Significance
Microbes have the genetic capacity to produce large numbers of specialized compounds, yet produce only a small fraction of these in the laboratory. Here we introduce a genetic platform that allows the efficient production of natural product molecules from uncharacterized gene collections. We used transformation-associated recombination in yeast to directly clone and express an orphan biosynthetic gene cluster for the production of the lipopeptide antibiotic taromycin A. With this direct cloning approach, a single genomic capture and expression vector was designed directly from next-generation sequencing data, which precisely captures genetic loci of interest and readily facilitates genetic manipulations. This study highlights a “plug-and-play” approach to cryptic biosynthetic pathways for the discovery and development of natural product drug candidates.
Keywords: biosynthesis, drug discovery, genome mining
Abstract
Recent developments in next-generation sequencing technologies have brought recognition of microbial genomes as a rich resource for novel natural product discovery. However, owing to the scarcity of efficient procedures to connect genes to molecules, only a small fraction of secondary metabolomes have been investigated to date. Transformation-associated recombination (TAR) cloning takes advantage of the natural in vivo homologous recombination of Saccharomyces cerevisiae to directly capture large genomic loci. Here we report a TAR-based genetic platform that allows us to directly clone, refactor, and heterologously express a silent biosynthetic pathway to yield a new antibiotic. With this method, which involves regulatory gene remodeling, we successfully expressed a 67-kb nonribosomal peptide synthetase biosynthetic gene cluster from the marine actinomycete Saccharomonospora sp. CNQ-490 and produced the dichlorinated lipopeptide antibiotic taromycin A in the model expression host Streptomyces coelicolor. The taromycin gene cluster (tar) is highly similar to the clinically approved antibiotic daptomycin from Streptomyces roseosporus, but has notable structural differences in three amino acid residues and the lipid side chain. With the activation of the tar gene cluster and production of taromycin A, this study highlights a unique “plug-and-play” approach to efficiently gaining access to orphan pathways that may open avenues for novel natural product discoveries and drug development.
Since the first genome of the bacterial pathogen Haemophilus influenzae Rd was revealed by shotgun sequencing in 1995 (1), the number of deposited genome sequences has grown exponentially, with >700 in the year 2012 alone (2). This rapid expansion of genomic information has benefited from increased throughput, improved fidelity, and lower costs associated with next-generation sequencing technologies (2, 3). Whereas initial genome sequencing efforts focused on microbial pathogens, bacterial genome sequencing projects have since increasingly involved industrial and environmental microbes. Consequently, the sequence analysis of pharmaceutically important actinobacteria, such as the soil-dwelling genus Streptomyces (4), has revealed their remarkable capacity to biosynthesize large numbers of structurally diverse antibiotics and other specialized metabolites (5). A mathematical model of known streptomycete antibiotics predicts that this genus has the capacity to produce as many as 100,000 secondary metabolites, yet only a small fraction of these have been discovered to date (6, 7). Thus, genomics is beginning to guide the discovery of chemical entities that have previously gone unnoticed, representing a modern approach to natural product drug discovery by connecting genes to molecules (8).
Currently available methods to maximize genetic resources for chemical production include targeted cultivation, whole pathway heterologous expression, and genetic manipulation of regulatory genes (9, 10). Synthetic biology approaches also can be used to refactor orphan gene clusters by optimizing promoters, transcriptional regulation, ribosome-binding sites, and even codon use (11). Examples of successful regulatory gene manipulation to elicit the production of new natural products from silent biosynthetic gene clusters in WT strains have been reported in bacteria and fungi (12, 13); however, this approach requires the de novo development of genetic protocols optimized for each native producer strain. In contrast, the heterologous expression of cryptic pathways in well-investigated biosynthetic host strains has some clear advantages, owing to the wide variety of available genetic tools to manipulate regulatory genes. In fact, many biosynthetic studies of actively expressed secondary metabolite pathways have been explored through mutagenesis and/or heterologous expression of cosmid/fosmid clones selected from genomic libraries (14). Next-generation sequencing technology does not require large insert clonal libraries (3), which have served as a resource for previous heterologous biosynthetic studies. Nonetheless, time-consuming cosmid/fosmid library construction and screening is still routinely practiced in the biosynthetic community.
A further complication with the cosmid/fosmid approach involves its size-selective bias of genomic fragments in the range of 30–40 kb, which is often inadequate to completely capture biosynthetic gene clusters, which can range in size up to 100 kb. Although innovative methods have been developed to stitch together overlapping clones into a functional single clone harboring the complete gene cluster (15), this approach can be very time-consuming (16, 17). Even with bacterial artificial chromosomes capable of cloning large genomic DNA fragments up to 200 kb, the need to identify desired clones that stably maintain the complete cluster remains. Recently reported approaches to cloning targeted gene clusters directly from genomic DNA have used RecE-mediated homologous recombination (18, 19), oriT-directed capture (20), or transformation-associated recombination (TAR) (21–23) to facilitate functional expression experiments.
TAR cloning in Saccharomyces cerevisiae takes advantage of yeast’s natural in vivo homologous recombination activity and has been applied to capture and express large biosynthetic gene clusters from environmental DNA samples (24–26). We adapted this TAR cloning strategy to capture and refactor a transcriptionally silent natural product biosynthetic gene cluster from actinomycetes. With this TAR cloning approach, a single genomic capture and expression vector can be designed directly from next-generation sequencing data that precisely captures genetic loci of interest and readily facilitates genetic manipulations. Here we report the construction of a TAR cloning vector that we used to capture, activate, and express a 67-kb nonribosomal peptide synthetase (NRPS) biosynthetic gene cluster from the marine actinomycete Saccharomonospora sp. CNQ-490, allowing discovery of the lipopeptide antibiotic taromycin A.
Results and Discussion
Design and Validation of the pCAP01 Gene Cluster Capture Vector.
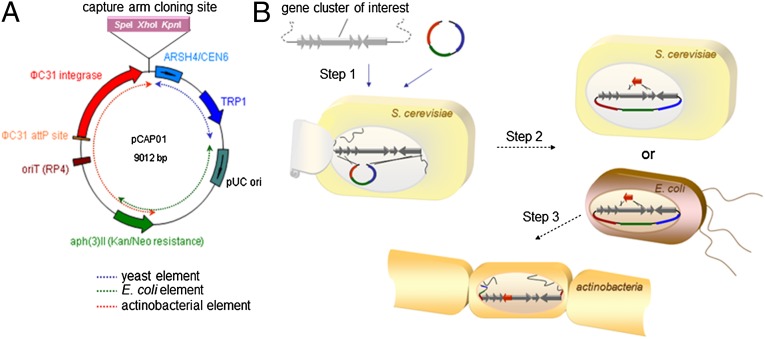
We designed the S. cerevisiae/Escherichia coli shuttle-actinobacterial chromosome integrative capture vector pCAP01 around the SuperCos1 cosmid, which is routinely used in heterologous biosynthesis experiments (27) (Fig. 1A). pCAP01 is maintained as a single copy in yeast cells to avoid unintended multiple recombination events during TAR cloning. The vector functions at multiple copies in E. coli through the action of the pUC ori (28) derived from SuperCos1, which provides sufficient plasmid DNA without induction, in contrast to a bacterial artificial chromosome-based TAR system (pTARa) reported previously (24–26). In general, a low copy vector is associated with higher stability when larger DNA constructs are used. However, we previously observed that SuperCos1-based cosmids equipped with pUC ori could stably carry >50-kb inserts in conventional E. coli cloning hosts (17), and we capitalized on this feature when designing the vector. Whereas the pTARa vector is equipped with the apramycin resistance gene (aac(3)IV, Aprr) for selection of both E. coli and Streptomyces recombinants (24–26), pCAP01 has the aph(3)II gene, providing neomycin/kanamycin resistance (Neor/Kanr) in both E. coli and actinobacteria.
Fig. 1.
Design and strategy of TAR-based cloning and expression. (A) Physical map of the gene cluster capture vector pCAP01. The vector consists of three elements that allow direct cloning and manipulation of pathways in yeast (blue), maintenance and manipulation in E. coli (green), and chromosomal integration and expression of cloned pathways in heterologous actinobacteria (red). For the construction of a pathway-specific capture vector, homology arms corresponding to both ends of the pathway are introduced into the capture arm cloning sites. (B) The procedure for TAR-based natural product discovery involves three steps. In step 1, transformation-associated recombination in yeast involves homologous recombination between the linearized pathway specific capture vector and cointroduced genomic DNA fragments to yield a circular construct that can be grown on selective media. In step 2, the cloned pathway can be manipulated using either in vivo yeast recombination or E. coli-based genetic manipulations, such as λ-Red recombination. Finally, in step 3, through conjugal DNA transfer from E. coli, the cloned and manipulated pathway is integrated into the chromosome of an actinomycete host strain for small-molecule expression studies.
Because the aac(3)IV gene is known to impart cross-resistance to aminoglycoside antibiotics such as kanamycin (29), many of the commonly used aminoglycoside resistance markers are not compatible with pTARa. In contrast, use of the aph(3)II gene makes pCAP01 compatible with conventional genetic manipulations in actinobacteria (30). The vector was designed to allow site-specific integration of the cloned gene cluster into chromosomes of heterologous actinobacterial cells via intergeneric conjugal DNA transfer from E. coli with the functions of the φC31 integration elements on the vector backbone (31). In addition, this vector system is compatible with a broad range of actinobacteria as heterologous hosts, owing to the relaxed directionality of the φC31 integration element on the vector backbone (32, 33). Our TAR-based natural product discovery approach is shown in schematic form in Fig. 1B.
We validated the function of pCAP01 by direct cloning of the 30-kb marinopyrrole (mpy) biosynthetic gene cluster from Streptomyces sp. CNQ418 and its heterologous expression in Streptomyces coelicolor M512 (SI Appendix, Fig. S12). Whereas we previously cloned the mpy cluster from two overlapping SuperCos1 cosmid clones by λ-Red recombination-mediated assembly and cluster minimization, a process that took months of labor (17), we accomplished the TAR cloning of mpy in just a fraction of this time (SI Appendix, Methods). With the successful completion of this proof-of-principle experiment, we set out to TAR clone and functionally express a larger, transcriptionally silent biosynthetic operon of unknown function from a nonstreptomycete.
Discovery and TAR Cloning of the Taromycin Biosynthesis (tar) Gene Cluster.
Bioinformatic analysis of the 4.9-Mb draft genome sequence of Saccharomonospora sp. CNQ490 revealed 19 conspicuous biosynthetic gene clusters, indicating diverse secondary metabolic capacity. Despite considerable efforts to explore the natural product chemistry of this marine actinomycete, only one compound, the novel alkaloid lodopyridone, has been reported to date (34). Such a disconnection of natural product biosynthetic potential and expressed chemistry is commonly observed in actinomycetes (5), suggesting that many of its pathways may be transcriptionally silent (35).
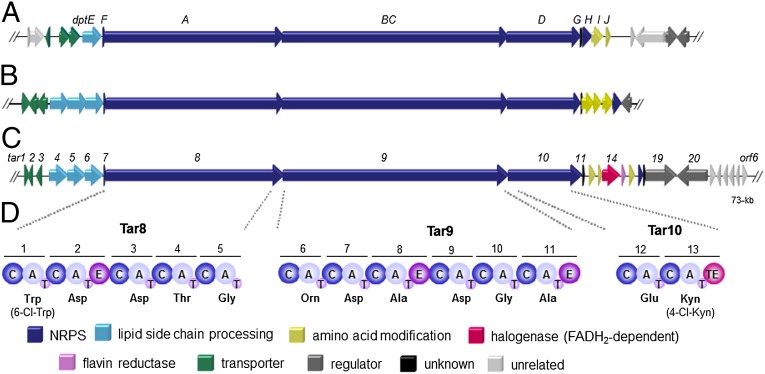
Of the three NRPS loci maintained in the Saccharomonospora sp. CNQ490 genome, the 67-kb NRPS cluster (designated the tar locus) encoded on contig-9 (B126DRAFT_scaffold 9.10, 14,775–81,515 nt) caught our attention owing to its sequence similarity with the daptomycin biosynthesis (dpt) gene cluster (36). The clinically approved lipopeptide antibiotic daptomycin is particularly important in treating β-lactam–resistant staphylococcal and enterococcal infections. Daptomycin was isolated from Streptomyces roseosporus NRRL11379, which, to the best of our knowledge, is the sole reported bacterial source of this antibiotic (37). The tar and dpt clusters, as well as an uncharacterized gene cluster from Saccharomonospora viridis DSM43017 (GenBank accession no: NC_013159), are organized similarly, yet have several notable differences (Fig. 2 and SI Appendix, Table S2). The 20-ORF tar locus maintains the dpt genes that encode the three NRPS megasynthetases. Furthermore, the enzymes required for biosynthesis of the nonproteinogenic amino acid residues 3-methyl-glutamic acid (3mGlu) (38, 39) and kynurenine (Kyn) are also present, as is the machinery required for incorporation of the fatty acid side chain, daptomycin export, and resistance.
Fig. 2.
Gene organization of lipopeptide biosynthetic gene clusters. Examples include the daptomycin (dpt) gene cluster in Streptomyces roseosporus NRRL11379 (A), an orphan lipopeptide biosynthetic gene cluster in Saccharomonospora viridis DSM43017 (B), and the taromycin (tar) biosynthetic gene cluster in Sacharomonospora sp. CNQ-490 (C). (D) The modular architecture of the taromycin NRPS assembly line and the predicted amino acid substrate specificity of each module A domain. A, adenylation domain; C, condensation domain; E, epimerization domain; T, thiolation domain; TE, thioesterase domain.
Four additional biosynthesis genes are uniquely harbored in the tar gene cluster, suggesting key structural differences of the tar lipopeptide compared with daptomycin. In particular, the tar5 and tar6 genes encode acyl-CoA dehydrogenases, which may desaturate the lipid side chain. Moreover, tar15 and tar14 code for flavin reductase and FADH2-dependent tryptophan (Trp) halogenase enzymes, respectively, suggesting that the tar lipopeptide may be halogenated at Trp and/or Trp-derived Kyn residues.
Owing to the projected novelty of the tar lipopeptide and its predicted structural relatedness to daptomycin, we initially interrogated Saccharomonospora sp. CNQ490 for the production of a halogenated derivative of daptomycin. All of our attempts to identify a lipopeptide in the native strain were unsuccessful, however. Further inspection of the tar cluster revealed two transcriptional regulators, Tar19 and Tar20, which were respectively identified as a Streptomyces antibiotic regulatory protein (SARP) and a LuxR transcription regulator. Given that LuxR-type regulators are known to act as transcription activators and/or repressors (40), we suspected that expression of the tar gene cluster might be suppressed by the Tar20 LuxR-type regulator, accounting for the absence of a lipopeptide from native strain cultures.
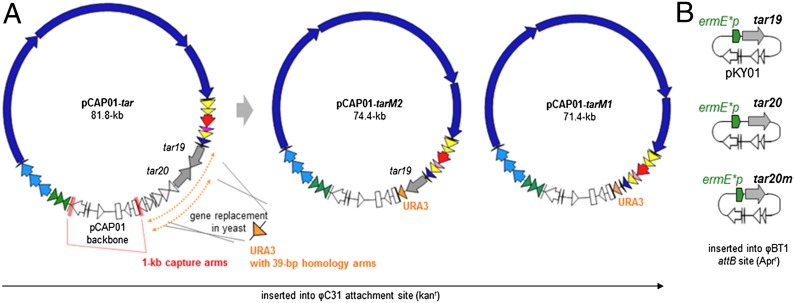
To activate the orphaned tar gene cluster, we targeted a 73-kb genomic region containing the 67-kb tar gene cluster for TAR cloning and genetic refactoring. Whereas previous reports showed that short 40- to 70-bp capture arms were effective in TAR cloning (21–23), we used a recent modification (41) of longer 1-kb capture arms that corresponded to tar1 (including its putative promoter region) and orf6 to generate the tar pathway-specific capture vector. After transformation of S. cerevisiae VL6-48 spheroplasts with the linearized vector and XbaI restriction enzyme-digested genomic DNA, positive clones were identified by PCR and confirmed by restriction mapping to give pCAP01-tar (Fig. 3). We observed that the 81.8-kb pCAP01-tar vector was stable after propagation through E. coli (SI Appendix, Fig. S2). Although we have not yet explored the upper size limits for gene cluster insertions, the size of the 73-kb tar insert covers most secondary metabolic biosynthetic loci. In cases where greater size capacity is required, this cosmid-based vector might not be singularly appropriate, however. In those cases, we envisage using a complementary cosmid-based vector in conjunction with pCAP01 or engaging the bacterial artificial chromosome-based pTARa system (24–26).
Fig. 3.
Physical maps of the TAR-cloned tar gene cluster and the pKY01-based complementation vectors. (A) The 73-kb genomic region containing the tar gene cluster was directly cloned in yeast, yielding pCAP01-tar. The tar regulatory genes tar19/tar20 and unrelated genes beyond the cluster border on pCAP01-tar (dotted arrows) were replaced with the URA3 auxotrophic marker gene in yeast, generating regulatory gene-deficient tar gene cluster expression constructs pCAP01-tarM1 and pCAP01-tarM2. The three constructs were introduced into the φC31 attachment site on the chromosome of the heterologous host S. coelicolor M1146. (B) Both of the eliminated regulatory genes and the codon redressed tar20 luxR regulator gene were introduced separately into the expression mutants using the φBT1 integrative expression vector pKY01. Color codes for the genes are the same as in Fig. 2.
Genetic Manipulation of the tar Regulatory Genes.
Because expression of the tar gene cluster is likely suppressed by the Tar20 LuxR-type regulator, we eliminated the tar20 gene and all unnecessary genes beyond the putative biosynthetic gene cluster boundary (tar20 to orf6) from pCAP01-tar. We similarly targeted the tar19 SARP regulator gene to explore its function. Normally, we use λ-Red recombination-mediated PCR targeting in E. coli for gene inactivation/deletion (42). Although this approach is compatible with the pCAP01 vector, we have observed unintended recombination events in target clones that carry repetitive sequences, such as those residing in modular NRPS and polyketide synthase genes.
To avoid this potential issue with the tar cluster and to further expand our TAR cloning strategy, we directly manipulated the tar regulatory genes by in vivo yeast recombination-mediated PCR targeting (43). We replaced the targeted regions containing the tar19 and/or tar20 genes with a PCR-amplified URA3 auxotrophic marker gene flanked by 39-nt homology arms. After introduction of the URA3 cassettes into S. cerevisiae VL6-48 carrying pCAP01-tar, plasmids were extracted from colonies grown on uracil-deficient media. Restriction mapping of the plasmids propagated in E. coli confirmed that the region tar19-orf6 and tar20-orf6 on pCAP01-tar were successfully replaced, yielding pCAP01-tarM1 (∆tar19-20) and pCAP01-tarM2 (∆tar20), respectively (Fig. 3).
Expression of the tar Gene Cluster and Characterization of Taromycin A.
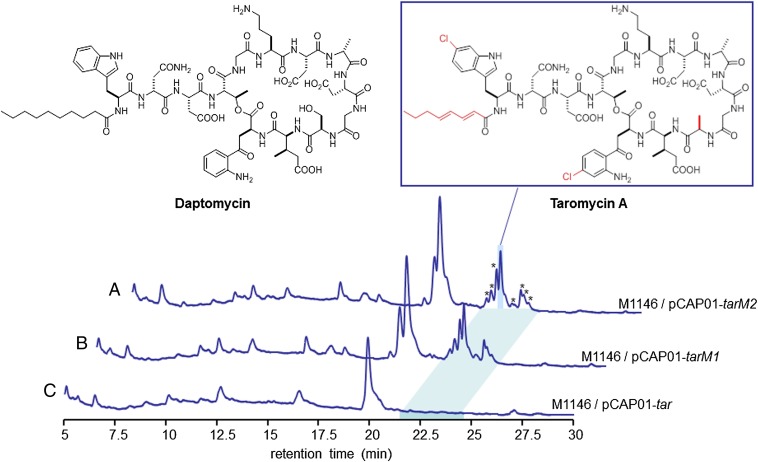
For the heterologous production of tar lipopeptide, we chose S. coelicolor, a model actinomycete commonly used as a biosynthetic host. S. coelicolor also produces the lipopeptide molecule calcium-dependent antibiotic (CDA), which, like daptomycin, contains a 3mGlu residue (44). We introduced pCAP01-tar and its two derivatives, pCAP01-tarM1 and pCAP01-tarM2, by conjugative transfer from E. coli ET12567/pUB307 (45) into the genome minimized S. coelicolor strain M1146 (46), which lacks four biosynthetic pathways known to be involved in the production of common endogenous antibiotics in this strain, including the cda locus. Although our initial attempt to heterologously produce a unique metabolite in S. coelicolor M1146 through the expression of pCAP01-tar was unsuccessful, both mutants produced a series of unique chlorinated lipopeptides, as revealed by HPLC-MS analysis (Fig. 4 and SI Appendix, Fig. S3).
Fig. 4.
HPLC analysis of the taromycins produced heterologously by S. coelicolor mutants and structures of daptomycin and taromycin A. Extracts from S. coelicolor M1146/pCAP01-tarM2 (∆tar20 luxR) (A), S. coelicolor M1146/pCAP01-tarM1 (∆tar19 sarp, ∆tar20 luxR) (B), and S. coelicolor M1146/pCAP01-tar (intact) (C) were analyzed by C18 reversed-phase HPLC. UV absorption at 254 nm was monitored. *Taromycin A derivatives. The taromycin structure is elucidated in SI Appendix, Tables S3–S5 and Figs. S8–S11. Structural differences between taromycin A and daptomycin are highlighted in red.
To determine the chemical structure of the predominant heterologously expressed lipopeptide, we cultured 4 L of S. coelicolor M1146/pCAP01-tarM1 to yield a ∼60-mg mixture of at least eight closely related lipopeptide derivatives, from which we purified 4 mg of taromycin A. Inspection of taromycin A’s mass spectral fragments, together with its molecular formula, C70H91N17O25Cl2, revealed by high-resolution MS, suggests that taromycin A specifically differs from daptomycin in the Trp, Kyn, and lipid residues, and by the substitution of an Ala for a Ser residue at position 11 (Fig. 4). Comprehensive MS and NMR analyses placed each of the two chlorine atoms in the two aromatic amino acid residues as 6-Cl-Trp and 4-Cl-Kyn, and aided construction of the planar structure of taromycin A carrying an octa-2,4-dienoyl lipid side chain (SI Appendix, Fig. S9). Although the residue 6-Cl-Trp has been reported as a residue in other peptides, including microsclerodermin D (47), to the best of our knowledge, taromycin A’s 4-Cl-Kyn residue has not been previously reported in nature.
With just a single halogenase encoded in the tar cluster, the tryptophan chlorinase Tar14 apparently catalyzes the synthesis of 6-Cl-Trp. This residue can be either incorporated directly by module 1 or further transformed by oxidative cleavage to 4-Cl-Kyn, which is then assimilated by module 13. We assigned taromycin A’s absolute stereochemistry by Marfey analysis and by direct comparison with the daptomycin hydrolysate (SI Appendix, Table S5). Taromycin A and daptomycin share the same amino acid configuration for all common residues. The unique Ala residue at position 11 in taromycin A is configured D, which is configurationally consistent with daptomycin’s d-Ser11 residue and biosynthetically consistent with the domain organization of the tar NPRS module 11, which contains an epimerase domain. The 6-Cl-Trp and 4-Cl-Kyn residues were found to be configured L, consistent with the stereochemistry observed for the respective nonhalogenated residues Trp and Kyn in daptomycin (SI Appendix, Table S5).
Taromycin A was screened against bloodstream isolates of methicillin-resistant Staphylococcus aureus (MRSA) O325, daptomycin-sensitive strain Enterococcus faecalis 613 and its isogenic daptomycin-resistant strain 613D, as well as daptomycin-sensitive strain Enterococcus faecium 447 and its isogenic resistant strain 447D. The bioactivity of taromycin A is highly dependent on the presence of calcium in the growth medium, as previously observed with daptomycin and CDA (48). Our assays show that taromycin A has moderate bioactivity, with a minimum inhibitory concentration of 6–100 μM, against all strains tested at the optimal calcium concentration of 50 mg/mL (SI Appendix, Table S6). In addition to taromycin A, the expression of the tar locus generated further analogs that, based on multistage MS analyses (SI Appendix, Fig. S3), appear to differ largely in the nature of the lipid side chain, which in the case of daptomycin was shown to be critical for its antibiotic potency (49). Structures and bioactivity of the full suite of taromycin derivatives will be reported in due course.
Analysis of the tar Regulatory System.
Production of the taromycins in S. coelicolor M1146/pCAP01-tarM2 strongly suggests that the Tar20 LuxR-type transcriptional regulator functions as a negative regulator of the tar gene cluster. To further clarify this observation, we reintroduced the tar20 gene back into S. coelicolor M1146/pCAP-tarM1 (∆tar19-20) and S. coelicolor M1146/pCAP-tarM2 (∆tar20) under control of the strong constitutive ermE* promoter using a second integrative plasmid, pKY01 (Fig. 3) (17). Contrary to our expectation, the constitutive overexpression of the native tar20 did not suppress taromycin biosynthesis (SI Appendix, Figs. S4 and S5). Further inspection revealed that, surprisingly, the tar20 gene contains five rare TTA codons known to limit expression in high guanine-cytosine content Streptomyces strains (50). We suspected that Tar20 could be expressed as a truncated, nonfunctional protein under control of the strong ermE* promoter. To address this codon issue, we generated the codon-redressed tar20 gene by PCR-mediated mutagenesis in which we replaced the TTA codons with CTG codons that similarly code for leucine (SI Appendix, Fig. S6). Complementation with the refactored tar20 gene did indeed significantly reduce taromycin production (SI Appendix, Fig. S7), clearly demonstrating that the Tar20 LuxR-type regulator functions as a repressor in taromycin biosynthesis.
Although SARP regulators are known to be transcriptional activators, the activity of the Tar19 SARP regulator does not appear to control taromycin synthesis. Whereas SARP regulators recognize heptameric repeats (TCGAGXX) spaced by 4–15 nt and located 8 nt upstream of the −10 region of a promoter to activate expression of downstream genes (51, 52), inspection of the tar locus revealed no such sequences. Furthermore, neither deletion nor overexpression of the tar19 gene resulted in a change in taromycin production level, suggesting that the Tar19 SARP regulator is not involved in taromycin biosynthesis under normal laboratory growth conditions (SI Appendix, Figs. S4 and S5).
Conclusion
As the pace and ease of DNA sequencing continues to improve, new synthetic biology methods are emerging to more efficiently explore genome-scale topics. With the realization that many microbial genomes possess a bounty of natural product biosynthetic gene clusters of mostly unknown functions, we have further developed TAR cloning (21–24) for the efficient capture, refactoring, and expression of large biosynthetic loci for the exploration of new chemical entities. Here we have described the TAR-directed cloning and functionalization of a large NRPS biosynthetic locus that codes for synthesis of the unique lipopeptide antibiotic taromycin A from a marine Saccharomonospora isolate. Our study validates the power of direct cloning for pathway-driven natural product discovery and offers a “plug-and-play” approach to rapidly gain access to orphan biosynthetic pathways from a wide range of genome resources that may be involved in the production of pharmaceutically important drug leads.
Methods
Strains.
All strains used in this study are listed in SI Appendix, Table S1. Culture conditions are described in SI Appendix, Methods.
Bioinformatics Analysis of the tar Gene Cluster.
The draft genome of Saccharomonospora sp. CNQ-490 was sequenced at the Joint Genome Institute (project ID: 407426). The bioinformatics program antiSMASH (http://antismash.secondarymetabolites.org/) (53) was initially used to analyze the whole genome sequence. The sequence of the orphan 67-kb NRPS gene cluster tar encoded on contig-9 (B126DRAFT_scaffold 9.10; 14,775–81,515 nt) was further analyzed and annotated using FramePlot 3.0 (54) (http://watson.nih.go.jp/∼jun/cgi-bin/frameplot-3.0b.pl) and BLAST (http://blast.ncbi.nlm.nih.gov). The amino acid specificity codes for adenylation domains in NRPS enzymes were analyzed using NRPSpredictor2 (http://nrps.informatik.uni-tuebingen.de/Controller?cmd=SubmitJob).
Preparation of Genomic DNA Fragments for TAR.
Saccharomonospora sp. CNQ-490 was grown in A1 liquid media as reported previously (34). Genomic DNA was isolated from midlog phase cells by standard procedures (30). Approximately 200 μg of genomic DNA was digested with 200 U of XbaI, which does not cut the tar gene cluster, in an overnight reaction at 37 °C. The digested genomic DNA fragments were precipitated and cleaned with 70% ethanol. The resulting DNA pellet was dissolved in 200 μL of 20% Tris-EDTA buffer (2 mM Tris⋅HCl and 0.2 mM EDTA; pH 8.0).
Construction of the Gene Cluster Capture Vector pCAP01.
The yeast element consisting of ARSH4/CEN6 (replication origin) and TRP1 auxotrophic marker from a yeast centromeric plasmid pRS314 (American Type Culture Collection 77143), the E. coli element pUC ori from SuperCos1 (Stratagene), and the Streptomyces elements consisting of the φC31 integrase gene (int) and its attachment site (attP), origin of DNA transfer (oriT), and the aph(3)II gene (Kan/Neo resistance) from the pSET152 derivative pLAE101 were assembled in S. cerevisiae strain VL6-48 (American Type Culture Collection MYA-3666), yielding the gene cluster capture vector pCAP01. Detailed information is provided in SI Appendix, Methods.
Direct Cloning of the tar Gene Cluster Using TAR.
The tar pathway-specific capture vector was constructed by introducing two PCR-amplified 1-kb homology arms corresponding to flanking regions of the tar gene cluster (tar1 and orf6) into pCAP01. Spheroplast cells of S. cerevisiae VL6-48 were transformed with the linearized tar pathway-specific capture vector and enzymatically fragmented genomic DNA (23). Desired transformants were selected on synthetic tryptophan dropout agar (SD-Trp agar; SI Appendix, Methods) and identified by PCR. Direct cloning of the tar cluster was confirmed by restriction mapping to give pCAP01-tar. More detailed information is provided in SI Appendix, Methods.
Heterologous Expression of the tar Gene Cluster.
pCAP01-tar and its derivatives, which have the φC31 integrase (int) gene with its attachment site (attP), were introduced into E. coli ET12567 (31) and then transferred to S. coelicolor M1146 (46) by triparental intergeneric conjugation facilitated by E. coli ET12567/pUB307 (45). Kanr exconjugants were routinely precultivated in trypticase soy broth containing kanamycin (50 μg/mL) and nalidixic acid (25 μg/mL) at 30 °C for 3 d. A portion (10 mL) of the precultures were inoculated into 1 L of modified MP production medium (17) and grown for 4–5 d at 30 °C in a 2.8-L flask with rotary shaking. Solid-phase extraction of the culture supernatants was performed with Amberlite XAD resin, and the methanol extracts from the resin were analyzed by reversed-phase HPLC-MS. Detailed information, including the recipe for the medium and the analytical conditions for HPLC, is provided in SI Appendix, Methods.
Other Methods.
Other methods, including genetic manipulation of the tar regulatory genes, structural elucidation of taromycins, and antimicrobial bioassay, are described in SI Appendix, Methods.
Nucleotide Sequence Accession Number.
The nucleotide sequence of the 76,640-bp genomic region containing the tar loci in Saccharomonospora sp. CNQ-490 has been deposited in the GenBank database (accession no. KF301601).
Supplementary Material
Acknowledgments
We thank P. R. Jensen and W. Fenical for providing Saccharomonospora sp. CNQ-490, B. M. Dunggan for assistance with NMR, and T. A. M. Gulder, A. C. Ross, and P. A. Jordan for helpful discussions. This work was supported by grants from the National Institutes of Health (GM085770, to B.S.M.; GM097509, to B.S.M. and P.C.D.; AI057153, to V.N.; and Instrument Grant S10-OD010640-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The nucleotide sequence of the 76,640-bp genomic region containing the tar loci in Saccharomonospora sp. CNQ-490 has been deposited in the GenBank database (accession no. KF301601).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319584111/-/DCSupplemental.
References
- 1.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269(5223):496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Bertelli C, Greub G. Rapid bacterial genome sequencing: Methods and applications in clinical microbiology. Clin Microbiol Infect. 2013;19(9):803–813. doi: 10.1111/1469-0691.12217. [DOI] [PubMed] [Google Scholar]
- 3.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 4.Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26(11):1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Streptomyces? Arch Microbiol. 2001;176(5):386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 7.Baltz RH. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J Ind Microbiol Biotechnol. 2006;33(7):507–513. doi: 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- 8.Challis GL. Genome mining for novel natural product discovery. J Med Chem. 2008;51(9):2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- 9.Bode HB, Müller R. The impact of bacterial genomics on natural product research. Angew Chem Int Ed Engl. 2005;44(42):6828–6846. doi: 10.1002/anie.200501080. [DOI] [PubMed] [Google Scholar]
- 10.Van Lanen SG, Shen B. Microbial genomics for the improvement of natural product discovery. Curr Opin Microbiol. 2006;9(3):252–260. doi: 10.1016/j.mib.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Medema MH, Breitling R, Takano E. Synthetic biology in Streptomyces bacteria. Methods Enzymol. 2011;497:485–502. doi: 10.1016/B978-0-12-385075-1.00021-4. [DOI] [PubMed] [Google Scholar]
- 12.Bergmann S, et al. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3(4):213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 13.Laureti L, et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA. 2011;108(15):6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gust B. Chapter 7: Cloning and analysis of natural product pathways. Methods Enzymol. 2009;458:159–180. doi: 10.1016/S0076-6879(09)04807-1. [DOI] [PubMed] [Google Scholar]
- 15.Wolpert M, Heide L, Kammerer B, Gust B. Assembly and heterologous expression of the coumermycin A1 gene cluster and production of new derivatives by genetic engineering. ChemBioChem. 2008;9(4):603–612. doi: 10.1002/cbic.200700483. [DOI] [PubMed] [Google Scholar]
- 16.Wenzel SC, et al. Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via red/ET recombineering. Chem Biol. 2005;12(3):349–356. doi: 10.1016/j.chembiol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Yamanaka K, Ryan KS, Gulder TA, Hughes CC, Moore BS. Flavoenzyme-catalyzed atropo-selective N,C-bipyrrole homocoupling in marinopyrrole biosynthesis. J Am Chem Soc. 2012;134(30):12434–12437. doi: 10.1021/ja305670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian X, et al. Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering. ChemBioChem. 2012;13(13):1946–1952. doi: 10.1002/cbic.201200310. [DOI] [PubMed] [Google Scholar]
- 19.Fu J, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30(5):440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 20.Kvitko BH, McMillan IA, Schweizer HP. An improved method for oriT-directed cloning and functionalization of large bacterial genomic regions. Appl Environ Microbiol. 2013;79(16):4869–4878. doi: 10.1128/AEM.00994-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larionov V, Kouprina N, Solomon G, Barrett JC, Resnick MA. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc Natl Acad Sci USA. 1997;94(14):7384–7387. doi: 10.1073/pnas.94.14.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouprina N, et al. Functional copies of a human gene can be directly isolated by transformation-associated recombination cloning with a small 3′ end target sequence. Proc Natl Acad Sci USA. 1998;95(8):4469–4474. doi: 10.1073/pnas.95.8.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3(3):371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 24.Kim JH, et al. Cloning large natural product gene clusters from the environment: Piecing environmental DNA gene clusters back together with TAR. Biopolymers. 2010;93(9):833–844. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Z, Kim JH, Brady SF. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA-derived type II PKS gene cluster. J Am Chem Soc. 2010;132(34):11902–11903. doi: 10.1021/ja104550p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Z, Kallifidas D, Brady SF. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc Natl Acad Sci USA. 2011;108(31):12629–12634. doi: 10.1073/pnas.1103921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens DC, Hari TP, Boddy CN. The role of transcription in heterologous expression of polyketides in bacterial hosts. Nat Prod Rep. 2013;30(11):1391–1411. doi: 10.1039/c3np70060g. [DOI] [PubMed] [Google Scholar]
- 28.Lin-Chao S, Chen WT, Wong TT. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol Microbiol. 1992;6(22):3385–3393. doi: 10.1111/j.1365-2958.1992.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 29.Magalhaes ML, Blanchard JS. The kinetic mechanism of AAC3-IV aminoglycoside acetyltransferase from Escherichia coli. Biochemistry. 2005;44(49):16275–16283. doi: 10.1021/bi051777d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keiser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
- 31.MacNeil DJ, et al. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene. 1992;111(1):61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 32.Bierman M, et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 33.Lechner A, Eustáquio AS, Gulder TA, Hafner M, Moore BS. Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem Biol. 2011;18(12):1527–1536. doi: 10.1016/j.chembiol.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maloney KN, et al. Lodopyridone, a structurally unprecedented alkaloid from a marine actinomycete. Org Lett. 2009;11(23):5422–5424. doi: 10.1021/ol901997k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8(2):208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 36.Miao V, et al. Daptomycin biosynthesis in Streptomyces roseosporus: Cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology. 2005;151(Pt 5):1507–1523. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- 37.Baltz RH. Antibiotic discovery from actinomycetes: Will a renaissance follow the decline and fall? SIM News. 2005;55:186–196. [Google Scholar]
- 38.Nguyen KT, et al. A glutamic acid 3-methyltransferase encoded by an accessory gene locus important for daptomycin biosynthesis in Streptomyces roseosporus. Mol Microbiol. 2006;61(5):1294–1307. doi: 10.1111/j.1365-2958.2006.05305.x. [DOI] [PubMed] [Google Scholar]
- 39.Mahlert C, Kopp F, Thirlway J, Micklefield J, Marahiel MA. Stereospecific enzymatic transformation of alpha-ketoglutarate to (2S,3R)-3-methyl glutamate during acidic lipopeptide biosynthesis. J Am Chem Soc. 2007;129(39):12011–12018. doi: 10.1021/ja074427i. [DOI] [PubMed] [Google Scholar]
- 40.Pompeani AJ, et al. The Vibrio harveyi master quorum-sensing regulator LuxR, a TetR-type protein, is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol. 2008;70(1):76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karas BJ, Tagwerker C, Yonemoto IT, Hutchison CA, 3rd, Smith HO. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Synth Biol. 2012;1(1):22–28. doi: 10.1021/sb200013j. [DOI] [PubMed] [Google Scholar]
- 42.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100(4):1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25(2):451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hojati Z, et al. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol. 2002;9(11):1175–1187. doi: 10.1016/s1074-5521(02)00252-1. [DOI] [PubMed] [Google Scholar]
- 45.Flett F, Mersinias V, Smith CP. High-efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1997;155(2):223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 46.Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4(2):207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt EW, Faulkner DJ. Microsclerodermins C–E, antifungal cyclic peptides from the lithistid marine sponges Theonella sp. and Microscleroderma sp. Tetrahedron. 1998;54:3043–3056. [Google Scholar]
- 48.Jung D, Rozek A, Okon M, Hancock RE. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol. 2004;11(7):949–957. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 49.Debono M, et al. Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: The synthesis and evaluation of daptomycin (LY146032) J Antibiot (Tokyo) 1988;41(8):1093–1105. doi: 10.7164/antibiotics.41.1093. [DOI] [PubMed] [Google Scholar]
- 50.Ueda Y, Taguchi S, Nishiyama K, Kumagai I, Miura K. Effect of a rare leucine codon, TTA, on expression of a foreign gene in Streptomyces lividans. Biochim Biophys Acta. 1993;1172(3):262–266. doi: 10.1016/0167-4781(93)90212-v. [DOI] [PubMed] [Google Scholar]
- 51.Wietzorrek A, Bibb M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25(6):1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 52.Arias P, Fernández-Moreno MA, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181(22):6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blin K, et al. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(Web Server issue):W204-–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishikawa J, Hotta K. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol Lett. 1999;174(2):251–253. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.