos1 (residues 213–638) was expressed as an OSS-TEV fusion in insect cells. The protein was affinity-purified using streptactin beads followed by anion exchange and size-exclusion chromatography. Details can be found in SI Appendix, Materials and Methods.
Crystallization and Structure Determination.
Dos1WD crystals were grown at 17 °C using the hanging-drop vapor-diffusion method. Details of the crystallization and structure determination are provided in SI Appendix, Materials and Methods. Additional information can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Chandni Ashar and Amanda Epstein for technical support, the members of the L.J. and R.A.M. laboratories for their support, Janet Partridge and Cem Kuscu for helpful discussions, and Annie Héroux for help at the National Synchrotron Light Source, which is supported by Department of Energy, Office of Basic Energy Sciences. We thank the Proteomic shared resources at Cold Spring Harbor Laboratory, which is supported by Cancer Center Support Grant 5P30CA045508 and the Protein Core Facility at Columbia University. T.S. was supported by a Human Frontiers in Research Program fellowship. This work was supported by The Robertson Research Fund of Cold Spring Harbor Laboratory. L.J. and R.A.M. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4O9D).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313096111/-/DCSupplemental.
References
- 1.Black JC, Whetstine JR. Chromatin landscape: Methylation beyond transcription. Epigenetics. 2011;6(1):9–15. doi: 10.4161/epi.6.1.13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper BJ, Lowe BR, Partridge JF. Centromeric heterochromatin assembly in fission yeast: Balancing transcription, RNA interference and chromatin modification. Chromosome Res. 2012;20(5):521–534. doi: 10.1007/s10577-012-9288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bühler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14(11):1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 4.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 5.Thon G, et al. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005;171(4):1583–1595. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, et al. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol. 2005;15(16):1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 7.Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7(10):1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 8.Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19(14):1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong EJ, Villén J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2(3):106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 10.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15(4):381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 11.Shanker S, et al. Continuous requirement for the Clr4 complex but not RNAi for centromeric heterochromatin assembly in fission yeast harboring a disrupted RITS complex. PLoS Genet. 2010;6(10):e1001174. doi: 10.1371/journal.pgen.1001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayne EH, et al. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140(5):666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 14.Jackson S, Xiong Y. CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34(11):562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angers S, et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443(7111):590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 16.He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20(21):2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higa LA, et al. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8(11):1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 18.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23(5):709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Fischer ES, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147(5):1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 20.Buscaino A, et al. Raf1 is a DCAF for the Rik1 DDB1-like protein and has separable roles in siRNA generation and chromatin modification. PLoS Genet. 2012;8(2):e1002499. doi: 10.1371/journal.pgen.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135(2):272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3(12):1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 23.Scrima A, et al. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135(7):1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He C, et al. Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification. Proc Natl Acad Sci USA. 2013;110(21):E1879–E1888. doi: 10.1073/pnas.1212155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9(2):218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17(1):105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2(3):202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26(1):131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Tang X, et al. Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc Natl Acad Sci USA. 2012;109(9):3287–3292. doi: 10.1073/pnas.1116455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cockell M, Renauld H, Watt P, Gasser SM. Sif2p interacts with Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr Biol. 1998;8(13):787–790. doi: 10.1016/s0960-9822(98)70304-5. [DOI] [PubMed] [Google Scholar]
- 31.Cerna D, Wilson DK. The structure of Sif2p, a WD repeat protein functioning in the SET3 corepressor complex. J Mol Biol. 2005;351(4):923–935. doi: 10.1016/j.jmb.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 32.Schuetz A, et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25(18):4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13(8):698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 34.Han Z, et al. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell. 2006;22(1):137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 35.Ruthenburg AJ, et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13(8):704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerrero-Santoro J, et al. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 2008;68(13):5014–5022. doi: 10.1158/0008-5472.CAN-07-6162. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22(3):383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 39.Kapetanaki MG, et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA. 2006;103(8):2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbas T, et al. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40(1):9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oda H, et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40(3):364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Centore RC, et al. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40(1):22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braun S, et al. The Cul4-Ddb1(Cdt)² ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144(1):41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475(7355):244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5(6):460–468. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479(7371):135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez M, He H, Sun S, Li C, Li F. Cell cycle-dependent deposition of CENP-A requires the Dos1/2-Cdc20 complex. Proc Natl Acad Sci USA. 2013;110(2):606–611. doi: 10.1073/pnas.1214874110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salyan ME, et al. A general liquid chromatography/mass spectroscopy-based assay for detection and quantitation of methyltransferase activity. Anal Biochem. 2006;349(1):112–117. doi: 10.1016/j.ab.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 49.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.