Significance
This paper describes the use of natural variation to investigate a cell-autonomous function in the experimentally well-studied model Schizosaccharomyces pombe. We determined the genome structure of a natural isolate with four chromosomes, rather than the three present in the laboratory strain and in all other natural isolates studied to date. Two of the centromeres in this variant lack adjacent heterochromatin but assemble their kinetochores normally, demonstrating that kinetochore and heterochromatin assembly are autonomous in this organism.
Keywords: yeast, microbial diversity, epigenesis
Abstract
Kinetochores in multicellular eukaryotes are usually associated with heterochromatin. Whether this heterochromatin simply promotes the cohesion necessary for accurate chromosome segregation at cell division or whether it also has a role in kinetochore assembly is unclear. Schizosaccharomyces pombe is an important experimental system for investigating centromere function, but all of the previous work with this species has exploited a single strain or its derivatives. The laboratory strain and most other S. pombe strains contain three chromosomes, but one recently discovered strain, CBS 2777, contains four. We show that the genome of CBS 2777 is related to that of the laboratory strain by a complex chromosome rearrangement. As a result, two of the kinetochores in CBS 2777 contain the central core sequences present in the laboratory strain centromeres, but lack adjacent heterochromatin. The closest block of heterochromatin to these rearranged kinetochores is ∼100 kb away at new telomeres. Despite lacking large amounts of adjacent heterochromatin, the rearranged kinetochores bind CENP-ACnp1 and CENP-CCnp3 in similar quantities and with similar specificities as those of the laboratory strain. The simplest interpretation of this result is that constitutive kinetochore assembly and heterochromatin formation occur autonomously.
The fission yeast Schizosaccharomyces pombe is a key system for the experimental study of centromere function. To date, all of this work has involved the strain 968h90 and its derivatives 972h− and 975h+, each of which contains three chromosomes. Each of these chromosomes contains a kinetochore embedded in heterochromatin (1) and resembles the centromeres of many metazoan organisms; consequently they have been intensively studied. These studies have demonstrated that heterochromatin is necessary for the formation of new kinetochores (2–4), but that once formed, kintetochores (5) can be maintained on a circular plasmid in the absence of heterochromatin. In evolution, novel centromeres form on regions of chromosomes lacking heterochromatin but subsequently acquire it (6, 7), an order of events at odds with the conclusions of published studies of S. pombe. Thus, the relationship between heterochromatin and kinetochore assembly in S. pombe requires investigation using new approaches.
A study of 88 independent natural isolates of S. pombe showed that this fission yeast is karyotypically variable, and that one Japanese strain, CBS 2777, contains four chromosomes (8). In the present study, we determined the genome sequence and organization of CBS 2777 and found that it is related to the karyotype of the laboratory strains by a complex rearrangement. We show that two of the centromeres in CBS 2777 lack flanking heterochromatic repeated sequences. This DNA is not detectably associated with siRNA and does not show any detectable enrichment of Rad21 (cohesin) binding. Despite these differences in flanking DNA, the rearranged centromeres in CBS 2777 bind the constitutive centromere proteins Cnp1 (CENP-A) and Cnp3 (CENP-C) similarly to the unrearranged centromere. The simplest interpretation of these results is that constitutive heterochromatin and constitutive kinetochore assembly occur autonomously.
Although the difference in genome organization between the laboratory strains and CBS 2777 is complex and resembles the rearrangements seen in fungal plant pathogens (9) and in some types of cancer cells (10), it can be explained by a sequence of simple events.
Results
CBS 2777 Genome Structure.
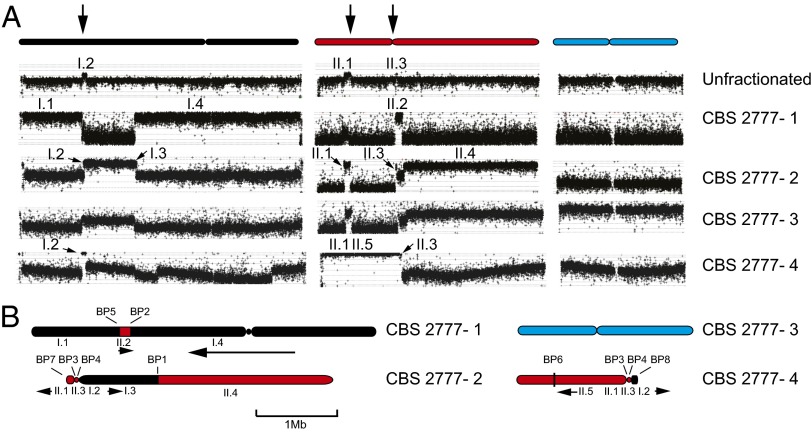
We used array-based comparative genome hybridization (CGH) to identify the components of the individual chromosomes of CBS 2777 (for which Arabic numerals are used, with the 972h− chromosomes designated by Roman numerals; Fig. 1A) and PCR combined with agarose gel analysis to link them together (SI Appendix, Figs. S1–S5). We found that CBS 2777 chromosomes 1, 2, and 4 are composed of sequences normally found on 972h− chromosomes I and II (Fig. 1B). Sequencing across breakpoints indicated that the breaks were typically reciprocal and conservative and characterized by little or no homology (SI Appendix).
Fig. 1.
Genome organization of S. pombe strain CBS 2777. (A) Sequence content of size- fractionated chromosomes of CBS 2777. Either unfractionated CBS 2777 DNA or DNA from each of the four chromosomes that had been size-fractionated by pulsed-field gel electrophoresis was analyzed by CGH using Agilent 4 × 44K ChIP-on-ChIP arrays (G4810A-015424). The results are aligned with respect to the three laboratory strain (972h−) chromosomes listed at the top of the panel and color-coded black, red, and blue, respectively. Chromosome 3 of CBS 2777 is not detectably rearranged, but the DNA used in the CGH was contaminated with CBS 2777 chromosome 2 as predicted, given the small difference in their respective sizes. Segments of the 972h− chromosomes present in the CBS 2777 chromosomes are labeled according to their chromosomal origin using Roman numerals. Segment II.5 extends from II:569,645 to the imr2 left repeat of chromosome II and corresponds to an inversion to which the CGH is insensitive. The three vertical arrows at the top of the figure refer to the three regions of the laboratory strain karyotype that are duplicated around the centromeres of CBS 2777 chromosomes 2 and 4. (B) Arrangement of the sequences identified in A on the CBS 2777 chromosomes. The arrows below the chromosomes indicate whether the respective segment is in the same orientation as in the 972h− genome as drawn at the top or inverted (running right to left). Each of the eight breakpoints (BP 1–8) defining the CBS 2777 karyotype is indicated.
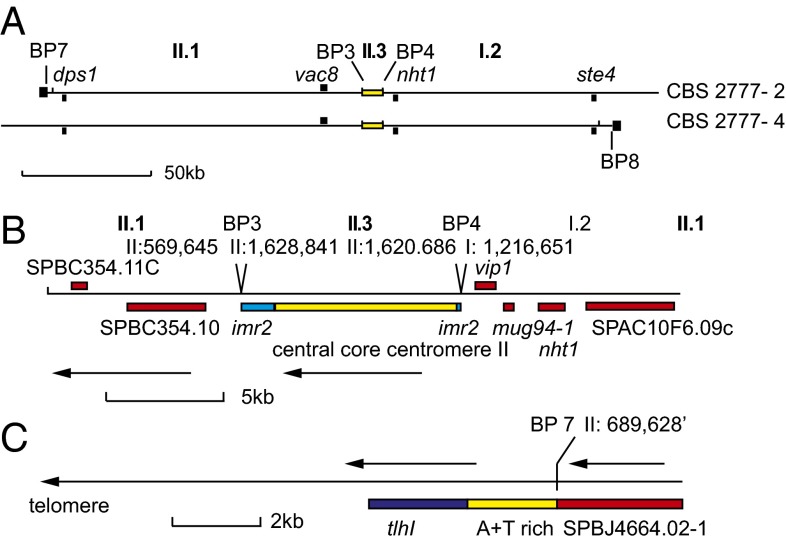
The central core of the centromere of chromosome II was duplicated in the genome of CBS 2777. One copy was present on both chromosomes 2 and 4, and each was flanked by two segments of DNA, termed I.2 and II.1, each ∼100 kb long, that are normally present in the centromere-distal regions of the short arms of the laboratory strain chromosomes (Fig. 2 A and B). Neither segment is associated with large amounts of heterochromatin in the laboratory strain. In CBS 2777, one copy of each segment is terminated by sequences (Fig. 2C) that include the telomeric helicase gene (tlh1 in S. pombe) (11), a feature common to the telomeres of most fungi and yeast (Fig. 2C). Thus, these telomeric ends define 210 kb that is held in common between chromosomes 2 and 4 and that accounts for the duplicated sequences seen in the CGH of unfractionated CBS 2777 DNA (Fig. 1A).
Fig. 2.
Organization of the centromeric and telomeric sequences of chromosomes 2 and 4 in S. pombe strain CBS 2777. (A) Sequences held in common between the centromeres of CBS 2777 chromosomes 2 and 4. Breakpoints 7 and 8 are at the left and right ends of CBS 2777 chromosomes 2 and 4, respectively, and are adjacent to the black squares. The genes used in the silencing and structural analyses (described in SI Appendix) are indicated in the upper part of the panel. The yellow rectangle indicates the position of the central core sequences of chromosome 2. The black squares at the ends of the two idiograms represent the telomeres. (B) Sequence organization around the centromere central core sequences on the centromeres of CBS 2777 chromosomes 2 and 4. Genes are indicated in red, imr repeats are in blue, and the centromere central core sequences are in yellow. The sequence position at breakpoints 3 and 4 are indicated with respect to the 972h− genome reference sequence. The arrows indicate that the sequence is inverted with respect to the 972h− genome reference assembly. (C) Sequence organization around the telomere at breakpoint 7. A similar arrangement of the tlh1 gene is seen at breakpoint 8. Details of the sequences and breakpoints are provided in SI Appendix.
This map of the CBS 2777 genome was confirmed by DNA sequencing (SI Appendix), which identified the common pericentric inversion of chromosome I (8) but no further rearrangements. The short-read sequencing method that we used would not have been sensitive to rearrangements within repeated sequences longer than 300 bp, and thus we cannot exclude the presence of inversions with such breakpoints.
Along with 972h−, S. pombe type strain NCYC 132 and the Kambucha strain (YFS 276) have been sequenced as well (12). We wanted to know how recently the rearrangement occurred, and so evaluated the relationships between CBS 2777 and these three strains. The percentage of single nucleotide divergence (coding and noncoding) between 972h− and the S. pombe Kambucha strain, S. pombe type strain NCYC 132, and CBS 2777 were 0.317%, 0.437%, and 0.447%, respectively. This indicates that CBS 2777 is as closely related to 972h− as the S. pombe type strain (NCYC 132), but is slightly more divergent than the Kambucha strain (YFS 276). The extent of polymorphism among these four strains was not uniform across the genome, but rather showed a segmental pattern (SI Appendix, Fig. S6), suggesting that all four strains are mosaics that recently descended from a small pool of relatively diverse natural founder strains, as has been observed for Saccharomyces cerevisiae industrial isolates (13). The two other Japanese strains, CBS 2775 and 2776 (8), were isolated at the same time and in the same location as CBS 2777 and have the same genotype, but contain three chromosomes. The close relationships among these three strains also suggest that the chromosome rearrangement in CBS 2777 is of recent origin.
Cnp1 and Cnp3 Binding at the Rearranged Centromeres in S. pombe CBS 2777.
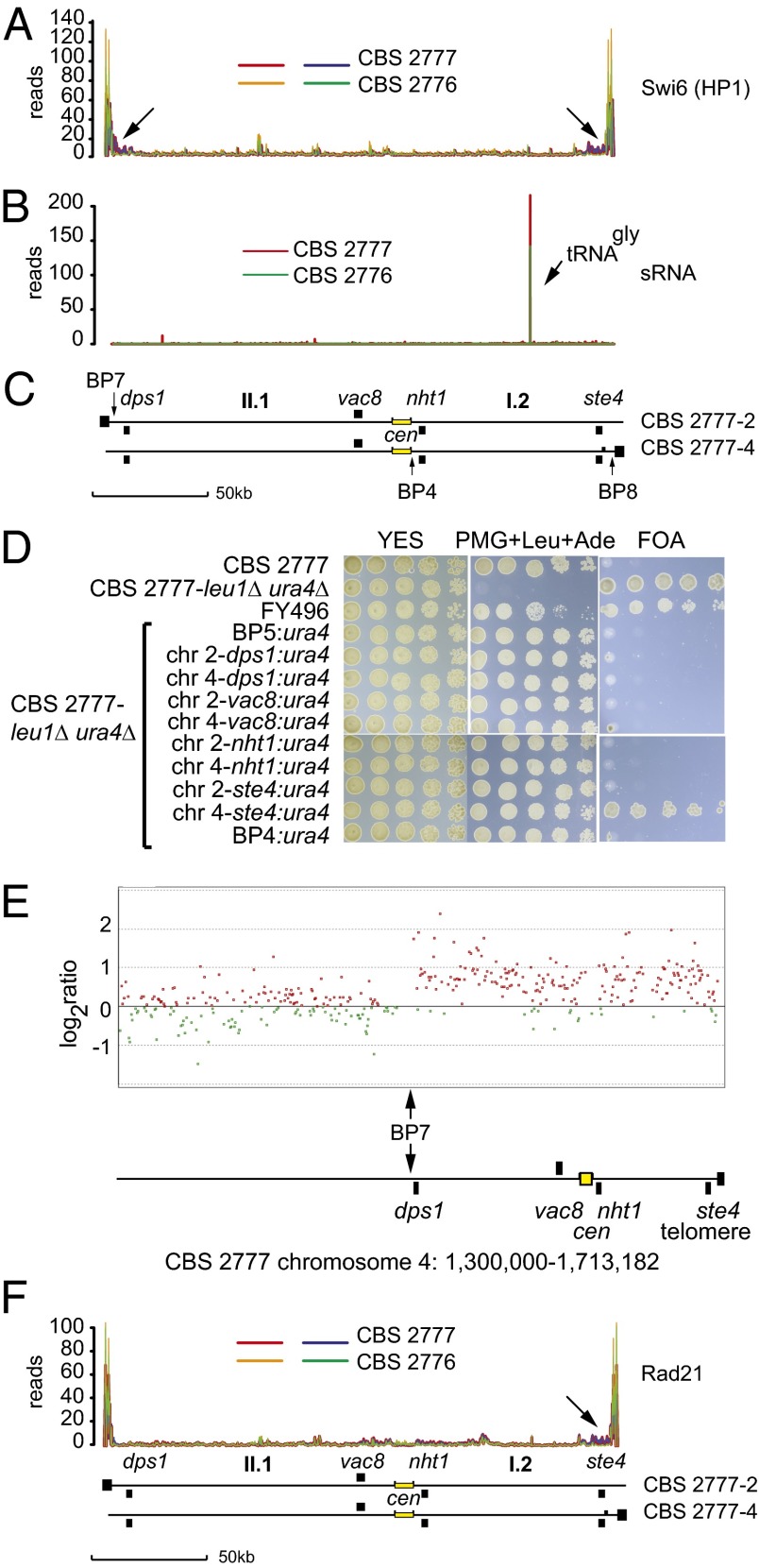
Our structural analysis of the CBS 2777 genome showed that chromosomes 2 and 4 contain centromeric central core sequences, but that these are adjacent to sequences not associated with heterochromatin in the laboratory strain. Heterochromatin has been suggested to promote kinetochore assembly, and so it was of interest to compare the binding of the centromere components Cnp1 and Cnp3 to the chromosomes of CBS 2777 and CBS 2776, an isogenic natural isolate with the three chromosome karyotype characteristic of the laboratory strain (8). To do so, we constructed monomeric GFP amino terminus tagged Cnp1 (CENP-A) strains as described previously (14) and strains in which Cnp3 (CENP-C) was tagged at the carboxyl terminus with the PK9 epitope, and carried out sequencing of DNA coimmunoprecipitated with the respective tagged protein (ChIP-seq). We found that the centromere central core and flanking sequences were the only major sites of binding of both of these proteins on the rearranged chromosomes 2 and 4 of CBS 2777 (Fig. 3A). Minor sites of binding either at the tdh1 gene on chromosome 2 (indicated by arrows in Fig. 3A) or at breakpoint 6 on chromosome 4 almost certainly arose as a result of ectopic mapping of ChIP-seq reads (Fig. 3). Thus, based on these data, we conclude that despite the absence of adjacent dgdh repeated sequences, the central core sequences are the functional centromeres on these chromosomes.
Fig. 3.
Kinetochore assembly around the centromeric DNA of CBS 2777 chromosomes 2 and 4. (A) Localization of the kinetochores on chromosomes 2 and 4 of CBS 2777 by ChIP-seq of epitope-tagged Cnp3 and Cnp1. The sequences mapping to BP6 on chromosome 4 are derived from the fragment of the imr2 element left behind at this position as a result of the rearrangement and are assumed to be artifacts of the sequence mapping that cannot discriminate between reads originating from this copy of the imr2 and the copy at the centromere. The arrows in the chromosome 2 panels illustrate peaks at a position corresponding to the tdh1 gene. Tdh1 is the paralog of the gpd3 gene present at breakpoint 3, and we assume that the reads also have been assigned ectopically. The ChIP-seq experiments for each tagged protein were duplicated, and these are color-coded. Only those reads in which the mates map within 180 bp and 250 bp of one another were used in this mapping. (B) Localization of Cnp1 and Cnp3 across the centromeres of CBS 2777 chromosomes 2 and 4 by ChIP-seq of epitope-tagged Cnp3 and Cnp1. (Inset) Color-coding. The arrow indicates the peak of binding indicated in D. (C) Localization of Cnp1 and Cnp3 across the unrearranged centromere 2 of CBS 2776 by ChIP-seq of epitope-tagged Cnp3 and Cnp1. (Inset) Color-coding of the respective analyses. (D) Comparison of the binding of Cnp1 and Cnp3 to centromere-adjacent DNA in CBS 2777 and to the corresponding sequences in CBS 2776. The results show the numbers of uniquely mapping reads derived from the respective sequences in each of the two strains, annotated according to the laboratory strain coordinates along chromosome II. (Upper) The arrow indicates the peak just beyond the breakpoint 3 sequences at chromosome II, 569645, which is also indicated in B.
We wished to know whether these central core sequences bind similar amounts of these key kinetochore components as the unrearranged central core sequence on chromosome II of CBS 2776. Thus, we counted the number of reads in the Cnp1 and Cnp3 ChIP-seq experiments that were specifically derived from the centromeric loci of chromosomes 2 and 4 in CBS2777 and from the centromere of the chromosome II of CBS 2776. Comparing these counts, after correcting for differences in the efficiency of ChIP between the two strains, showed that the total number derived from the centromeric loci of chromosomes 2 and 4 in CBS2777 was increased by an estimated factor of 1.84-fold for Cnp1 and 1.41-fold for Cnp3. The pattern of Cnp1 and Cnp3 binding to the central core sequences of centromere II in both CBS 2777 and CBS 2776 was similar as well (Fig. 3C). Given the interexperimental variability of the ChIP-seq technique, these results suggest that the presence of adjacent heterochromatic DNA does not play a significant role in determining Cnp1 and Cnp3 binding at the centromere. Comparing the levels of Cnp1 and Cnp3 binding to the rearranged centromeres (Fig. 3A) and to the unrearranged centromeres (SI Appendix, Fig. S7) within CBS 2777 confirmed that all of the centromeres bound similar amounts of both proteins regardless of the presence of heterochromatin.
The ChIP-seq data also demonstrate that Cnp1 and Cnp3 bind to DNA beyond the laboratory strain centromere sequences adjacent to breakpoints 3 and 4 and spread into the flanking sequences (Fig. 3 B and D, with arrows indicating corresponding peaks). The spreading of both proteins is correlated and extends as far as 50 kb from the boundary with the imr2 sequences (Fig. 3D). Similar spreading was seen on both sides, suggesting that in some cells, the functional centromere may include as much as 100 kb of DNA.
Lack of Heterochromatin Adjacent to the Rearranged Centromeres in S. pombe CBS 2777.
Centromeric dgdh repeats are associated with the presence of heterochromatin (15), and thus the Cnp1 and Cnp3 ChIP-seq results suggest that heterochromatin plays no significant direct role in kinetochore maintenance in S. pombe cells proliferating in the natural environment. To test this idea, we needed to exclude the possibility that heterochromatin was forming ectopically on the sequences II.1 and I.2 in CBS 2777. We did so through four different experimental approaches. In the first approach, we tagged the heterochromatin protein Swi6 (HP1) at the amino terminus with GFP and compared binding across the I.2 and II.1 sequences in the two strains by ChIP-seq. Swi6 demonstrated the same amount and distribution of binding to the I.2 and II.1 sequences immediately adjacent to the centromeres in CBS 2777 as to the unrearranged sequences in CBS 2776 (Fig. 4A), but with increased binding in CBS 2777 around the dps1 gene (arrows in Fig. 4A), which was subtelomeric on CBS 2777 chromosome 2, and around the ste4 gene, which was subtelomeric on CBS2777 chromosome 4. Although the I.2 and II.1 sequences are considered euchromatic and distinct from the heterochromatic sequences found at centromeres and telomeres, Fig. 4A clearly shows low levels of Swi6 binding across the entire interval in both strains.
Fig. 4.
Absence of heterochromatin or an increase in Rad21 binding around the centromeres of CBS 2777 chromosomes 2 and 4. (A) Comparison of the binding of Swi6 (HP1) to the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 in strains CBS 2777 and CBS 2776 as measured by ChIP-seq on GFP epitope-tagged Swi6 (HP1). The data from the duplicate experiments of the CBS 2777 are shown in red and blue, and those from CBS 2776 are shown in green and orange. Arrows indicate the increased binding of Swi6 to dps1 and ste4 in CBS 2777. (B) Small RNAs derived from the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 in strains CBS 2777 and CBS 2776. (C) Organization of the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 aligned with respect to the data shown in A and B. (D) Absence of gene silencing in the pericentric DNA of CBS 2777 chromosomes 2 and 4. Silencing activity was assayed by testing the growth of URA+ cells on 5-FOA. Cells were grown to saturation in YES and inoculated in serial fivefold dilutions starting from an initial concentration of ∼7.5 × 103 on agar plates of the indicated composition. In FY496 (27), the ura4 gene was placed into the imr1L repeat of 972h−; this strain served as a positive control for the silencing seen in CBS 2777. (E) Absence of repression of gene expression around the acrocentric centromeres of CBS 2777 chromosome 4. Total RNA was extracted from either CBS 2777 or CBS 2776 cells, and cRNA probes were prepared and labeled with CY3 or CY5, respectively, and analyzed by competitive hybridization to a custom 4 × 44K gene expression microarray. The results were processed and analyzed using Agilent Genespring software. Red and green points mark genes with greater or lesser expression, respectively, in CBS 2777 compared with CBS 2776. The region around the centromere of CBS 2777 chromosome 4 is illustrated. The vertical arrow marks the junction with the region duplicated on chromosome 2 defined by breakpoint 7. (F) Comparison of the binding of Rad21 to the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 in strains CBS 2777 and CBS 2776 as measured by ChIP-seq on PK9 epitope-tagged Rad21. The arrow indicates the increase in binding of Rad21 to ste4 in CBS 2777. The traces in F are aligned with those in A and B, and this is indicated by the idiogramatic representation (C) of the centromeric region of chromosomes 2 and 4 placed below the three sets of traces.
Our second approach was to analyze the small RNA molecule content of CBS 2777 and CBS 2776 by RNA sequencing. No detectable siRNA was derived from sequences I.2 and II.1 in either CBS 2776 or CBS 2777 (Fig. 4B). In addition, there were no qualitative or quantitative differences between the total siRNA derived across the genome of the two strains (SI Appendix, Fig. S8), demonstrating that siRNA formation occurs in a position-independent manner.
In our third experiment, we constructed a CBS 2777 leu1Δ ura4Δ strain and introduced a ura4 transgene into breakpoint 5 on chromosome 1, into breakpoint 4 at the edge of the CNP1 and CNP3 binding region, into the vac8 and nht1 loci that are centromere-proximal on chromosomes 2 and 4, and into the dps1 and ste4 loci that are centromere-distal but telomere-proximal on chromosomes 2 and 4 (Fig. 4C). We then assayed heterochromatin-associated gene silencing by the ability to proliferate in the presence of the antimetabolite 5-fluoro-orotic acid (5-FOA). The positive control in these experiments was the strain FY496 (15), in which a ura4 transgene is placed in the imr1 repeat on the centromere of chromosome 1 of the laboratory strain. The ura4 gene showed no detectable silencing when placed immediately adjacent to the centromere in breakpoint 4 or in either of the centromere-proximal loci vac8 or nht1. In contrast, the ura4 transgene was silenced when placed into the subtelomeric copy of ste4 present on chromosome 4. The specific silencing of this subtelomeric copy of the ste4 chromosome 4 locus is consistent with the observation that ste4 is one of the few noncentromeric loci to bind components of the ClrC complex, which is responsible for heterochromatin formation (16). Why the apparent increased binding of Swi6 around breakpoint 7 sequences is not accompanied by silencing at the dps 1 locus on CBS 2777 chromosome 2 requires further experimental investigation.
In our final experiment, we compared the levels of gene expression across the centromeric interval held in common between chromosomes 2 and 4, using two-color microarrays with the isogenic unrearranged strain CBS 2776 as the source of control mRNA. Our findings showed that the levels of gene expression across the interval in CBS 2777 were higher, in most cases approximately twofold more, compared with those in the isogenic CBS 2776 (Fig. 4E). Thus, we conclude that the DNA flanking the centromeres on CBS 2777 chromosomes 2 and 4 is not heterochromatic compared with the DNA flanking centromere II of the laboratory strain (SI Appendix, Fig. S9).
No Increase in Binding of Rad21 Cohesin to DNA Adjacent to the Rearranged Centromeres in S. pombe CBS 2777.
Although we had excluded the possibility that the rearranged centromeres in CBS 2777 were associated with the centromere adjacent heterochromatin, it remained possible that they were recruiting cohesin to the flanking DNA by a heterochromatin-independent pathway, and this cohesin was playing a role in the assembly of the kinetochores. To exclude this possibility, we tagged the Rad21 subunit of cohesin with the PK-9 epitope as described previously (17), and then compared binding across the I.2 and II.1 sequences of the two proteins in the two strains by ChIP-seq. We found that Rad21 has the same amount and distribution of binding to the I.2 and II.1 sequences around the centromeres in CBS 2777 as to the unrearranged sequences in CBS 2776 (Fig. 4F). The overall pattern of Rad21 binding was similar to that of Swi6 binding, and both proteins showed increased binding to the ste4 gene, which was subtelomeric on CBS 2777 chromosome 4 (indicated by the arrow in Fig. 4F). However, there was no evidence of increased binding of Rad21 to the dps1 gene, which was subtelomeric on CBS 2777 chromosome 2 and bound increased amounts of Swi6. Fig. 4B clearly shows low levels of Rad21 binding across the whole interval in both strains.
Higher-resolution analysis of the regions of highest binding revealed a correlation between Rad21 binding and Swi6 binding (SI Appendix, Figs. S10 and S11). This pattern of irregular but tightly correlated low-level binding of Swi6 and Rad21 was not exceptional but was observed over the entire euchromatic portion of the S. pombe genome (CBS 2777 chromosome 4 shown in SI Appendix, Fig. S12), with a correlation coefficient of 0.96 (to 2 dp) over the 1-Mb to 3-Mb interval of CBS 2776 chromosome 1, suggesting that Swi6 is associated with cohesin binding across the entire genome as well as at heterochromatin (18, 19).
Discussion
Our analysis of the CBS 2777 chromosome 2 and 4 centromeres showed that constitutive kinetochore assembly in S. pombe does not depend on the presence of heterochromatin in cis. This supports the idea that kinetochore assembly is likely to be similar to how it occurs in other single-celled eukaryotes that have regional centromeres but lack heterochromatin, such as the pathogenic budding yeast Candida albicans (20), and is consistent with the observation that during evolution in metazoan organisms, new centromeres form on DNA lacking heterochromatin (6, 7).
The fact that CBS 2777 is a viable natural isolate does not imply that the centromeres of chromosomes 2 and 4 of CBS 2777 function as accurately as those of the laboratory strain 972h−, however. In optimal conditions, CBS 2777 proliferates more slowly than any of 87 other strains of S. pombe studied earlier (8). This increase in doubling time is consistent with the observation that cells of the laboratory strain deleted for heterochromatin components, such as Swi6, show an excess of lagging chromosomes at cell division. However, preliminary work has shown that any increase in the duration of mitosis in CBS 2777 is too small to account for the increase in doubling time, and thus the effect of the rearrangement on the cell cycle is likely complex. Nevertheless, the decreased rate of proliferation of CBS 2777 under optimal conditions implies that the CBS 2777 rearrangement is maladaptive and hence raises the question of how CBS 2777 arose in the environment. It may have done so simply as a result of genetic drift in a small subpopulation. In the diploid budding yeast S. cerevisiae, genetic drift is a powerful factor driving gene diversification (21). Although S. pombe is almost exclusively haploid, genetic drift also could be a factor in this diversification if S. pombe exists in small subpopulations, as we suggested earlier (8) based on the extensive karyotypic diversity. Another possible explanation is that soft selection on CBS 2777 proliferation is weak in nature, with its fitness determined predominantly by viability and hard selection.
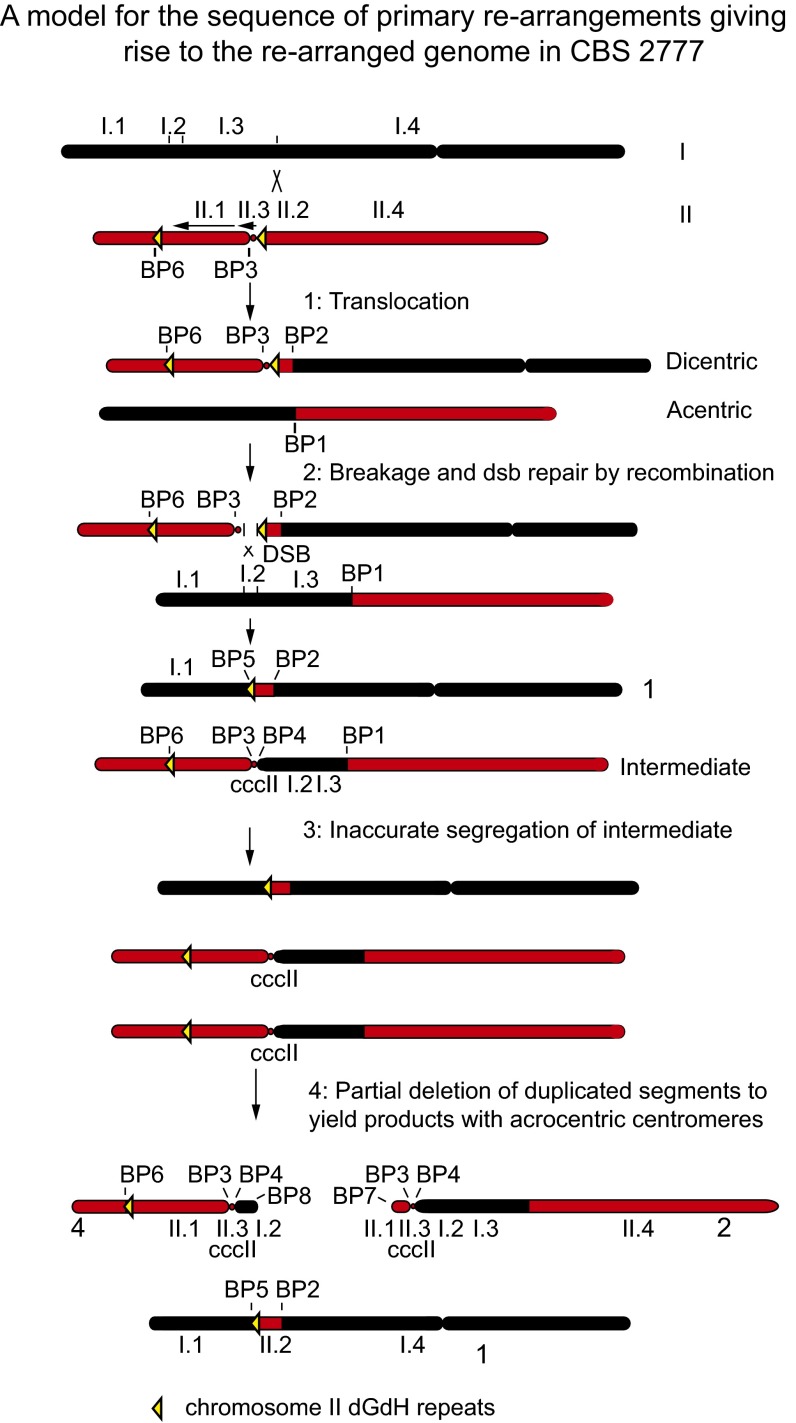
The sequence arrangement in CBS 2777, although complex, can be explained as arising from a series of elementary chromosome rearrangements (Fig. 5). We first assume that chromosome II of the immediate ancestor of CBS 2777 contained an inversion of the central core of the centromere and a paracentric inversion of the short arm between breakpoints designated 3 and 6 in the structure shown in Fig. 1. The first step in the rearrangement was a translocation between the ancestral chromosome I and a copy of the ancestral chromosome II, which generated a dicentric chromosome and an acentric derivative chromosomes and breakpoints 1 and 2. These two derivative chromosomes then entered cell division, which led to breakage of the dicentric chromosome within the right-hand imr2 repeat of the centromere II-derived sequences. The broken ends then triggered nonhomologous DNA repair from the chromosome I-derived part of the acentric chromosome, which in turn generated chromosome 1 of CBS 2777 and an intermediate chromosome containing a metacentric copy of the central core of chromosome II. If this chromosome then segregated inaccurately at cell division because of an absence of nearby heterochromatin to yield a daughter cell with two copies of the intermediate chromosome, then the daughter would be effectively disomic for the chromosome II-derived sequences.
Fig. 5.
Model for the sequence of elementary rearrangements giving rise to the rearranged genome in CBS 2777. See Discussion for details.
Experimental observations suggest that cells with two copies of chromosome II sequences would have lower fitness than monosomic cells in the native environment (22). Correspondingly, we suggest that one of the copies of the intermediate chromosome lost most of the short arm to yield CBS 2777 chromosome 2, and that the other copy lost most of the long arm to yield CBS 2777 chromosome 4. The deletions define breakpoints 7 and 8 (Fig. 1B) and place the central core sequences close enough to telomeres to provide the cohesion necessary for their accurate segregation at cell division and reduce the extent of aneuploidy, such that the cells now outgrow the initial rearrangement. The mechanism by which these deletions occur is unknown, but the sequence beyond breakpoints 7 and 8 that includes the tlh1 gene differ by only four residues, suggesting that both deletions occurred through a sequence-specific mechanism of exchange of subtelomeric DNA.
Although the model set out in Fig. 5 accounts for the CBS 2777 rearrangement, it does not explain why sequences 1.2 and II.1 in CBS 2777 are both ∼100 kb long. A possible explanation is that the pericentromeric DNA may have an architectural function. The centromeres that we have discovered in CBS 2777 should allow investigation of this and other aspects of centromere function without the apparently unnecessary complication of adjacent heterochromatin.
Materials and Methods
Microarray and PCR Analysis of the Rearranged Karyotype.
For microarray analysis, chromosomal DNA was size-fractionated by pulsed-field gel electrophoresis as described previously (8). After electroelution from the gel into dialysis tubing, the DNA was concentrated using butan-2-ol, and then amplified using the Qiagen REPL1-g Mini Kit before labeling using the Agilent DNA ULS labeling kit (5190-0419). Labeled DNA was then purified and hybridized to the Agilent S. pombe 4 × 44k ChIP-on-ChIP array (G4810), using unfractionated reciprocally labeled 972h− DNA as a competitor. Arrays were scanned with an Agilent scanner, and the data were analyzed using the Agilent Genomic Workbench 5.0.14. Oligonucleotide primers were designed on the basis of the reference strain sequence at each end of the segments indicated in Fig. 1A and then linked to one another using conventional PCR for breakpoints 1–6. For breakpoints 7 and 8, the ends of segments I.2 and II.1 were linked to the tlh1 gene using the Expand dNTP Kit (Roche). Breakpoints 3 and 4 were linked similarly. Details of the results, primer sequences (SI Appendix, Table S1), and sequences across the breakpoints are presented in the SI Appendix.
Strain Construction and Yeast Culture.
The ura4 and leu1 genes in CBS 2777 were deleted using sequence targeting and a kanMX6 cassette of Bahler et al. (23) flanked by ΦC31 integrase attP and attB sites. A codon-optimized ΦC31 integrase expression vector (24) was used to enable recycling of the kanMX6 cassette. A cassette containing the ura4 gene and an Asc I site was then targeted to the dps1, vac8, nht1, and ste4 genes on chromosomes 2 and 4 of the CBS 2777 leu1Δura4Δ strain. Silencing was assayed using 5-FOA, as described at www-bcf.usc.edu/∼forsburg/plasmids.html. S. pombe was otherwise grown in yeast extract with five supplements (YES) medium, and was handled and stored as described previously (8, 23). CNP1 was tagged at the amino terminus with mGFP as described previously (14), CNP3 was tagged at the carboxyl terminus with the PK-9 epitope, Swi6 was tagged with GFP using the EGFP-pAL2U plasmid (a gift of Jun-ichi Nakayama, Nagoya City University, Nagoya City, Japan), and Rad 21 was tagged with PK-9 as described previously (17). Tagging constructs were assembled from constituent fragments using SLiCE (25). All tagging and targeting used plasmid constructs, because CBS 2777 transforms ∼50-fold less efficiently than the laboratory strain.
Targeting sequences were at least 500 bp long and, with the exception of the Swi6 GFP-tagging plasmid EGFP-pAL2U, were isogenic with CBS 2777 and CBS 2776. A standard S. pombe lithium acetate transformation procedure was modified to use 108 cells and ∼200 μg of linearized DNA in each transformation. All tagging was carried out using leu1Δ ura4Δ derivatives of either CBS 2776 or CBS 2777.
ChIP.
ChIP was performed according to the method of Saitoh et al. (26) using either the anti-GFP antibody (AB 290; Abcam) at 1,000-fold dilution or the anti–PK-9 antibody (MCA 4764; AbD Serotec) at 500-fold dilution and protein A-conjugated Dynabeads (Invitrogen). Control experiments in which no antibody was used established that the ChIP was antibody- and tag-specific. The ChIP was monitored by quantitative PCR.
DNA Sequencing and Analysis.
Genome sequencing of the CBS 2777 was provided by the Genome Analysis Centre, Norwich, UK. Libraries were constructed using the Illumina DNA TruSeq protocol, with a true insert size of 359 bp (mean size) or 479 bp including Illumina adapters (60 bp on either end of the fragments). The library was sequenced on a single lane of an Illumina HiSEq 2000 sequencing system using 100-bp paired-end reads, which resulted in 84.3 million pairs of reads. The Q30 quality scores were up to base 85 for read one and up to base 70 for read two. The 20 ChIP samples, including controls and inputs, were multiplexed and sequenced in a single lane of the Illumina HiSEq 2000 using 50-bp paired-end reads by the project manager at the High-Throughput Genomics Wellcome Trust Centre for Human Genetics, Oxford, UK. Details of the sequence analysis and bioinformatics are provided in SI Appendix.
Microarray Expression Analysis.
Total RNA was extracted from either CBS 2777 or CBS 2776 cells with hot phenol. The size and integrity were checked by agarose gel electrophoresis. cRNA probes were prepared and labeled with CY3 or CY5 using the Agilent Low-Input Quick Amp Labeling Kit. The probes were purified, fragmented, and analyzed by competitive hybridization to a custom 4 × 44K gene expression microarray (design ID 033946; courtesy of Jurg Bähler and colleagues, University College, London). The results were scanned with an Agilent microarray scanner, processed, and analyzed using Agilent Genespring software.
Supplementary Material
Acknowledgments
We thank Christian Rudolph for discussion, Robin Allshire for strains; Jun-ichi Nakayama for the Swi6 GFP tagging plasmid EGFP-pAL2U; Frank Uhlmann and Wolfgang Zachariae for the details of the tagging of Rad21 with PK-9; Jian-qui Wu and Valerie Coffman for details of their Cnp1 mGFP tag, plasmids, and advice; Jurg Bähler for microarray design; and Danielle Fletcher and Darren Marjenberg (both of Agilent) for help with microarray data analysis. This work was supported by Nottingham University and by a Genetics Society summer studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The unannotated sequence data is available at http://www.nottingham.ac.uk/~plzloose/cbs2777/index.php.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216934111/-/DCSupplemental.
References
- 1.Allshire R, Pidoux A. Centromeres. Curr Biol. 2001;11(12):R454. doi: 10.1016/s0960-9822(01)00278-0. [DOI] [PubMed] [Google Scholar]
- 2.Steiner NC, Clarke L. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 1994;79(5):865–874. doi: 10.1016/0092-8674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 3.Ishii K, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321(5892):1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 4.Kagansky A, et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324(5935):1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319(5859):94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomiento M, Jiang Z, D’Addabbo P, Eichler EE, Rocchi M. Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol. 2008;9(12):R173. doi: 10.1186/gb-2008-9-12-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piras FM, et al. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 2010;6(2):e1000845. doi: 10.1371/journal.pgen.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown WR, et al. A geographically diverse collection of Schizosaccharomyces pombe isolates shows limited phenotypic variation but extensive karyotypic diversity. G3 (Bethesda) 2011;1(7):615–626. doi: 10.1534/g3.111.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croll D, Zala M, McDonald BA. Breakage-fusion-bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLoS Genet. 2013;9(6):e1003567. doi: 10.1371/journal.pgen.1003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen KR, Ibarra PT, Thon G. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 2006;34(1):78–88. doi: 10.1093/nar/gkj415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhind N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332(6032):930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195(4):563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76(1):157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15(4):381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt CK, Brookes N, Uhlmann F. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 2009;10(5):R52. doi: 10.1186/gb-2009-10-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard P, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294(5551):2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 19.Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4(1):89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 20.Ketel C, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5(3):e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zörgö E, et al. Life history shapes trait heredity by accumulation of loss-of-function alleles in yeast. Mol Biol Evol. 2012;29(7):1781–1789. doi: 10.1093/molbev/mss019. [DOI] [PubMed] [Google Scholar]
- 22.Niwa O, Tange Y, Kurabayashi A. Growth arrest and chromosome instability in aneuploid yeast. Yeast. 2006;23(13):937–950. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- 23.Bähler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14(10):943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z, et al. Site-specific recombination in Schizosaccharomyces pombe and systematic assembly of a 400-kb transgene array in mammalian cells using the integrase of Streptomyces phage phiBT1. Nucleic Acids Res. 2008;36(1):e9. doi: 10.1093/nar/gkm1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Werling U, Edelmann W. SLiCE: A novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40(8):e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90(1):131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 27.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9(2):218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.