Abstract
High-throughput DNA sequencing significantly contributed to diagnosis and prognostication in patients with myelodysplastic syndromes (MDS). We determined the biological and prognostic significance of genetic aberrations in MDS. In total, 944 patients with various MDS subtypes were screened for known/putative mutations/deletions in 104 genes using targeted deep sequencing and array-based genomic hybridization. In total, 845/944 patients (89.5%) harbored at least one mutation (median, 3 per patient; range, 0–12). Forty-seven genes were significantly mutated with TET2, SF3B1, ASXL1, SRSF2, DNMT3A, and RUNX1 mutated in >10% of cases. Many mutations were associated with higher risk groups and/or blast elevation. Survival was investigated in 875 patients. By univariate analysis, 25/48 genes (resulting from 47 genes tested significantly plus PRPF8) affected survival (P<0.05). The status of 14 genes combined with conventional factors revealed a novel prognostic model (‘Model-1') separating patients into four risk groups (‘low', ‘intermediate', ‘high', ‘very high risk') with 3-year survival of 95.2, 69.3, 32.8, and 5.3% (P<0.001). Subsequently, a ‘gene-only model' (‘Model-2') was constructed based on 14 genes also yielding four significant risk groups (P<0.001). Both models were reproducible in the validation cohort (n=175 patients; P<0.001 each). Thus, large-scale genetic and molecular profiling of multiple target genes is invaluable for subclassification and prognostication in MDS patients.
Keywords: next-generation sequencing, molecular markers, myelodysplastic syndromes, prognostic score
Introduction
Myelodysplastic syndromes (MDS) are a heterogeneous group of myeloid neoplasms characterized by varying degrees of cytopenias and a predisposition to acute myeloid leukemia.1 With conspicuous clinical and biological heterogeneity in MDS, an optimized choice of treatment based on accurate diagnosis and risk stratification in individual patients is central to the current therapeutic strategy.2 In fact, relying on conventional cytogenetics, cytomorphology and peripheral blood parameters, the International Prognostic Scoring System (IPSS) and other models2, 3, 4 have been successfully used for these purposes, and the recent revision of IPSS (IPSS-R) further improved prognostication.5
Meanwhile, our knowledge about the molecular pathogenesis of MDS has dramatically improved during the past 10 years mainly through the identification of major mutational targets.6 These mutations not only involved previously well-defined pathways such as signal-transducing molecules and transcription factors, but also affected newly identified pathways, including epigenetic regulators such as TET2,7 IDH1/2,8, 9 DNMT3A,10 ASXL111 and EZH2,12, 13 and multiple components of the RNA-splicing machinery,14, 15, 16 and the list of gene mutations implicated in MDS pathogenesis17, 18 is still growing. A recent study described oncogenic mutations in 78% of patients with MDS or closely related myeloid neoplasms.19 Further studies are warranted to clarify how to integrate this increased knowledge of gene mutations in our understanding of MDS pathogenesis and into clinical practice, which is even more emphasized by the recent advance of high-throughput genomics. Bejar et al.20, 21 recently suggested that the mutational status of multiple gene targets could better predict the clinical outcome in MDS. According to ELN (European Leukemia Network) recommendations, mutation analysis of candidate genes that can allow a conclusive diagnosis and a reliable prognostic evaluation is already suggested in patients with MDS.22
The purpose of the present study was to provide a solid basis for applying the knowledge of this group of gene mutations to medical practice and to further understanding of MDS pathogenesis, using revolutionized technologies for genome analysis.23, 24 We investigated a large cohort of 944 patients with different MDS subtypes for genetic lesions, in which gene mutations and copy number alterations in a total of 104 known/putative gene targets were comprehensively analyzed using massively parallel sequencing. This set of information about genetic lesions was utilized to build novel prognostic models that outperformed existing models, employing a conventional training/validation-cohort design. Analyses of spectrum of mutations and their mutational burdens and correlation of different mutations also provided novel insights into intratumoral heterogeneity and hierarchy of mutations, together with their association to phenotypes.
Patients and methods
Patients
A total of 944 adult patients with different MDS subtypes were enrolled in this study, whose bone marrow and peripheral blood samples had been sent for diagnosis to the MLL Munich Leukemia Laboratory between August 2005 and August 2011 (Table 1). The training cohort consisted of 730 patients, the validation cohort of 214 patients. In the training cohort (n=730 patients), follow-up data was available in 670 cases, information on treatment in 648 cases (supportive care: n=504/648, 77.8% azacytidine or lenalidomide: n=62/648, 9.6% cytoreductive treatment only: 22/648, 3.4% acute myeloid leukemia-induction therapy: n=60/648, 9.3%). In the independent validation cohort (n=214 patients), data on follow-up was available in 205, on therapy in 192 patients with a similar distribution of therapeutic strategies (supportive care: n=148/192, 77.1% azacytidine or lenalidomide: n=18/192, 9.4% cytoreductive treatment: n=11/192, 5.7% acute myeloid leukemia-induction therapy: n=15/192; 7.8%). Non-amplified genomic DNA from 53 healthy individuals and paired tumor/normal DNA from an additional 16 MDS cases were also analyzed and used for performance of our data analysis pipelines. All samples were investigated by cytomorphology, chromosome banding analysis, and in part, fluorescence in situ hybridization (Supplementary Table S1), and standard molecular analyses. All patients gave their written informed consent with genetic analyses and with research studies. The study was approved by the internal review boards of the MLL and the University of Tokyo and was following the Declaration of Helsinki.
Table 1. Demographic data, peripheral blood parameters, MDS subtypes, results of cytogenetics and risk classification according to IPSS/IPSS-R for the total cohort.
| Parameter | Total cohort (n=944) |
|---|---|
| Patient numbers (ratios, ranges or percentages) | |
| Demographic characteristics | 944 |
| Males:females (ratio) | 580:364 (1.6) |
| Median age (years) | 72.8 (23.3–90.8) |
| <50 | 46 (4.9%) |
| 50–<60 | 81 (8.6%) |
| 60–<70 | 240 (25.4%) |
| 70–<80 | 423 (44.8%) |
| ⩾80 | 154 (16.3%) |
| Cases with follow-up | 875 |
| Median follow-up in months | 32.3 |
| Median OS in months | 54.5 |
| MDS subtypes (WHO, 2008) | 944 |
| RA | 41 (4.3%) |
| RARS | 81 (8.6%) |
| RARS-T | 28 (3.0%) |
| RCMD | 195 (20.7%) |
| RCMD-RS33 | 183 (19.4%) |
| RAEB-1 | 191 (20.7%) |
| RAEB-2 | 188 (19.9%) |
| MDS with isolated 5q-deletion | 37 (3.9%) |
| Cytogenetics | 944 |
| Normal karyotype | 648 (68.6%) |
| Aberrant karyotype | 296 (31.4%) |
| IPSS risk group | 848 |
| Low | 324 (38.2%) |
| Intermediate-1 | 319 (37.6%) |
| Intermediate-2 | 171 (20.2%) |
| High | 34 (4.0%) |
| IPSS-R risk group | 848 |
| Very low | 122 (14.4%) |
| Low | 340 (40.1%) |
| Intermediate | 203 (23.9%) |
| High | 130 (15.3%) |
| Very high | 53 (6.3%) |
Abbreviations: IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndrome; OS, overall survival.
Procedures
In total, 104 known or putative mutational gene targets in MDS were examined for mutations in all 944 cases from the cohort using massively parallel sequencing (Illumina, San Diego, CA, USA) of SureSelect (Agilent, Santa Clara, CA, USA) captured target sequences (Supplementary Tables S2 and S3 and Methods in the Supplementary Information). All sequencing data were analyzed using our in-house pipeline, through which high-probability oncogenic mutations were called by eliminating sequencing/mapping errors and known/possible SNPs based on the available databases and frequencies of variant reads (Supplementary Figures S1 and S2 and Methods in the Supplementary Information). Genomic copy number status was calculated by directly enumerating corresponding sequencing reads in each exon, and also assessed by array-based comparative genomic hybridization (aCGH) in 494 (76.2%) out of 648 samples with normal karyotypes. Genome-wide methylation status was measured in 191 patients using Infinium 450 K arrays (Illumina) (Supplementary Information).
Impact of genetic lesions on overall survival (OS) was investigated by Cox regression, in which the Least Absolute Shrinkage and Selection Operator (lasso)25, 26 was used for selecting variables. The linear predictor from the Cox regression was then used to assign the patients into discrete risk groups. Prognostic models were constructed in a training set and confirmed using an independent validation cohort and their performance was compared to the IPSS-R using the Akaike information criterion and J-tests.27 Detailed information describing mutation/copy number calling, cytomorphology, chromosome banding analysis, array comparative genomic hybridization (aCGH), risk prediction model building, global methylation studies and other statistical analyses are given in the Supplementary Information.
Results
Targeted sequencing
The mean depth of the targeted resequencing study in 104 genes was 1066-fold (257–2306 reads) across the entire cohort (n=944). More than 99% of the target sequences (that is, 357 861 bp nucleotides including six bases from a splice junction) were analyzed with >30 independent reads. After excluding sequencing/mapping errors and known or possible polymorphisms, a total of 2764 single nucleotide variants and insertions/deletions (indels) were called in 96 genes as high-probability somatic changes (Figure 1a and Supplementary Figure S3). For validation, we sequenced randomly selected 300 variants (Supplementary Table S4) using independent sequencing platforms (Sanger sequencing and/or Illumina MiSeq), of which 299 were successfully confirmed (99.7%). Our pipeline generated only 10 false-positive calls, when applied to the analysis of DNA from 53 healthy individuals (Supplementary Information). The accuracy of our pipeline was also assessed by using paired tumor/normal DNA from 16 MDS cases, in which 74 of 75 (98.7%) mutations called by our pipeline were confirmed as being true somatic mutations using normal DNA. Finally, further cross-validation was performed for a total of 1207 gene loci among 446 MDS cases, in which an overall concordance of 99.4% congruent results was observed (Supplementary Information).
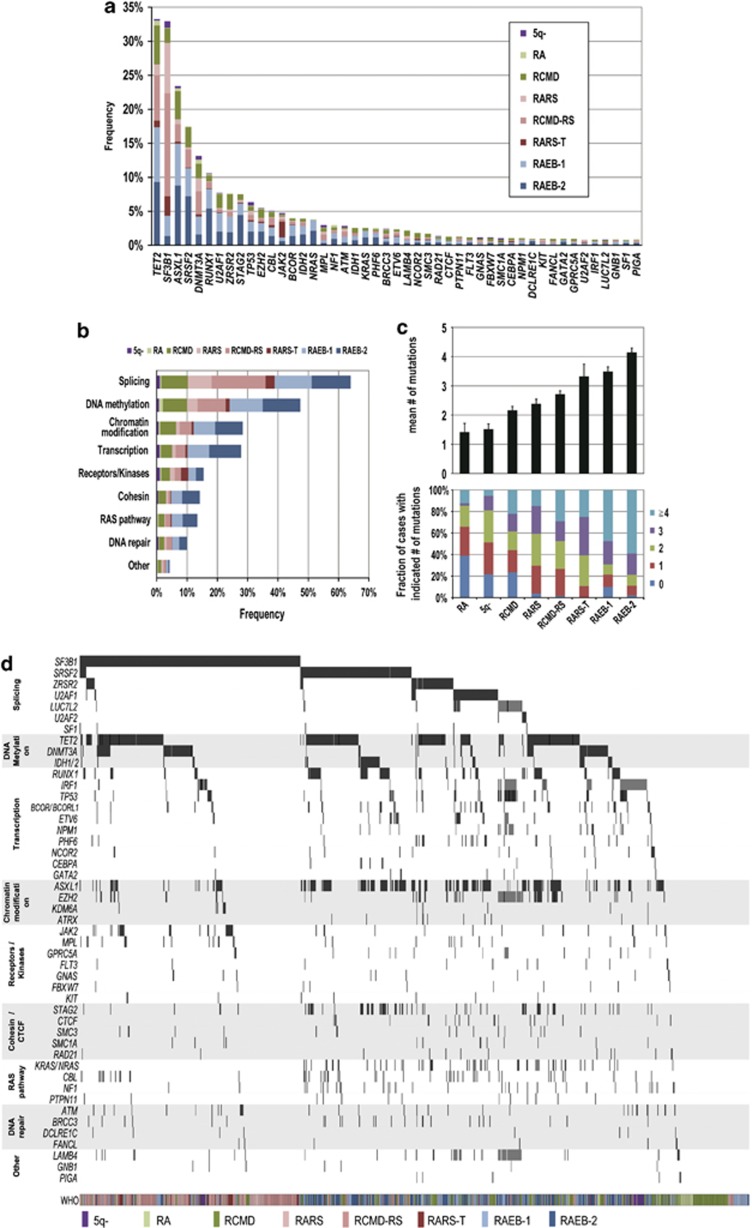
Figure 1.
Significantly mutated genes in MDS. (a) Frequency of mutations in 47 significantly mutated genes in 944 cases with different WHO subtypes, which are shown in indicated colors. (b) Frequency of gene mutations involved in common functional pathways, which are defined in Supplementary Table S3. (c) Number of gene mutations detected in different MDS subtypes. (d) Distribution of mutations/deletions of significantly mutated genes in 944 MDS cases.
Frequency and spectrum of gene mutations
In total, 845 of the 944 patients (89.5%) harbored at least one mutation with a median of 3 (0–12) mutations per sample, in which 574 (67.9%) cases had a normal karyotype (Supplementary Figure S4). In 19 cases gene deletions without mutations were observed. Sixty-eight genes were mutated in >0.5% of the cases and 47 genes were considered as statistically significantly mutated (q<0.01) after calculating type I errors for 104 genes following the method of Benjamini and Hochberg28 (Supplementary Table S5 and Methods in the Supplementary Information). The six most frequently mutated genes were TET2, SF3B1, ASXL1, SRSF2, DNMT3A and RUNX1 (mutated in >10% of the cases). Less common mutations (2–10%) involved U2AF1, ZRSR2, STAG2, TP53, EZH2, CBL, JAK2, BCOR, IDH2, NRAS, MPL, NF1, ATM, IDH1, KRAS, PHF6, BRCC3, ETV6 and LAMB4 (Figure 1a). Most of the significantly mutated genes were previously well documented either in MDS or other myeloid malignancies. However, some were newly identified or re-confirmed as recurrently mutated genes, including BRCC3,14 FANCL, LUC7L2,29 STAG2 and other cohesin components17 such as RAD21, SMC3 and SMC1A,30 or CTCF, GPRC5A, LAMB4 and IRF1.31, 32
Mutated genes were grouped into several functional pathways, which were hypothesized to characterize MDS pathogenesis (Supplementary Figure S5). Amongst these, the most frequent target was RNA splicing, with mutations observed in as many as 64% of cases, followed by genes associated with DNA methylation, chromatin modification, transcription, RAS/signal transduction pathways, cohesin and DNA repair pathways (Figure 1b). The mean number and the spectrum of mutations closely correlated with WHO subtypes. SF3B1 mutations were frequently confirmed in MDS subtypes with increased ring sideroblasts (240/292, 82.2%) and also in isolated del(5q) (9/37, 24.3%). Except for SF3B1, DNMT3A, JAK2 and MPL mutations, the majority of common mutations occurred more frequently in high-risk WHO subtypes (RAEB-1/-2), than in RA/RARS cases (Figure 1a). Similarly, the mean number of mutations tended to be higher in high-risk subtypes (Figure 1c). Notably, even in RA and RCMD with normal karyotype, gene mutations were found in 133/183 cases (72.7%). Some tumor suppressor genes were also identified as common targets of gene deletions as revealed by conventional cytogenetics, aCGH, and/or digital copy number analyses, which included EZH2, LUC7L2, ETV6, TET2, IRF1, TP53 and PHF6 (Supplementary Figures S6 and S7). Combined, these data provide a landscape of genetic alterations in MDS (Figure 1d).
Correlation of gene mutations
Statistically significant correlations (q<0.01) were found across 82 combinations of significantly mutated genes, indicating the presence of functional interactions among different mutations in MDS pathogenesis. These included correlations, such as positive correlations among mutations/deletions in STAG2, IDH2, ASXL1, RUNX1 and BCOR, and mutually exclusive relationships between SRSF2 mutations and mutations/deletions of DNMT3A, EZH2, IRF1 and between ASXL1 and DNMT3A (Figure 2a). Reflecting lower number of coexisting mutations in SF3B1-mutated cases, SF3B1 mutations largely showed negative correlations with common mutational targets other than DNMT3A and JAK2 mutations. ASXL1 mutations were more likely to coexist with other mutations, except for significantly negative correlations with SF3B1, DNMT3A and IRF1 mutations. TET2 mutations showed positive correlations with SRSF2 and ZRSR2 mutations.
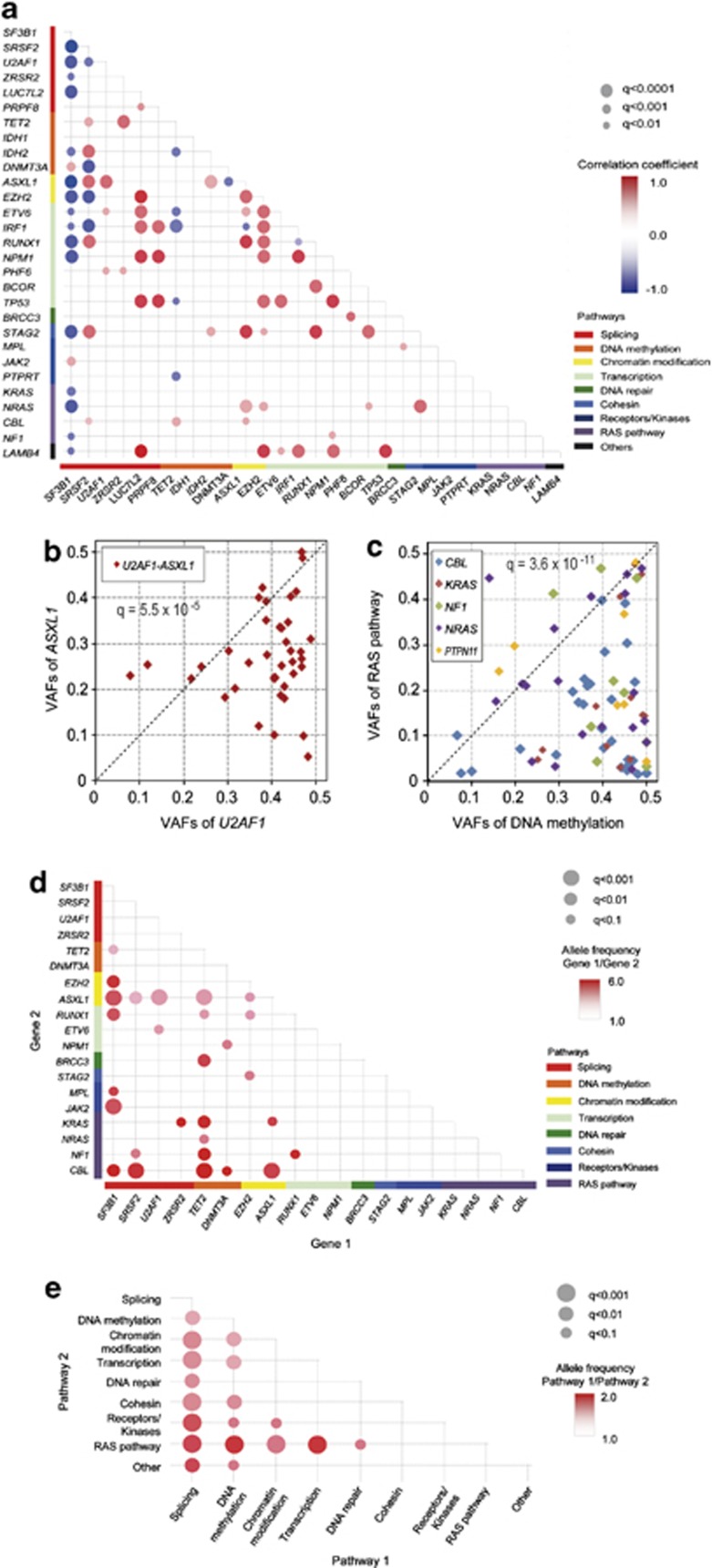
Figure 2.
Comparison of mutation loads between major gene targets in MDS. (a) Correlations between major genetic lesions, where the correlation coefficients are indicated by a color gradient and show diagonal plots of variant allele frequencies (VAFs) between ASXL1 and U2AF1 mutations (b) and between mutations in RAS pathway genes (CBL, KRAS, NF1, NRAS and PTPN11) and DNA methylation-related genes (TET2, IDH1/2 and DNMT3A) (c), in which copy number-adjusted VAF values between indicated mutations or mutations of indicated functional pathways were compared using paired T-tests. Comparison was made exhaustively between all possible combinations of commonly mutated genes (>2.5% of mutation rates) (d) or gene pathways (e) with adjustment for multiple testing. Significance (q-values) and difference (relative difference in VAFs) is indicated by the size of circles and color gradient, as indicated, respectively.
Intratumoral heterogeneity and hierarchy of mutations
Accurate determination of variant allele frequencies (VAFs) by deep sequencing allowed for investigating substructures within the tumor population in terms of gene mutations, relying on a modified χ2 test for heterogeneity (Supplementary Figure S8 and Methods in the Supplementary Information). Intratumoral heterogeneity was recognized in as many as 456 cases (48.3%), even though the small number of gene mutations available for evaluation was thought substantially to underestimate the real frequency. The number of observed intratumoral subpopulations tended to correlate with the number of detected mutations and therefore, advanced WHO subtypes and risk groups with poorer prognosis (Supplementary Figure S9).
Mean VAFs showed significant variations among major gene targets (Supplementary Figure S10). Specifically, when VAFs of coexisting genes and/or pathway genes were compared between major gene targets (N=47) or target pathways (N=9), significant difference was demonstrated in 28 out of 224 gene pairs and 18 out of 36 pathway pairs investigated (Figures 2b–e, Supplementary Table S6). These results suggested clonogenic hierarchy among these common mutations during clonal evolution. For example, mutations in genes on DNA methylation and RNA splicing pathways tended to have higher mutation burden than those in other genes, suggesting earlier origins of these mutations (Figures 2c and d). Also, mutations of genes involved in chromatin modifications and transcriptions tended to have higher VAFs than RAS pathway mutations (Figure 2e). Similarly, RARS-T cases were characterized by a coexisting JAK2 mutation, in which all but one JAK2 mutations showed significantly lower mutation burden than the corresponding SF3B1 mutation, strongly suggesting that most RARS-T cases evolved from RARS/RCMD-RS with an additional acquisition of a JAK2 mutation (Supplementary Figure S11).
Development of a novel prognostic model including molecular markers
The impact of these genetic lesions on clinical outcomes was initially investigated in 875 patients, in which follow-up data were available. In univariate analysis, 25 out of 48 genes (resulting from 47 genes tested significantly plus PRPF8) affected OS (P<0.05) (Supplementary Figure S12), and only SF3B1 mutations were associated with a better clinical outcome. Next, to evaluate the combined effect of these multiple gene mutations/deletions, together with common clinical/cytogenetic variables used for IPSS-R, OS was modeled by a conventional Cox regression. For this purpose, complete follow-up data and other clinical/cytogenetic data were available in 786 patients, of which 611 were used as a training set, while the remaining 175 were used for validation (Supplementary Information). To reduce the initial set of putative explanatory variables, we included only 32 genes in the regression analysis that were altered in at least 15 patients of the training set, and then performed variable selection using lasso (Methods in the Supplementary Information).
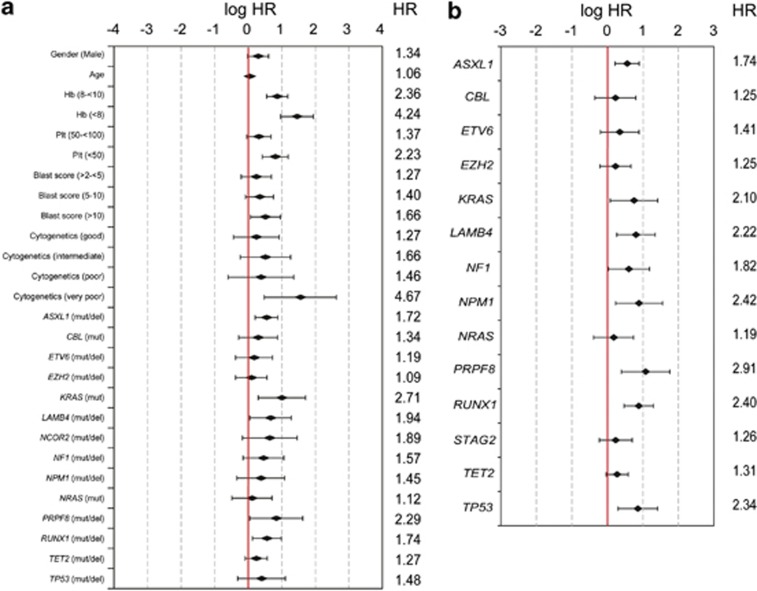
A total of 14 genes, together with age, gender and, using the categories of the IPSS-R, white blood cell counts, hemoglobin, platelet counts, bone marrow blast count and cytogenetic score were finally selected for the Cox regression in a proportional hazard model (Figure 3a and Supplementary Tables S7 and S8). Based on the linear predictor of the regression model, we constructed a prognostic model (Model-1), in which patients were classified into four risk groups showing significantly different OS (‘low', ‘intermediate', ‘high' and ‘very high risk') with significantly differing 3-year survival of 95.2, 69.3, 32.8 and 5.3%, respectively (P<0.001; Figure 4a). The model was largely reproducible in the validation cohort of 175 patients (P<0.001; Figure 4d) and also in the 463 patients who were treated with supportive care only (P<0.001; Supplementary Figure S13). When the variables for a new lasso model were limited to genes only (results of Cox regression in a proportional hazard model shown in Figure 3b), a prognostic model (gene-only model; Model-2) was generated based on 14 selected genes yielding four significant risk groups (P<0.001; Figure 4b), which was also statistically reproduced in the validation set (P<0.001; Figure 4e and Supplementary Table S9). Significantly, 13 of the 14 genes selected for Model-1 were again selected for Model-2.
Figure 3.
Illustration of hazard ratios for Model-1 and Model-2. Hazard ratios (HRs, given in numbers) as well as logHR and their 95% confidential intervals (given as chart) for parameters used for Model-1 including clinical and genetic variables (a) and for Model-2 including only genetic variables (referring to the training cohort) (b) are plotted. For a, baseline level for the analysis was the respective IPSS-R risk category with the least risk score (hemoglobin: ⩾10 g/dl, platelets score: ⩾100 × 109/l, blast score: ⩽2%, cytogenetic score very good).
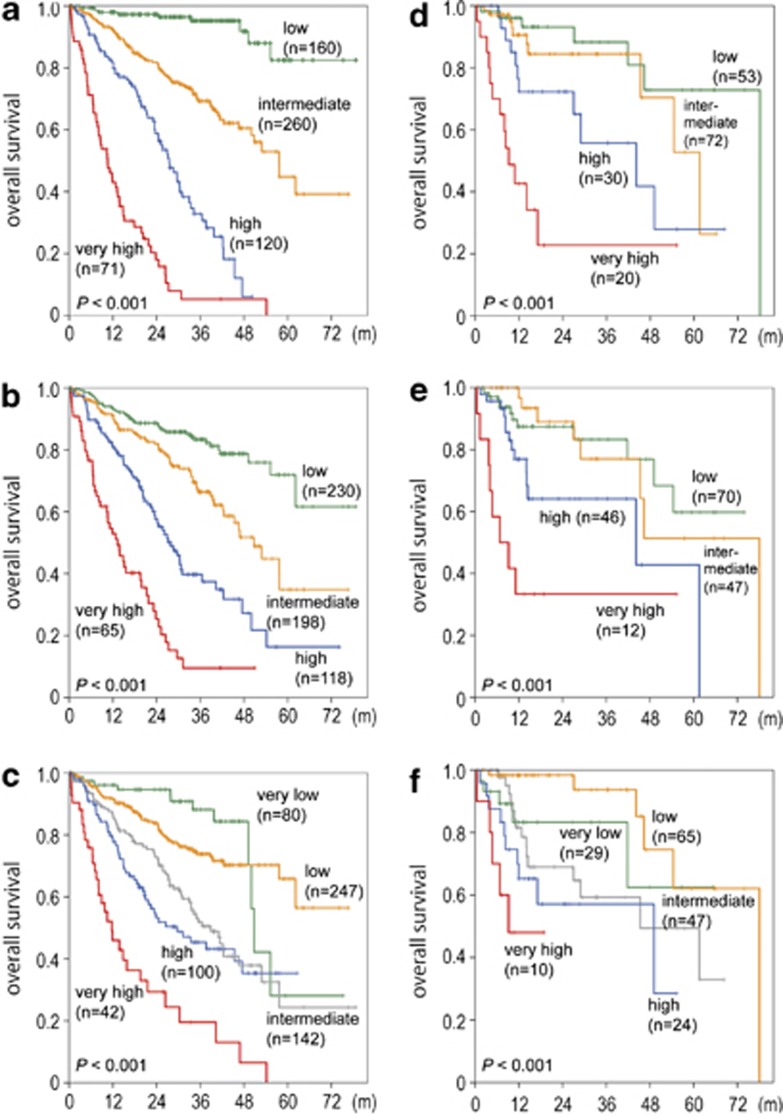
Figure 4.
Development of a novel prognostic risk classification. (a) Kaplan–Meier estimates of OS in months (m) for four groups according to Model-1 in the training cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 95.2, 69.3, 32.8 and 5.3%, respectively. (b) Kaplan–Meier estimates of OS for four groups according to Model-2 in the training cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 83.3, 66.4, 39.7 and 9.5%, respectively. (c) Kaplan–Meier estimates of OS for five groups according to IPSS-R in the training cohort. Three-year OS for very low, low, intermediate, high and very high-risk groups amounts to 88.2, 73.9, 51.9, 45.3 and 19.6%, respectively. (d) Kaplan–Meier estimates of OS for four groups according to Model-1 in the validation cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 88.3, 84.4, 55.7 and 22.8%, respectively. (e) Kaplan–Meier estimates of OS for four groups according to Model-2 in the validation cohort. Three-year OS for low, intermediate, high and very high-risk groups amounts to 83.3, 77.0, 64.1 and 33.3%, respectively. (f) Kaplan–Meier estimates of OS for five groups according to IPSS-R in the validation cohort. Three-year OS was 83.3, 93.7, 59.3 and 57.1% for the very low, low, intermediate and high-risk groups. For the very high-risk group, the median OS was 9.2 months, as 3-year OS was not applicable.
These results demonstrated that the mutation/deletion status of a set of genes could be used to build a clinically relevant prognostic system as independent variables from clinical parameters. When applied to the validation cohort (Figures 4d–f), Model-1 was shown to outperform the IPSS-R (Figure 4f; see also Figure 4c for training cohort according to IPSS-R) and Model-2 (Figure 4e). The superiority of Model-1 was also indicated by the J-test: adding the predictions from Model-1 to the IPSS-R model significantly improved the model (P<0.001), whereas the predictions from the IPSS-R did not substantially improve Model-1. As expected, when comparing Model-2 to the IPSS-R, the J-test indicates that both models could be improved by information from the other model (Supplementary Table S10; see also Supplementary Information and Supplementary Figure S21A–E). The results did not change remarkably when the validation cohort was reduced to only samples with supportive treatment (see Supplementary Information).
We investigated possible correlations of the before mentioned genes with WHO classification, risk categories in IPSS/IPSS-R and Model-1/2, karyotypes, demographic and clinical data within the training cohort (Supplementary Tables S11 and S12). ASXL1 (P=0.021) and TET2 (P=0.007) mutations were significantly associated with male sex. Patients with TET2 mutations (P<0.001) showed a higher mean age as compared with patients without the respective mutations. Most of the above mutations were associated with higher risk groups and/or with MDS subtypes with elevated blast counts (Supplementary Figures S14-S17).
Discussion
High-throughput deep sequencing of the 104 gene targets across 944 MDS patients, combined with genome-wide copy number analyses, revealed a large-scale comprehensive landscape of genetic lesions in MDS with a broad spectrum of gene mutations and deletions. Combined with copy number abnormalities, genetic lesions were demonstrated in as many as 864/944 (91.5%) of MDS cases. This even exceeds the frequency of gene mutations in another large-scale gene sequencing study in MDS published recently.19 It should be emphasized that there was an extremely high concordance rate between the results of NGS and other molecular diagnostic techniques that were used for secondary validation in part of the patients in our cohort. Given the deep sequencing depths uniformly achieved over the entire target sequences, we could expect highly sensitive detection of mutations, except for false-negative detection of some indels, especially those having large insertion/deletion size. This is also true for other mutations that had very high (>0.45) allelic burdens and were previously unreported and for mutations existing in very small subclones only (further details are given in Supplementary Information). Also, the lack of germline controls and the technical limitations in the current sequencing devices/chemistry may have produced some false positives, even after extensive exclusion of known SNPs and possible sequencing errors.
A paucity of known gene mutations was a characteristic feature of RA and other low-risk MDS and correlated with better clinical outcome, whereas higher numbers of gene mutations were observed in advanced WHO subtypes. Heterogeneity of tumor population in MDS17 has now been confirmed to be quite common in MDS and more prominent in advanced WHO subtypes and high-risk prognostic groups, which were also associated with increasing numbers of gene mutations and intratumoral subpopulations. Taken together, accumulating numbers of gene mutations and increasing intratumoral heterogeneity characterize the clonal evolution of MDS and are associated with a worse prognosis. The presence of a hierarchy of mutations was another important finding, which not only advances our understanding of MDS pathogenesis, but also may suggest a clinical relevance in molecular monitoring of disease progression. The gene mutations found in our study were found to show significant correlations, many of which were confirming recently published data,19 for example, the positive correlation of TET2 and ZRSR2 mutations, or the negative correlation of ASXL1 and SF3B1 mutations.
However, probably the most significant finding of the current study was the feasibility of testing mutations in multiple genes (>100 candidates), based on which we can improve the way to predict the prognosis of patients with MDS to enhance making clinical decisions. In fact, correlation between multiple genetic lesions and clinical outcome was so close to allow for building novel, clinically applicable prognostic models for MDS. Thus, further combining the 14 prognostic genes with conventional risk factors, such as age, gender and the parameters used in the IPSS-R, our model successfully discriminated four significant risk groups and better predicted patients' OS than gene-only model (Model-2) and IPSS-R. This combined model succeeded to discriminate four risk groups (‘low', ‘intermediate', ‘high' and ‘very high risk') with significantly differing 3-year survival 95.2, 69.3, 32.8 and 5.3% (P<0.001), respectively. It should be pointed out that >77% of patients from both the training and the validation cohort in our study had received supportive treatment only, but part of the patients had received therapy, for example, with demethylating agents. However, we were able to confirm the prognostic power of our prognostic Model-1 separately in part of the patients from the training cohort who received supportive treatment only.
In conclusion, comprehensive molecular genetic profiling using deep-sequencing is a feasible19 and highly promising approach to contribute to diagnostic accuracy, pathogenesis, biologic subclassification and finally prognostication and risk stratification in patients with MDS. Considering the decrease of costs for this technology and therefore its availability for large numbers of patients in the near future, molecular profiling of multiple target genes is feasible and soon will be integrated in individualized therapeutic decision making for patients with MDS.
Acknowledgments
We thank Melanie Zenger and Marita Staller for performing array CGH analyses and all co-workers from the MLL Munich Leukemia Laboratory. We are grateful to all physicians for providing and caring for patients as well as collecting data. This work was supported by Grant-in-Aids from the Ministry of Health, Labor and Welfare of Japan and KAKENHI (23249052, 22134006, 24221011 and 21790907), by the Industrial Technology Research Grant Program from NEDO (08C46598a), by Project for Development of Innovative Research on Cancer Therapeutics (P-Direct) and by the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST).
Author contributions
TH, YN, VG, YO, UB, TA, HPK, WK and SO performed study design and interpretation of data. YN, VG, SS, MS, AK, KY, SM, AyK and AlK performed molecular studies. YN, YO, AR, NN, YuiS, YusS, KC, HT, HUK, MD and SM performed bioinformatics. Methylation analyses were done by GN, HUK and HA. HUK, MD, AR and NN performed risk models and analyzed methylation data. Patient samples were characterized by TH, SS, AlK, CH and WK. TH, YN, VG, YO, UB, TA, WK and SO wrote the manuscript. TH, WK and SO supervised the study. All authors contributed to writing of the manuscript and approved the final version.
TH, SS, CH and WK declare equity ownership of MLL Munich Leukemia Laboratory GmbH. VG, UB, TA, AR, NN and AlK are employed by MLL Munich Leukemia Laboratory GmbH. All other authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Brunning R, Orazi A, Germing U, Le Beau MM, Porwit A, Baumann I, et al. Myelodysplastic syndromesIn: Swerdlow S, Campo E, Lee Harris N, Jaffe ES, Pileri SA, Stein H, et al(eds). 4 edn,WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 200887–93.
- Cazzola M, la Porta MG, Travaglino E, Malcovati L. Classification and prognostic evaluation of myelodysplastic syndromes. Semin Oncol. 2011;38:627–634. doi: 10.1053/j.seminoncol.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Greenberg P, Cox C, Le Beau MM, Fenaux P, Morel P, Sanz G, et al. International Scoring System for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- Malcovati L, Germing U, Kuendgen A, Della Porta MG, Pascutto C, Invernizzi R, et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J Clin Oncol. 2007;25:3503–3510. doi: 10.1200/JCO.2006.08.5696. [DOI] [PubMed] [Google Scholar]
- Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, et al. Revised International Prognostic Scoring System (IPSS-R) for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Mardis ER. New strategies and emerging technologies for massively parallel sequencing: applications in medical research. Genome Med. 2009;1:40. doi: 10.1186/gm40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Kato M, Kawahata R, Matsubara A, Takita J, et al. A nonsense mutation of IDH1 in myelodysplastic syndromes and related disorders. Leukemia. 2011;25:184–186. doi: 10.1038/leu.2010.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Trouplin V, Roquain J, Adelaide J, Carbuccia N, Esterni B, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151:365–375. doi: 10.1111/j.1365-2141.2010.08381.x. [DOI] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Shao J, Ding L, White B, Kandoth C, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013;27:1275–1282. doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes Blood 2013. e-pub ahead of print 12 September 2013;doi: 10.1182/blood-2013-08-518886 [DOI] [PMC free article] [PubMed]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Hellstrom-Lindberg E, Bowen D, Ades L, Cermak J, del Canizo C, et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European LeukemiaNet. Blood. 2013;122:2943–2964. doi: 10.1182/blood-2013-03-492884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J Royal Statist Soc B. 1996;58:267–288. [Google Scholar]
- Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- Davidson R, Mackinnon JG. Several tests for model specification in the presence of alternative hypotheses. Econometrica. 1981;49:781–793. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu KS, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willman CL, Sever CE, Pallavicini MG, Harada H, Tanaka N, Slovak ML, et al. Deletion of IRF-1, mapping to chromosome 5q31.1, in human leukemia and preleukemic myelodysplasia. Science. 1993;259:968–971. doi: 10.1126/science.8438156. [DOI] [PubMed] [Google Scholar]
- Boultwood J, Fidler C, Lewis S, MacCarthy A, Sheridan H, Kelly S, et al. Allelic loss of IRF1 in myelodysplasia and acute myeloid leukemia: retention of IRF1 on the 5q- chromosome in some patients with the 5q- syndrome. Blood. 1993;82:2611–2616. [PubMed] [Google Scholar]
- Brunning RD, Bennett JM, Flandrin G, Matutes E, Head D, Vardiman J, et al. Myelodysplastic syndromesIn: Jaffe ES, Harris N, Stein H, Vardiman J, edsWorld Health Organization of Tumors, Pathology & Genetics, Tumors of haematopoietic and lymphoid tissues IARC Press: Lyon, France; 200161–73. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.