Significance
Polycyclic aromatic hydrocarbons (PAHs) constitute a class of well-characterized environmental pollutants. Benzo[a]pyrene (BP), a typical and extensively studied PAH, can be metabolically converted to mutagenic reactive intermediates in vivo to form the major 10S (+)-trans-anti-BP diol epoxide (BPDE)-N2-dG (BPDE-dG) lesions and thus impede DNA replication and enhance mutagenesis. DNA polymerase κ (Polκ) is the only known DNA polymerase in mouse or human cells that can bypass the BPDE-dG adduct in an error free manner, thereby reducing mutation risk. Here we show that a structural gap and the N-clasp of Polκ are essential for accurate translesion synthesis of the bulky adduct.
Keywords: translesion DNA synthesis, polycyclic aromatic hydrocarbons
Abstract
DNA polymerase κ (Polκ) is the only known Y-family DNA polymerase that bypasses the 10S (+)-trans-anti-benzo[a]pyrene diol epoxide (BPDE)-N2-deoxyguanine adducts efficiently and accurately. The unique features of Polκ, a large structure gap between the catalytic core and little finger domain and a 90-residue addition at the N terminus known as the N-clasp, may give rise to its special translesion capability. We designed and constructed two mouse Polκ variants, which have reduced gap size on both sides [Polκ Gap Mutant (PGM) 1] or one side flanking the template base (PGM2). These Polκ variants are nearly as efficient as WT in normal DNA synthesis, albeit with reduced accuracy. However, PGM1 is strongly blocked by the 10S (+)-trans-anti-BPDE-N2-dG lesion. Steady-state kinetic measurements reveal a significant reduction in efficiency of dCTP incorporation opposite the lesion by PGM1 and a moderate reduction by PGM2. Consistently, Polκ-deficient cells stably complemented with PGM1 GFP-Polκ remained hypersensitive to BPDE treatment, and complementation with WT or PGM2 GFP-Polκ restored BPDE resistance. Furthermore, deletion of the first 51 residues of the N-clasp in mouse Polκ (mPolκ52–516) leads to reduced polymerization activity, and the mutant PGM252–516 but not PGM152–516 can partially compensate the N-terminal deletion and restore the catalytic activity on normal DNA. However, neither WT nor PGM2 mPolκ52–516 retains BPDE bypass activity. We conclude that the structural gap physically accommodates the bulky aromatic adduct and the N-clasp is essential for the structural integrity and flexibility of Polκ during translesion synthesis.
Benzo[a]pyrene (BP) is one of the best characterized polycyclic aromatic hydrocarbons generated during incomplete fuel combustion, in tobacco smoke, and in cooked food. The most mutagenic and tumorigenic metabolite of BP in vivo is the (+)-7R,8S,9S,10R-BP dihydrodiol epoxide [(+)-anti-BPDE] (1), which reacts readily with the exocyclic amino groups of guanine residues in DNA to form the major 10S (+)-trans-anti-BPDE-N2-dG adduct (2) (abbreviated as BPDE-dG hereafter). This adduct typically impedes normal DNA replication. Translesion DNA synthesis (TLS), which provides one mode of DNA damage tolerance, uses specialized bypass polymerases to synthesize DNA opposite and beyond a variety of replication-blocking lesions, thus avoiding replication fork collapse (3). The Y-family DNA polymerases are specialized for TLS. To date, four of them, namely DNA polymerase (Pol) η, Polι, Polκ, and REV1, have been identified in mammals (4–6). Polκ is the only one with homologs in bacteria (DinB; also known as Pol IV in Escherichia coli), archaea [DNA polymerase IV (Dpo4) in Sulfolobus solfataricus and DinB homologue (Dbh) in Sulfolobus acidocaldarius] and all eukaryotes. The mouse and human POLK gene are expressed ubiquitously, with highest levels in testis (7).
Human Polκ can accurately incorporate dCMP opposite the BPDE-dG adduct and bypasses the lesion in an almost error-free manner (8–10). In agreement with in vitro analyses, Polκ-deficient mouse embryonic fibroblasts (MEFs) and embryo stem cells are sensitive to killing by BP and exhibit increased BPDE-induced mutagenesis (11–14). E. coli DinB can also correctly incorporate dCMP opposite some N2-dG adducts, including N2-furfuryl-dG (15) and N2-(1-carboxyethyl)-2′-dG (16) with greater catalytic proficiency than opposite normal dG. Similarly, BPDE-dG (15, 17) is incorporated with high accuracy but reduced efficiency. Although phylogenetically related to Polκ, the enzyme Dpo4 possesses UV-lesion bypass activity akin to that of Polη (18), but bypasses BPDE-dG lesions inefficiently and induces mutations by a −1 frame-shift mechanism (19).
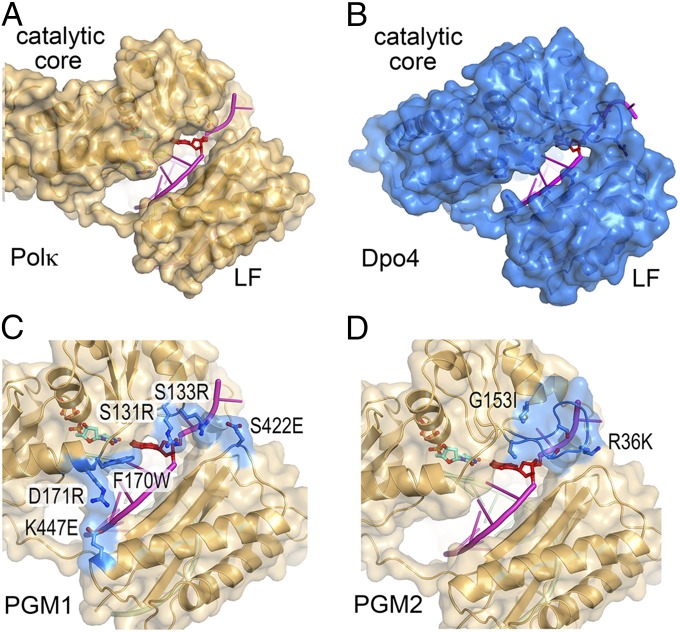
The crystal structure of Dpo4 in ternary complex with BPDE-dG and dNTP (19) shows that the BPDE-dG lesion at the template position is looped out and the downstream normal base is used to direct nucleotide incorporation. Similar loop-out of the adducted base is observed during primer extension immediately after the lesion (19) and results in −1 frameshift mutation. The looped out bulky minor groove adduct is accommodated in a structural gap between the catalytic core (composed of palm, finger, and thumb domains) and the little finger (LF) domain. A ternary complex crystal structure of human Polκ with native DNA has been reported (20). As the DNA adduct is absent in the Polκ structure, it is not known how a BPDE-dG lesion is accommodated and how dCTP is selected by Polκ. The structures of Dpo4 and human Polκ are similar in the polymerase domain (21, 22). Both contain a structural gap between the catalytic core and LF domain (Fig. 1 A and B). Interestingly, the gap is larger in Polκ than in Dpo4. The gap between the LF and the catalytic core is virtually absent in human and yeast Polη (23, 24), which are specialized to bypass cyclobutane pyrimidine dimers and depend on close interactions between the catalytic core and LF to stabilize the UV radiation-induced lesion. The uniquely large structural gap in Polκ has been hypothesized to be essential for accommodating minor groove adducts, such as BPDE-dG, and hence for efficient bypass (19, 25, 26). In addition, Polκ has a 100-residue N-terminal extension, designated the N-clasp, which is absent in the archaeal homolog Dpo4 (22). This N-clasp has been shown to encircle a normal primer/template pair and connects and fastens the flexible LF domain to the catalytic core (20). Human Polκ (hPolκ) with a 90-residue (residues 91–559) or a 67-residue truncated N-clasp (residues 68–526) has diminished catalytic activity compared with the native catalytic domain (residues 1–526) or the domain without the first 18 residues (residues 19–526) (20, 22).
Fig. 1.
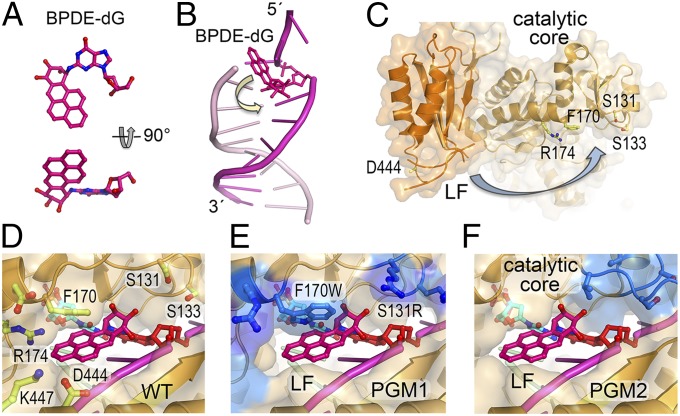
Design of PGM1 and PGM2 mutant mPolκ. (A) A back view of the structure of hPolκ–DNA–dNTP complex structure (PDB ID code 2OH2). Polκ is shown in gold ribbon diagram and covered with a semitransparent molecular surface. The residues to be mutated are shown in ball-and-stick model. The DNA template strand is shown in magenta and the template base is highlighted in red opposite the incoming dNTP (cyan). (B) Structure of Dpo4–DNA–dNTP complex (PDB ID code 2AGQ). After superposition with Polκ, Dpo4 is shown in blue with the corresponding residues along the structural gap highlighted in ball-and-stick models. (C) Models of PGM1 and (D) PGM2 mutant mPolκ. The mutated residues are highlighted in blue and labeled according to mPolκ residue number. The corresponding human residues are exactly one number higher (Fig. S1).
Previous studies have investigated the fidelity of Y-family polymerases by mutating the steric gate and residues surrounding the incoming nucleotide, e.g., Phe13 and Tyr79 of DinB (15, 27), Phe12 in Dbh (28), Tyr112 of Polκ (29), and Tyr11 of UmuC (30, 31). Similarly, TLS efficiency and specificity have been examined after swapping the LF domain (32), the finger domain (33), or the linker between the thumb and LF among Y-family members (34). In terms of lesion specificity, amino acid substitution of residue Arg332 in the LF of Dpo4 (35) and Leu508 in the LF of hPolκ (36) have been shown to alter the efficiency of 8-oxo-G bypass. Studies of BPDE-dG bypass are limited. Phe171 in hPolκ located along the structural gap and predicted to stack with (26) or proximal to the minor groove adduct was replaced by Ala (37). Interestingly, the F171A mutant form of hPolκ and other alanine substitutions along the predicted BP binding pocket, which enlarge the structural gap, do not alter BPDE-dG bypass efficiency by Polκ (37).
To understand the molecular mechanism for bypass synthesis of BPDE-dG adducts by Polκ, we elected to probe the structural gap in Polκ by reducing its size (Fig. 1C). Two mouse Polκ (mPolκ) variants [Polκ Gap Mutant (PGM) 1 and PGM2] were generated and compared for their ability to synthesize DNA opposite BPDE-modified dG templates as well as undamaged DNA. Polκ-deficient MEFs stably complemented with WT, PGM1, and PGM2 GFP-Polκ were established to validate the function of Polκ in vivo. To explore the effect of the N-clasp on DNA synthesis by Polκ mutants, we generated N-terminal 51-aa deleted WT and PGM mutant forms of mPolκ (Δ51) and examined their DNA synthetic and TLS activities. Collectively, our results offer a molecular mechanism for lesion-bypass specificity by Polκ and uncover a key role of the N-clasp during TLS.
Results
Design of PGM1 and PGM2 Variants of mPolκ.
Structural comparisons of hPolκ and Dpo4, each in complex with DNA primer/template and a dNTP, reveal different sizes of the gap between the catalytic core and LF domains (Fig. 1 A and B). A shorter loop connecting the secondary structures in the Polκ finger domain compared with Dpo4 enlarges the gap in Polκ on the 5′ side (i.e., downstream) of the template base. Smaller amino acid side chains in the finger and LF domains of Polκ than those in Dpo4 along the crevice between the catalytic core and the LF further enlarge the gap in Polκ on the 3′ side (i.e., upstream) of the template base. The first 540 residues of human and mouse Polκ, including the entire polymerase catalytic domain, are 95% identical (Fig. S1). For the ease of cell-based complementation assays of Polκ activity, we chose to generate mutations in mPolκ. All residues discussed in this manuscript, including the ones that determine the gap size, are strictly conserved between human and mouse Polκ. However, most residue numbers between the two Polκs differ by one because the eighth residue (Cys8) of hPolκ is absent in mouse Polκ. For the structure-based models, mouse residue numbers are used hereafter for consistency (Fig. S1).
The PGM1 Polκ mutant was designed to reduce the gap size along its entire length by increasing the sizes of amino acid side chains, e.g., S131R, S133R, F170W, and D171R, and by creating charge–charge interaction pairs across the gap, e.g., S422E opposite S131R and S133R, K447E opposite D171R (Fig. 1C). In the PGM2 Polκ mutant, a 3-residue loop connecting β2 and β3 (Ser131–Met132–Ser133) in the finger domain was replaced by the corresponding 10-residue loop in Dpo4 (Val32–Gly41). To graft the Dpo4 loop onto Polκ, two additional mutations were introduced, i.e., Gly153 (G153I) to mimic the corresponding residue in Dpo4 (Ile61) and Arg36 in the grafted Dpo4 loop was mutated to Lys (R36K) to avoid potential steric clashes (Fig. 1D). The result of the loop replacement closes one end of the gap downstream of the template base. Our prediction was that these Polκ mutants would retain WT-like activity when replicating normal DNA, and show reduced activity when bypassing BPDE-dG (Fig. 2A) adducts.
Fig. 2.
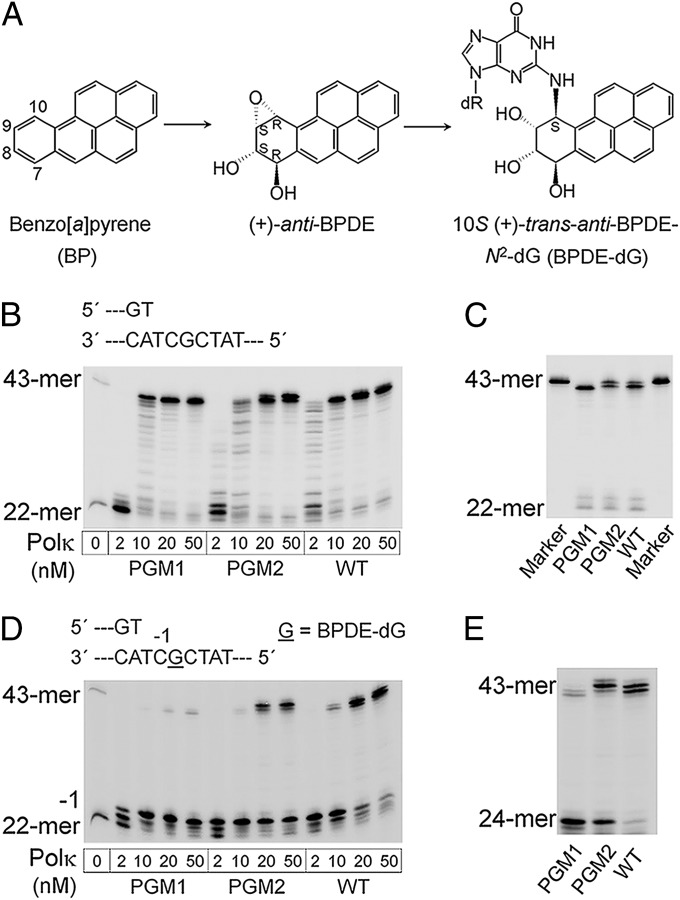
Primer extension opposite normal and BPDE-dG templates by full-length WT, PGM1, and PGM2 mouse Polκ. (A) Structures of BP, its (+)-(7R,8S,9S,10R) dihydrodiol epoxide metabolite [(+)-anti-BPDE], and the BPDE-dG [10S (+)-trans-anti-[BP]-N2-dG (2)] adduct in DNA. (B) Reactions were carried out in the presence of all four dNTPs for 5 min at 37 °C with increased concentrations of Polκ as shown below each track. (C) Analysis of extension product lengths over normal dG template by PGM1, PGM2, and WT mPolκ. The 43-mer marker is in a separate lane. The reactions were catalyzed by 10 nM Polκ at 37 °C for 15 min. (D) Running-start primer extension experiments on BPDE-dG template. Reactions were carried out with increasing concentrations of Polκ at 37 °C for 15 min. The underlined “G” represents the BPDE-dG lesion. The 22-mer primer was readily extended to the two nucleotides 3′ of the lesion and stalled at template position −1, immediately before the lesion. (E) Comparison of primer extension products length catalyzed by PGM1, PGM2, and WT mPolκ by using the BPDE-dG template. The assays included 50 nM of each Polκ at 37 °C for 30 min.
Primer Extension by Full-Length WT, PGM1, and PGM2 mPolκ Proteins.
Full-length WT, PGM1, and PGM2 Polκ proteins were purified to near homogeneity by using FPLC (Fig. S2). Like WT Polκ, PGM1 and PGM2 readily extended the primer to the end of the normal DNA template at enzyme concentrations greater than 10 nM (Fig. 2B). In the case of PGM1, a single extension product corresponding to a 42-mer was observed, whereas for WT and PGM2 Polκ, a fully extended 43-mer was observed in addition to the 42-mer products (Fig. 2C).
In the presence of a BPDE-dG, WT Polκ readily extended the 22-mer primer by 2 nt and paused immediately before the adduct (“−1”). When the polymerase concentration was increased from 2 to 50 nM, essentially all primers were extended to the end or near the end of the template strand (Fig. 2D). As predicted, extension of the 22-mer primer was inefficient by PGM1 or PGM2 as a result of the strong pause at the −1 position. Notably, primer extension by PGM1 was strongly blocked by the bulky adduct, whereas stalling of PGM2 was less pronounced (Fig. 2 D and E and Figs. S3 and S4). These data confirmed that mutations introduced to PGM1 or PGM2 Polκ led to a significant decrease in bypass efficiency. In addition, extension product length varied with different Polκ forms (Fig. 2E). WT generated 42- and 43-mer full extension products; PGM1 predominantly produced 41- and a little 42-mer; and PGM2 extended the primer to the full-length 43-mer with minor bands corresponding to the 42- and 44-mers.
Steady-State Kinetic Measurements of Full-Length WT and Mutant Polκ.
Kinetic parameters Vmax and Km were measured first with WT and mutant Polκ for single dNTP insertion opposite normal dG (Table S1). The two mutant proteins behaved like WT on native DNA, with less than threefold reduction of catalytic efficiency of dCTP incorporation (Table S2). Both WT and PGM1 are accurate in dCTP incorporation opposite dG, with misincorporation frequencies in the order of 10−2 to 10−3 (Table S1). However, PGM2 was less accurate compared with WT or PGM1, and had a 10-fold higher frequency in misincorporation of dTTP opposite dG (Table S1).
Vmax and Km were also determined for WT and both Polκ mutants with respect to nucleotide insertion opposite the BPDE-dG adduct (Table 1). Compared with WT Polκ, the modifications in PGM1 and PGM2 led to decreases in the efficiency (Vmax/Km) of dCTP insertion opposite a damaged dG by 24- and seven fold (Table S2), respectively, which is consistent with the observation that PGM1 is more severely blocked by BPDE-dG than PGM2 in the running-start TLS assay (Fig. 2D).
Table 1.
Steady-state kinetic parameters for nucleotide incorporation opposite the BPDE-dG adduct by WT, PGM1, and PGM2 mPolκ
| Polκ/dNTP | Vmax, nM⋅min−1 | Km, nM | Vmax/Km, min−1 | Misincorporation frequency |
| WT | ||||
| A | 0.85 ± 0.065 | (1.68 ± 0.017) × 105 | 5.06 × 10−6 | 5.50 × 10−2 |
| C | 1.04 ± 0.091 | (1.13 ± 0.334) × 104 | 9.20 × 10−5 | 1 |
| T | 0.40 ± 0.011 | (2.96 ± 0.288) × 105 | 1.35 × 10−6 | 1.47 × 10−2 |
| PGM1 | ||||
| A | 0.57 ± 0.018 | (4.99 ± 0.163) × 105 | 1.14 × 10−6 | 2.99 × 10−1 |
| C | 1.06 ± 0.055 | (2.78 ± 0.305) × 105 | 3.81 × 10−6 | 1 |
| T | 0.56 ± 0.0069 | (1.49 ± 0.132) × 106 | 3.76 × 10−7 | 9.87 × 10−2 |
| PGM2 | ||||
| A | 0.80 ± 0.016 | (6.72 ± 0.269) × 105 | 1.19 × 10−6 | 8.88 × 10−2 |
| C | 1.17 ± 0.072 | (8.71 ± 1.23) × 104 | 1.34 × 10−5 | 1 |
| T | 0.21 ± 0.0050 | (1.36 ± 0.230) × 105 | 1.54 × 10−6 | 1.15 × 10−1 |
Data are expressed as mean ± SD obtained from three independent experiments. No apparent incorporation of dGMP was observed even when high concentrations of dNTP (2.5 mM) were used by WT and mutant proteins. Thus, steady-state kinetic parameters were determined only for dATP, dCTP, and dTTP incorporation.
The single dNTP insertion across the BPDE-dG lesion by WT Polκ is accurate (Table 1). The misincorporation frequencies are decreased by two orders of magnitude compared with that of dCTP incorporation and are comparable to the misincorporation frequencies with undamaged DNA template (Table S1). The decreased efficiency of dATP or dTTP misincorporation by WT Polκ mainly results from an increased Km compared with dCTP incorporation. PGM1 and PGM2 Polκ show reduced fidelity compared with WT, even though the absolute efficiencies of misincorporation (Vmax/Km) by PGM1 and PGM2 are lower than that of WT (Table 1). The Km of misincorporation by PGM1 and PGM2 relative to the correct dCTP is not increased as much as with WT protein. In particular, PGM1 prefers to misincorporate dATP, whereas PGM2 favors the misincorporation of dTTP (Table 1).
Clonogenic Assay upon BPDE Treatment.
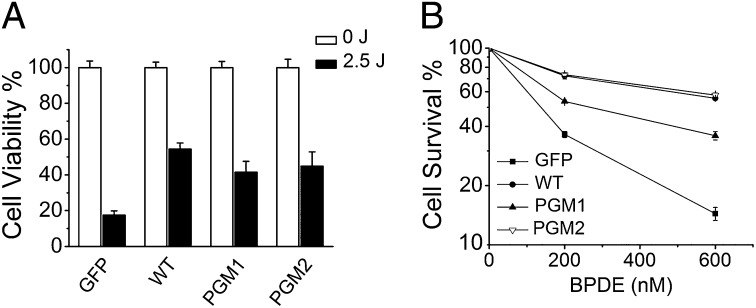
It was previously shown that Polκ-deficient MEFs are abnormally sensitive to UV radiation (38, 39) and BPDE treatments (13, 14), and complementation with GFP-Polκ can rescue cellular hypersensitivity. We thus determined whether Polκ-deficient MEFs complemented with WT, PGM1, or PGM2 GFP fusion proteins showed altered sensitivity to BPDE treatment. Polκ-deficient MEFs expressing GFP-tagged WT, PGM1, or PGM2 were generated by lentivirus infection (Fig. S5). Expression of WT, PGM1, or PGM2 GFP-Polκ increased the UV resistance of Polκ-deficient MEFs compared with that observed with expression of GFP alone (Fig. 3A), indicating that WT, PGM1, and PGM2 GFP-Polκ are functional in Polκ-deficient MEFs.
Fig. 3.
PGM1 GFP-Polκ has a defect in complementation of BPDE hypersensitivity of Polκ-deficient MEFs. The Polκ-deficient MEFs stably complemented with GFP or WT, PGM1, and PGM2 GFP-Polκ were treated with UV (A) or BPDE (B) as described in SI Materials and Methods. Surviving fractions are expressed as a percentage of mock-treated cells. Values are the mean of three independent experiments (±SE).
We performed a clonogenic assay to examine the effects of the Polκ mutants on cell survival upon BPDE treatment (Fig. 3B). Expression of WT GFP-Polκ, but not GFP alone, rescued the BPDE hypersensitivity of Polκ-deficient MEFs, in accordance with the notion that Polκ is required for survival after BPDE exposure. Interestingly, the killing curves were similar between cells complemented with WT or PGM2 GFP-Polκ, whereas cells complemented with PGM1 exhibited less efficient rescue, suggesting that PGM2 but not PGM1 is capable of restoring BPDE resistance. This cell-based observation is consistent with the result obtained in vitro that bypass synthesis over the BPDE-dG lesion is more efficient by PGM2.
Effect of the N-Clasp on the Catalytic Activity of Polκ with Reduced Gap Size.
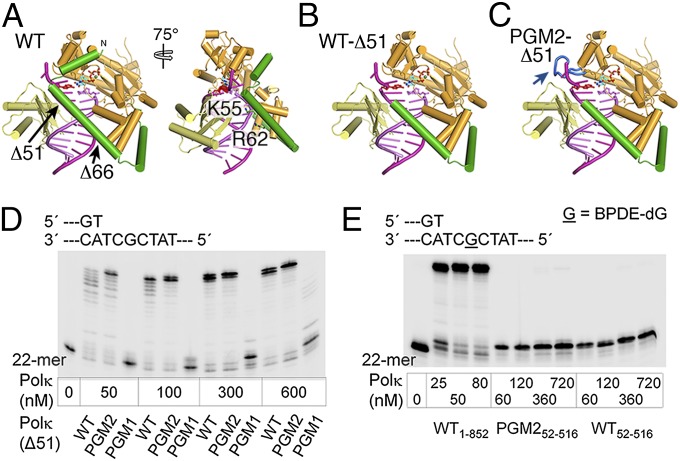
It has been shown that the N-clasp of Polκ encircles the substrate DNA, thereby enabling DNA synthesis (20). As PGM1 and PGM2 have increased interactions between the catalytic core and LF domain and a reduced structural gap, we asked whether the N-clasp of Polκ is still required for the protein structure and catalytic activity of PGM1 and PGM2. In the structure of the hPolκ–DNA complex (Fig. 4A), the first 51 residues in the N-clasp appear to bridge the catalytic core with the LF, and, in the second part of the N-clasp (52–67 aa), Lys55 and Arg62 interact with the DNA primer directly (Fig. 4A). We therefore deleted residues 1 to 51 and 1 to 66 of WT, PGM1, and PGM2, creating the mPolκ52–516 and mPolκ67–525 variants, and examined their activity during normal DNA synthesis and TLS. Deletion of the first 66 aa of mPolκ resulted in nearly complete loss of catalytic activity of PGM167–525, and significantly reduced the activities of PGM2 and WT mPolκ67–525 in normal DNA synthesis (Fig. S6). Interestingly, restoring 15 residues to the N-clasp (mPolκ52–516) rescued the DNA polymerase activities of WT and mutant Polκ (mPolκ67–525) to different degrees (Fig. 4 D and E). PGM252–516 manifested much enhanced activity relative to WT52–516, and at enzyme concentrations greater than 50 nM, PGM252–516 exhibited full extension activity. However, the normal DNA synthesis ability of PGM152–516 remained lowest, and no complete extension could be detected even at 600 nM enzyme concentration. Surprisingly, removal of the first 51 aa resulted in complete loss of TLS activity of PGM2 and WT mPolκ52–516 (Fig. 4E). Strong primer stalling was observed in the vicinity of the adduct even at a 70:1 protein/DNA molar ratio.
Fig. 4.
N-terminal truncated Polκ and their activities in normal and TLS. (A) A front view of the hPolκ–DNA–dNTP complex structure (PDB ID code 2OH2). Residues 32 to 51 of the N-clasp (green) connect the catalytic core (bright orange) with LF (pale yellow), and, in the second part of the N-clasp (52–66 aa), Lys55 and Arg62 directly interact with the primer strand. (B) Deletion of residues 1 to 51 in the N-clasp is predicted to result in loss of hydrophobic interactions of the N-clasp with the finger and LF domains. (C) In PGM2, the requirement of 1 to 51 aa to connect the catalytic core and LF is predicted to be substituted by the long loop inserted (blue) in the finger domain. (D) DNA synthesis activities over normal dG template. Reactions were carried out with 20 nM DNA substrate and increasing concentration of mPolκ52–516 at 37 °C for 15 min. (E) Primer extension on BPDE-dG damaged template. A total of 10 nM DNA was incubated with different concentrations of PGM2 and WT mPolκ52–516 at 37 °C for 15 min. The reaction with WT Polκ is described in Fig. 2D.
Discussion
A structural gap of different sizes between the polymerase catalytic core and LF has been observed among different Y family polymerases (23, 25) and predicted to play an influential role in lesion bypass specificity and efficiency. In the present study, based on the structural superposition of Y-family Polκ and Dpo4, we engineered the gap in Polκ to closely resemble that found in Dpo4 and showed that the BPDE-dG bypass efficiency and accuracy by these Polκ mutants are indeed reduced.
In PGM1 Polκ, a collection of six amino acid replacements reduced the gap along its entire length (Fig. 1C). In contrast, in PGM2, replacement of a three-residue loop in the finger domain by the corresponding 10-residue loop in Dpo4 closed one end of the gap in Polκ around the template base (Fig. 1D). Studies that used the in vitro TLS system reveal that PGM1 and PGM2 retain activities comparable to those of WT on normal DNA. However, mutations in PGM1 Polκ strongly inhibit its TLS ability on a BPDE-dG template, leading to extensive primer pausing in the vicinity of the adducted dG, whereas mutations in the PGM2 moderately hinder BPDE bypass (Fig. 2D). The differences in polymerase stalling that result from amino acid substitutions around the BPDE-binding sites are mirrored in the different catalytic efficiencies of dCTP incorporation opposite the BPDE-dG adduct (Table S2). Kinetic analysis of PGM1, PGM2, and WT Polκ reveals that the reduction of catalytic efficiency stems from an increase in Km of dCTP binding with little change in Vmax (Table S2). Molecular modeling of WT and mutant proteins suggest that the reduced gap size perturbs binding of the adduct and the template base and thus may cause the increase in Km (Fig. 5).
Fig. 5.
A molecular model of BPDE accommodation and bypass by Polκ. (A) Atomic structure of the (+)-trans-anti-BPDE-dG determined by NMR (40, 41). (B) The BPDE adduct is situated in the minor groove and points toward the 5′ end at a template and primer junction (PDB ID code 1AXO). (C) In the apo-hPolκ68–517 structure (PDB ID code 1T94), the LF is flexible and far removed from the catalytic core. (D) Both DNA and Polκ need to undergo conformational changes as indicated by arrows in B and C, to accommodate the BPDE-dG lesion in the minor groove for TLS. The adduct is predicted to point toward the 3′ end of the template strand and form van der Waals contacts with Phe170. (E) In the PGM1 complex, the aromatic rings of BPDE would clash with F170W and the sugar moiety of BPDE would clash with S131R. (F) In the PGM2 complex, the sugar moiety of BPDE would clash with the loop transplanted from Dpo4 (shown in blue). The rest of the large structural gap likely remains intact.
The NMR structures of DNA containing the BPDE-dG lesion (40, 41) show that the BP ring whether in the middle of a duplex or at the template and primer junction always points toward the 5′ end of the modified strand (Fig. 5 A and B). The molecular dynamic simulation studies of Polκ bypassing a BPDE-dG lesion shows that the BP ring must point toward the 3′ end to avoid steric collisions with the finger domain, and the presence of the adduct does not perturb the prereactant or transition states of the reaction pathway (26, 42). In our model of mPolκ and the BPDE-DNA complex, the lesion is situated as proposed by Jia et al. (26), and WT Polκ readily accommodates the minor groove BPDE-dG adduct. The BP moiety points toward the 3′ end of the template strand and forms van der Waals contacts with Phe170 (Fig. 5D). In the PGM1 complex (Fig. 5E), the predicted narrowed gap restricts the flexibility of the template strand, and the aromatic rings of BP clash with F170W, suggesting that the aromatic adduct may be shifted and thereby promote misincorporation. Alternatively, the lesion base may be looped out and the next base downstream of the adduct may serve as the template to direct primer extension, resulting in a −1 frame shift and a shortened product (19) (Fig. 2 D and E). However, incorporation of dGTP opposite the adduct by PGM1 is rarely observed even at extremely high nucleotide concentrations (Table 1); instead, the misincorporation frequency of dATP is increased relative to dCTP. We suspect that narrowing of the groove where the BPDE is accommodated may destabilize the binding of damaged template to PGM1 and lead to increased misincorporation of dATP by the A-rule (43, 44). In the modeled structure of PGM2 (Fig. 5F), only the sugar moiety of BPDE clashes with the loop transplanted from Dpo4, whereas the rest of the large structural gap remains intact. The engineered long loop probably stabilizes a blunt DNA duplex in the active site to allow template-independent nucleotide addition, as observed in Dpo4 (45), thereby resulting in the longer DNA product. PGM2 manifests increased misincorporation frequency of dTTP in particular (Table 1). We suspect that the templating dG and particularly BPDE-dG base may shift toward the minor groove, thus facilitating a dG:dTTP wobble base pair.
To compare the function of these Polκ variants in cells after BPDE treatment, we reconstituted Polκ−/− MEFs with WT, PGM1, and PGM2 GFP fusion proteins. We observed that cells complemented with PGM1 retained hypersensitivity to BPDE treatment and that complementation with WT or PGM2 restored the BPDE resistance. As Polκ is required for TLS opposite BP adducts in vivo (11), the impaired ability of PGM1 Polκ to restore BPDE resistance is most likely a result of an inability to efficiently catalyze TLS over BPDE-dG lesions. For the PGM2 complex (Fig. 5F), the remaining large structural gap in between the core and LF provides space for the BPDE adduct to reorient and fit, which may explain why PGM2 can restore BPDE resistance in Polκ-deficient cell lines. These data further confirm that mutations in the BPDE binding sites modulate the bypass function of Polκ across minor-groove adducts. Notably, the structural gap size in Polκ appears to have little consequence in Polκ’s role in survival from UV radiation exposure.
Residues 1 to 51 of the N-clasp unique to Polκ appear to bridge the catalytic core and LF domain across the large structural gap. To examine whether this part of the N-clasp is required for the structural integrity in the case of PGM1 and PGM2, in which the gap size is reduced, we made N-terminal–deleted mPolκ and tested its polymerase activities (Fig. 4 and Fig. S6). We find that the N-clasp1–51 is required for even normal DNA synthesis. However, its role of connecting the catalytic core and LF domain can be partially replaced by the engineered long loop in the finger domain that interacts with the LF, as demonstrated in PGM252–516 (Fig. 4D) and in the rest of the Y-family polymerases (34). The higher activity of PGM252–516 than that of Polκ52–516 supports our prediction that the Dpo4 loop transplanted into PGM2 qualitatively serve the same function as the N-clasp in normal DNA synthesis, although the N-clasp is more effective than the loop insertion in PGM252–516 (Fig. S7). The N-clasp1–51 forms hydrophobic interactions with the finger (Ile35, Ile38, and Ala42) and LF (Phe48 to Phe464). The resulting interface is strong, even at high salt concentrations, and yet flexible, which may enable N-clasp1–51 to counteract against undesirable interactions and facilitate substrate binding and DNA synthesis by Polκ as demonstrated by the active full-length PGM1 and the inactive PGM152–516 (Fig. 2B and Fig. 4D). The complete failure of TLS to bypass BPDE-dG by Polκ52–516 indicates that N-clasp1–51 has a second role in addition to linking the catalytic core and LF. In the presence of the N-clasp Polκ possibly undergoes conformational changes itself (Fig. 5C), and induces changes in the BPDE-dG adduct after initial association, for example, from pointing toward the 5′ end as observed in solution by NMR to pointing toward the 3′ end to fit into the active site for the adduct to template dCTP incorporation. The second role of the N-clasp in TLS is irreplaceable, and deletion of the first 51 residues leads to failure of BPDE bypass (Fig. 4E). Dpo4 has no N-clasp but still can bypass a BPDE-dG adduct with lower efficiency and reduced accuracy than Polκ (19, 46, 47). This may reflect the strong linker between the thumb and LF domains of Dpo4 (34).
Collectively, these results imply that the structure gap and N-clasp of Polκ are essential for BPDE-dG bypass but not for normal DNA synthesis. With the reduced size of the structural gap, PGM1 or PGM2 Polκ has lower efficiency and accuracy in bypassing the BPDE-dG adduct. Finally, our data reinforce the notion that accurate replicative bypass of polycyclic aromatic G-adducts in DNA may be a primary function of Polκ, and that defective translesion synthesis of such lesions in DNA may generate predisposition to tumors associated with exposure to polycyclic hydrocarbons.
Materials and Methods
A full description of the present study materials and methods is provided in SI Materials and Methods.
Molecular Modeling.
Crystal structure of Dpo4 [Protein Data Bank (PDB) ID code 2AGQ] (48) and human Polκ (PDB ID code 2OH2) (20) complexed with DNA and dNTP were superimposed by using COOT (49). The major differences that determine the size of gap were located. To reduce its size, in Polκ PGM1, six residues along the gap were replaced with larger side chains. In PGM2, the short loop between β2 and β3 in Polκ was replaced by the corresponding loop in Dpo4 (Val32–Phe33–Ser34–Gly35–Arg36–Phe37–Glu38–Asp39–Ser40–Gly41). To avoid close contact, Arg36 of Dpo4 was replaced by Lys when transplanted to Polκ. Gly154 of Polκ was also replaced by Ile to enhance the stability of the transplanted long loop between β2 and β3. Structural models are displayed by using PyMOL. BPDE-dG is modeled after the NMR structure (PDB ID code 1AXO) (41) and the molecular simulation (26, 42). Details of protein purification, primer extension assay, steady-state kinetic analyses, and cell survival assay are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Dr. N. Geacintov for providing the BPDE modified oligodeoxynucleotide and advice on fidelity assay, Drs. S. Broyde and L. Lior-Hoffmann for the molecular simulated models of Polκ-BPDE-dG complex, Dr. H. Wang for BPDE reagents, Dr. T. Ogi for advice on establishment of stable cell lines, X. Zhang for assistance in cell culture, and Drs. Y. Zhang and P. Fischhaber for helpful discussion. This work was supported by Chinese National 973 Project Grants 2013CB945000, 2011CB944302, and 2012CB944702; Natural Science Foundation of China Grants 30970588, 31170730, 81371415, 31300658, and 31100557; National Institutes of Health Intramural Program DK036146-07; the Chinese Academy of Sciences “One-Hundred-Talent Program”; and the State Key Laboratory of Biomembrane and Membrane Biotechnology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324168111/-/DCSupplemental.
References
- 1.Brookes P, Osborne MR. Mutation in mammalian cells by stereoisomers of anti-benzo[a] pyrene-diolepoxide in relation to the extent and nature of the DNA reaction products. Carcinogenesis. 1982;3(10):1223–1226. doi: 10.1093/carcin/3.10.1223. [DOI] [PubMed] [Google Scholar]
- 2.Geacintov NE, et al. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: Principles, patterns, and diversity. Chem Res Toxicol. 1997;10(2):111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg E, et al. DNA Repair and Mutagenesis. 2nd Ed. Washington, DC: ASM; 2006. [Google Scholar]
- 4.Ohmori H, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8(1):7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 5.Yang W. Damage repair DNA polymerases Y. Curr Opin Struct Biol. 2003;13(1):23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 6.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlach VL, et al. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci USA. 1999;96(21):11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: errOr-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair (Amst) 2002;1(7):559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 9.Rechkoblit O, et al. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277(34):30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, et al. Effects of base sequence context on translesion synthesis past a bulky (+)-trans-anti-B[a]P-N2-dG lesion catalyzed by the Y-family polymerase pol kappa. Biochemistry. 2003;42(8):2456–2466. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- 11.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA. 2002;99(24):15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avkin S, et al. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: The role of DNA polymerase kappa. J Biol Chem. 2004;279(51):53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 13.Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280(23):22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 14.Stancel JNK, et al. Polk mutant mice have a spontaneous mutator phenotype. DNA Repair (Amst) 2009;8(12):1355–1362. doi: 10.1016/j.dnarep.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439(7073):225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 16.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc Natl Acad Sci USA. 2008;105(25):8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen X, et al. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J Biol Chem. 2002;277(7):5265–5274. doi: 10.1074/jbc.M109575200. [DOI] [PubMed] [Google Scholar]
- 18.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): An archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 2001;29(22):4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer J, et al. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc Natl Acad Sci USA. 2007;104(38):14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lone S, et al. Human DNA polymerase kappa encircles DNA: Implications for mismatch extension and lesion bypass. Mol Cell. 2007;25(4):601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell. 2001;107(1):91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 22.Uljon SN, et al. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure. 2004;12(8):1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Biertümpfel C, et al. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465(7301):1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silverstein TD, et al. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465(7301):1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Woodgate R. What a difference a decade makes: Insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104(40):15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia L, Geacintov NE, Broyde S. The N-clasp of human DNA polymerase kappa promotes blockage or error-free bypass of adenine- or guanine-benzo[a]pyrenyl lesions. Nucleic Acids Res. 2008;36(20):6571–6584. doi: 10.1093/nar/gkn719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarosz DF, Cohen SE, Delaney JC, Essigmann JM, Walker GC. A DinB variant reveals diverse physiological consequences of incomplete TLS extension by a Y-family DNA polymerase. Proc Natl Acad Sci USA. 2009;106(50):21137–21142. doi: 10.1073/pnas.0907257106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31(14):4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niimi N, et al. The steric gate amino acid tyrosine 112 is required for efficient mismatched-primer extension by human DNA polymerase kappa. Biochemistry. 2009;48(20):4239–4246. doi: 10.1021/bi900153t. [DOI] [PubMed] [Google Scholar]
- 30.Kuban W, et al. Escherichia coli UmuC active site mutants: Effects on translesion DNA synthesis, mutagenesis and cell survival. DNA Repair (Amst) 2012;11(9):726–732. doi: 10.1016/j.dnarep.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaisman A, et al. Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res. 2012;40(13):6144–6157. doi: 10.1093/nar/gks233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boudsocq F, et al. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J Biol Chem. 2004;279(31):32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 33.Kirouac KN, Ling H. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase iota. EMBO J. 2009;28(11):1644–1654. doi: 10.1038/emboj.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson RC, Jackson MA, Pata JD. Y-family polymerase conformation is a major determinant of fidelity and translesion specificity. Structure. 2013;21(1):20–31. doi: 10.1016/j.str.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rechkoblit O, et al. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4(1):e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irimia A, Eoff RL, Guengerich FP, Egli M. Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase kappa opposite the 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J Biol Chem. 2009;284(33):22467–22480. doi: 10.1074/jbc.M109.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sassa A, et al. Phenylalanine 171 is a molecular brake for translesion synthesis across benzo[a]pyrene-guanine adducts by human DNA polymerase kappa. Mutat Res. 2011;718(1-2):10–17. doi: 10.1016/j.mrgentox.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Schenten D, et al. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32(11):3152–3160. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8(6):640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 40.Cosman M, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci USA. 1992;89(5):1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng B, et al. Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction. Biochemistry. 1997;36(45):13769–13779. doi: 10.1021/bi970069s. [DOI] [PubMed] [Google Scholar]
- 42.Lior-Hoffmann L, et al. Preferred WMSA catalytic mechanism of the nucleotidyl transfer reaction in human DNA polymerase κ elucidates error-free bypass of a bulky DNA lesion. Nucleic Acids Res. 2012;40(18):9193–9205. doi: 10.1093/nar/gks653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 44.Strauss BS. The “A” rule revisited: Polymerases as determinants of mutational specificity. DNA Repair (Amst) 2002;1(2):125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- 45.Fiala KA, et al. Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. J Mol Biol. 2007;365(3):590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perlow-Poehnelt RA, Likhterov I, Scicchitano DA, Geacintov NE, Broyde S. The spacious active site of a Y-family DNA polymerase facilitates promiscuous nucleotide incorporation opposite a bulky carcinogen-DNA adduct: elucidating the structure-function relationship through experimental and computational approaches. J Biol Chem. 2004;279(35):36951–36961. doi: 10.1074/jbc.M404332200. [DOI] [PubMed] [Google Scholar]
- 47.Xu P, Oum L, Geacintov NE, Broyde S. Nucleotide selectivity opposite a benzo[a]pyrene-derived N2-dG adduct in a Y-family DNA polymerase: A 5′-slippage mechanism. Biochemistry. 2008;47(9):2701–2709. doi: 10.1021/bi701839q. [DOI] [PubMed] [Google Scholar]
- 48.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24(17):2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.