Significance
Quorum sensing is a mechanism for intra- and/or interspecies microbial communication of cell density information. It can regulate group behaviors, like virulence, biofilm formation, and competence. Candida albicans is a major opportunistic fungal pathogen of humans that resides in the gastrointestinal tract and mucosal membranes as a commensal microbiota. Its capacity to switch between yeast and hyphal morphologies is a key virulence factor. The morphologic transition is inhibited by the sesquiterpine alcohol farnesol, a quorum-sensing molecule in C. albicans. Here, we show that farnesol controls hyphal initiation through the N-end rule pathway-mediated protein degradation. This regulation provides molecular insights into how C. albicans cells communicate in the host microbiota to regulate the transition between commensal and pathogenic growth states.
Abstract
Candida albicans is the most common cause of invasive fungal infections in humans. Its ability to undergo the morphological transition from yeast to hyphal growth forms is critical for its pathogenesis. Hyphal initiation requires the activation of the cAMP-PKA pathway, which down-regulates the expression of NRG1, the major repressor of hyphal development. Hyphal initiation also requires inoculation of a small amount of C. albicans cells from overnight culture to fresh medium. This inoculation releases the inhibition from farnesol, a quorum-sensing molecule of C. albicans, that accumulated in the spent medium. Here, we show that farnesol inhibits hyphal initiation mainly through blocking the protein degradation of Nrg1. Through screening a kinase mutant library, we identified Sok1 as the kinase required for Nrg1 degradation during inoculation. SOK1 expression is transiently activated on inoculation during hyphal initiation, and overexpression of SOK1 overcomes the farnesol-mediated inhibition of hyphal initiation. Screening a collection of transcription factor mutants, the homeodomain-containing transcription repressor Cup9 is found to be responsible for the repression of SOK1 expression in response to farnesol inhibition. Interestingly, farnesol inhibits Cup9 degradation mediated by the N-end rule E3 ubiquitin ligase, Ubr1. Therefore, hyphal initiation requires both the cAMP-PKA pathway-dependent transcriptional down-regulation of NRG1 and Sok1-mediated degradation of Nrg1 protein. The latter is triggered by the release from farnesol inhibition of Cup9 degradation and consequently, derepression of SOK1 transcription. Neither pathway alone is sufficient for hyphal initiation.
Quorum sensing is the regulation of gene expression and group behavior in response to changes in cell population density. Quorum-sensing molecules were initially discovered in bacteria. They can mediate the sensing of cell density and control group behavior, like virulence, competence, conjugation, antibiotic production, and biofilm formation (1). Much less is known about quorum-sensing molecules, their functions, and their mechanisms of sensing in fungi. For the human commensal and pathogenic fungus Candida albicans, dense cultures display a reduced propensity for the yeast-to-hyphal switch. This reduced propensity is because of the accumulation of a sesquiterpine alcohol farnesol that can inhibit hyphal formation (2). Farnesol can also affect the expression of virulence genes and biofilm formation (3, 4). C. albicans produces farnesol from an intermediate of the sterol biosysthesis pathway, farnesyl pyrophosphate (5). At concentrations of 10–250 μM, farnesol inhibits hyphal initiation but does not suppress hyphal elongation (6). Farnesol-dependent inhibition of hyphal formation involves the histidine kinase Chk1 (7), the conserved Ras, and the adenylate cyclase Cyr1 pathway (8). Recently, down-regulation of Nrg1, a major repressor of the hyphal transcriptional program, was found to be essential for hyphal initiation (9). The cAMP-protein kinase A (PKA) pathway is required for down-regulation of NRG1 expression during initiation (9). Furthermore, the Nrg1 protein is degraded during hyphal initiation, when C. albicans cells from dense cultures are inoculated into fresh medium; adding farnesol to the medium prevents Nrg1 degradation and hyphal initiation (9).
Ubiquitin-mediated protein turnover is a key regulatory mechanism for proteins with concentration that must be dynamically regulated during cellular signaling, development, or stress responses. Enzymatic conjugation of ubiquitin to proteins that contain primary degradation signals is mediated by the E1 Ub-activating enzyme and Ub-conjugating (E2) enzyme. The transfer of ubiquitin from E2 to a lys residue of an appropriate substrate protein requires an E3 ubiquitin ligase, which is responsible for the initial recognition of a substrate’s degradation signal (degron) (10). Different degrons are recognized by different E3s. The mammalian genome encodes at least 1,000 distinct E3s. The first E3 identified and cloned was Ubr1, which recognizes proteins’ N-terminal degradation signals (the N-end rule) (11). The RING-type E3 ligase contains types 1 and 2 substrate binding sites that recognize the unmodified basic (Arg, Lys, or His) and bulky hydrophobic (Leu, Phe, Tyr, Trp, or Ile) N-terminal residues, respectively (12, 13). In addition, Ubr1 also contains binding sites that recognize internal degrons of the Cup9 transcriptional repressor (14). Degradation of specific proteins by the N-end rule pathway can be regulated in response to changes in many physiological signals, including heme, nitric oxide, amino acids, and short peptides (14–17), and it is important in many cellular functions, such as the fidelity of chromosome segregation, the regulation of DNA repair, and peptide import (15, 18, 19). For example, dipeptides accelerate their uptake by activating the Ubr1-dependent degradation of Cup9, a transcriptional repressor of a dipeptide transporter (15).
In this report, we show that Cup9 is a transcriptional repressor of SOK1 expression in C. albicans and that Sok1 is required for the degradation of Nrg1 and hyphal initiation. Farnesol sensing during the yeast-to-hyphal transition is mostly through Ubr1-mediated degradation of Cup9. This study provides an example of a regulated proteolysis by the N-end rule pathway in response to quorum-sensing molecules.
Results
Release from Farnesol Inhibition Induces Nrg1 Degradation Independent of the cAMP-PKA Pathway.
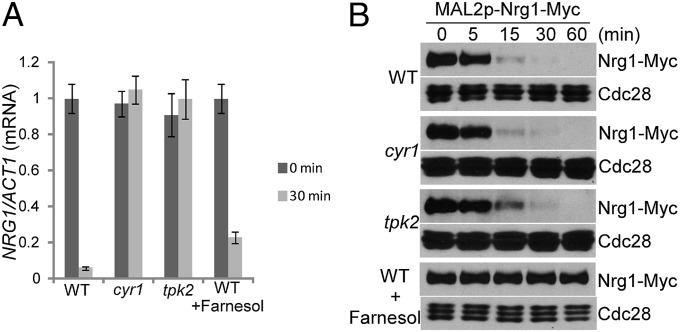
Hyphal initiation requires rapid down-regulation of the Nrg1 protein through reducing NRG1 expression and activating Nrg1 degradation. The decrease in NRG1 expression was dependent on the cAMP-PKA pathway, because Cyr1 or Tpk2 was required for reduced NRG1 expression during hyphal initiation (Fig. 1A) (9). The preferential requirement of Tpk2 for NRG1 transcriptional down-regulation is consistent with the distinct but overlapping roles reported for Tpk1 and Tpk2 in hyphal development (20). Farnesol is thought to affect hyphal initiation by inhibiting the Ras1-Cyr1 pathway (8, 21, 22). Consistent with this notion, we detected about 23% of the NRG1 transcript left after 30 min in farnesol-containing medium at 37 °C, which was higher than without farnesol (Fig. 1A). However, the expression level of NRG1 was still dramatically reduced in the presence of farnesol (Fig. 1A). This result suggests that, in addition to being involved in cAMP-mediated regulation of NRG1 transcription, farnesol inhibits hyphal initiation, mainly through a different mechanism. Indeed, Nrg1 was unstable, which was shown by promoter shutdown experiments, when cells from overnight cultures were inoculated into fresh medium (Fig. 1B). Disruption of cAMP-PKA signaling did not affect Nrg1 protein stability, but adding farnesol blocked Nrg1 degradation (Fig. 1B). Therefore, Nrg1 transcription down-regulation requires the activation of the cAMP-PKA pathway, whereas release from farnesol inhibition during inoculation triggers the degradation of Nrg1. The two independent pathways are both required for rapid clearing of Nrg1 to initiate hyphal development.
Fig. 1.
Nrg1 degradation on release from farnesol inhibition is not regulated by the cAMP-PKA pathway. (A) Transcriptional down-regulation of NRG1 during hyphal initiation is regulated by the cAMP-PKA pathway but not farnesol. Overnight culture of WT and the indicated mutant cells was inoculated at a 1:100 ratio into fresh YPD media with or without 100 µM farnesol at 37 °C. NRG1 mRNA levels were determined by quantitative RT-PCR. The signal obtained from ACT1 mRNA was used as a loading control for normalization. The 0 h normalized value of NRG1/ACT1 for the WT cells was set to be 1.00. The data show the average of three independent quantitative RT-PCR experiments, with error bars representing the SEM. (B) Nrg1 stability is monitored by MAL2 promoter shutdown. WTs or the indicated mutant cells carrying Nrg1-Myc under the MAL2 promoter were inoculated from overnight cultures into fresh YPD media at 30 °C with and without 100 µM farnesol.
Sok1 Kinase Is Required for Nrg1 Degradation During Inoculation.
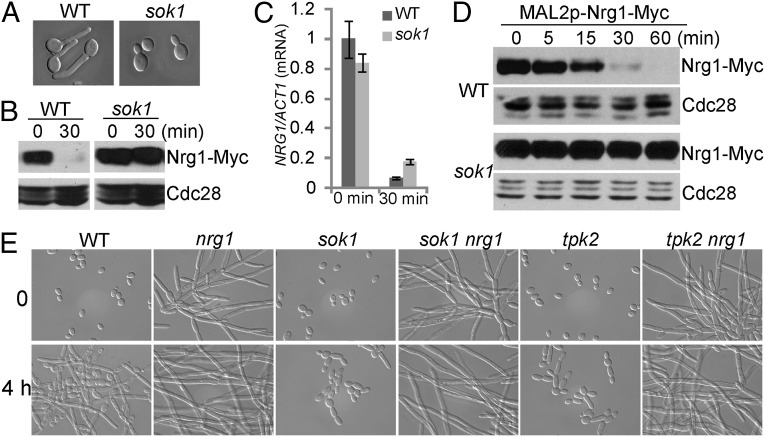
To uncover how farnesol regulates Nrg1 degradation, we screened a kinase mutant library of C. albicans (23) for mutants defective in hyphal initiation; 5 of 80 mutants showed defects to varying extents in hyphal initiation when cells were inoculated into fresh medium at 37 °C, but only tpk2 and sok1 mutants showed no decrease in levels of Nrg1. As shown in Fig. 2 A and B, germ-tube formation was completely blocked, and the Nrg1 protein level remained high in an sok1 mutant during hyphal initiation. In Saccharomyces cerevisiae, Sok1 is involved in cAMP-PKA–mediated signaling, and overexpression of Sok1 suppresses the growth defect of mutants lacking PKA activity (24). However, in C. albicans, Sok1 is not required for the cAMP-PKA–dependent transcriptional down-regulation of NRG1, because the level of NRG1 expression was significantly decreased in the sok1 mutant (Fig. 2C). Therefore, we predicted that Nrg1 degradation during inoculation could be blocked in the sok1 mutant. Indeed, Nrg1 was much more stable in the sok1 mutant when cells were inoculated into a fresh medium compared with Nrg1 stability in WT cells (Fig. 2D). We suggest that Sok1 is involved in farnesol-mediated regulation of Nrg1 degradation.
Fig. 2.
Sok1 is required for Nrg1 degradation during hyphal initiation. Overnight cultures of WT and sok1 mutant cells carrying Nrg1-Myc were inoculated into YPD + 10% serum at 37 °C for (A) 1 h for cell morphology analysis or (B) 30 min for Western analysis. (C) Sok1 is not required for the transcriptional down-regulation of NRG1 during hyphal initiation. NRG1 mRNA levels were determined as described in Fig. 1A. (D) Overnight cultures of WT or sok1 mutant cells carrying Nrg1-Myc under the MAL2 promoter were inoculated into fresh YPD medium at 30 °C. The protein stability of Nrg1 is monitored by MAL2 promoter shutdown as described in Fig. 1B. (E) Deletion of NRG1 in an sok1 or tpk2 mutant results in constitutive filamentous growth similar to the nrg1 single mutant. Overnight culture of WT and the indicated mutant cells was inoculated at a 1:100 ratio into fresh YPD media at 37 °C. Cells were collected at 0 and 4 h for cell morphology analysis.
Sok1 is required for Nrg1 degradation, and Tpk2 is required for down-regulating NRG1 expression; both kinases are required for hyphal initiation. To determine whether Nrg1 degradation or down-regulation of NRG1 expression are the major functions of Sok1 and Tpk2 in hyphal development, NRG1 was deleted in the sok1 or tpk2 mutant. Both the sok1 nrg1 and tpk2 nrg1 double mutants behaved like the nrg1 single mutant. They grew as elongated hyphae under a yeast growth condition and had no defect in hyphal induction (Fig. 2E). Therefore, the major functions of Sok1 and Tpk2 in hyphal development are to down-regulate Nrg1.
SOK1 Expression Is Activated on Release from Farnesol Inhibition During Inoculation, and Ectopic SOK1 Expression Promotes Hyphal Initiation Without Inoculation.
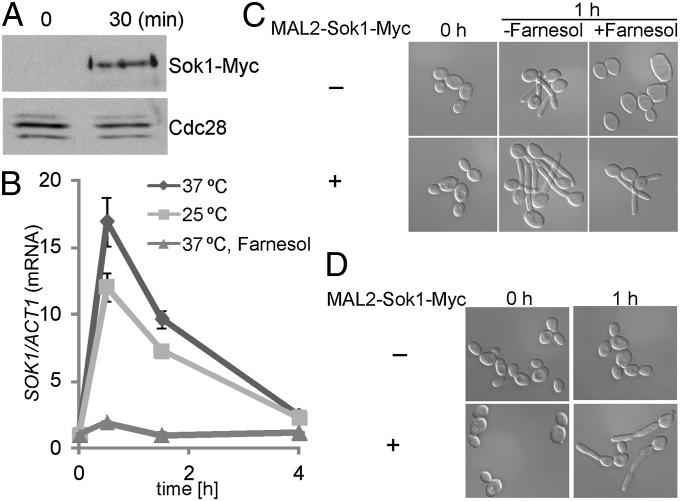
We next examined whether Sok1 protein levels changed in response to a drop in cell density by inoculation. The protein level of Sok1-myc under its endogenous promoter increased at 30 min after inoculation (Fig. 3A). The increase likely reflected transcriptional regulation of SOK1 by farnesol, because Sok1 stability was not affected by farnesol (Fig. S1). As expected, SOK1 expression was activated when cells were inoculated to fresh media, regardless of temperature, but decreased afterward (Fig. 3B). In contrast, the brief increase in SOK1 expression was not observed when farnesol was added into the medium. Our results indicate that release from farnesol inhibition is critical for the transient expression of SOK1 during inoculation.
Fig. 3.
SOK1 transcription is activated during inoculation for accelerated Nrg1 degradation. (A) An overnight culture of WT cells carrying Sok1-Myc was inoculated into YPD medium at 37 °C for 30 min for Western analysis. (B) Quantitative RT-PCR analysis of SOK1 expression. An overnight culture of WT cells was diluted into prewarmed YPD media at 25 °C or 37 °C in the presence or absence of 100 µM farnesol. The 0 h normalized value of SOK1/ACT1 for the WT cells was set to be 1.00. (C) Constitutive expression of SOK1 induces germ-tube formation in the presence of farnesol. WT cells with or without the MAL2-driven Sok1-Myc were grown in yeast extract–peptone plus 2% maltose (YPM) medium at 30 °C overnight and inoculated into prewarmed YPM medium in the presence or absence of 100 µM farnesol at 37 °C. (D) Constitutively expressed SOK1 promotes germ-tube formation in cells grown in log phase. Cells of WT with or without Sok1-Myc under the MAL2 promoter were grown in YPM medium at 30 °C overnight; they were diluted into YPM medium at 30 °C for 3 h (displayed as 0 h) and then transferred to 37 °C for 1 h (displayed as 1 h).
Because Sok1 was required for Nrg1 degradation during hyphal initiation and SOK1 expression level increased during inoculation, we examined whether overexpression of SOK1 could overcome farnesol inhibition in germ-tube formation. Ectopic expression of SOK1 in a WT strain could initiate hyphal growth at 37 °C, even in the presence of farnesol (Fig. 3C). Furthermore, hyphal development could be induced to a certain extent by a shift in temperature to 37 °C in cells growing in log phase without inoculation when SOK1 was constitutively expressed (Fig. 3D). Hyphae initiated from unbudded G1 cells gave rise to germ tubes without constriction at the base of germ tube, whereas hyphae initiated from budded cells (non-G1 cells) generated elongated buds with a constriction (25). Our results suggest that release from farnesol inhibition promotes Nrg1 degradation through transient activation of SOK1 expression for hyphal initiation.
Cup9 Represses SOK1 Expression in Response to Farnesol Inhibition.
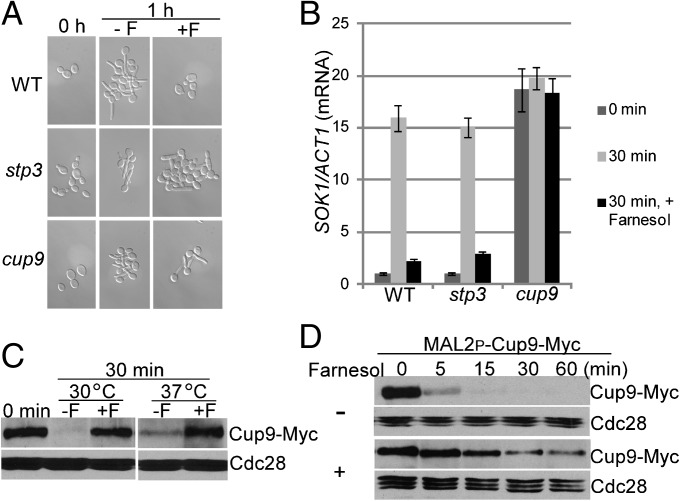
To identify transcription factors responsible for farnesol-mediated regulation of SOK1 expression, we first screened a KO library of 165 transcription factor genes in C. albicans (26) for mutants defective in hyphal initiation. Seven mutants could not induce germ-tube formation when inoculated into yeast extract peptone dextrose (YPD) medium at 37 °C, and none of them showed defects in activation of SOK1 expression (Fig. S2). Therefore, we reasoned that, instead of a transcriptional activator of SOK1 expression, SOK1 expression may be repressed by a transcriptional repressor in the presence of farnesol. When released from farnesol inhibition during inoculation, the repressor is inactivated, leading to derepression of SOK1 expression. We then screened the same mutant library of transcription factors for mutants that could form germ tubes even in the presence of farnesol. Only 2 of 165 mutants exhibited better hyphal initiation in farnesol-containing medium compared with WT cells (Fig. 4A). However, there was a dramatic difference between the ratios of germ-tube formation in these two mutants in response to farnesol. Over 90% of cup9 mutant cells formed germ tubes in the presence of farnesol, whereas only about 25% of stp3 mutant cells were elongated under the same growth conditions (Fig. 4A). The cup9 mutant has previously been reported to exhibit hyperfilamentation in other screens using this mutant library (26–28). We next examined the expression levels of SOK1 in these two mutants during inoculation in the presence or absence of farnesol. As shown in Fig. 4B, SOK1 was constitutively expressed in the cup9 mutant, regardless of farnesol. In contrast, deletion of STP3 had no effect on SOK1 expression (Fig. 4B). Our data suggest that SOK1 expression is derepressed during inoculation through inhibiting the transcriptional repressor Cup9.
Fig. 4.
Cup9 is stabilized in response to farnesol to repress SOK1 expression. (A) Overnight cultures of WT and the stp3 or cup9 mutant cells were inoculated into YPD media at 37 °C for 1 h in the presence or absence of 100 µM farnesol for cell morphology analysis. (B) SOK1 expression is derepressed in the presence of farnesol in the cup9 mutant but not the stp3 mutant. Quantitative RT-PCR analysis of SOK1 expression in WT and stp3 or cup9 mutants. Cells were diluted into prewarmed YPD medium at 37 °C in the presence or absence of 100 µM farnesol. SOK1 mRNA levels were determined as described in Fig. 3B. The 0 h normalized value of SOK1/ACT1 for the WT was set to be 1.00. (C) WT cells carrying Cup9-Myc were inoculated into YPD media at 30 °C or 37 °C in the presence or absence of 100 µM farnesol for 30 min for Western analysis. The white space indicates noncontiguous lanes from the Western blot. (D) Cup9 is stabilized by farnesol on inoculation. Cup9 stability is monitored by MAL2 promoter shutdown. A Western blot of WT cells carrying Cup9-Myc under the MAL2 promoter inoculated from an overnight culture into fresh YPD media at 37 °C with and without 100 µM farnesol. F, farnesol.
Chk1, a histidine kinase, was reported to be important for inhibition of hyphal development by farnesol. The addition of farnesol blocks hyphal formation by WT cells but not chk1 null mutants (7). However, farnesol could still repress SOK1 expression in a chk1 mutant, like in WT cells or the stp3 mutant (Fig. S3). Therefore, Chk1 is unlikely to function through the same farnesol-sensing pathway as Cup9 in the regulation of SOK1 expression.
Cup9 Is Stabilized by Farnesol.
Cup9 is a homeodomain-containing transcriptional repressor in S. cerevisiae (29). The farnesol-mediated regulation could be at the level of Cup9 localization, expression, or stability. We first examined Cup9 localization by indirect immunofluorescence. Cup9 was tagged at its C terminus with 13Myc, and its expression was under the control of the MAL2 promoter. Cup9-Myc displayed nuclear localization under all conditions with or without farnesol (Fig. S4). We then examined whether Cup9 protein levels changed during inoculation. The protein level of Cup9-myc under its endogenous promoter was high in cells from overnight cultures (Fig. 4C). It decreased rapidly at 30 min on inoculation, regardless of temperature in the absence of farnesol (Fig. 4C). In the presence of farnesol, the protein levels of Cup9 were largely unchanged (Fig. 4C), suggesting that release from farnesol inhibition is important for the down-regulation of the Cup9 protein. We next determined that CUP9 transcription was not regulated by farnesol (Fig. S5). Therefore, the rapid decrease in Cup9 levels during inoculation likely reflects the regulation of Cup9 protein stability by farnesol. With the MAL2 promoter shutoff assay, we found that Cup9 was extremely unstable in the absence of farnesol, because it disappeared at around 15 min after inoculation into YPD medium, regardless of temperature (Figs. 4D and 5A). Adding farnesol stabilized Cup9 (Figs. 4D and 5A). When the MAL2 promoter was shut off for 20 min before inoculating cells into fresh medium to release farnesol inhibition, Cup9 protein completely disappeared by 5 min (Fig. S6). Together, our data suggest that Cup9 is rapidly degraded on loss of farnesol inhibition. The rapid response to release from farnesol also indicates that Cup9 degradation may be directly controlled by farnesol instead of a network of actions brought on by farnesol. These results suggest that Cup9 is stabilized in the presence of farnesol to repress SOK1 expression.
Fig. 5.
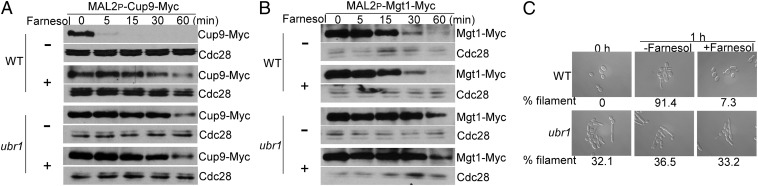
Ubr1 is critical for Cup9 degradation. (A) A Western blot of WT and ubr1 mutant cells carrying Cup9-Myc under the MAL2 promoter inoculated from overnight cultures into fresh YPD media at 30 °C with and without 100 µM farnesol. (B) Farnesol does not affect Ubr1-mediated Mgt1 degradation. Promoter shutoff experiments were performed as described in A. (C) Overnight culture of WT and ubr1 mutant cells was inoculated into YPD with or without 100 µM farnesol at 37 °C for 1 h for cell morphology analysis.
Cup9 Degradation Requires the E3 Ubiquitin Ligase Ubr1.
In S. cerevisiae, Cup9 degradation is blocked in the ubr1 mutant (15). To determine whether a similar regulation exists in C. albicans, we constructed a ubr1 null mutant by sequential gene disruption. Compared with WT cells, Cup9 was much more stable in the ubr1 mutant when saturated cells were inoculated into fresh medium without farnesol (Fig. 5A). Cup9 is likely a direct target of the Ubr1 E3 ubiquitin ligase, because the N-terminal residue of Cup9 is Lys, an N-end rule pathway degron for the type 1 binding site of Ubr1 (12, 13, 30, 31). The stability of Cup9 in the ubr1 mutant in the absence of farnesol was reminiscent of its stability in WT cells in the presence of farnesol (Fig. 5A). Furthermore, adding farnesol could not make Cup9 more stable in the ubr1 mutant, suggesting that the effect of farnesol on Cup9 stability was mediated through Ubr1. To determine if farnesol acts through regulating Ubr1 or specific targets of Ubr1, such as Cup9, we examined if any other Ubr1-mediated protein degradation is inhibited by farnesol. Mgt1, an O6-alkylguanine-DNA alkyltransferase, is degraded by Ubr1 in S. cerevisiae (18). We found that the degradation of C. albicans Mgt1 also required Ubr1 (Fig. 5B). However, unlike Cup9, the Ubr1-mediated degradation of Mgt1 was not affected by farnesol (Fig. 5B). These results suggest that farnesol is specifically involved in the Ubr1-mediated degradation of Cup9 but does not regulate protein degradation of all Ubr1 targets.
Because Cup9 was constitutively stabilized in the ubr1 mutant, the mutant should be impaired in hyphal initiation. Consistent with this prediction, the ratio of elongated cells in the ubr1 mutant did not increase significantly after cells were inoculated into YPD medium at 37 °C (Fig. 5C), indicating that Ubr1 is important for temporal dynamics of hyphal initiation. Unexpectedly, about 32% of the ubr1 mutant cells were elongated in overnight cultures at 30 °C (Fig. 5C). The higher tendency to develop filaments in the ubr1 mutant could mean that stability of additional proteins is also under the control of Ubr1 and that some of the other Ubr1 targets are involved in hyphal development. It is also possible that Ubr1 directly controls the degradation of Nrg1; thus, the ubr1 mutant is defective in hyphal initiation. To examine this possibility, we performed a promoter shutoff experiment to assess Nrg1 protein degradation in the ubr1 mutant. As shown in Fig. S7, Nrg1 protein degradation was blocked in the ubr1 mutant when saturated cells were inoculated into fresh medium. However, when SOK1 was constitutively expressed, Nrg1 was degraded rapidly, even in the ubr1 mutant (Fig. S7), indicating that Ubr1 indirectly controls Nrg1 degradation for hyphal initiation through the regulation of Cup9 stability. Our data show that Ubr1-mediated Cup9 degradation is activated on inoculation for hyphal initiation and inhibited by addition of farnesol. This dynamic change in Cup9 degradation and subsequently, Nrg1 protein level is limited to a short time period during inoculation, because similar levels of Nrg1 protein were detected from overnight cultures of the cup9 or sok1 mutant compared with the level of Nrg1 in WT cells (Fig. S8).
Discussion
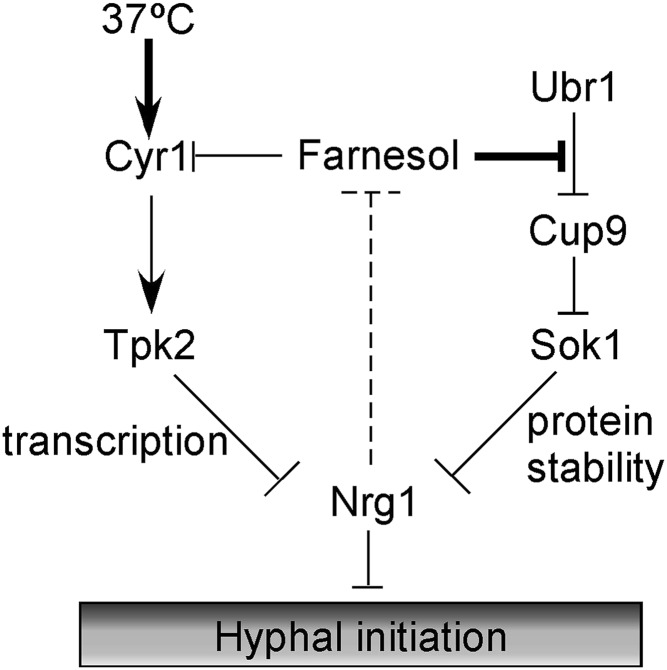
Farnesol is a natural product that is made endogenously from isoprene compounds in plants, animals, and fungi. The most known biological function of farnesol is as a quorum-sensing molecule that inhibits hyphal initiation (2), and this function seems to be specific to C. albicans (32). Because the ability to develop hyphae is a key virulence attribute of C. albicans (33, 34), uncovering molecular mechanisms involved in farnesol sensing and inhibition of hyphal initiation is important for understanding its role in pathogenesis. We have previously shown that transcriptional down-regulation of NRG1 expression by the activation of the cAMP-PKA pathway is essential for hyphal initiation (9). In this study, we identified a cAMP-PKA–independent farnesol-sensing pathway that is essential for hyphal initiation. During inoculation, when cells are released from farnesol inhibition, the Cup9 transcriptional repressor is degraded, allowing the expression of Sok1 and subsequent induction of Nrg1 protein degradation. In the presence of farnesol, Cup9 is stable, thereby repressing Sok1 expression; the repression of Sok1 expression, in turn, blocks Nrg1 degradation and hyphal initiation (Fig. 6). There may be an internal feedback loop in the system to balance between Nrg1 level and farnesol production, because the nrg1 mutant produces about 19 times more farnesol than the WT strain (Fig. 6, dashed line) (35). Together with our previous report (9), our data show that the temporary clearing of Nrg1 protein during hyphal initiation is achieved by two independent regulations: the cAMP-dependent transcriptional down-regulation of NRG1 and the degradation of Nrg1 protein, which is triggered by the release from farnesol inhibition during inoculation. Both pathways are required for the rapid clearing of Nrg1 protein, and neither one is sufficient for hyphal initiation. This requirement for the activation of both pathways explains previous genetic studies that suggest that the Ras1-cAMP pathway and Tup1-Nrg1 function in the farnesol-sensing pathway during hyphal induction (8, 21, 35, 36). Our study provides the underlying mechanism for how the different signals (e.g., nutritional signal vs. cell density signal) are integrated in the regulation of hyphal initiation in C. albicans.
Fig. 6.
A schematic diagram depicting the two pathways involved in down-regulation of Nrg1 protein during hyphal initiation. NRG1 transcriptional down-regulation requires the activation of the cAMP-PKA pathway, whereas Nrg1 protein degradation requires release from farnesol inhibition.
How do cells sense farnesol? Other than inhibition of hyphal initiation in C. albicans, many studies have shown that farnesol has growth-inhibitory and apoptosis-inducing effects on both mammalian cells and fungi (37–40). Several signaling pathways or cellular processes have been implicated in the apoptosis effects of farnesol, but a common underlying mechanism for how farnesol is sensed by mammalian or fungal cells is still elusive. Our study shows that Cup9 protein stability is regulated by farnesol. Cup9 is stable in the presence of farnesol but rapidly degraded on release from farnesol inhibition during inoculation in a Ubr1-dependent manner. The rapid response to release from farnesol inhibition in Cup9 degradation indicates a possible direct action of farnesol on Ubr1-mediated Cup9 degradation. This function of farnesol is specific to Cup9, because farnesol does not affect the degradation of another Ubr1 target. In S. cerevisiae, degradation of Cup9, the transcriptional repressor of the di- and tripeptide transporter PTR2, is also dependent on Ubr1 (14, 15). Ubr1 is the only E3 of the N-end rule pathway in S. cerevisiae. It contains at least three substrate binding sites in the N-terminal one-half of the protein. The type 1 and type 2 sites are specific for basic and bulky hydrophobic N-terminal residues, respectively. The third binding site recognizes an internal (non–N-degron) degradation signal in Cup9, denoted i (12, 13, 41). The i site is autoinhibited by the C-terminal domain Ubr1. Binding of the Arg-Ala and/or Leu-Ala dipeptide to type 1 and/or type 2 sites of Ubr1 induces a conformational change, which unmasks the i site and allows Cup9 binding with Ubr1, leading to Cup9 degradation (14, 15, 41). Therefore, dipeptides with destabilizing N-terminal residues can bind to Ubr1 and activate Cup9 degradation; consequently, they derepress the expression of PTR2 and increase the cell’s capacity to import more peptides, resulting in a positive feedback. However, dipeptides did not seem to affect C. albicans Cup9 stability or germ-tube formation (Fig. S9), indicating a difference between S. cerevisae and C. albicans Cup9 proteins in their interactions with Ubr1. The N-terminal residue of C. albicans Cup9 is Lys, an N-end rule degron for recognition by the type 1 site of Ubr1; however, S. cerevisiae Cup9 is recognized through the i site of Ubr1, and its N-terminal residue Asn is not an N-end rule degron. In addition, farnesol inhibition of Cup9 degradation in C. albicans is reminiscent to hemin inhibition of Ubr1 (16). Hemin can block the dipeptide-induced conformational change of Ubr1 and consequently, Cup9 degradation in S. cerevisiae. It is possible that farnesol adopts a similar mechanism to inhibit the binding of Ubr1 to Cup9 in C. albicans. Additional experiments are needed to uncover the molecular mechanism of the Ubr1-mediated Cup9 degradation by farnesol.
Living as a commensal, C. albicans must adapt and respond to environmental cues generated by the mammalian host and microbes comprising the natural microbiota. The human gastrointestinal (GI) tract is colonized with trillions of commensal microbes, including Candida species. Interestingly, C. albicans stays mostly as the yeast form in the GI tract (42). Farnesol may play a role, because the cup9 mutant displayed slightly increased colonization in the GI tract in a mouse model of intestinal colonization with pooled mutants (43). In addition to farnesol, other quorum-sensing molecules in the host’s gut microbiota may also regulate hyphal development of C. albicans. For example, the quorum-sensing molecule 3-oxo-C12-homoserine lactone, which is secreted by Pseudomonas aeruginosa, can also inhibit the yeast-to-hyphal transition (2, 22, 44). Other conditions unique to the host GI tract, such as high iron levels, can also contribute to the growth state of the commensal pathogen (45). Our study links the N-end rule pathway to hyphal development in C. albicans. Instead of being a regulator of di- and tripeptide transport, Cup9 is used to control the yeast–hyphal transition in response to the cell density cue (farnesol) in C. albicans. This study provides an example of how a commensal pathogen rewires its transcriptional circuits to adapt to varied host environments to become a successful commensal and a pathogen.
Materials and Methods
Media and Growth Conditions.
Hyphal inductions were performed as follows. Strains were grown overnight in liquid YPD at 30 °C, pelleted, washed two times in PBS, resuspended in an equal volume of PBS, and diluted 1:100 in YPD medium with or without farnesol at 37 °C.
Promoter Shutdown Assays.
C. albicans strains containing Nrg1-Myc or Cup9-Myc under the regulation of the MAL2 promoter were grown in yeast extract–peptone plus 2 g/100 mL maltose at 30 °C overnight to induce the expression of Nrg1-Myc or Cup9-Myc. The overnight culture was diluted 1:100 into fresh YPD medium at 30 °C to shut off the promoter. Aliquots were collected after the times indicated.
Supplementary Material
Acknowledgments
We thank Drs. A. P. Mitchell and A. D. Johnson for providing the kinase deletion collection and the transcriptional factor deletion collection to the research community through the Fungal Genetics Stock Center. This work is supported by the National Institutes of Health Grants R01GM/AI55155 and R01AI099190 (both to H.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318690111/-/DCSupplemental.
References
- 1.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 2.Hornby JM, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67(7):2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao YY, et al. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Chemother. 2005;49(2):584–589. doi: 10.1128/AAC.49.2.584-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramage G, Saville SP, Wickes BL, López-Ribot JL. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 2002;68(11):5459–5463. doi: 10.1128/AEM.68.11.5459-5463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornby JM, Kebaara BW, Nickerson KW. Farnesol biosynthesis in Candida albicans: Cellular response to sterol inhibition by zaragozic acid B. Antimicrob Agents Chemother. 2003;47(7):2366–2369. doi: 10.1128/AAC.47.7.2366-2369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosel DD, Dumitru R, Hornby JM, Atkin AL, Nickerson KW. Farnesol concentrations required to block germ tube formation in Candida albicans in the presence and absence of serum. Appl Environ Microbiol. 2005;71(8):4938–4940. doi: 10.1128/AEM.71.8.4938-4940.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruppa M, et al. The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot Cell. 2004;3(4):1062–1065. doi: 10.1128/EC.3.4.1062-1065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol. 2008;67(1):47–62. doi: 10.1111/j.1365-2958.2007.06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011;9(7):e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 11.Bartel B, Wünning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia Z, et al. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J Biol Chem. 2008;283(35):24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tasaki T, et al. The substrate recognition domains of the N-end rule pathway. J Biol Chem. 2009;284(3):1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Z, Turner GC, Hwang CS, Byrd C, Varshavsky A. Amino acids induce peptide uptake via accelerated degradation of CUP9, the transcriptional repressor of the PTR2 peptide transporter. J Biol Chem. 2008;283(43):28958–28968. doi: 10.1074/jbc.M803980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405(6786):579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 16.Hu RG, Wang H, Xia Z, Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci USA. 2008;105(1):76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu RG, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437(7061):981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 18.Hwang CS, Shemorry A, Varshavsky A. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc Natl Acad Sci USA. 2009;106(7):2142–2147. doi: 10.1073/pnas.0812316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410(6831):955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 20.Bockmühl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol Microbiol. 2001;42(5):1243–1257. doi: 10.1046/j.1365-2958.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- 21.Hogan DA, Muhlschlegel FA. Candida albicans developmental regulation: Adenylyl cyclase as a coincidence detector of parallel signals. Curr Opin Microbiol. 2011;14(6):682–686. doi: 10.1016/j.mib.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Hall RA, et al. The quorum-sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans. Eukaryot Cell. 2011;10(8):1034–1042. doi: 10.1128/EC.05060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6(2):e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward MP, Garrett S. Suppression of a yeast cyclic AMP-dependent protein kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally regulated mouse gene. Mol Cell Biol. 1994;14(9):5619–5627. doi: 10.1128/mcb.14.9.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazan I, Sepulveda-Becerra M, Liu H. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol Biol Cell. 2002;13(1):134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5(12):e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan G, et al. Bcr1 plays a central role in the regulation of opaque cell filamentation in Candida albicans. Mol Microbiol. 2013;89(4):732–750. doi: 10.1111/mmi.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su C, Lu Y, Liu H. Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Mol Biol Cell. 2013;24(3):385–397. doi: 10.1091/mbc.E12-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrd C, Turner GC, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17(1):269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 31.Varshavsky A, Bachmair A, Finley D. The N-end rule of selective protein turnover: Mechanistic aspects and functional implications. Biochem Soc Trans. 1987;15(5):815–816. doi: 10.1042/bst0150815. [DOI] [PubMed] [Google Scholar]
- 32.Nickerson KW, Atkin AL, Hornby JM. Quorum sensing in dimorphic fungi: Farnesol and beyond. Appl Environ Microbiol. 2006;72(6):3805–3813. doi: 10.1128/AEM.02765-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao F, et al. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol Biol Cell. 2006;17(1):295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo H-J, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 35.Kebaara BW, et al. Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction. Eukaryot Cell. 2008;7(6):980–987. doi: 10.1128/EC.00357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langford ML, Atkin AL, Nickerson KW. Cellular interactions of farnesol, a quorum-sensing molecule produced by Candida albicans. Future Microbiol. 2009;4(10):1353–1362. doi: 10.2217/fmb.09.98. [DOI] [PubMed] [Google Scholar]
- 37.Joo JH, Jetten AM. Molecular mechanisms involved in farnesol-induced apoptosis. Cancer Lett. 2010;287(2):123–135. doi: 10.1016/j.canlet.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semighini CP, Hornby JM, Dumitru R, Nickerson KW, Harris SD. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol Microbiol. 2006;59(3):753–764. doi: 10.1111/j.1365-2958.2005.04976.x. [DOI] [PubMed] [Google Scholar]
- 39.Shirtliff ME, et al. Farnesol-induced apoptosis in Candida albicans. Antimicrob Agents Chemother. 2009;53(6):2392–2401. doi: 10.1128/AAC.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, et al. Farnesol-induced apoptosis in Candida albicans is mediated by Cdr1-p extrusion and depletion of intracellular glutathione. PLoS One. 2011;6(12):e28830. doi: 10.1371/journal.pone.0028830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc Natl Acad Sci USA. 2002;99(22):14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White SJ, et al. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3(12):e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez JC, Kumamoto CA, Johnson AD. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol. 2013;11(3):e1001510. doi: 10.1371/journal.pbio.1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54(5):1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 45.Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 2011;10(2):118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.