Significance
We investigated the possible role of TANK-binding kinase 1 (TBK1) protein in tamoxifen resistance and found that phosphorylation by TBK1 at the Ser-305 site stabilized estrogen receptor α (ERα) and modulated its transcriptional activity. Ectopic expression of TBK1 rendered breast cancer cells resistant to tamoxifen. TBK1 inhibition sensitized breast cancer cells to tamoxifen-induced cell death. The expression of TBK1 was increased in subjects with breast cancer and was positively correlated with ERα, ERα Ser-305, and cyclin D1 expression. Subjects with tumors that highly expressed TBK1 had poor responsiveness to tamoxifen treatment. Therefore, TBK1 is potentially a unique predictive marker of tamoxifen resistance and a potential therapeutic target for breast cancer.
Abstract
Resistance to antiestrogens is one of the major challenges in breast cancer treatment. Although phosphorylation of estrogen receptor α (ERα) is an important factor in endocrine resistance, the contributions of specific kinases in endocrine resistance are still not fully understood. Here, we report that an important innate immune response kinase, the IκB kinase-related TANK-binding kinase 1 (TBK1), is a crucial determinant of resistance to tamoxifen therapies. We show that TBK1 increases ERα transcriptional activity through phosphorylation modification of ERα at the Ser-305 site. Ectopic TBK1 expression impairs the responsiveness of breast cancer cells to tamoxifen. By studying the specimens from patients with breast cancer, we find a strong positive correlation of TBK1 with ERα, ERα Ser-305, and cyclin D1. Notably, patients with tumors highly expressing TBK1 respond poorly to tamoxifen treatment and show high potential for relapse. Therefore, our findings suggest that TBK1 contributes to tamoxifen resistance in breast cancer via phosphorylation modification of ERα.
TANK-binding kinase 1 (TBK1) and IκB kinase ε (IKKε) are two IKK-related serine/threonine kinases that display 64% sequence identity and trigger the antiviral response of interferons (IFN) through NF-κB activation and interferon regulatory transcription factor (IRF) 3/7 phosphorylation (1–3). In addition to the proposed roles of IKK-related kinases in controlling transcription factors NF-κB and IRF, the involvement of TBK1 and IKKε in AKT-induced oncogenic transformation has been demonstrated in a recent study (4). TBK1 is identified as a Ras-like (Ral) B effector in the Ral guanine nucleotide exchange factor pathway that is required for Ras-induced transformation (5). IKKε acts downstream of the PI3K-AKT pathway and cooperates with activated MEK to promote cellular transformation (6). IKKε has also been identified recently as a breast cancer oncogene that is frequently amplified or overexpressed in human breast cancer, and the phosphorylation of ERα by IKKε contributes to tamoxifen resistance in breast cancer (7–9). Interestingly, TBK1 is also highly expressed in breast cancer (10), and knocking down TBK1 diminishes the viability of MCF-7 cells (9). However, the exact role of TBK1 in breast cancer remains unclear.
Estrogen receptor α (ERα) is a nuclear receptor that exerts a profound influence on the initiation and progression of breast cancer by regulating cell transformation, proliferation, and metastasis (11–13). For ERα-positive patients with breast cancer, targeting the ER signaling pathway with tamoxifen, a selective ER modulator, is efficacious in both prevention and treatment of breast cancer (14). Unfortunately, a substantial proportion of patients are intrinsically resistant to this therapy, and a significant number of patients with advanced disease eventually develop acquired resistance to the treatment (15–18). ERα is a key determinant of breast cancer susceptibility to endocrine therapy. Recent studies demonstrate that ERα phosphorylation may have had a significant impact on ERα signaling and its response to endocrine therapies (19, 20). For instance, ERα phosphorylation at Ser-118 has been suggested to be involved in protein turnover and directly associated with tamoxifen sensitivity (21–23). However, the contributions and mechanisms of specific kinase-mediated ERα phosphorylation in endocrine resistance are not fully known.
Here, we investigated the possible role of TBK1 protein in tamoxifen resistance and found that phosphorylation by TBK1 at the Ser-305 site stabilized ERα and modulated its transcriptional activity. Although ubiquitin-like domain (ULD)–mutated TBK1 failed to activate IFN-β promoters, it retained the ability to phosphorylate ERα, induce ERα transactivational activity, and modulate breast cancer cell growth. Moreover, ectopic expression of TBK1 rendered breast cancer cells resistant to tamoxifen. Suppressing TBK1 with its pharmacological inhibitor BX795 sensitized breast cancer cells to tamoxifen-induced cell death. Administration of BX795 in conjunction with tamoxifen achieved synergistic inhibitory effects on tumors. The expression of TBK1 was increased in patients with breast cancer and was positively correlated with ERα, ERα S305, and cyclin D1 expression. Notably, patients with tumors highly expressing TBK1 responded poorly to tamoxifen treatment and showed a high potential for relapse. Therefore, TBK1 is potentially a unique predictive marker of tamoxifen resistance and a therapeutic target for breast cancer.
Results
TBK1 Regulating the Transcriptional Activity of ERα Is Independent of Its Role in the Innate Immune Response.
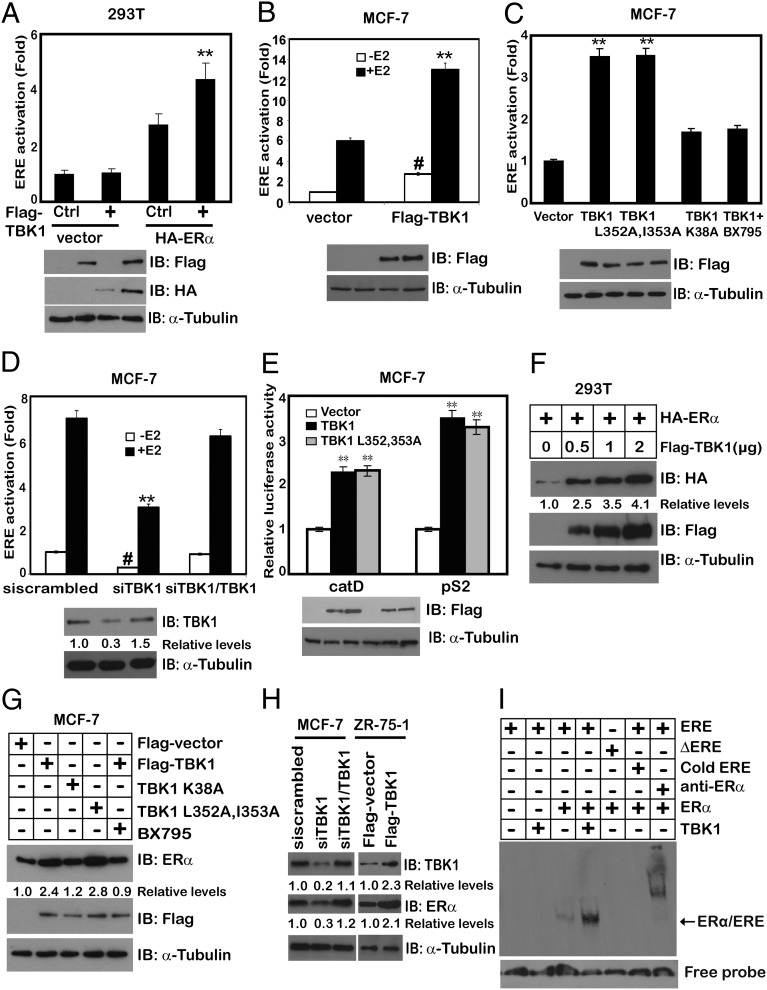
To define the exact role of TBK1 in breast tumor growth, we investigated whether TBK1 regulates estrogen signaling. TBK1 overexpression in cell lines, including ERα-negative 293T (Fig. 1A), ERα-positive ZR-75-1 (Fig. S1A), MCF-7 (Fig. 1 B and C and Fig. S1B), and BT474 (Fig. S1C) cell lines, increased the transcription of a luciferase reporter containing the estrogen-responsive element (ERE). TBK1 enhanced the transcriptional activity of ERα regardless of the presence of 17β-estradiol (E2) (Fig. 1B), suggesting that the activation of ERα by TBK1 is both ligand-dependent and -independent. This effect required TBK1 kinase activity, because expression of the kinase-inactive TBK1 K38A mutant failed to increase ERα transcriptional activity (Fig. 1C and Fig. S1A). Specifically, inhibition of TBK1 by TBK1 siRNA oligos (Fig. 1D) reduced the transcriptional activity of ERα, which was rescued by TBK1 reintroduction in siTBK1 cells (Fig. 1D). A similar result was also achieved by using TBK inhibitor BX795 (24) (Fig. 1C). ULD-mutated TBK1 (TBK1 L352A, I353A) failed to activate IFN-β, IRF3, and NF-κB promoters (Fig. S1 D–F) but retained its ability to induce ERα transactivation activity (Fig. 1C). Consistent with this, overexpression of TBK1 and TBK1 L352A, I353A increased the expression of ERα target genes catD and pS2, as shown by luciferase reporter assay (Fig. 1E) and quantitative PCR (Fig. S1G). Enhancement of cyclin D1, Bcl-xL, c-fos, and catD by TBK1 overexpression was also observed at the protein level by immunoblotting (Fig. S1H).
Fig. 1.
TBK1 regulates the transcriptional activity of ERα independent of its role in the innate immune response. The 293T cells (A) or MCF-7 cells (B) were transfected with the ERE-Luc reporter, ERα, and TBK1 with or without E2. The Luc activity was measured 24 h later and normalized for transfection efficiency. #P < 0.01 and **P < 0.01 vs. control (Ctrl) Flag vector without E2 or with E2, respectively. IB, immunoblot. (C) MCF-7 cells were cotransfected with ERE-Luc together with TBK1, TBK1 L352A, I353A, or TBK1 K38A. Cells were treated with or without 1 μM BX795 for 2 h. The Luc activity was measured 24 h later and normalized for transfection efficiency. (D) MCF-7 cells were transfected with TBK1 siRNA oligos (siTBK1) and siRNA-resistant TBK1 (siTBK1/TBK1) expression plasmid. The Luc activity was measured 24 h later and normalized for transfection efficiency. (E) MCF-7 cells were transfected with pS2-Luc or catD-Luc together with TBK1 or TBK1 L352A, I353A. The Luc activity was measured 24 h later and normalized for transfection efficiency. (F) 293T cells were transfected with HA-ERα and different doses of Flag-TBK1. Whole-cell lysates were analyzed by immunoblotting with anti-Flag or anti-HA antibody. α-Tubulin was used as an equal loading control. (G) MCF-7 cells were transfected with Flag-TBK1 or its mutants in the presence or absence of 1 μM BX795 for 2 h. Whole-cell lysates were analyzed by immunoblotting with anti-Flag or anti-ERα antibody. α-Tubulin was used as an equal loading control. (H) MCF-7 cells were transfected with siTBK1 and siTBK1/TBK1 expression plasmid. ZR-75-1 cells were transfected with Flag-TBK1. Whole-cell lysates from those treatments were analyzed by immunoblotting with anti-TBK1 or anti-ERα antibody. α-Tubulin was used as an equal loading control. (I) EMSA was performed using biotin-labeled ERE probe and nuclear proteins extracted from 293T cells transfected with Flag-ERα and Myc-TBK1. For competition experiments, a 100-fold molar excess of unlabeled ERE was incubated with the labeled probe. The biotin-labeled mutant ERE probe (∆ERE) was used as a negative control. Supershifts were performed using specific anti-ERα antibody. Cell-based studies were performed at least three independent times with comparable results. The numbers below some Western blots indicate the relative levels determined by software-based quantification of the representative experiment shown (also Fig. S1). Data represent mean ± SEM. The Student t test was used for statistical analysis (**P < 0.01).
To investigate how TBK1 increased the transactivational activity of ERα, we examined the effect of TBK1 on ERα protein expression. Indeed, overexpression of TBK1 and TBK1 L352A, I353A, together with ERα in ERα-negative 293T cells, led to increased ERα expression (Fig. 1G). Both of them also led to up-regulation of endogenous ERα expression in ERα-positive MCF-7 breast cancer cells (Fig. 1F and Fig. S1I). In contrast, TBK1 K38A mutation or BX795 treatment was incapable of up-regulating endogenous ERα expression (Fig. 1G). Consistently, knocking down TBK1 with specific siRNA decreased ERα expression in WT MCF-7 but not in the parental cells expressing siRNA-resistant TBK1. On the contrary, TBK1 overexpression in ZR-75-1 increased ERα expression (Fig. 1H). Furthermore, TBK1 increased the binding of ERα to the ERE sequence (Fig. 1I). Therefore, we suggest that TBK1 regulates estrogen signaling and that a TBK1-mediated antiviral response is dispensable for TBK1-mediated up-regulation of ERα transactivational activity.
TBK1 Interacts with ERα.
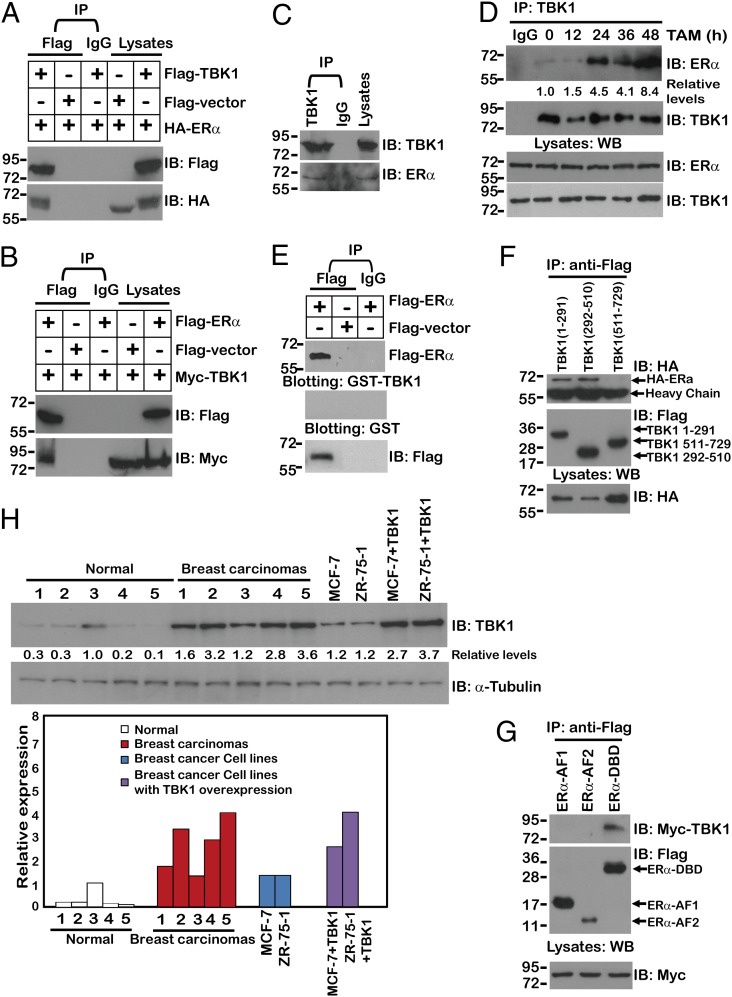
Based on our findings that TBK1 regulated ERα expression and its transactivational activity, we tested whether TBK1 physically interacts with ERα. Indeed, coimmunoprecipitation experiments showed that exogenous TBK1 protein associated with exogenous ERα (Fig. 2 A and B). Importantly, endogenous TBK1 interacted with endogenous ERα in the absence of tamoxifen (Fig. 2C), but the interaction between TBK1 and ERα increased considerably at 24 and 48 h in the presence of tamoxifen treatment (Fig. 2D). To rule out indirect binding mediated by other components in the cell lysate, Far-Western immunoblotting was conducted, and the results showed that ERα bonded to TBK1 directly (Fig. 2E). As a control, Flag-ERα did not bind to IgG (Fig. 2E).
Fig. 2.
TBK1 interacts with ERα. (A and B) 293T cells were cotransfected with the indicated plasmids, and anti-Flag or IgG immunoprecipitates (IP) were analyzed by immunoblotting with anti-Myc or anti-Flag antibody. Lysates from MCF-7 cells (C) or from MCF-7 cells treated with tamoxifen (TAM) for the indicated times (D) were subjected to immunoprecipitation with anti-TBK1 or IgG, fractionated by SDS/PAGE, and subsequently analyzed by immunoblotting with anti-ERα. (E) Anti-Flag or IgG immunoprecipitates prepared from cells transfected with Flag-ERα or Flag vector-expressing plasmids were subjected to SDS/PAGE and blotted onto nitrocellulose membrane. The nitrocellulose membrane was incubated with soluble GST-TBK1 or IgG for 2 h and then analyzed with anti-GST, anti-IgG, or anti-Flag antibody. (F and G) 293T cells were cotransfected with the indicated plasmids, and anti-Flag immunoprecipitates were analyzed by immunoblotting with anti-Myc or anti-Flag antibody. WB, Western blot. (H) Immunoblotting analysis for TBK1 expression in two breast cancer cell lines (MCF-7 and ZR-75-1) transfected with different doses of TBK1 and in tumors from five cases of breast cancer. Fold changes are referenced to values obtained with the normal sample with the highest TBK1 expression. All coimmunoprecipitation experiments were performed independently two to three times with comparable results. The numbers below some Western blots indicate relative levels determined by software-based quantification of the representative experiment shown.
To explore the molecular basis of the interaction between TBK1 and ERα further, we defined the domains of TBK1 and ERα required for their interaction. The results of coimmunoprecipitation experiments showed that the N terminus (1–510 aa) of TBK1 and the DNA binding domain (180–282 aa) of ERα were indispensable for the interaction (Fig. 2 F and G).
To evaluate the possible physiological relevance of using TBK1-overexpressing breast cancer cells as a biochemical study model, the relative abundance of TBK1 protein was compared between breast cancer cell lines with ectopic TBK1 expression and patient-derived primary breast cancer tissue samples. We observed a two- to threefold increase in TBK1 expression in the TBK1 transfectants from MCF-7 and ZR-75-1 cells, respectively, compared with their parental cells (Fig. 2H). The ranges of the levels of overexpressed TBK1 in cell lines were comparable to those in breast carcinomas (Fig. 2H), indicating that overexpression of TBK1 in breast cancer cell lines could mimic the possible physiological effect exerted by the increased TBK1 expression in breast carcinomas.
TBK1 Mediates Phosphorylation of ERα at the Serine-305 Site.
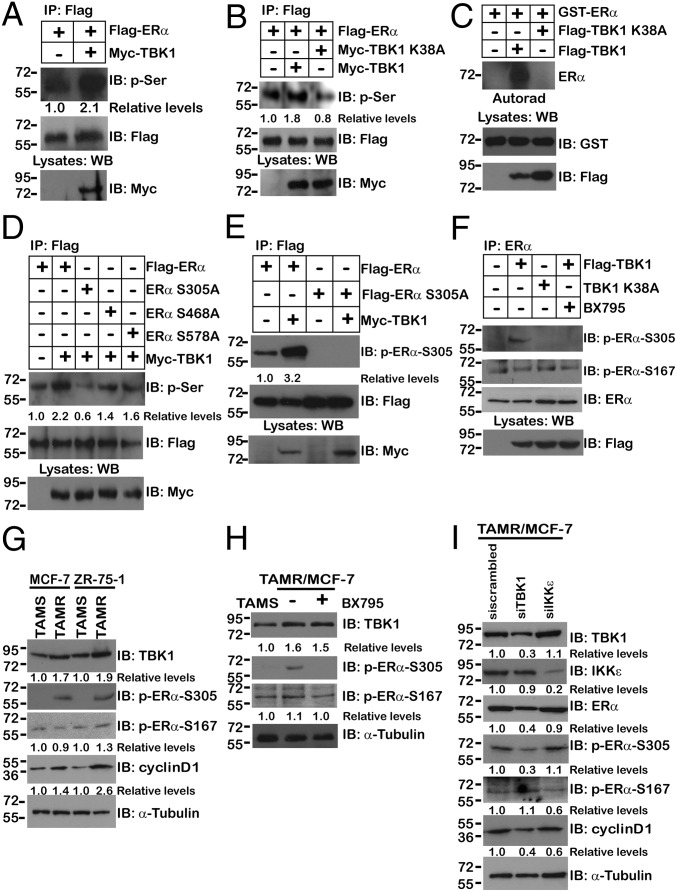
The association between ERα and TBK1 suggests that ERα could be a new substrate for this serine kinase. Analysis of the precipitates demonstrated that ERα was phosphorylated by WT TBK1 but not by the TBK1 K38A mutant both in vivo (Fig. 3 A and B) and in vitro (Fig. 3C). Examination of the amino acid sequence of ERα showed that three of 46 serine residues are putative targets for phosphorylation by TBK1 (i.e., Ser-305, Ser-468, Ser-578). We thus generated a single mutation to replace the serine residue in the serine–lysine motifs with alanine. Although overexpression of TBK1 led to partially reduced phosphorylation in ERα S468A and ERα S578A, TBK1 overexpression-induced phosphorylation was completely abolished in the ERα mutant in which Ser-305 was mutated to alanine (S305A) (Fig. 3D).
Fig. 3.
TBK1 mediates phosphorylation of ERα at the Ser-305 site. (A and B) 293T cells were cotransfected with the indicated plasmids. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti–p-Ser or anti-Flag antibody. (C) Recombinant GST-ERα was incubated with Flag-TBK1 or Flag-TBK1 K38A immunoprecipitates from transfected 293T cells in the presence of [γ-32P]-ATP. The reaction products were analyzed by SDS/PAGE and autoradiography (Autorad). (D) 293T cells were cotransfected with Myc-TBK1 and Flag-ERα or Flag-ERα mutants. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti–p-Ser or anti-Flag antibody. (E) 293T cells were cotransfected with Myc-TBK1 and Flag-ERα or Flag-ERα S305A. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti–p-ERα–S305 or anti-Flag antibody. (F) MCF-7 cells were transfected with Flag-TBK1 or Flag-TBK1 K38A. Cells were treated with or without 1 μM BX795 for 2 h. Anti-ERα immunoprecipitates were analyzed by immunoblotting with anti–p-ERα–S305, anti–p-ERα–S167, or anti-Flag antibody. (G) Cell lysates from TAMR or tamoxifen-sensitive (TAMS) MCF-7 or ZR-75-1 were analyzed by immunoblotting with anti-TBK1, anti–p-ERα–S305, anti–p-ERα–S167, or anti-cyclin D1 antibodies. α-Tubulin was used as an equal loading control. (H) TAMR/MCF-7 cells were treated with 1 μM BX795 in cell culture medium for 2 h. Whole-cell lysates were analyzed by immunoblotting with anti-TBK1, anti–p-ERα–S305, or anti–p-ERα–S167 antibodies. α-Tubulin was used as an equal loading control. (I) TAMR/MCF-7 cells were transfected with siTBK1 or siIKKε RNAi oligos. Whole-cell lysates were analyzed by immunoblotting with anti-TBK1, anti-IKKε, anti-ERα, anti–p-ERα–S305, anti–p-ERα–S167 antibody, or anti-cyclin D1 antibody. α-Tubulin was used as an equal loading control. The numbers below some Western blots indicate relative levels determined by software-based quantification of the representative experiment shown. All coimmunoprecipitation experiments and immunoblotting assays were performed independently two to three times with comparable results (also Fig. S2).
To confirm the phosphorylation of ERα Ser-305 in vivo, we used a monoclonal antibody specific for the Ser-305–phosphorylated peptide. This antibody specifically recognizes the overexpressed WT form of ERα but not S305A. Our results showed that the signal for S305 phosphorylation was significantly enhanced by overexpressed TBK1, whereas such an increase in phosphorylation was greatly blunted in experiments with ERα S305A, the TBK1 K38A mutation, or BX795 (Fig. 3 E and F). Moreover, the signal for S167 phosphorylation was not induced by overexpressed TBK1 (Fig. 3F). These findings demonstrated that ERα was phosphorylated by TBK1 mainly at Ser-305.
To determine whether BX795 specifically inhibits TBK1 but not IKKε, the phosphorylation status of ERα S167 and S305 (the two presumed substrates of IKKε and TBK1, respectively) was analyzed in the presence of different dosages of BX795 (7, 24). The results indicated that TBK1 overexpression led to increased ERα S305 phosphorylation, which was reduced by ∼85% in the presence of 1 μM BX795. In contrast, IKKε-mediated ERα S167 phosphorylation was resistant to 1 μM BX795 treatment (Fig. S2A). In addition, dose–response experiments indicated that the half-maximal inhibitory concentration (IC50) of BX795-mediated TBK1 inhibition was about 0.3 μM, whereas the IC50 of IKKε was about 2 μM (Fig. S2B). Therefore, we concluded that the observed responses under treatment with 1 μM BX795 are a direct consequence of TBK1 inhibition.
To explore the role of TBK1-mediated ERα phosphorylation in acquired tamoxifen resistance further, the profiles of TBK1 expression in tamoxifen-resistant (TAMR) MCF-7 and TAMR ZR-75-1 cells were analyzed. The protein expressions of TBK1, ERα S305, and cyclin D1 were significantly up-regulated in TAMR/MCF-7 and TAMR/ZR-75-1 cells (Fig. 3 G and H). However, ERα S167 expression remained unchanged between TAMR and tamoxifen-sensitive cells. More importantly, the up-regulation of ERα S305 induced by longtime tamoxifen treatment was nearly abolished by 1 μM BX795 (Fig. 3H). Furthermore, knockdown of TBK1 with siRNA in TAMR/MCF-7 cells significantly decreased the protein levels of ERα, ERα S305, and cyclin D1. By contrast, although knocking down IKKε caused a significant reduction of ERα S167 phosphorylation, the expression of ERα and ERα S305 remained unchanged (Fig. 3I). Overall, these results suggest that TBK1 mediates tamoxifen resistance by regulating phosphorylation of ERα at Ser-305.
TBK1 Increases ERα Transcriptional Activity Primarily Through Phosphorylation of ERα at Ser-305.
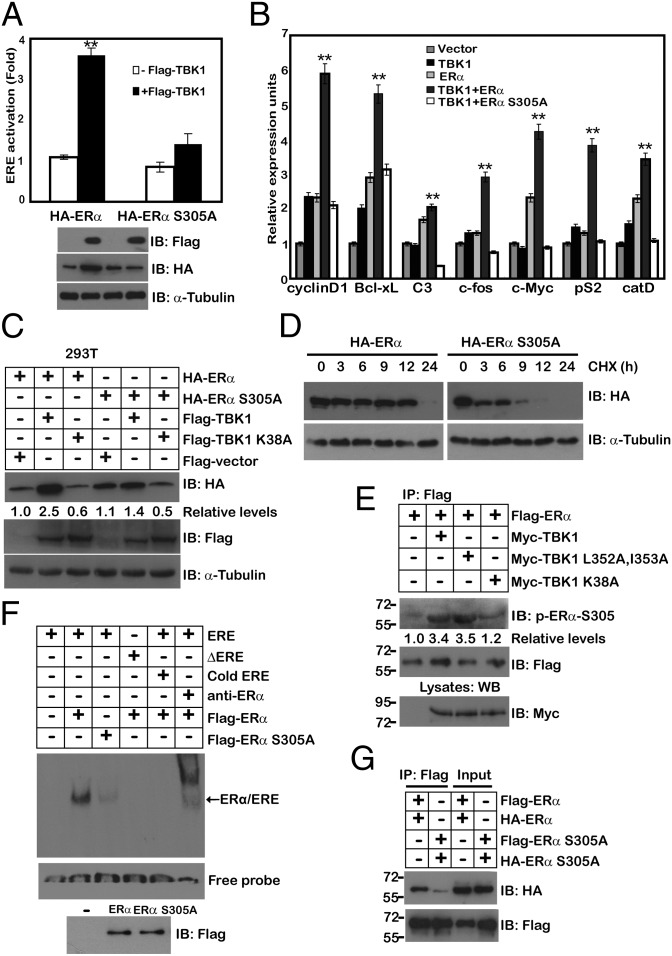
We then determined the effect of ERα phosphorylation at Ser-305 by TBK1 on estrogen signaling. Ectopic expression of TBK1 enhanced the transcriptional activity of ERα, as well as the expression of its target genes in the presence of WT ERα, but not ERα S305A mutant (Fig. 4 A and B). Furthermore, WT TBK1, but not its kinase-inactive mutant counterpart or its chemical inhibitor, induced an increase in ERα expression (Fig. 4C). Real-time PCR analysis showed no significant difference in the transcriptional level of ERα under TBK1 overexpression (Fig. S3A), indicating that TBK1 up-regulated ERα at a posttranscriptional level. Cotransfection of TBK1 significantly increased the protein level of WT ERα, but not that of the ERα S305A mutant (Fig. 4C and Fig. S3B). Indeed, the estimated t1/2 of WT ERα (longer than 12 h) was significantly longer than that of ERα S305A (shorter than 3 h; Fig. 4D and Fig. S3C). Immunoblotting experiments revealed that TBK1 and TBK1 L352A, I353A also effectively increased the endogenous ERα S305 phosphorylation level, whereas cotransfection of TBK1 K38A failed to do so (Fig. 4E). These results suggest that the effect of TBK1 on the ERα protein level is mainly caused by phosphorylation on the Ser-305 site. Interestingly, the binding of ERα S305A to the ERE sequence was nearly abrogated, compared with that of WT ERα (Fig. 4F). ERα binds DNA as either a homodimer or heterodimer with ERβ. The inability of the receptor to dimerize would result in the loss of DNA binding. To investigate the effect of the ERα S305A mutant on dimerization of ERα, we next examined the ability of WT ERα and the ERα S305A mutant to coimmunoprecipitate within themselves. Unlike ERα, ERα S305A nearly failed to associate with ERα S305A (Fig. 4G).
Fig. 4.
TBK1 increases ERα transcriptional activity primarily through phosphorylation of ERα at Ser-305. (A) ZR-75-1 cells were cotransfected with ERE-Luc and TBK1 together with ERα or ERα S305A. The Luc activity was measured 24 h later and normalized for transfection efficiency. (B) MCF-10A cells were cotransfected with TBK1 together with ERα or ERα S305A, and semiquantitative RT-PCR was performed using the primers specific for ERα-responsive genes, including cyclin D1, Bcl-xL, C3, c-fos, c-Myc, pS2, and catD. (C) HA-ERα or its mutants were cotransfected with Flag-TBK1 or Flag-TBK1 K38A in 293T cells. Whole-cell lysates were analyzed by immunoblotting with anti-Flag or anti-HA antibody. α-Tubulin was used as an equal loading control. (D) 293T cells were transfected with the expression vector encoding HA-ERα or HA-ERα S305A. After 24 h, both cell types were treated with cycloheximide (50 μM), and the level of ERα was monitored by immunoblotting using anti-HA antibody. α-Tubulin was used as an equal loading control. (E) MCF-7 cells were transfected with Myc-TBK1 or its mutants. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti–p-ERα–S305 or anti-Flag antibody. (F) 293T cells were transfected with Flag-ERα or Flag-ERα S305A. EMSA was performed as described in Fig. 1I. (G) 293T cells were cotransfected with Flag-ERα or Flag-ERα S305A and HA-ERα or HA-ERα S305A. After 48 h, cell lysates were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-Flag and anti-HA antibodies. Cell-based studies were performed at least three independent times with comparable results. The numbers below some Western blots indicate relative levels determined by software-based quantification of the representative experiment shown (also Fig. S3). Data represent mean ± SEM. The Student t test was used for statistical analysis (**P < 0.01).
TBK1 Has a Major Role in the Resistance of Breast Cancer Cells to Tamoxifen Treatment.
Next, we determined the effect of TBK1-ERα interaction on breast cancer cell growth. TBK1 knockdown cells grew slower than those transfected with scrambled siRNA control, and this phenotype was rescued by TBK1 and TBK1 L352A, I353A reexpression, but not by TBK1 K38A reexpression (Fig. S4A). WT ERα-transfected cells grew faster than those transfected with ERα S305A mutant (Fig. S4B). These results indicate that TBK1 regulation of cell growth is mediated by ERα phosphorylation. TBK1 knockdown MCF-7 cells also displayed a greatly inhibited migratory capacity for wound healing. The observed effects were rescued by TBK1 and TBK1 L352A, I353A reexpression but not by TBK1 K38A reexpression (Fig. S4C). We next examined if cancer cell lines selectively sensitive to siRNA-mediated TBK1 depletion were also selectively sensitive to BX795. A total of 13 cell lines were used: Eight of them were breast cancer cell lines and three were positive for ERα. We found that 1 μM BX795 was toxic to 100% of the ERα-positive breast cancer cell lines (three of three cell lines; P ≤ 0.005), 20% of ERα-negative breast cancer cell lines (one of five cell lines), and 20% of non-breast cancer cell lines (one of five cell lines; Fig. S4D).
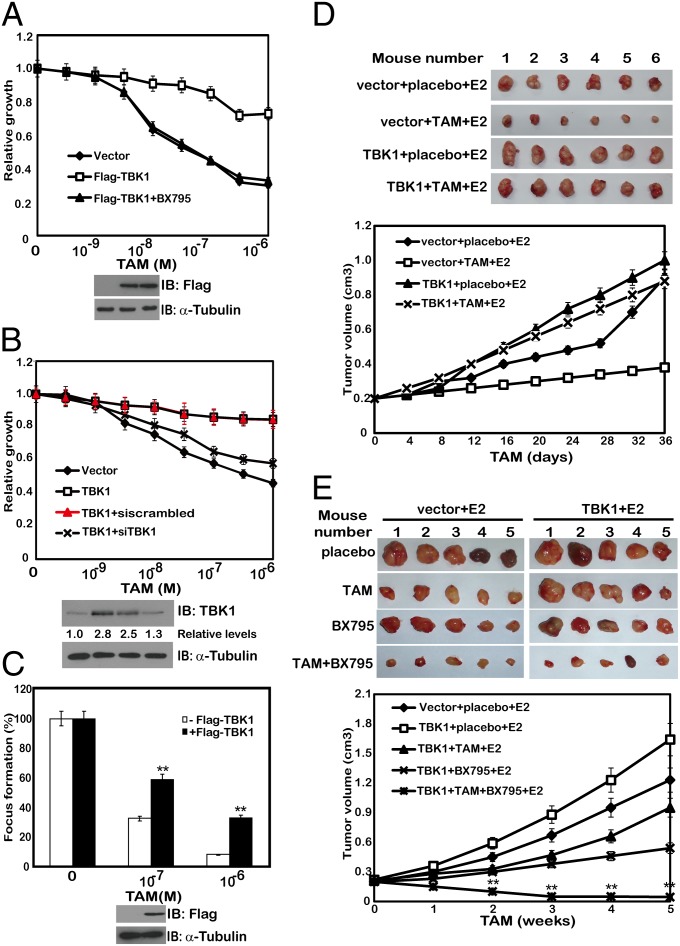
Based on the results that phosphorylation of ERα on Ser-305 by TBK1 involved tamoxifen resistance, we proposed that TBK1 is also implicated in the resistance to tamoxifen-induced growth inhibition and cell death. To prove this hypothesis, MCF-7 cells stably transfected with a plasmid encoding TBK1 were treated with different doses of tamoxifen and cell growth was measured under different conditions. TBK1 overexpression rendered reduced sensitivity of the MCF-7 and ZR-75-1 cells to tamoxifen treatment (Fig. 5A and Fig. S5A). In contrast, TBK1 knockdown cells displayed increased sensitivity to tamoxifen-mediated cell growth inhibition (Fig. S5B). To rule out nonspecific effects of TBK1 overexpression on tamoxifen sensitivity, we knocked down the expression of TBK1 in the TBK1 stably transfected MCF-7 cells and found that knockdown of TBK1 restored the sensitivity of the cells to tamoxifen (Fig. 5B). Moreover, TBK1-induced tamoxifen resistance could be relieved by BX795 (Fig. 5A). TBK1-overexpressing cells were apparently more resistant to tamoxifen-induced cell death, as measured by colony formation assay, whereas the control cells remained susceptible (Fig. 5C). To verify these results, we performed xenograft experiments and confirmed that tamoxifen effectively inhibited the tumor growth in BALB/c nude mice implanted with control tumors but not in the mice with TBK1-overexpressing tumors (Fig. 5D). These results suggest that TBK1 has a major role in the resistance of breast cancer cells to tamoxifen therapies.
Fig. 5.
TBK1 has a major role in the resistance of breast cancer cells to tamoxifen treatment. (A) MCF-7 cells transfected with TBK1 were treated with a range of concentrations of tamoxifen in the presence or absence of 1 μM BX795. Cell viability was assessed 7 d after the tamoxifen treatment. (B) MCF-7 cells transfected with Flag-TBK1 were transfected with scrambled siRNA (siscrambled) or siTBK1 and were treated with a range of concentrations of tamoxifen. Cell viability was assessed 7 d after the tamoxifen treatment. (C) MCF-7 cells stably transfected with a plasmid encoding TBK1 were treated with a range of concentrations of tamoxifen. Colony formation was assessed 21 d after the tamoxifen treatment. (D) Volume of xenograft tumors was derived from control or TBK1-overexpressing ZR-75-1 cells upon treatment with placebo or tamoxifen (n = 6 in each group). Tumor sizes were measured using a caliper at the indicated time points. (E) Volume of xenograft tumors was derived from control or TBK1-overexpressing ZR-75-1 cells upon treatment with the indicated drugs (n = 5 in each group). Tumor sizes were measured using a caliper at the indicated time points. Cell-based studies were performed at least three independent times with comparable results (also Figs. S4–S6 and Table S1). Data represent mean ± SEM. The Student t test was used for statistical analysis (**P < 0.01).
Because inhibition of TBK1 by siRNA or BX795 increased the sensitivity of breast cancer cells to tamoxifen treatment, we next investigated the potential synergistic effect of BX795 and tamoxifen. A well-established mathematical model for studying multidrug interactions was applied to the analysis (25). Our results showed that the combined inhibition of tamoxifen and BX795 achieved considerable synergy, as shown in the xenograft experiment, in which we overexpressed TBK1 in MCF-7 cells. The calculated combination index value was significantly lower than 1 (Fig. 5E, Fig. S6, and Table S1), indicating that BX795 and tamoxifen could act synergistically in breast cancer therapies.
TBK1 Expression Is Elevated in Breast Cancer Tumors.
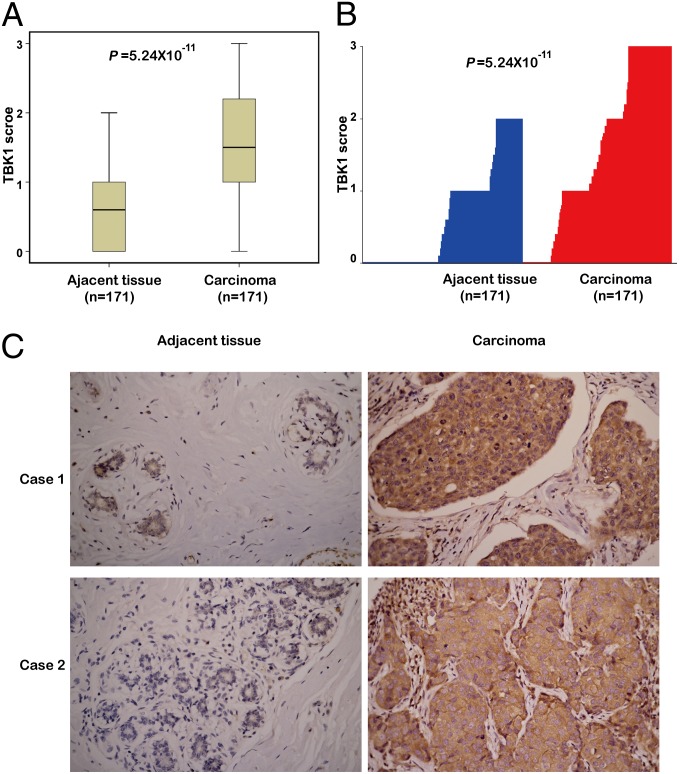
To investigate the potential clinical role of TBK1 in breast cancer, we collected 473 breast cancer tissue samples and 171 paired adjacent normal tissue samples from three hospitals (Table S2). To determine TBK1 expression in these samples, we performed immunohistochemistry staining on all the samples with an anti-TBK1 antibody. We confirmed the specificity of the antibody by immunoblotting of lysates from MCF-7 cells transfected with TBK1 siRNA control (Fig. S7A); immunoblotting and immunohistochemical staining of tumors isolated from mice inoculated with MCF-7 cells transfected with TBK1, IKKε, TBK1 siRNA, or IKKε siRNA (Fig. S7 B and C); and immunohistochemical staining of breast cancer samples incubated with anti-TBK1 antibody preincubated with their respective antigens (Fig. S7D). Immunohistochemical staining of 171 breast cancer tissues and paired adjacent normal tissues, using TBK1 antibody with confirmed specificity, showed that TBK1 expression was significantly higher in breast tumor tissues compared with that in matched adjacent normal tissues (Fig. 6 A–C).
Fig. 6.
TBK1 expression is elevated in breast tumors. (A) TBK1 expression scores are shown as box plots, with the horizontal lines representing the median; the upper and lower parts of the boxes representing the 25th and 75th percentiles, respectively; and the vertical bars representing the range of data. We compared TBK1 scores in breast cancer tissues (n = 171) with those in matched adjacent normal tissues (n = 171) using the Mann–Whitney U test. (B) Plot of TBK1 scores in each carcinoma and adjacent normal tissues. (C) Representative images from immunohistochemical staining of TBK1 in tumors and adjacent normal tissues from two cases of breast cancer (also Fig. S7 and Table S2). (Magnification: 400×.)
TBK1 Is Positively Correlated with ERα, ERα Ser-305, and Cyclin D1 in Patients with Breast Cancer.
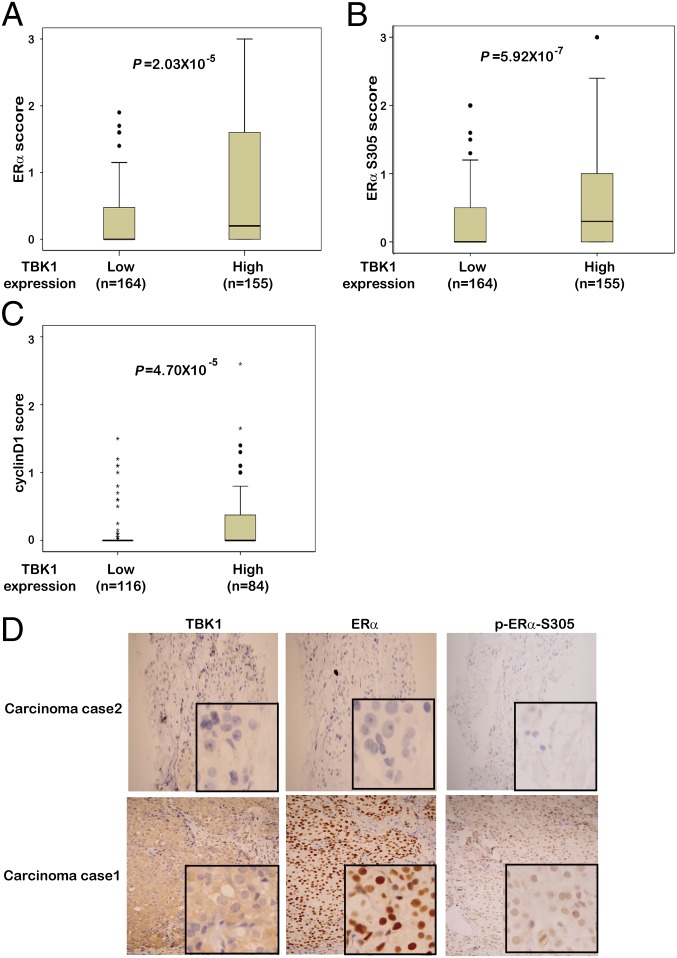
Spearman correlation scores of TBK1 expression with ERα and ERα Ser-305 were determined in 319 breast cancer samples from two hospitals (Table S2), whereas Spearman correlation scores of TBK1 expression with cyclin D1 were assessed in the 200 patients from Beijing (Table S2). The results suggested that TBK1 had a significant positive correlation with ERα expression (P = 1.53 × 10−6, r = 0.265; Fig. 7D and Figs. S8B and S9), ERα Ser-305 expression (P = 6.14 × 10−7, r = 0.275; Fig. 7D and Fig. S9), and cyclin D1 expression (P = 2.02 × 10−7, r = 0.358; Fig. S9). To understand the correlation of TBK1 with ERα, ERα Ser-305, and cyclin D1 better, we divided breast cancer samples into two groups on the basis of TBK1 levels defined by their expression scores. The differences in the expression of ERα, ERα Ser-305, and cyclin D1 between the two groups were analyzed using the Mann–Whitney U test. The protein expression in each group was represented by its median expression score (Fig. 7 A–C). The median expression scores of ERα in tumors with high TBK1 expression were higher than those in tumors with lower TBK1 expression (Fig. 7A). Similarly, the expression of ERα Ser-305 and cyclin D1 showed statistical differences between the two groups (Fig. 7 B and C).
Fig. 7.
TBK1 is positively correlated with ERα, ERα Ser-305, and cyclin D1 in patients with breast cancer. Box plots of ERα (A), ERα Ser-305 (B), and cyclin D1 (C) expression in breast cancer from 319 (A and B) and 200 (C) cases. The cases were divided into two groups based on TBK1 expression scores in the tumors, representing low (0 ≤ scores ≤ 1.5) and high (1.5 < scores ≤ 3.0) expression of TBK1. Any outliers are marked with a circle, and extreme cases are marked with an asterisk. Data were analyzed by the Mann–Whitney U test. (D) Representative images from immunohistochemical staining of ERα and ERα Ser-305 in tumors from TBK1 high-expression and low-expression groups. (Magnification: 200×.) The boxed areas are magnified images (also Figs. S8 and S9). (Magnification: 400×.)
TBK1 Predicts the Clinical Outcome of Tamoxifen Therapy.
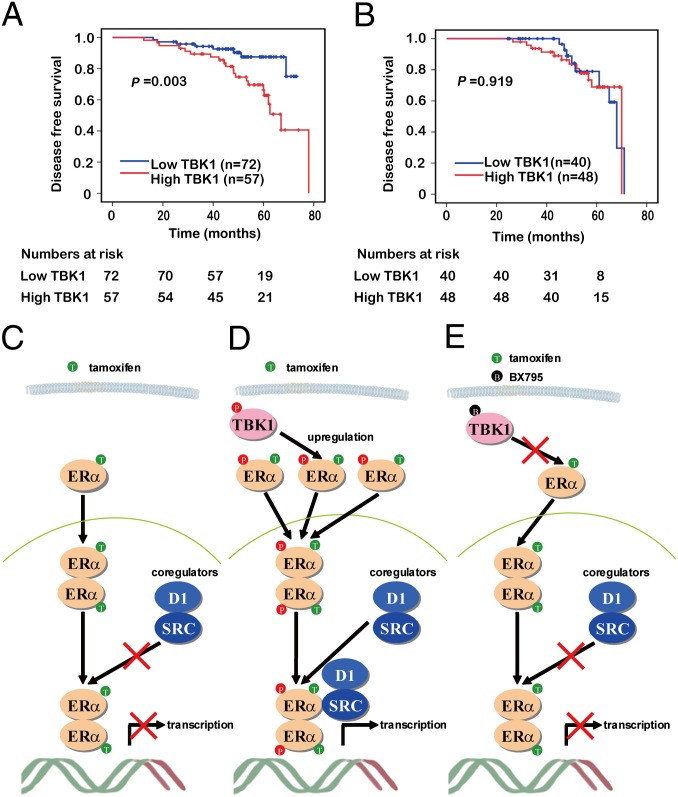
To evaluate the clinical association of TBK1 up-regulation of ERα transcriptional activity, we carried out a survival analysis in 217 subjects using Kaplan–Meier curves. The 217 subjects were divided into two groups according to whether they received tamoxifen treatment. Subsequently, subjects were dichotomized into TBK1-low and TBK1-high groups according to the cutoff value (low: 0 ≤ TBK1 score ≤ 1.5; high: 1.5 < TBK1 score ≤ 3). For the subjects who received tamoxifen (n = 129), those with tumors highly expressing TBK1 (n = 57) showed significantly poorer disease-free survival (DFS) than those with tumors with low TBK1 expression (n = 72; P = 0.003; Fig. 8A). In contrast, the remaining subjects, who did not receive tamoxifen therapy, had no significant differences in their DFS regardless of the TBK1 amounts in the tumors (n = 88; P = 0.919; Fig. 8B). Our results show that patients with tumors that highly express TBK1 did not respond to tamoxifen treatment as effectively as those with tumors with low TBK1 expression, suggesting a potential crucial role of TBK1 in the clinical resistance of breast cancer to hormone therapies.
Fig. 8.
TBK1 predicts the clinical outcome of tamoxifen therapy. Kaplan–Meier estimates of DFS of the subjects who received adjuvant tamoxifen therapy (A) and did not receive tamoxifen therapy (B). Comparison was made between groups with high TBK1 expression (scores >1.5) and low TBK1 expression (scores ≤1.5). Marks on graph lines represent censored samples. P value refers to two-sided log-rank tests. (C–E) Model for TBK1-mediated resistance to tamoxifen. The ERα-complex binds to its cognate ERE recognition site in the promoter of estrogen-responsive genes. Transcription is mediated through the subsequent recruitment of a number of coactivators. In tamoxifen nonresistant cells (C), binding of tamoxifen to ERα prevents the recruitment of coactivator SRC-1 and impedes ERα-mediated transcription. In tamoxifen-resistant cells (D), up-regulation of TBK1, as observed in many breast cancer patients, prevents tamoxifen-mediated inhibition of ERα transactivation by phosphorylation of ERα at serine-305. Inhibition of TBK1 activity by BX795 resumed tamoxifen-mediated inhibition of ERα transactivation. (E) Phosphorylation of Ser-305 of ERα by TBK1 then controls the switch from inhibition to growth stimulation by tamoxifen. P, phosphorylation; T, tamoxifen.
Discussion
Various mechanisms underlie tamoxifen resistance, including kinase activities that result in phosphorylation of ERα (19, 20, 26–28). The coactivator recruitment, subcellular localization, receptor dimerization, ligand binding, and posttranslational modifications of ERα are regulated through the phosphorylation of certain individual residues (29). Elucidating the regulation mechanism of ERα phosphorylation may thus provide new therapeutic targets for overcoming tamoxifen resistance (30). The present study revealed a key role of the TBK1 protein in tamoxifen resistance of breast cancer. First, ectopic TBK1 expression induced ERα transactivational activity, enhanced ERα DNA binding, increased the expression of endogenous ERα target genes, and impaired the responsiveness of breast cancer cells to tamoxifen through phosphorylation modification of ERα at the Ser-305 site. Second, ULD-mutated TBK1 failed to activate IFN-β promoter and retained the ability to phosphorylate ERα, induce ERα transactivational activity, and modulate the growth of breast cancer cells. Third, expression of TBK1 positively correlated with ERα, ERα S305, and cyclin D1 protein levels in patients with breast cancer. Fourth, subjects with tumors that highly expressed TBK1 responded poorly to tamoxifen treatment and displayed a high potential for relapse. Therefore, our findings raise the possibility that TBK1 is a crucial determinant of resistance to tamoxifen therapies in breast cancer independent of its roles in innate immunity.
ERα can be phosphorylated on multiple amino acid residues throughout the whole protein and within all major structural domains (23, 28, 29, 31–34). Phosphorylation has been suggested to be involved in protein turnover via a proteasome-mediated mechanism. For instance, phosphorylation on S118 and S167 protects ERα from proteasomal degradation (30). However, how phosphorylation on other sites may affect receptor turnover is not clear and is underexplored. In this report, we found that TBK1 kinase activity was required for the up-regulated phosphorylation of ERα at S305 and that the t1/2 of ERα S305A mutant was significantly shorter than that of WT ERα, suggesting that Ser-305 is a phosphorylation site protecting ERα from proteasomal degradation. Several lines of evidence suggest that increased ERα protein levels play crucial roles in breast cancer tumorigenesis via stimulation of cell division and tumor growth. Thus, our results may suggest a unique mechanism by which elevated TBK1 expression in breast cancer contributes to tumorigenesis by up-regulating the protein level of ERα, which may differ from the mechanism of TBK1 in Ras-induced oncogenic transformation in lung cancer.
In general, ERα phosphorylation is associated with the clinical outcome in patients who have breast cancer (23). For example, a higher level of ERα S167 and/or S118 phosphorylation is often associated with a better clinical outcome in patients on tamoxifen therapy (22, 35). In contrast, detection of ERα S305 is more likely to be associated with increased aggressiveness of tumors. Identifying the kinase that is responsible for S305 induction is therefore of vital importance. PKA and PAK1 are the possible candidates (28, 36). Although only a positive correlation of S305 phosphorylation with nuclear PAK1 expression is found, PAK1 itself does not lead to S305 phosphorylation directly (36–38). Obviously, additional kinases must exist for inducing S305 phosphorylation. One notable finding of this study is to show the importance of TBK1 in ERα S305 phosphorylation. In support of our notion, PAK1 is reported to be upstream of IKKε and TBK1 in the viral activation of IRF3 (39); therefore, modification of ERα S305 by PAK1 may be mediated by TBK1. Still, we cannot exclude additional mechanisms for inducing transcriptional activation mediated by TBK1, because TBK1 K38A mutant appears to induce ERE activation compared with vector control (Fig. 1C). We also noticed that overexpression of TBK1 led to reduced phosphorylation in ERα S468A and ERα S578A, suggesting that the combination of phosphorylation sites within ERα, rather than any individual site, may be more important for affecting function and responding to tamoxifen therapies. Therefore, further studies evaluating the contributions of S468 and S578 on receptor turnover and transcriptional activity would be of great interest.
The molecular links between chronic inflammation and cancer have begun to emerge only recently (40). There is now compelling evidence showing that the transcription factor NF-κB plays a key role in cancer development and progression (41). Thus, a recent suggestion that the IKK-related kinases TBK1 and IKKε also regulate the proliferation and survival of cancer cells is not totally unexpected (9). However, our data showed that TBK1 exerted its oncogenic effect in breast cancer cells through the modulation of ERα signaling and that ULD-mutated TBK1 failed to activate IFN-β, IRF3, and NF-κB promoters but retained the ability to phosphorylate ERα, induce ERα transactivational activity, and stimulate breast cancer cell growth. These findings have an impact on the understanding of the complex relationship between innate immune effectors and the signaling events that drive tumor formation.
Although TBK1 and IKKε share 64% homology in amino acid sequences and activate the same substrates, it seems that they differ in the way that they regulate ERα signaling (1, 7, 42). IKKε has recently been identified as a breast cancer oncogene that is frequently amplified or overexpressed in human breast cancer (9). IKKε phosphorylates ERα at Ser-167, a site most often associated with a better clinical outcome in patients on tamoxifen therapy (7, 35). Here, we show that TBK1 phosphorylates ERα at Ser-305, a site more likely to be associated with increased aggressiveness of tumors. Many ERα-positive tumors are resistant to tamoxifen without any prior exposure, and many of the tumors that initially respond to tamoxifen can acquire resistance during and after tamoxifen therapy (23). Most acquired tamoxifen resistance (70–80%) occurs and exhibits an estrogen-independent phenotype even with the presence of ERα (43). Actually, we noticed increased TBK1, ERα, and ERα S305 expression in breast cancer cells with acquired tamoxifen resistance, and the up-regulation of ERα and ERα S305 required TBK1 regardless of the presence of E2. Consistent with this, the clinical data and xenograft experiments presented here suggest that patients with breast tumors that highly express TBK1 are less likely to benefit from hormonal therapy compared with those with tumors that show low TBK1 expression. Recent studies suggest that TBK1 is highly expressed in breast cancer, even though the role of TBK1 in breast cancer remains unclear. Previous reports (28) showed that phosphorylation of Ser-305 affects the stability of ERα conformation upon antiestrogen binding rather than the binding properties per se. Therefore, phosphorylation at Ser-305 blocks the conversion of ERα into its inactive conformation by tamoxifen and leads to a specific TAMR proliferation. Consistent with this finding, we found that Ser-305 phosphorylation affected not only ERα stability but its dimerization ability, which is of vital importance for the transcriptional activity of ERα. Taken together, our data revealed a positive feedback loop of TBKI-ERα-ERα S305 signaling and identified the TBK1-ERα-ERα S305 axis as a unique signaling cascade for transcription activation of ERα. Because administration of BX795 together with tamoxifen achieved a synergistic effect on tumor suppression (Fig. 8 C–E), TBK1 might form a unique therapeutic target for overcoming both intrinsic and acquired tamoxifen resistance in breast cancers.
Materials and Methods
Full experimental procedures and any associated references are available in SI Materials and Methods.
Ethics Statement.
All animals were handled in strict accordance with the Guide for the Care and Use of Laboratory Animals (44) and the principles for the utilization and care of vertebrate animals (45), and all animal work was approved by the Institutional Animal Care Committee of Beijing Institute of Biotechnology.
The use of all patient tissue specimens was carried out according to the laws and regulations of China.
Plasmids and SiRNA.
The reporter constructs ERE-Luc, catD-Luc, and pS2-Luc and the expression vectors for ERα, Flag-tagged ERα, and HA-tagged ERα have been described previously (46). Other mammalian expression vectors encoding Flag-, Myc-, or HA-fusion proteins tagged at the amino terminus were constructed by inserting PCR-amplified fragments into pcDNA3 (Invitrogen) or pIRESpuro2 (Clontech). Plasmids encoding GST fusion proteins were generated by cloning PCR-amplified sequences into pGEX4T-1 (Amersham Pharmacia Biotech). The cDNA target sequence of siRNA for TBK1 is 5′-AAGCGGCAGAGUUAGGUGAAdT-3′, and it was inserted into pSUPER.retro RNAi vector (Oligoengine).
Cell Culture, Transfection, and Luciferase Reporter Assay.
The 293T embryonic kidney cells, MCF-7, ZR-75-1, and BT474 breast cancer cells were routinely cultured in DMEM (Invitrogen) containing 10% (vol/vol) FBS (HyClone). For hormone treatment experiments, cells were cultured in medium containing phenol red-free DMEM supplemented with 10% (vol/vol) charcoal/dextran-treated FBS (HyClone). TAMR/MCF-7 and TAMR/ZR-75-1 were derived from WT MCF-7 or ZR-75-1 cells by continuous exposure to 1 μM tamoxifen diluted in 0.1% ethanol. Cells were maintained as monolayers in a humidified atmosphere containing 5% (vol/vol) CO2 at 37 °C, and culture medium was replaced every the other day. The medium for matched control cells contained 0.1% ethanol. Lipofectamine 2000 reagent was used for transfection following the manufacturer’s protocol (Invitrogen). Stable cell lines were selected in 500 μg/mL G418 or 1 μg/mL puromycin for ∼2 mo. Pooled clones or individual clones were screened by standard immunoblot protocols and produced similar results. A luciferase reporter assay was performed as described previously.
Western Blotting and Immunoprecipitation.
Cell extracts were prepared, immunoprecipitated, and analyzed as previously described (46). An aliquot of the total lysate [5% (vol/vol)] was included as a control for the interaction assay. Immunoprecipitation was performed with anti-Flag M2 Affinity Gel (A2220; Sigma–Aldrich), anti-Myc (A5598; Sigma–Aldrich), anti-ERα (catalog no. sc-7207; Santa Cruz Biotechnology), anti-HA (H9658; Sigma–Aldrich), anti-TBK1 (3296-1; Epitomics), antiphosphoserine (61-8100; Zymed), anti–phospho-ERα-Ser305 (07-962 and 05-922R; Millipore), anti–phospho-ERα-Ser167 (catalog no. sc-101676; Santa Cruz Biotechnology), anti-cyclin D1 (2261-1; Epitomics), IKKε (07-580; Millipore), or anti–α-tubulin (T6074; Sigma–Aldrich) antibody. The antigen/antibody complexes were visualized by chemiluminescence. When necessary, figures were cropped using Photoshop software (Adobe). Band density was analyzed using Image-Quant software (Amersham).
In a direct binding assay, immunoprecipitates were separated by SDS/PAGE and then blotted onto nitrocellulose membranes. Membranes were subsequently incubated with purified GST-fusion proteins for 2 h at room temperature. The GST fusion proteins binding to nitrocellulose were probed with anti-GST antibody.
In Vitro Kinase Assay.
Purified GST-ERα (2 μg) was incubated with Flag-TBK1 or Flag-TBK1(K38A) immunoprecipitates from transfected 293T cells in kinase buffer [20 mM Hepes (pH 7.5), 75 mM KCl, 10 mM MgCl2, and 10 mM MnCl2] containing 2.5 mCi of [γ-32P]-ATP for 30 min at 37 °C. The reaction products were analyzed by SDS/PAGE and autoradiographed.
Statistics.
Statistical analysis was performed using SPSS 17.0 (SPSS, Inc.) and R 2.13.0 (www.r-project.org). We determined the correlation between TBK1 expression and that of ERα, ERα Ser-305, and cyclin D1, respectively, by the Spearman correlation test. The Mann–Whitney U test was used to compare immunohistochemistry scores between groups. CompSyn software (www.combosyn.com) was used to assess synergistic effects between BX795 and tamoxifen. Estimation of DFS was performed using the Kaplan–Meier analysis, and differences between curves were compared using log-rank tests. All statistical tests were two-sided, and P values <0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported, in part, by the Basic Research Program of China (Grants 2012CB518900 and 2010CB529403), the National Natural Science Foundation of China (Grants 31300637, 31170029, 31207911, 31270800, 81071771, 30725035, 30930103, and 31100960), and the National Science and Technology Major Program (Grant 2011ZX09102-010-02).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316255111/-/DCSupplemental.
References
- 1.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4(5):491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 2.McWhirter SM, et al. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA. 2004;101(1):233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451(7179):725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 4.Xie X, et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc Natl Acad Sci USA. 2011;108(16):6474–6479. doi: 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien Y, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127(1):157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 6.Guo JP, Coppola D, Cheng JQ. IKBKE protein activates Akt independent of phosphatidylinositol 3-kinase/PDK1/mTORC2 and the pleckstrin homology domain to sustain malignant transformation. J Biol Chem. 2011;286(43):37389–37398. doi: 10.1074/jbc.M111.287433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Guo JP, et al. IKKepsilon phosphorylation of estrogen receptor alpha Ser-167 and contribution to tamoxifen resistance in breast cancer. J Biol Chem. 2010;285(6):3676–3684. doi: 10.1074/jbc.M109.078212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Qin B, Cheng K. Silencing of the IKKε gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast Cancer Res. 2010;12(5):R74. doi: 10.1186/bcr2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 10.Korherr C, et al. Identification of proangiogenic genes and pathways by high-throughput functional genomics: TBK1 and the IRF3 pathway. Proc Natl Acad Sci USA. 2006;103(11):4240–4245. doi: 10.1073/pnas.0511319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17(5):1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 12.Nadji M, Gomez-Fernandez C, Ganjei-Azar P, Morales AR. Immunohistochemistry of estrogen and progesterone receptors reconsidered: Experience with 5,993 breast cancers. Am J Clin Pathol. 2005;123(1):21–27. doi: 10.1309/4wv79n2ghj3x1841. [DOI] [PubMed] [Google Scholar]
- 13.Osborne CK, Schiff R, Fuqua SA, Shou J. Estrogen receptor: Current understanding of its activation and modulation. Clin Cancer Res. 2001;7(12) Suppl:4338s–4342s. discussion 4411s–4412s. [PubMed] [Google Scholar]
- 14.Jordan VC, Gapstur S, Morrow M. Selective estrogen receptor modulation and reduction in risk of breast cancer, osteoporosis, and coronary heart disease. J Natl Cancer Inst. 2001;93(19):1449–1457. doi: 10.1093/jnci/93.19.1449. [DOI] [PubMed] [Google Scholar]
- 15.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11(4):643–658. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- 16.Shou J, et al. Mechanisms of tamoxifen resistance: Increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96(12):926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 17.Clarke R, et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22(47):7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 18.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 19.Kato S, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270(5241):1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 20.Le Goff P, Montano MM, Schodin DJ, Katzenellenbogen BS. Phosphorylation of the human estrogen receptor. Identification of hormone-regulated sites and examination of their influence on transcriptional activity. J Biol Chem. 1994;269(6):4458–4466. [PubMed] [Google Scholar]
- 21.Likhite VS, Stossi F, Kim K, Katzenellenbogen BS, Katzenellenbogen JA. Kinase-specific phosphorylation of the estrogen receptor changes receptor interactions with ligand, deoxyribonucleic acid, and coregulators associated with alterations in estrogen and tamoxifen activity. Mol Endocrinol. 2006;20(12):3120–3132. doi: 10.1210/me.2006-0068. [DOI] [PubMed] [Google Scholar]
- 22.Murphy LC, Niu Y, Snell L, Watson P. Phospho-serine-118 estrogen receptor-alpha expression is associated with better disease outcome in women treated with tamoxifen. Clin Cancer Res. 2004;10(17):5902–5906. doi: 10.1158/1078-0432.CCR-04-0191. [DOI] [PubMed] [Google Scholar]
- 23.Murphy LC, Seekallu SV, Watson PH. Clinical significance of estrogen receptor phosphorylation. Endocr Relat Cancer. 2011;18(1):R1–R14. doi: 10.1677/ERC-10-0070. [DOI] [PubMed] [Google Scholar]
- 24.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: A distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284(21):14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 26.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2(2):101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 27.Jordan VC, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol. 2007;25(36):5815–5824. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 28.Michalides R, et al. Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer Cell. 2004;5(6):597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 29.Williams CC, et al. Identification of four novel phosphorylation sites in estrogen receptor alpha: Impact on receptor-dependent gene expression and phosphorylation by protein kinase CK2. BMC Biochem. 2009;10:36. doi: 10.1186/1471-2091-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Romancer M, et al. Cracking the estrogen receptor’s posttranslational code in breast tumors. Endocr Rev. 2011;32(5):597–622. doi: 10.1210/er.2010-0016. [DOI] [PubMed] [Google Scholar]
- 31.Arnold SF, Vorojeikina DP, Notides AC. Phosphorylation of tyrosine 537 on the human estrogen receptor is required for binding to an estrogen response element. J Biol Chem. 1995;270(50):30205–30212. doi: 10.1074/jbc.270.50.30205. [DOI] [PubMed] [Google Scholar]
- 32.Britton DJ, et al. A novel serine phosphorylation site detected in the N-terminal domain of estrogen receptor isolated from human breast cancer cells. J Am Soc Mass Spectrom. 2008;19(5):729–740. doi: 10.1016/j.jasms.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen D, Pace PE, Coombes RC, Ali S. Phosphorylation of human estrogen receptor alpha by protein kinase A regulates dimerization. Mol Cell Biol. 1999;19(2):1002–1015. doi: 10.1128/mcb.19.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, Bai W. Regulation of estrogen receptor nuclear export by ligand-induced and p38-mediated receptor phosphorylation. Mol Cell Biol. 2002;22(16):5835–5845. doi: 10.1128/MCB.22.16.5835-5845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang J, et al. Phosphorylation of estrogen receptor-alpha at Ser167 is indicative of longer disease-free and overall survival in breast cancer patients. Clin Cancer Res. 2007;13(19):5769–5776. doi: 10.1158/1078-0432.CCR-07-0822. [DOI] [PubMed] [Google Scholar]
- 36.Kok M, et al. PKA-induced phosphorylation of ERα at serine 305 and high PAK1 levels is associated with sensitivity to tamoxifen in ER-positive breast cancer. Breast Cancer Res Treat. 2011;125(1):1–12. doi: 10.1007/s10549-010-0798-y. [DOI] [PubMed] [Google Scholar]
- 37.Holm C, et al. Phosphorylation of the oestrogen receptor alpha at serine 305 and prediction of tamoxifen resistance in breast cancer. J Pathol. 2009;217(3):372–379. doi: 10.1002/path.2455. [DOI] [PubMed] [Google Scholar]
- 38.Bostner J, Skoog L, Fornander T, Nordenskjöld B, Stål O. Estrogen receptor-alpha phosphorylation at serine 305, nuclear p21-activated kinase 1 expression, and response to tamoxifen in postmenopausal breast cancer. Clin Cancer Res. 2010;16(5):1624–1633. doi: 10.1158/1078-0432.CCR-09-1733. [DOI] [PubMed] [Google Scholar]
- 39.Ehrhardt C, et al. Rac1 and PAK1 are upstream of IKK-epsilon and TBK-1 in the viral activation of interferon regulatory factor-3. FEBS Lett. 2004;567(2-3):230–238. doi: 10.1016/j.febslet.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 40.Vendramini-Costa DB, Carvalho JE. Molecular link mechanisms between inflammation and cancer. Curr Pharm Des. 2012;18(26):3831–3852. doi: 10.2174/138161212802083707. [DOI] [PubMed] [Google Scholar]
- 41.Karin M, Greten FR. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 42.Hiscott J. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18(5-6):483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Robertson JF. Oestrogen receptor: A stable phenotype in breast cancer. Br J Cancer. 1996;73(1):5–12. doi: 10.1038/bjc.1996.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Institute of Laboratory Animal Resources . Guide for the Care and Use of Laboratory Animals. Washington, DC: National Academy Press; 1996. [Google Scholar]
- 45.US Office of Science and Technology Policy Laboratory animal welfare: U.S. government principles for the utilization and care of vertebrate animals used in testing, research, and training. Fed Regist. 1985;50(97):20864–20865. [PubMed] [Google Scholar]
- 46.He X, et al. c-Abl regulates estrogen receptor alpha transcription activity through its stabilization by phosphorylation. Oncogene. 2010;29(15):2238–2251. doi: 10.1038/onc.2009.513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.