Summary
Trehalose synthase (TreS) was thought to catalyze flux from maltose to trehalose, a precursor of essential trehalose mycolates in mycobacterial cell walls. However, we now show, using a genetic approach, that TreS is not required for trehalose biosynthesis in Mycobacterium smegmatis, whereas two alternative trehalose-biosynthetic pathways (OtsAB and TreYZ) are crucial. Consistent with this direction of flux, trehalose levels in Mycobacterium tuberculosis decreased when TreS was overexpressed. In addition, TreS was shown to interconvert the α anomer of maltose and trehalose using 1H and 19F-nuclear magnetic resonance spectroscopies using its normal substrates and deoxyfluoromaltose analogs, with the nonenzymatic mutarotation of α/β-maltose being slow. Therefore, flux through TreS in mycobacteria flows from trehalose to α-maltose, which is the appropriate anomer for maltose kinase of the GlgE α-glucan pathway, which in turn contributes to intracellular and/or capsular polysaccharide biosynthesis.
Graphical Abstract
Highlights
► Flux through trehalose synthase (TreS) in mycobacteria is from trehalose to maltose ► The appropriate α anomer is formed by TreS for maltose kinase of the GlgE pathway ► The specificity of TreS for α-maltose is retained with deoxyfluoro analogs ► TreS supports cytosolic/capsular α-glucan but not trehalose mycolate biosynthesis
It is currently thought that trehalose synthase contributes to the biosynthesis of trehalose mycolates destined for mycobacterial cell walls. Miah et al. show that flux through this enzyme is in the reverse direction from trehalose to maltose and that the appropriate α anomer is generated for the GlgE α-glucan pathway.
Introduction
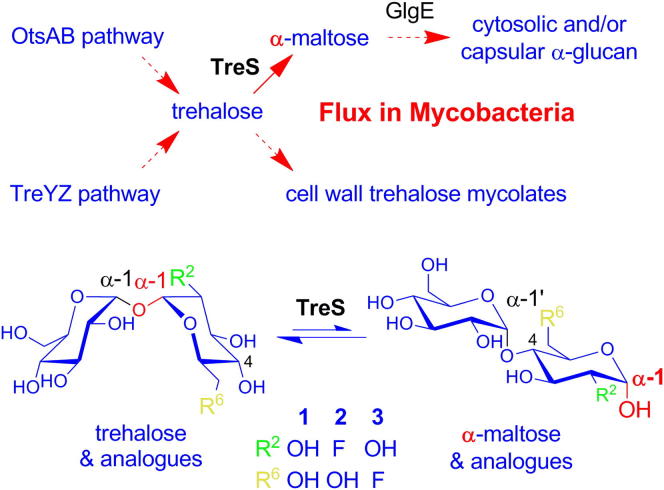
Trehalose (α-D-glucopyranosyl-(1→1)-α-D-glucopyranoside) is a nonreducing disaccharide that has many roles in biology (Argüelles, 2000; Elbein et al., 2003; Paul et al., 2008). For example, it is a precursor for cell wall trehalose mycolates (Figure 1A) that are essential for the growth and virulence of mycobacteria, such as Mycobacterium tuberculosis (Takayama et al., 2005), the causative agent of the globally widespread human disease tuberculosis (Dye, 2006). For this reason, trehalose has attracted attention in the development of imaging agents (Backus et al., 2011; Swarts et al., 2012) and drugs (Lin et al., 2007; Rose et al., 2002; Wang et al., 2004) to help diagnose and treat tuberculosis. It has been widely thought that there are three pathways responsible for the biosynthesis of trehalose in mycobacteria (Figure 1A): the OtsAB, TreYZ, and trehalose synthase (TreS) pathways (Avonce et al., 2006; Elbein et al., 2003). In vitro experiments have shown that all three pathways could, in principle, operate in mycobacteria (De Smet et al., 2000). In addition, genetic experiments appeared to show that all pathways are capable of synthesizing trehalose de novo in the fast-growing avirulent species Mycobacterium smegmatis (Woodruff et al., 2004). By contrast, it has been reported that the OtsAB pathway is dominant in trehalose biosynthesis in Mycobacterium tuberculosis and that TreS could have a role only in late-stage pathogenesis in infected mice (Murphy et al., 2005). This implies Mycobacterium smegmatis is not an appropriate model organism with respect to the metabolism of trehalose in Mycobacterium tuberculosis. The picture is complicated further by evidence that only the OtsAB and TreYZ pathways, but not the TreS pathway, appear to be important in the de novo biosynthesis of trehalose and trehalose mycolate formation in the related actinomycete, Corynebacterium glutamicum (Tzvetkov et al., 2003; Wolf et al., 2003).
Figure 1.
Metabolism of Trehalose in Mycobacteria and Proposed Mechanism of TreS
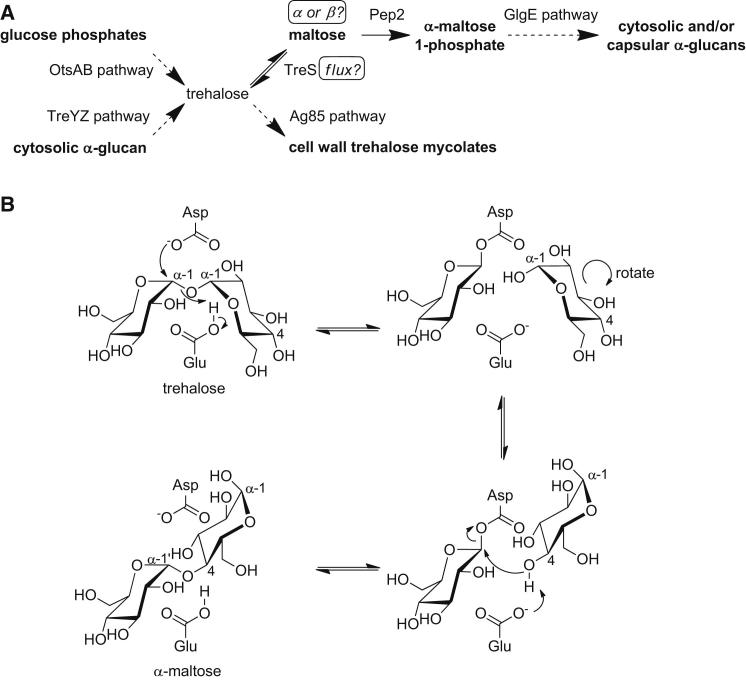
(A) All known metabolic pathways associated with trehalose in mycobacteria are shown, except for its hydrolysis by trehalase to form glucose as a carbon source for growth (Carroll et al., 2007). The questions addressed by this work are indicated in boxes.
(B) Proposed catalytic mechanism of TreS with the most likely relative orientations of the glucose rings of trehalose and maltose. Hydrolysis would be expected to occur when water attacks the glucosyl-enzyme intermediate, generating a second glucose molecule with an α anomeric configuration.
TreS is a maltose α-D-glucosylmutase (EC 5.4.99.16) that interconverts maltose (α-D-glucopyranosyl-(1,4)-D-glucopyranose) and trehalose (Nishimoto et al., 1995; Pan et al., 2004). Therefore, an oft ignored puzzle has been the lack of an obvious and significant source of maltose in a mycobacterium either from its own metabolism or its environment. This is compounded by the lack of a maltose transporter in Mycobacterium tuberculosis (Kalscheuer et al., 2010a). However, we recently discovered an alternative route for α-glucan biosynthesis, the GlgE pathway, which is widespread among bacteria and involves the consumption of trehalose by TreS (Chandra et al., 2011; Kalscheuer et al., 2010b). These observations prompt the question as to whether TreS contributes to the biosynthesis of either trehalose mycolate, α-glucans, or both in mycobacteria.
There are no reports of experimental evidence defining the anomeric configuration of maltose that TreS produces. The configuration could have consequences for metabolic flux, because the nonenzymatic mutarotation of maltose is so slow that the half-life of anomeric equilibration is of the order of tens of minutes (Bailey et al., 1967). Furthermore, although maltose mutarotase enzymes are known, they appear to be rare and have only been detected in higher plants (Bailey et al., 1967) and Lactobacillus brevis (Shirokane and Suzuki, 1995). TreS is a glycoside hydrolase GH13_3 (Stam et al., 2006) family member according to the CaZy database (Cantarel et al., 2009). Thus, it is predicted to have a (β/α)8 fold, defining an active site containing an Asp nucleophile and a Glu proton donor that catalyze an α-retaining double-displacement reaction mechanism (Figure 1B). In support of this mechanism, evidence for the glucosyl-enzyme intermediate involving Asp230 in the Mycobacterium smegmatis enzyme has been reported recently (Zhang et al., 2011). One would therefore predict that α-maltose is utilized and produced by TreS, but evidence to support this notion is currently lacking.
We now show, using a genetic approach, that flux through TreS is from rather than to trehalose in both Mycobacterium smegmatis and Mycobacterium tuberculosis. Therefore, TreS supplies intermediates for the GlgE α-glucan pathway rather than the trehalose mycolate pathway, both potential targets for imaging agents and therapeutic inhibitors. Furthermore, we show using 1H-nuclear magnetic resonance (NMR) spectroscopy, supported by 19F-NMR spectroscopy and deoxyfluoro substrate analogs, that the appropriate α anomer of maltose is formed for maltose kinase of the GlgE pathway. These findings have implications for the study and targeting of trehalose-dependent pathways in mycobacteria and other bacteria.
Results
Characterization of Trehalose Auxotrophs of Mycobacterium Smegmatis
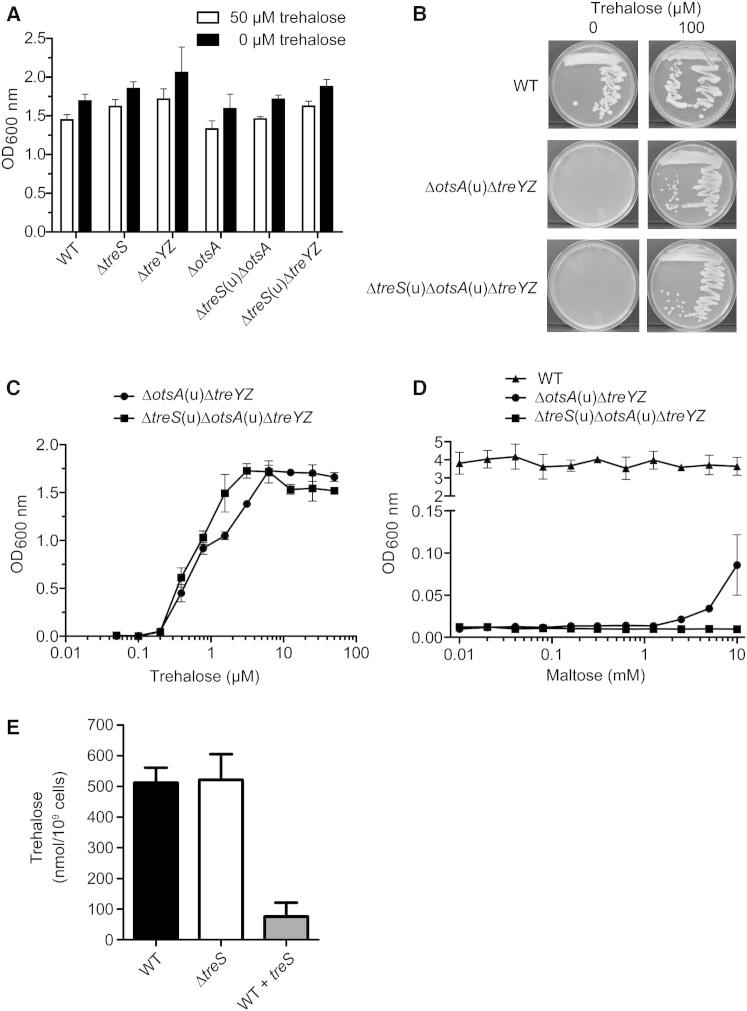
In order to definitively assess the contribution of TreS for the de novo biosynthesis of essential trehalose in mycobacteria, we generated site-specific gene deletion mutants in Mycobacterium smegmatis in the three reported trehalose biosynthetic pathways by targeting the genes treS, otsA, and treY1-treY2-treZ, both individually and in combination (Figure S1 available online). Inactivation of treS alone or in combination with either otsA or treYZ did not lead to any detectable growth defect in the absence of exogenous trehalose (Figure 2A). In contrast, we found that not only the ΔtreS(u)ΔotsA(u)ΔtreYZ triple mutant but also the ΔotsA(u)ΔtreYZ double mutant strictly required trehalose supplementation for growth on solid medium (Figure 2B) and in liquid culture (Figure 2C), despite the double mutant possessing an intact treS gene. Supplementation with exogenous trehalose in the μM range was sufficient to support growth of these two trehalose auxotrophs (Figure 2C), showing that TreS does not contribute significantly to the de novo production of trehalose in Mycobacterium smegmatis. In the absence of trehalose, exogenous maltose in the mM range could partially restore growth of the ΔotsA(u)ΔtreYZ but not the ΔtreS(u)ΔotsA(u)ΔtreYZ mutant (Figure 2D). These data indicate that TreS could be capable of synthesizing trehalose in mycobacteria but that the biosynthesis of maltose is severely limited in the growth conditions tested.
Figure 2.
The TreS Pathway Does Not Contribute to the De Novo Biosynthesis of Trehalose in Mycobacteria
(A) Trehalose growth requirements in liquid culture of Mycobacterium smegmatis gene deletion mutants (see Figure S1 for how they were generated). Cultures were incubated for 48 hr at 37°C in Middlebrook 7H9 medium containing 0.5% (v/v) glycerol and 10% (v/v) albumin-dextrose-saline (ADS) enrichment containing either 0 or 50 μM trehalose. Values are means of triplicates ± SD.
(B) Trehalose growth requirements on solid media of the trehalose-auxotrophic Mycobacterium smegmatis mutants ΔotsA(u)ΔtreYZ and ΔtreS(u)ΔotsA(u)ΔtreYZ. Cells were cultivated for 72 hr on Middlebrook 7H10 agar containing 0.5% (v/v) glycerol and 10% (v/v) ADS enrichment in the presence or absence of 100 μM trehalose.
(C) Trehalose growth requirements in liquid culture of the trehalose-auxotrophic Mycobacterium smegmatis mutants ΔotsA(u)ΔtreYZ and ΔtreS(u)ΔotsA(u)ΔtreYZ. Growth conditions were essentially as described for (A) with 0–50 μM trehalose.
(D) Maltose supplementation of trehalose auxotrophic Mycobacterium smegmatis gene deletion mutants. Growth conditions were essentially as described for (A) without trehalose but with 0–10 mM maltose. Values are means of triplicates ± SD.
(E) Effect of treS deficiency and overexpression on intracellular trehalose concentration in Mycobacterium tuberculosis. Cultures of Mycobacterium tuberculosis H37Rv wild-type, the ΔtreS mutant and a treS overexpressing strain (wild type [WT] + treS) were incubated for 14 days in Middlebrook 7H9 medium containing 0.5% (v/v) glycerol and 10% (v/v) oleic acid-albumin-dextrose-catalase enrichment before enzymatically determining trehalose concentrations. Values are means of sextuplicates ± SD.
TreS Consumes Trehalose in Mycobacterium Tuberculosis
In order to assess the direction of flux through TreS in Mycobacterium tuberculosis, we analyzed intracellular trehalose levels in Mycobacterium tuberculosis wild-type, the ΔtreS mutant and treS-overexpressing strains (Figure 2E). Deletion of treS had no influence on the intracellular trehalose concentration, precluding the ability to distinguish between TreS having a role in trehalose formation and consumption. However, the intracellular trehalose level in the treS-overexpressing strain was dramatically reduced, which indicates that TreS has a role in the consumption of trehalose in Mycobacterium tuberculosis, a direction of flux consistent with that in Mycobacterium smegmatis.
TreS Interconverts Trehalose with the α Anomer of Maltose According to 1H-NMR Spectroscopy
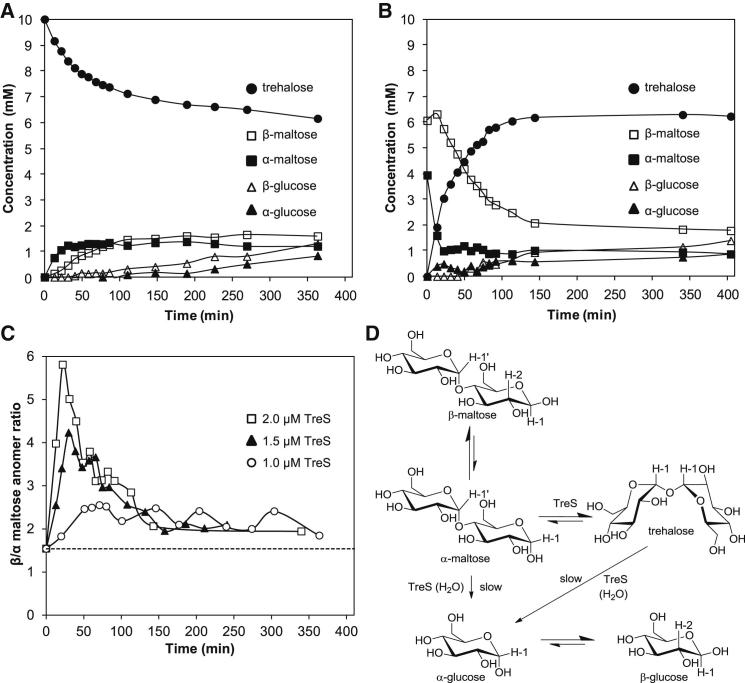
With a view to establishing whether TreS interconverts the α anomer of maltose, we explored the use of 1H-NMR spectroscopy. A spectrum of a reaction mixture generated from maltose by TreS allowed each component to be detected (Figure S2A), including glucose, a known product of hydrolysis (Nishimoto et al., 1996; Pan et al., 2004; Zhang et al., 2011), and the anomers of each reducing sugar. Since anomers mutarotate nonenzymatically, their equilibration upon dissolution of crystalline materials in citrate buffer at 25°C was monitored using 1H-NMR spectroscopy (Figure S2B). The rate constants determined for the mutarotation of α-glucose to β-glucose and the reverse reaction were 0.0316 ± 0.0004 and 0.0192 ± 0.0004 min−1 and, for maltose, 0.0286 ± 0.0002 and 0.0187 ± 0.0001 min−1, each respectively (Figure S2C). These rate constants were consistent with the literature (Bailey et al., 1967; Stults et al., 1987) and the expected dominance of the β anomers.
The conversion of trehalose into maltose by TreS in citrate buffer as a function of time was monitored using 1H-NMR spectroscopy (Figure 3A). The α anomer of maltose was formed 5.4-fold more rapidly than its β anomer, suggesting TreS generates the α anomer. A low level of glucose was also produced through hydrolysis, as observed previously. At longer times, the ratio between trehalose and α/β-maltose was 2.2:1 (at 25°C and pH 6.7), which is reasonably similar to the equilibrium position of 4.6:1 determined from the free energies of hydrolysis of these disaccharides (at 25°C and pH 5.65) (Syson et al., 2011; Tewari and Goldberg, 1991; Tewari et al., 2008) and of 3.2:1 determined from the kinetics of Mycobacterium smegmatis TreS (Zhang et al., 2011). The expected equilibrium positions between the anomers of both maltose and glucose were also approached at longer times.
Figure 3.
Mycobacterium Tuberculosis TreS Interconverts the α Anomer of Maltose
(A) The conversion of trehalose (10 mM) into maltose by TreS (2 μM) according to 1H-NMR spectroscopy (see Figures S2A–S2C for representative spectra and nonenzymic mutarotation controls). Each component was quantified by signal integration using citrate as an internal standard. Reaction mixtures contained 10% D2O to assist spectrum acquisition without significant risk of introducing solvent kinetic and/or equilibrium isotope effects. This necessitated solvent suppression and the introduction of experimentally determined correction factors for the cosuppression of resonances that were close to the solvent resonance.
(B) The conversion of pre-equilibrated α/β-maltose (10 mM) into trehalose by TreS (2 μM) according to 1H-NMR spectroscopy. Note that enzyme was added immediately after the t = 0 data were acquired, resulting in a small and reproducible change in the apparent concentration of starting materials at the second recorded time point. See Figure S2D for the fit of the β-maltose curve (Hoops et al., 2006). The times taken to produce 2 mM trehalose and consume 50% of the α/β-maltose were 14 and 55 min, respectively.
(C) Time courses of the ratios between the β and α anomers of maltose with different TreS concentrations during the conversion of pre-equilibrated α/β-maltose. The broken line indicates the equilibrium between the two anomers in these conditions.
(D) Proposed reaction scheme to account for TreS-catalyzed reactions. Protons used to quantify components of the reaction mixtures by 1H-NMR spectroscopy (Figure S2A) are indicated.
As TreS formed trehalose from pre-equilibrated α/β-maltose (α:β anomeric ratio of 1:1.5), a rapid depletion of α-maltose was immediately apparent (Figure 3B). By contrast, the consumption of β-maltose was significantly slower and conformed to a single exponential function (Figure S2D) with a rate of 0.019 min−1 that was consistent with the rate constant for the mutarotation described above (0.0187 ± 0.0001 min−1). This implied that TreS does not utilize the β anomer. When the experiment was repeated with lower TreS concentrations, the rate of consumption of α-maltose decreased, while that of the β anomer remained similar. Thus, the transient increase in the β/α ratio of the maltose anomers was less pronounced at lower enzyme concentrations (Figure 3C), providing further evidence that TreS converts only the α anomer of maltose into trehalose, as expected. While the initial formation of glucose through hydrolysis appeared to be more rapid from maltose than trehalose, this most likely reflected the more rapid initial consumption of maltose. This implies the equal probability of either α-maltose or trehalose being hydrolyzed. It is also noteworthy that the α anomer of glucose was produced more rapidly from maltose than its β anomer, suggesting that α-glucose is the product of hydrolysis.
The Specificity of TreS for the α Anomer of Maltose Is Retained with Deoxyfluoro Analogs According to 19F-NMR Spectroscopy
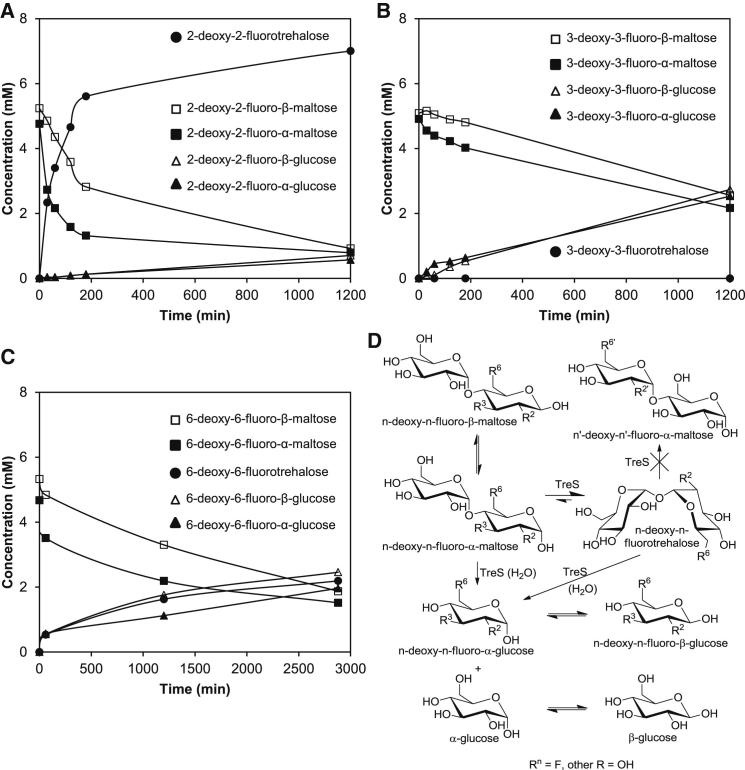
Deoxyfluorotrehalose analogs have been shown to label Mycobacterium tuberculosis cells and exhibit weak antimycobacterial activity (Backus et al., 2011). Since Mycobacterium tuberculosis has a trehalose transporter (Kalscheuer et al., 2010a) and TreS has been reported to utilize 2-fluoro-2-deoxymaltose as a substrate (Zhang et al., 2011), the ability of TreS to convert 2-, 3-, and 6-deoxyfluoromaltose analogs (Tantanarat et al., 2012) was monitored using 19F-NMR spectroscopy (Figure 4). The 2-deoxy-2-fluoro and 6-deoxy-6-fluoro compounds were converted to the corresponding deoxyfluorotrehalose analogs ∼2-fold and ∼180-fold less efficiently than the normal substrate (Figures 4A and 4C). The hydrolysis of each analog to the corresponding deoxyfluoroglucose compounds was detected (Figures 4A–4C), particularly with the 3-fluoro-3-deoxy and 6-fluoro-6-deoxy compounds, with the former being exclusively hydrolyzed (Figure 4B). The expected concomitant formation of nonfluorinated glucose was detected using 1H-NMR spectroscopy (data not shown). The α anomer was consumed more rapidly than the β anomer with all three analogs (Figures 4A–4C), consistent with TreS only acting on α anomers. The mutarotation rates of the deoxyfluoro analogs are not known, but fitting the decay of the β anomer of the 2-deoxy-2-fluoro analog (Figure 4A) suggested a rate constant of ∼0.004 min−1, an order of magnitude slower than that for maltose.
Figure 4.
Mycobacterium Tuberculosis TreS Converts the α Anomers of Deoxyfluoromaltose Analogs
(A) The conversion of pre-equilibrated 2-deoxy-2-fluoro-α/β-maltose (10 mM) by TreS (2 μM) was monitored using 19F-NMR spectroscopy (see Figure S3A for spectra). The times taken to produce 2 mM 2-deoxy-2-fluorotrehalose and consume 50% of the maltose analog were 25 and 125 min, respectively, both ∼2-fold longer than with maltose.
(B) Corresponding data with 3-deoxy-3-fluoro-α/β-maltose (Figure S3B). The time taken to consume 50% of the maltose analog was 1,100 min, 20-fold longer than with maltose. No 3-deoxy-3-fluorotrehalose was detected.
(C) Corresponding data with 6-deoxy-6-fluoro-α/β-maltose (Figure S3C). The times taken to produce 2 mM 6-deoxy-6-fluorotrehalose and consume 50% of the maltose analog were 2,500 and 1,500 min, respectively, 180- and 27-fold longer than with maltose.
(D) Proposed reaction scheme to account for the conversion of deoxyfluoromaltoses by TreS.
In order to assess whether fluoro substitution at the three and six positions resulted in poor binding to TreS or slow conversion by TreS, the extent of conversion of 0.52 mM 2-deoxy-2-fluoromaltose was monitored in the presence and absence of 2.1 mM of each of the other two analogs (Figure S3D). Neither of the analogs gave inhibition, and there may indeed have been a modest stimulation of activity. Given that the Km for maltose is 8–10 mM with the Mycobacterium smegmatis enzyme (Pan et al., 2004; Zhang et al., 2011) and the Km for the 2-deoxy-2-fluoro analog would not be expected to be orders of magnitude lower than this, the lack of inhibition is consistent with fluoro substitution at the three and six positions, compromising the ability of maltose analogs to bind to TreS.
Discussion
We have shown that the direction of flux through TreS is from trehalose to maltose and that the source of trehalose is a combination of the OtsAB and TreYZ pathways in Mycobacterium smegmatis (Figure 2). The overexpression of TreS led to a decrease in trehalose levels, indicating the same direction of flux in Mycobacterium tuberculosis. Thus, TreS appears not to generate trehalose for trehalose mycolate biosynthesis but to convert trehalose into maltose for the GlgE pathway in mycobacteria (Elbein et al., 2010; Kalscheuer et al., 2010b). Although the equilibrium of the TreS-catalyzed reaction is not in line with the direction of flux, the ATP requirement of maltose kinase for the formation of α-maltose 1-phosphate provides most of the driving force through the GlgE pathway, as discussed elsewhere (Syson et al., 2011). Consistent with this, exogenously supplied 14C-labeled trehalose is rapidly and substantially converted to α-maltose 1-phosphate in a Mycobacterium smegmatis ΔglgE mutant, with maltose only being detected when the pep2 maltose kinase gene was inactivated (Kalscheuer et al., 2010b). The observed direction of flux contrasts with a previous study in Mycobacterium smegmatis (Woodruff et al., 2004). However, the authors did not employ a defined ΔotsAΔtreY double mutant to study the specific contribution of TreS in the de novo biosynthesis of trehalose but rather a surrogate strain (a ΔotsAΔtreSΔtreY triple mutant with a reconstituted treS gene constitutively expressed from an episomal multicopy plasmid) that likely exhibited a much higher treS expression level compared with the native gene. Moreover, as nonspecified culture conditions were used, it is unclear whether the medium was devoid of maltose, which might be sufficient to support growth in this genetic context in the absence of trehalose. In any case, our observations are consistent with those of others in Mycobacterium tuberculosis (Murphy et al., 2005) and Corynebacterium glutamicum (Tzvetkov et al., 2003; Wolf et al., 2003). Thus, we have shown that the metabolism of trehalose in Mycobacterium smegmatis is similar to that in Mycobacterium tuberculosis after all, allowing Mycobacterium smegmatis to be used as a model organism in this context. Furthermore, our Mycobacterium smegmatis strains, particularly the strains that are auxotrophic for trehalose, could be used to study the impact of trehalose analogs on specific pathways. It is also intriguing that TreS may be important in late-stage pathogenesis in mice (Murphy et al., 2005), implying a potential role of GlgE pathway-generated cytosolic and/or capsular α-glucan in this process, noting that the latter has been implicated in immune evasion (Sambou et al., 2008).
TreS generates the appropriate α anomer for maltose kinase (Drepper et al., 1996; Mendes et al., 2010) of the GlgE pathway (Kalscheuer et al., 2010b). Thus, the formation of α-maltose 1-phosphate is not limited by the mutarotation of maltose in mycobacteria. This is relevant to many other species, because the treS gene coexists with the other genes of the GlgE pathway in 14% of all sequenced bacterial genomes (Chandra et al., 2011). In a further 28% of genomes that possess the treS gene, one or more of the other GlgE pathway genes is missing. It is therefore not possible to rule out that flux through TreS favors the conversion of maltose to trehalose in organisms that have access to sufficient cytosolic maltose from intracellular or extracellular sources. If the maltose were generated by an enzyme such as β-amylase, the mutarotation of β-maltose could become an issue for flux through TreS to trehalose. Indeed, this work highlights that the mutarotation of any given reducing sugar should not be assumed to be fast compared with the metabolism of a specific sugar anomer.
It is now possible to propose schemes defining the anomeric configurations and origins of all species associated with the TreS-catalyzed reactions studied (Figures 3D and 4D). TreS interconverts the α anomer of maltose (Figures 3A–3C) as expected (Figure 1B), and specificity was also retained with deoxyfluoro analogs (Figures 4A–4C). The TreS enzyme is thought to sterically capture the glucose molecule that it liberates (Figure 1B) and transiently exclude access to the active site such that exogenously supplied glucose does not get incorporated into the products of TreS (Koh et al., 2003; Nishimoto et al., 1996; Zhang et al., 2011). Thus, TreS catalyzes an isomerization, whereby the noncovalently captured glucose molecule must rotate within an enclosed active site. We have now established that this glucose molecule retains its α configuration, whether it goes on to produce normal disaccharide products or is released in the hydrolytic side reaction. Therefore, TreS catalyzes the mutarotation of neither maltose nor glucose.
Unlike trehalose, the deoxyfluorotrehalose products are asymmetric and might have been converted to the corresponding n′-deoxy-n′-fluoromaltose analogs but were not (Figure 4D). This could have been due to the destabilization of oxocarbenium ion-like transition states associated with these reactions (Withers et al., 1988). Alternatively, other effects could be involved, such as reduced binding affinities. Indeed, neither of the 3-deoxy-3-fluoro and 6-deoxy-6-fluoromaltose analogs appeared to bind well to the enzyme. That 3-deoxy-3-fluoromaltose was exclusively hydrolyzed means that 3-deoxy-3-fluoroglucose is less able than a water molecule to attack the glucosyl-enzyme intermediate. This could be due to changes in the nucleophilicity of 3-deoxy-3-fluoroglucose or more likely to an inability to orient itself appropriately within the active site.
Significance
Our findings about the flux through TreS to supply the trehalose mycolate and α-glucan biosynthetic pathways (Figure 1A) have implications for the design and efficacy of inhibitors/imaging agents that target them (Backus et al., 2011; Swarts et al., 2012). It is now much clearer which enzymes need to be targeted to affect either one or both of these pathways. Furthermore, substrate analogs can now be designed as prodrugs for a given pathway that avoid detoxification by competing pathways. For example, when targeting the synthesis of essential trehalose mycolates (Backus et al., 2011; Swarts et al., 2012), it would be an advantage if trehalose analogs were not converted and deactivated by TreS. Indeed, we have observed the limited ability of TreS to tolerate relatively small modifications of its substrates. However, when targeting GlgE or GlgB (Kalscheuer and Jacobs, 2010), there is the challenge of a trehalose analog being tolerated by not only TreS but also the trehalose importer (Kalscheuer et al., 2010a) and maltose kinase. Therefore, the weak inhibition of the growth of Mycobacterium tuberculosis by either 2-deoxy-2-fluoro or 6-deoxy-6-fluoromaltose (Backus et al., 2011) could be due to a lack of either import, efficient processing, and/or inhibition of the GlgE/GlgB targets. Interestingly, labeling of Mycobacterium smegmatis using a 4-azido analog of trehalose is TreS-dependent (Swarts et al., 2012). This observation implies the tolerance of this analog through each step in the biosynthesis of capsular α-glucan rather than trehalose mycolates. Our work supports this interpretation and also shows that this observation in Mycobacterium smegmatis is likely to be relevant to Mycobacterium tuberculosis.
Experimental Procedures
All details about the Experimental Procedures used are given in the Supplemental Experimental Procedures.
Acknowledgments
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (Doctoral Training Grant [BB/F017294/1] and Institute Strategic Programme Grant [BB/J004561/1]), the John Innes Foundation, the Strategic Research Fund of the Heinrich-Heine-University Düsseldorf, and the Jürgen Manchot Foundation. We thank Karl Syson for practical assistance and useful discussion, Shirley Fairhurst for assistance with the NMR spectroscopy, and Krit Tantanarat for kindly providing deoxyfluoromaltose analogs.
Contributor Information
Rainer Kalscheuer, Email: rainer.kalscheuer@med.uni-duesseldorf.de.
Stephen Bornemann, Email: stephen.bornemann@jic.ac.uk.
Supplemental Information
References
- Argüelles J.C. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 2000;174:217–224. doi: 10.1007/s002030000192. [DOI] [PubMed] [Google Scholar]
- Avonce N., Mendoza-Vargas A., Morett E., Iturriaga G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006;6:109. doi: 10.1186/1471-2148-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus K.M., Boshoff H.I., Barry C.S., Boutureira O., Patel M.K., D’Hooge F., Lee S.S., Via L.E., Tahlan K., Barry C.E., 3rd, Davis B.G. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat. Chem. Biol. 2011;7:228–235. doi: 10.1038/nchembio.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.M., Fishman P.H., Pentchev P.G. Studies on mutarotases. I. Purification and properties of a mutarotase from higher plants. J. Biol. Chem. 1967;242:4263–4269. [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J.D., Pastuszak I., Edavana V.K., Pan Y.T., Elbein A.D. A novel trehalase from Mycobacterium smegmatis - purification, properties, requirements. FEBS J. 2007;274:1701–1714. doi: 10.1111/j.1742-4658.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- Chandra G., Chater K.F., Bornemann S. Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology. 2011;157:1565–1572. doi: 10.1099/mic.0.044263-0. [DOI] [PubMed] [Google Scholar]
- De Smet K.A.L., Weston A., Brown I.N., Young D.B., Robertson B.D. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology. 2000;146:199–208. doi: 10.1099/00221287-146-1-199. [DOI] [PubMed] [Google Scholar]
- Drepper A., Peitzmann R., Pape H. Maltokinase (ATP:maltose 1-phosphotransferase) from Actinoplanes sp.: demonstration of enzyme activity and characterization of the reaction product. FEBS Lett. 1996;388:177–179. doi: 10.1016/0014-5793(96)00554-6. [DOI] [PubMed] [Google Scholar]
- Dye C. Global epidemiology of tuberculosis. Lancet. 2006;367:938–940. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- Elbein A.D., Pan Y.T., Pastuszak I., Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- Elbein A.D., Pastuszak I., Tackett A.J., Wilson T., Pan Y.T. Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J. Biol. Chem. 2010;285:9803–9812. doi: 10.1074/jbc.M109.033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops S., Sahle S., Gauges R., Lee C., Pahle J., Simus N., Singhal M., Xu L., Mendes P., Kummer U. COPASI—a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R., Jacobs W.R., Jr. The significance of GlgE as a new target for tuberculosis. Drug News Perspect. 2010;23:619–624. doi: 10.1358/dnp.2010.23.10.1534855. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R., Weinrick B., Veeraraghavan U., Besra G.S., Jacobs W.R., Jr. Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2010;107:21761–21766. doi: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R., Syson K., Veeraraghavan U., Weinrick B., Biermann K.E., Liu Z., Sacchettini J.C., Besra G., Bornemann S., Jacobs W.R., Jr. Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an α-glucan pathway. Nat. Chem. Biol. 2010;6:376–384. doi: 10.1038/nchembio.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S., Kim J., Shin H.J., Lee D., Bae J., Kim D., Lee D.S. Mechanistic study of the intramolecular conversion of maltose to trehalose by Thermus caldophilus GK24 trehalose synthase. Carbohydr. Res. 2003;338:1339–1343. doi: 10.1016/s0008-6215(03)00172-1. [DOI] [PubMed] [Google Scholar]
- Lin F.L., van Halbeek H., Bertozzi C.R. Synthesis of mono- and dideoxygenated α,α-trehalose analogs. Carbohydr. Res. 2007;342:2014–2030. doi: 10.1016/j.carres.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes V., Maranha A., Lamosa P., da Costa M.S., Empadinhas N. Biochemical characterization of the maltokinase from Mycobacterium bovis BCG. BMC Biochem. 2010;11:21. doi: 10.1186/1471-2091-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy H.N., Stewart G.R., Mischenko V.V., Apt A.S., Harris R., McAlister M.S.B., Driscoll P.C., Young D.B., Robertson B.D. The OtsAB pathway is essential for trehalose biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 2005;280:14524–14529. doi: 10.1074/jbc.M414232200. [DOI] [PubMed] [Google Scholar]
- Nishimoto T., Nakano M., Ikegami S., Chaen H., Fukuda S., Sugimoto T., Kurimoto M., Tsujisaka Y. Existence of a novel enzyme converting maltose into trehalose. Biosci. Biotechnol. Biochem. 1995;59:2189–2190. [Google Scholar]
- Nishimoto T., Nakano M., Nakada T., Chaen H., Fukuda S., Sugimoto T., Kurimoto M., Tsujisaka Y. Purification and properties of a novel enzyme, trehalose synthase, from Pimelobacter sp. R48. Biosci. Biotechnol. Biochem. 1996;60:640–644. doi: 10.1271/bbb.60.640. [DOI] [PubMed] [Google Scholar]
- Pan Y.T., Koroth Edavana V., Jourdian W.J., Edmondson R., Carroll J.D., Pastuszak I., Elbein A.D. Trehalose synthase of Mycobacterium smegmatis: purification, cloning, expression, and properties of the enzyme. Eur. J. Biochem. 2004;271:4259–4269. doi: 10.1111/j.1432-1033.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- Paul M.J., Primavesi L.F., Jhurreea D., Zhang Y.H. Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 2008;59:417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- Rose J.D., Maddry J.A., Comber R.N., Suling W.J., Wilson L.N., Reynolds R.C. Synthesis and biological evaluation of trehalose analogs as potential inhibitors of mycobacterial cell wall biosynthesis. Carbohydr. Res. 2002;337:105–120. doi: 10.1016/s0008-6215(01)00288-9. [DOI] [PubMed] [Google Scholar]
- Sambou T., Dinadayala P., Stadthagen G., Barilone N., Bordat Y., Constant P., Levillain F., Neyrolles O., Gicquel B., Lemassu A. Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice. Mol. Microbiol. 2008;70:762–774. doi: 10.1111/j.1365-2958.2008.06445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokane Y., Suzuki M. A novel enzyme, maltose 1-epimerase from Lactobacillus brevis IFO 3345. FEBS Lett. 1995;367:177–179. doi: 10.1016/0014-5793(95)00524-d. [DOI] [PubMed] [Google Scholar]
- Stam M.R., Danchin E.G.J., Rancurel C., Coutinho P.M., Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng. Des. Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- Stults C.L.M., Wade A.P., Crouch S.R. Immobilized enzyme kinetic study of D-glucose mutarotation by flow injection analysis. Anal. Chem. 1987;59:2245–2247. doi: 10.1021/ac00145a007. [DOI] [PubMed] [Google Scholar]
- Swarts B.M., Holsclaw C.M., Jewett J.C., Alber M., Fox D.M., Siegrist M.S., Leary J.A., Kalscheuer R., Bertozzi C.R. Probing the mycobacterial trehalome with bioorthogonal chemistry. J. Am. Chem. Soc. 2012;134:16123–16126. doi: 10.1021/ja3062419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syson K., Stevenson C.E., Rejzek M., Fairhurst S.A., Nair A., Bruton C.J., Field R.A., Chater K.F., Lawson D.M., Bornemann S. Structure of Streptomyces maltosyltransferase GlgE, a homologue of a genetically validated anti-tuberculosis target. J. Biol. Chem. 2011;286:38298–38310. doi: 10.1074/jbc.M111.279315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Wang C., Besra G.S. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2005;18:81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantanarat K., Rejzek M., O’Neill E., Ruzanski C., Hill L., Fairhurst S.A., Limpaseni T., Field R.A. An expedient enzymatic route to isomeric 2-, 3- and 6-monodeoxy-monofluoro-maltose derivatives. Carbohydr. Res. 2012;358:12–18. doi: 10.1016/j.carres.2012.05.026. [DOI] [PubMed] [Google Scholar]
- Tewari Y.B., Goldberg R.N. Thermodynamics of hydrolysis of disaccharides. Lactulose, alpha-D-melibiose, palatinose, D-trehalose, D-turanose and 3-o-beta-D-galactopyranosyl-D-arabinose. Biophys. Chem. 1991;40:59–67. doi: 10.1016/0301-4622(91)85029-p. [DOI] [PubMed] [Google Scholar]
- Tewari Y.B., Lang B.E., Decker S.R., Goldberg R.N. Thermodynamics of the hydrolysis reactions of 1,4-β-d-xylobiose, 1,4-β-d-xylotriose, d-cellobiose and d-maltose. J. Chem. Thermodyn. 2008;40:1517–1526. [Google Scholar]
- Tzvetkov M., Klopprogge C., Zelder O., Liebl W. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology. 2003;149:1659–1673. doi: 10.1099/mic.0.26205-0. [DOI] [PubMed] [Google Scholar]
- Wang J., Elchert B., Hui Y., Takemoto J.Y., Bensaci M., Wennergren J., Chang H., Rai R., Chang C.W.T. Synthesis of trehalose-based compounds and their inhibitory activities against Mycobacterium smegmatis. Bioorg. Med. Chem. 2004;12:6397–6413. doi: 10.1016/j.bmc.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Withers S.G., Rupitz K., Street I.P. 2-Deoxy-2-fluoro-D-glycosyl fluorides. A new class of specific mechanism-based glycosidase inhibitors. J. Biol. Chem. 1988;263:7929–7932. [PubMed] [Google Scholar]
- Wolf A., Krämer R., Morbach S. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol. Microbiol. 2003;49:1119–1134. doi: 10.1046/j.1365-2958.2003.03625.x. [DOI] [PubMed] [Google Scholar]
- Woodruff P.J., Carlson B.L., Siridechadilok B., Pratt M.R., Senaratne R.H., Mougous J.D., Riley L.W., Williams S.J., Bertozzi C.R. Trehalose is required for growth of Mycobacterium smegmatis. J. Biol. Chem. 2004;279:28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- Zhang R., Pan Y.T., He S.M., Lam M., Brayer G.D., Elbein A.D., Withers S.G. Mechanistic analysis of trehalose synthase from Mycobacterium smegmatis. J. Biol. Chem. 2011;286:35601–35609. doi: 10.1074/jbc.M111.280362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.