Abstract
Multiple reaction monitoring (MRM) has recently become the method of choice for targeted quantitative measurement of proteins using mass spectrometry. The method, however, is limited in the number of peptides that can be measured in one run. This number can be markedly increased by scheduling the acquisition if the accurate retention time (RT) of each peptide is known.
Here we present iRT, an empirically derived dimensionless peptide-specific value that allows for highly accurate RT prediction. The iRT of a peptide is a fixed number relative to a standard set of reference iRT-peptides that can be transferred across laboratories and chromatographic systems.
We show that iRT facilitates the setup of multiplexed experiments with acquisition windows more than 4 times smaller compared to in silico RT predictions resulting in improved quantification accuracy. iRTs can be determined by any laboratory and shared transparently. The iRT concept has been implemented in Skyline, the most widely used software for MRM experiments.
Keywords: Mass spectrometry, multiplexing, proteomics methods, optimization, quantitative analysis
Introduction
Liquid chromatography mass spectrometry (LC-MS) is a powerful method to analyze peptides from complex biological samples. Many different workflows with different underlying mass-spectrometer layouts [1] have been devised as variants of LC-MS. They can be broadly categorized into discovery and targeted approaches. Discovery approaches, most notably data independent LC-MS/MS workflows [2, 3], aim to analyze as exhaustively as possible the peptide composition of samples.
Targeted approaches, with multiple reaction monitoring (MRM, also referred to as selected reaction monitoring or SRM) as the prototypic method, are increasingly used for mass spectrometry based quantification of selected proteins [4–6]. The targets for MRM experiments are defined on a rational basis and depend on the hypothesis to be tested in the experiment.
The upfront work required to carefully design assays for the targets, including the selection of suitable proteotypic peptides [7]and the identification of the fragment ions that provide the highest signal intensities [8], is compensated by more reproducible and much simpler measurement data and improved sensitivity compared with, for instance, LC-MS/MS data [9, 10].
In both approaches, discovery and targeted, proteins are digested into smaller peptides prior to analysis. The resulting peptide mixture is usually chromatographically separated in order to reduce the complexity of the sample [11]. Chromatographic separation adds a time dimension to the recorded data of the mass spectrometer, the selectivity which manifests itself as a specific retention time (RT) in a particular chromatographic system. RT can be used in various ways as an orthogonal property to the mass to charge dimension.
In applications of discovery LC-MS the RT information has been used to align LC peaks [12, 13] across acquisitions and to support peptide identification [14, 15].
In targeted LC-MS applications the RT can be directly used to set-up a measurement schedule. Peptides can be measured for only a small window of time with the center of the window at the point where the peptide is expected to elute from the chromatographic column, the RT. This scheduling increases the number of transitions that can be measured in one LC-MRM run, since at any given time, the instrument measures only a subset of the transitions in the method [16]. The duration of the measurement window used for scheduling is usually chosen based on the anticipated accuracy of the predicted peptide RTs. Generally, more accurate predicted RTs allow RT windows of shorter duration.
In all cases it is advantageous to be able to predict the RT with high accuracy; retrospectively in discovery proteomics or prospectively in targeted proteomics. The differences in the RT of peptides for a specific LC-setup, the RT variance, are composed of essentially three main factors: peptide-intrinsic properties, variance in the LC-system, and residual variance. Peptide intrinsic retention (or selectivity) for a defined resin and solvent is specific for each peptide (sequence) and structure [17] and determined by the physicochemical interaction of the peptide, the resin, and the solvent [18]. The setup of the chromatographic system (solvent gradient, column length, dead volumes in the LC system) affects all peptides consistently and has been theoretically described by a Linear Solvent Strength Model [19]. The residual variance is composed of variability in the LC-system, such as effects of varying sample concentrations (resulting in overloading), variations in pump pressure, or changes in the column over continued use.
Currently, two main approaches are used to predict the peptide RTs for a specific set up: preliminary empirical measurement and in silico prediction.
It is in the nature of discovery approaches that novel peptides are identified for which no prior empirical data is available. Here RT has to be predicted de-novo by using empirically derived physicochemical parameters, such as hydrophobicity, which tries to capture the peptide-intrinsic effects [20]. The hydrophobicity is then translated into real RT by a linear fit to a single calibration run to take the LC-setup into account. Several algorithms have been developed [21], where the most widely used RT prediction algorithm is SSRCalc [22].
In targeted proteomics the situation is fundamentally different. Here usually peptides are targeted for which empirical information is available. In principle there is no need for de-novo prediction if the empirical data is available. The complication with using empirical RT information is that the RTs that are empirically measured are only valid for the same specific experiment set up and need to be repeated every time a single parameter is changed. If a large number of targets are to be measured this method can require many sample injections to schedule a single experimental method. It also requires that the targets can be easily identified over the whole gradient. These limitations restrict the direct approach to experiments with very few targets.
Our hypothesis was that once a normalized RT value is empirically determined for a peptide, its measurement could be easily scheduled on virtually any LC system without again determining its RT empirically before an LC-MRM run. Such empirically determined RT values could be used as more accurate predictions of future RTs than prediction based on peptide sequence alone.
Here we present a novel method of RT prediction for targeted proteomics applications, named iRT, which achieves the scale of the in silico approach and the accuracy of an empirical measurement. The iRT score for a peptide can be stored as a single number that is specific and stable for the peptide across a wide range of LC configurations. This score is empirically determined and normalized relative to a set of synthetic standard peptides. We furthermore combined iRT with the recently developed on-the-fly RT recalibration setup [23]. In doing so, we found that the combination can cancel out residual RT variance for increased analytical robustness. Finally, we will release support for the iRT workflow, with a step-by-step tutorial in its use, in version 1.2 of the open source Skyline software (http://proteome.gs.washingteon.edu/software/skyline/) [24].
In summary, we present an open, portable and standardized RT scale, the iRT scale. We show that peptide RT prediction based on iRT increases the throughput, quality, and portability of LC-MRM experiments. We further simplified the adoption of iRT through support in a popular, freely-available software tool.
Materials & Methods
Selection of iRT-peptides and definition of iRT-C18 scale
Peptides were selected from a shotgun LC-MS/MS dataset from Leptospira interrogans. Altogether 30 peptides were selected based on their intensities, absence of amino acids that are prone to modifications (e.g. M, C) and a broad distribution of RTs spanning the whole gradient. In these selected peptides amino acids were conservatively exchanged (e.g. G->A, T->S) and these modified sequences were searched against the complete database of NCBI with Blast to verify that none of the modified sequences is identical to a known natural sequence. Synthetic peptides were ordered from JPT (Berlin, Germany) and MRM assays were developed as described below. For 20 peptides highly purified (purity > 99%) peptides were ordered from JPT and tested under various conditions (heat, freeze thaw cycles, drying down etc.). A final number of 11 peptides (iRT-peptides) were selected to constitute the reference peptides for a new iRT scale (see Table 1 and Figure 1d). The reference peptides were mixed into a yeast total lysate background (approx. 100fmol each in 1 μg total protein) and measured in LC-MRM with a 90 min linear gradient (5%–35% acetonitrile as organic modifier with 0.1% formic acid) on a C18 column (Magic C18 AQ resin 3μm particle size/300Å pore size; Michrom, Leonberg, Germany). RTs were extracted for all 11 peptides and the new scale was defined by setting the iRT value of peptide b to 0 and the iRT value of peptide l to 100 (see also equation 1 in the Results, and Supplementary Table 1). The iRT values of the remaining peptides were calculated by linear regression to these 2 fixpoints. The resulting iRT scale is defined as iRT-C18.
Table 1.
iRT-peptide sequences and corresponding iRTs. Asterisks indicate defined fix-points for the iRT-C18 scale.
| Sequence | Name | iRT |
|---|---|---|
| LGGNEQVTR | RT-pep a | −24.92 |
| GAGSSEPVTGLDAK | RT-pep b | 0.00* |
| VEATFGVDESNAK | RT-pep c | 12.39 |
| YILAGVENSK | RT-pep d | 19.79 |
| TPVISGGPYEYR | RT-pep e | 28.71 |
| TPVITGAPYEYR | RT-pep f | 33.38 |
| DGLDAASYYAPVR | RT-pep g | 42.26 |
| ADVTPADFSEWSK | RT-pep h | 54.62 |
| GTFIIDPGGVIR | RT-pep i | 70.52 |
| GTFIIDPAAVIR | RT-pep k | 87.23 |
| LFLQFGAQGSPFLK | RT-pep l | 100.00* |
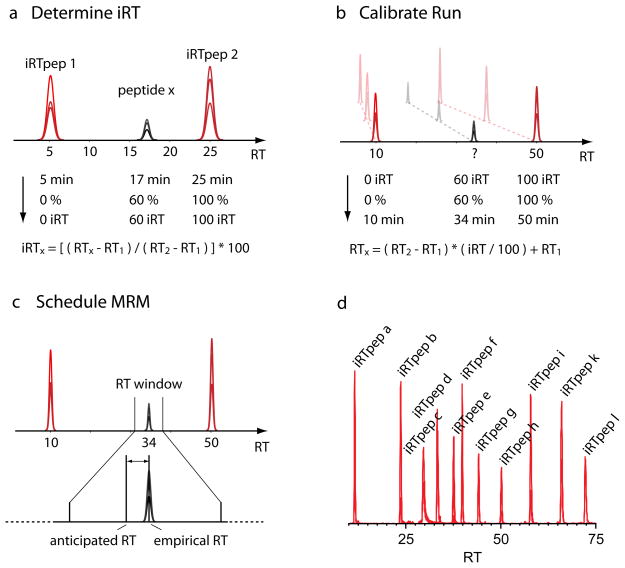
Figure 1.
iRT is a normalized retention time (RT) scale, independent of the chromatographic setup. (a) Schematic illustration of how the iRT is empirically determined for the case of LC-MRM. In practice, 11 reference peptides (iRT-peptides) are used and the transformation is based on a linear fit; for simplicity, in this illustration only two peptides are shown. A target peptide and the iRT-peptides are measured using liquid chromatography coupled to MRM (LC-MRM). The retention time of the target peptide (RTx) is normalized and hereby transformed into iRT using the coordinates of the iRT-peptides. Simplified, the iRT of the target peptide is expressed relative to the RTs of the iRT-peptides. iRT can be stored as a single stable number and transferred to any chromatographic setup using the iRT-peptides. (b) One calibration run is used to convert iRT to a new chromatographic setup. Depicted are three possible chromatographic setups with gradients of different length. (c) In order to increase the number of peptides measured in a single LC-MRM run, the measurement can be scheduled to the time point where the peptide elutes. The window width should be chosen such that the difference between anticipated and empirical RT never exceeds the window size. Hence, the more accurate the anticipated RT is, the smaller the RT window can be chosen. (d) LC-MRM measurement of the 11 iRT-peptides to define the iRT-C18 scale.
Hela cells
Hela cells were grown over 8 passages in High Glucose DMEM [12.43 g/l Dulbecco’s Modified Eagle’s Medium (Caisson Laboratories Inc., North Logan, UT, USA), 4.5 g/l D-(+)-Glucose anhydrous (Fluka, Buchs, Switzerland), 30 mg/l Glycine (Fluka)] supplemented with either light (Sigma-Aldrich) or heavy (13C 15N, Sigma-Aldrich) isotope-labeled lysine and arginine at 37°C and 5% CO2.
Cells were harvested at 80% confluency by trypsinization, washed three times with ice cold PBS (GIBCO (Invitrogen), Paisley, UK) and the cell number was determined using a Neubauer chamber. Hela cells were spun down at 300 ×g and resuspended in one cell pellet volume PBS. Two pellet volumes of 8 M Urea (Sigma-Aldrich, Buchs, Switzerland) containing 50 mM ammonium bicarbonate (Sigma-Aldrich) and 0.1% RapiGest (Waters, Baden, Switzerland) were thoroughly mixed with the resuspended cells. Subsequent to sonication (80% amplitude, 0.6 cycle, 1 min) cell debris was spun down at 16000×g.
The protein concentration of the lysate was measured by BCA assay (bicinchoninic acid, Thermo Scientific, Reinach, Switzerland). Proteins were reduced with 5 mM TCEP (tris(2-carboxyethyl)phosphine, Thermo Scientific) at 37°C for 15 min and alkylated with 10 mM iodoacetamide (Sigma-Aldrich) for 30 min in the dark. Proteins were first digested with lysyl endopeptidase (Wako Chemicals, Neuss, Germany) at an enzyme – substrate ratio of 1 to 50 (w/w) at 35°C for 2 hours. After dilution with 50 mM ammonium bicarbonate to 0.8M urea trypsin (Promega) was added at the same ratio. Tryptic digestion was carried out overnight at 37 °C. Peptides were acidified with 1% trifluoroacetic acid (TFA, Thermo Scientific) and purified by solid-phase extraction using C18 cartridges (Sep-Pak, Waters). The SPE eluate was evaporated to dryness and reconstituted in 3% acetonitrile (Thermo Scientific) and 0.2% formic acid (Sigma-Aldrich).
Sample generation for quantification experiment
Isotopically light and heavy SILAC [25] cell lysate digests were mixed at 1:1 and 5:1 ratios. The theoretical ratio between the two heavy-to-light ratios of the 1:1 and the 5:1 mixture equals five regardless the actual concentration of the individual peptides.
Selection of targets and setup of transition lists
Preselection of target peptides was carried out from a list of >500 peptides verified in Hela cells from the Biognosys repository of MRM assays (www.mrmbase.com). A set of 148 peptides (150 precursors), all with low SILAC background labeling (<10%), were selected for all scheduled MRM experiments.
Transitions for the target peptides were obtained from the Biognosys repository, including four light and four corresponding heavy transitions for each peptide where available (y- and b-ions as fragments) and assembled in a list containing 1232 transitions. Two precursors were included with two charge states (2+/3+). All samples were spiked with the iRT-C18 reference peptides (iRT-peptides), a mixture of eleven well-characterized synthetic peptides (RT-Kit; Biognosys, Schlieren, Switzerland). Three transitions to monitor each of the reference peptides (33 transitions) were included in the transition list.
LC-MRM
All experiments were carried out on a TSQ Vantage triple quadrupole mass spectrometer equipped with a nanoelectrospray source (Thermo Scientific, Reinach, Switzerland) coupled to an EASY-nLC nanoflow HPLC system (Thermo Scientific). Samples were separated on a nanoLC column prepared by packing a PicoFrit emitter (360μm outer diameter, 75μm inner diameter, and 10μm tip; New Objective; Woburn MA, USA) with 10cm of Magic C18 AQ resin (3μm particle size and 300Å pore size; Michrom; Leonberg, Germany). Gradient elution was performed using 1% acetonitrile/0.1% formic acid in water as solvent A and 97% acetonitrile/0.1% formic acid in water as solvent B according to the following program for iRT determination: 0–30min = 5–35%B, 30–32min = 35–100%B and 32–40min = 100%B followed by reequilibration to 5% B. Experiments for comparisons of SSCRCalc and iRT predictions were carried out using the same setup with a linear gradient of 0–90 minutes to 40%B. The flow rate was 300nl/min. The LC eluent was electrosprayed using an ionization voltage of 19kV. Q1 and Q3 resolutions were 0.7 Da and the cycle time was fixed to 2.5s. Collision energies were calculated by a linear regression specific to the TSQ Vantage instrument according to the manufacturer’s instructions[26]. Overall cycle time was 2.5s.
Scheduled MRM
iRT-peptides were used for a linear regression of RT by SSRCalc hydrophobicity and iRT respectively to calculate a linear predictor of RT. RTs of iRT-peptides were determined for the current chromatographic setup by measurement in unscheduled MRM-mode. For SSRCalc-based prediction, the hydrophobicity of each peptide was calculated by the SSRCalc algorithm [22] using the web-based interface at http://hs2.proteome.ca/SSRCalc/SSRCalcX.html, version 3.X for 100Å C18 column and 0.1%FA. This version turned out to predict RTs more accurately for our LC system than the version for 300 Å C18 columns and 0.1% TFA (data not shown). RTs for target peptides were predicted according to the following equation obtained from the regression of RT by hydrophicity of the eleven iRT-peptides: RT = 1.34*SSRCalc hydrophobicity-1.46 (R2=0.95). For iRT-based prediction, a similar linear regression of RT by iRT values of the eleven iRT-peptides produced the following equation used to predict RTs for the target peptides: RT = 0.38*iRT + 23.07 (R=0.998). Time windows for each peptide were set to 6min in a 90 minutes gradient. For quantification experiments time windows were set to 2, 4, 6, and 8 minutes respectively and both SILAC heavy/light 1:1 and heavy/light 5:1 sample mixtures were measured using all window sizes. Incomplete labeling of SILAC heavy cells was assessed by measurement of all peptides in the heavy SILAC cells (2min RT window) and all calculations for quantification experiment were corrected for this factor.
On-the-fly RT calibration
iRT-peptides were used as reference peptides for on-the-fly RT calibration. During the chromatographic run, RTs of the reference peptides are captured and a linear fit is used to assess the RT shifts occurring in the respective run. All time windows of the following peptides are subsequently adjusted so that target peptides are not missed due to minor changes in LC performance. It is important to know that at any given time, the instrument uses only the two last reference peptides to calculate the linear fit and ignores all preceding references. For example, a target peptide eluting at time 2.5 will have its RT window adjusted according to a linear fit through the RTs of reference peptides a and b. As soon as reference peptide c elutes, all windows for target peptides with higher RTs will be adjusted according to a linear fit through reference peptides b and c.
All light transitions from quantification experiments were monitored resulting in a list containing 632 peptides. Time windows for target peptides were set to 3min and to 6min for all reference peptides as well as for early-eluting peptides (before the third iRT-peptide) to compensate for the less stable calibration at early RTs (one-point calibration using only the first reference peptide). Xcalibur version 2.1.0) was run in iSRM mode and trigger values were set to 103 for reference peptides and to 107 for all target peptides as primary transitions so that no reference peptide was missed and target peptides were monitored constantly over the time of their adjusted RT window. 0.5μg, 1μg, and 2μg total protein were injected in triplicates.
Data analysis
Data analysis was carried out using mQuest, the scoring part of mProphet [27]. Data were manually checked and light/heavy ratios were calculated using apexsum values with removed outliers.
Plotting and statistics
All plots were generated and all statistical tests were carried out using the R package for statistics version 2.13.0 [28]. Boxplots were generated using the standard function with no further options. F-tests were performed to compare variances with confidence intervals of 0.95.
Skyline implementation
A complete workflow supporting the calculation of iRT values and their use in scheduling exported targeted methods was implemented in the Skyline software tool, as shown in Supplementary Figure 1. A single button click allows measured RTs to be converted to iRT values and stored in a SQLite file (iRT database). These files can be easily shared between labs and investigators using Skyline. Similarly, a new linear predictor for RT by iRT can be trained with a single click from a MRM run imported into Skyline. Finally, a scheduled method may be exported through the normal interface using the linear equation and stored iRT values.
Results
Definition of a dimensionless empirical measure for retention time
The goal of these experiments was to assess the viability of defining a universal prediction of RT for targeted proteomics applications based on empirical data. This requires previous observation of a peptide on an equivalent chromatographic system, where equivalence can be broadly defined as conservation of order of elution for a set of target peptides.
We refer to this general concept as iRT. The peptide specific value is the peptide-iRT and the concrete implementation of the scale is based on a concrete set of peptides. Several sets of standard peptides have been described [21, 29] and any of them can be used to define an iRT scale. We nonetheless developed a novel set that fulfills several criteria that we deem to be highly desirable for a standard set: The peptides are not occurring naturally, they are stable under various storage conditions, approximately balanced in their intensity, span a wide range of hydrophobicity, and are readily available as a premixed kit. Additionally we use more peptides (11) than most of the described sets. The optimum number was estimated based on a bootstrap simulation (see Supplementary Figure 4), where accuracy of prediction was measured as function of the number of peptides.
Based on the defined set of 11 peptides that is described here (iRT-peptides, see Methods, Figure 1d and Table 1) is referred to as iRT-C18 because it has been validated for C18 based chromatographic setups with acetonitrile as organic modifier (see Supplementary Figure 2). For simplicity we refer to the values in this iRT-C18 scale as iRT values. The use of different columns, ion-pairing agents or organic modifiers can affect the elution order of peptides [30, 31]. If different types of resins (e.g. C4) or organic modifiers (e.g. methanol) would be used the iRT-C18 scale could be replaced by another resin- or organic modifier-specific scale if the retention behavior is not linear to the iRT-C18 scale. Given this assignment, the formula for the one time determination of iRT for the 11 reference iRT-peptides on a chromatographic system with a linear gradient was:
| (1) |
The result is the fixed iRTs for 11 standard iRT-peptides given in Table 1.
The formula to determine an iRTx for a novel peptide based on the 11 iRT-peptides and a chromatographic system with a linear gradient is:
| (2) |
where m is the slope and n is the y-intercept from the linear regression of iRT by RT, using values for the reference peptides. The formula to predict RT based on the 11 iRT-peptides on a chromatographic system with a linear gradient is:
| (3) |
where m is the slope and n is the y-intercept from the linear regression of RT by iRT, using values for the reference peptides. Using the following approach, any empirical peptide RT can be converted into iRT: iRT-peptides and the peptides of interest are measured in an unscheduled LC-MRM run or in LC-MS/MS mode using a linear gradient. Linear regression of the iRT values for the 11 iRT-peptides (see Table 1) by their measured RTs is used to calculate m and n in equation 2. If the iRT for a peptide is known it can be scheduled on the current gradient using equation 3 by measuring the 11 iRT-peptides on the current chromatographic setup (see Figure 1). Linear regression of the measured RT values for the 11 iRT-peptides by their iRT values (see Table 1) is used to calculate m and n in equation 3. The ability to derive iRT values and schedule peptides based on iRT is now also implemented in Skyline (see Supplementary Figure 1). The iRT values calculated in Skyline are stored in a file which can be shared easily across experiments and labs. Supplementary Figure 2 shows the linearity iRT-peptide RTs across different mass spectrometers, LC systems and LC gradients. Alternatively, a web based tool for conversion of RT into iRT-C18 scale and vice versa is available at http://www.mrmbase.com/iRT.
If the chromatographic system is not set up with a linear gradient then the fit has to be chosen accordingly or the fit can be approximated with a number of linear fits in between neighboring standard iRT-peptides.
The determination of iRT can be performed for many peptides in a single run with virtually all mass spectrometric methods employing liquid chromatography. To demonstrate this we calculated the iRTs of 94 abundant peptides from 43 proteins measured with LC-MS/MS in human breast cancer tissue sample (data not shown). These peptides can serve as landmarks e.g. to monitor housekeeping proteins or to derive iRTs using equation 3 without using the 11 standard peptides (Supplementary Table 2). It should be noted, however, that these iRTs are based on MS/MS scan times, and are therefore less accurate than iRTs based on fully measured chromatographic peaks.
Comparison of prediction accuracy iRT vs. SSRCalc and quantification accuracy depending on window size
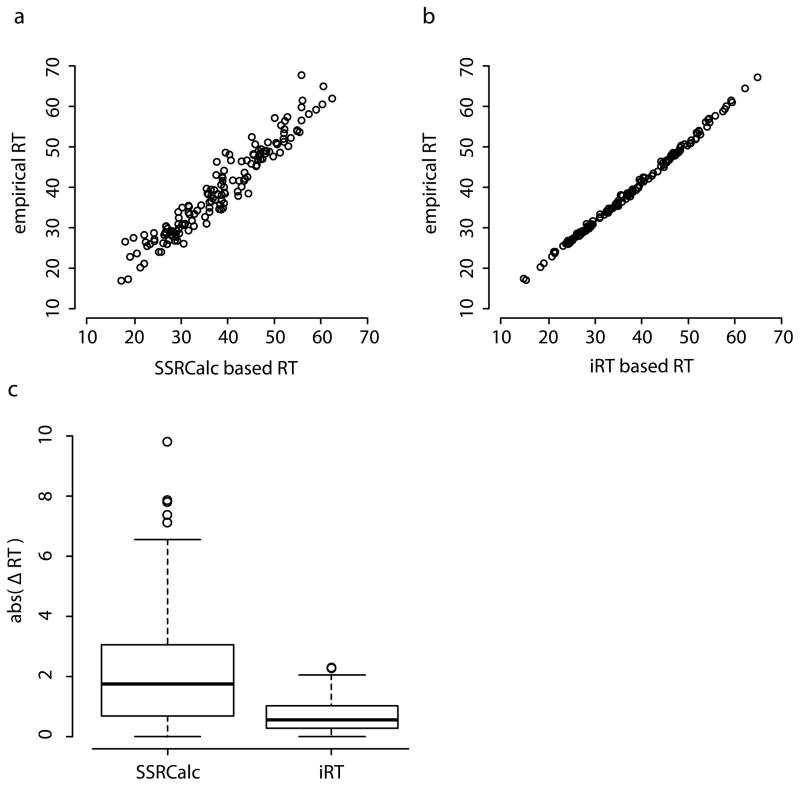
We selected 148 peptides (150 precursors) which were, in previous experiments, visible in a total cell lysate at various intensities and which were labeled close to completeness in SILAC. For these peptides, assays were developed as described in [27, 32] and stored in MRMbase (http://www.mrmbase.com/). The assays were used to calculate iRTs for each peptide and set up a scheduled transition list (assay panel) to measure all peptides in a single injection. We recorded the relationship between SSRCalc- and iRT-based predictions by measuring the endogenous peptides in a total cell lysate. Prediction of the RTs using the linear equation for iRT determined on a 30 min gradient from 0–35%B resulted in successful measurement of all 148 target peptides when using acquisition time windows of two minutes for a 30min 0–35%B gradient and six minutes for a 90min 0–40%B gradient. Correlation between the RT predicted with the SSRCalc linear equation and measured RT was 0.9246 (average deviation was 2.4min), and correlation between the RT predicted with the iRT linear equation and the actual measured RT was 0.9970 (average deviation was 0.8min; see Figure 2a, b). Even though the prediction using SSRCalc correlates strongly with measured RTs the overall variation of that approach was more than four times larger than the iRT approach. This variation effectively defines the minimum window size of 5.1min for the iRT based prediction and 21.9min for the in silico prediction for a 90min gradient (see Figure 2c). 75% percentiles were 3.4min for the in silico approach and 1.1min for iRT-based predictions.
Figure 2.
Accuracy of iRT and SSRCalc based retention time (RT) prediction based on the measurement of 148 peptides in a 90min linear gradient. (a) SSRCalc predicts the measured (empirical) RT with a good correlation of 0.92. (b) iRT values determined in an independent experiment in a 30min linear gradient predict RTs with a very high correlation (0.997) and low variance. (c) The absolute difference between empirical and anticipated retention time abs (ΔRT) is shown for predictions based on iRT and SSRCalc. iRT predictions are more accurate which allows choosing shorter RT windows without missing any targeted protein.
The results show that RT prediction using empirical data instead of a calculated parameter provides significantly better accuracy for targeted proteomics applications where target peptides have been observed before.
It is desirable to keep the window size as small as possible, because a smaller window allows the mass spectrometer to measure fewer transitions in each cycle. With fewer transitions per cycle, the dwell time for individual peptide increases and it can be expected that this produces an increase in signal-to-noise ratio.
We tested practical relevance of this relationship by measuring the same set of 150 precursors using window sizes of 2, 4, 6, and 8 minutes. The experiment was set up as a mix of light and heavy labeled cell lysates at a 1:5 ratio. This artificially introduced ratio served to simulate a 5x regulation. With this known ratio we tested the accuracy of quantification as a function of window size.
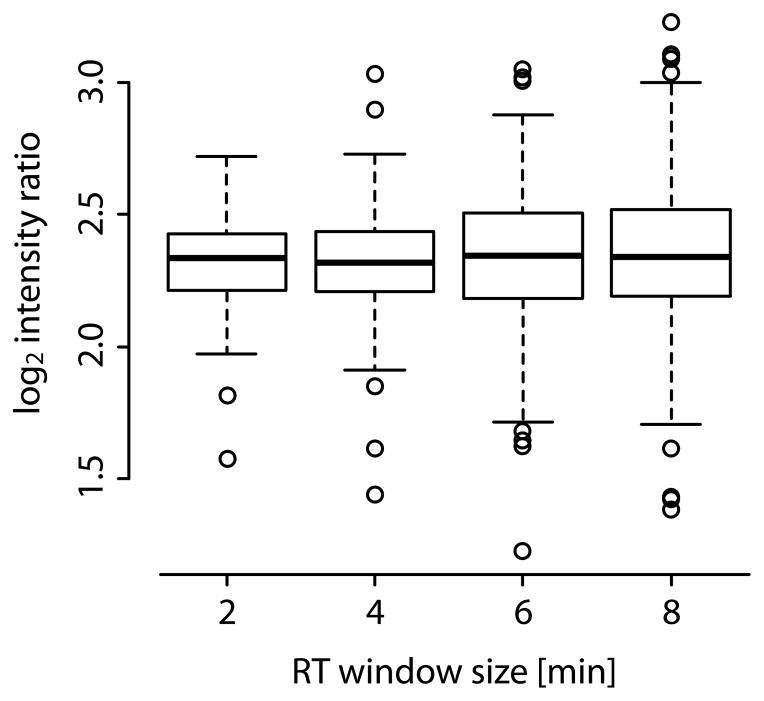
The mean values for the quantification of the 150 targeted precursors did not differ strongly from the predicted value of 5 between window sizes (from 5.02–5.19), but the mean variation increased from 12% CV to 23% CV with increasing window size (see Figure 3, Supplementary Figure 2 and Table 2).
Figure 3.
Quantification accuracy of 150 precursors (148 peptides) as a function of the retention time (RT) window size when using scheduled MRM. A log2 intensity ratio of 2.32 (5-fold difference) was expected based on the sample preparation. Generally, quantification was most accurate when using the smallest retention time windows of two minutes.
Table 2.
Quantification accuracy parameters for different RT window sizes in scheduled MRM. The theoretical (mean) value is 5 for all peptides.
| Window size | 2min | 4min | 6min | 8min |
|---|---|---|---|---|
| Median | 5.04 | 4.99 | 5.08 | 5.06 |
| Mean | 5.02 | 5.04 | 5.12 | 5.19 |
| Min | 2.97 | 2.70 | 2.33 | 2.60 |
| Max | 6.60 | 8.19 | 8.28 | 9.39 |
| CV (%) | 12 | 14 | 19 | 23 |
| F-test p value* | - | 0.04 | 1.7E-8 | 3.2E-13 |
F-test p value for variances between 2min window size and the respective window size.
This shows that accuracy of quantification can be significantly increased by a better prediction of RTs for a scheduled LC-MRM experiment (F-test p-value for 2min windows and 4min windows was 0.04).
On-the-fly calibration
We have shown that iRT provides highly accurate predictions, but the precision (i.e. the actual RT deviation) is also affected by the variance that occurs as systematic shifts from run to run. This variation is mainly introduced by differences in sample concentration, non-reproducible loading, or shifts introduced by the chromatographic system. This technical variance cannot be captured by any pre-run RT prediction and therefore affects the effective window size that has to be chosen in order to avoid losing data. On-the-fly RT calibration as recently introduced by Thermo Fisher [23] attempts to reduce this technical variance. Because this method also uses reference peptides to predict additional variance during acquisition, we tested it in combination with iRT to determine whether it allows shorter duration RT windows than iRT alone.
We tested this hypothesis by measuring a series of samples using the same assay panel as described above. Samples were repeatedly injected with varying concentrations, which is known to lead to shifts in RT.
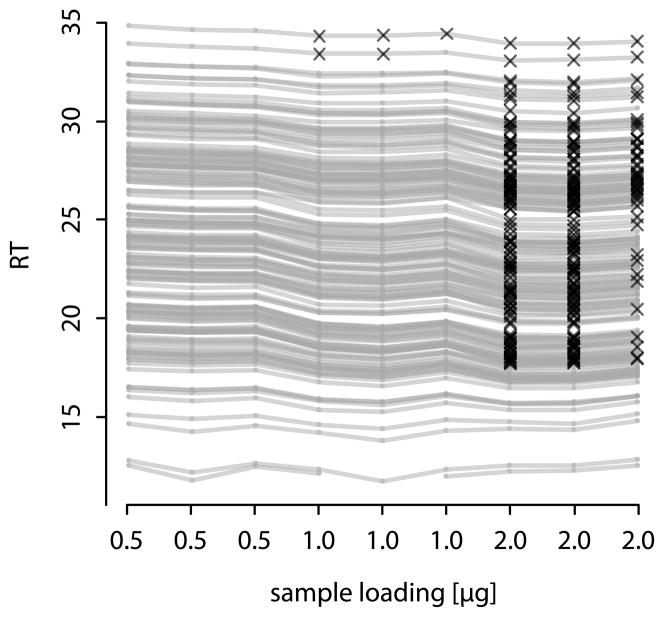
All peptides (except the earliest-eluting peptide in run 2 with 1μg loading) in the dataset were successfully detected using the on-the-fly RT calibration setup using iRT-peptides as references when setting RT windows to three minutes for target peptides and to six minutes for references as well as early-eluting peptides (eluting before the third iRT-peptide at RT approx. 18.5 when loading 0.5μg total protein). Figure 4 shows the RT shifts that are introduced by higher amounts of protein on the column where nearly all peptides elute at an earlier RT when more sample is loaded. Using small RT windows as in the on-the-fly-corrected setup, more than 80% of all target peptides would be missed when loading 2μg sample and not correcting for those RT shifts.
Figure 4.
RT shifts induced by different amounts of protein loaded on the column can be compensated using on-the-fly RT calibration. Grey lines represent peptide retention times when varying the sample load on the column. Black crosses indicate peptides that would have been missed without using on-the-fly RT calibration. With 2μg protein loaded on the column 111, 122, and 49 peptides would elute outside the retention time window and hence be missed in the three technical replicates. The earliest eluting peptide was missed once even using on-the-fly RT calibration in the second run of the 1μg load.
This experiment furthermore demonstrates higher accuracy of iRT based RT prediction compared to direct empirical prediction e.g. by using RTs measured in the first injection. We calculated iRT values for each peptide by equation (3) based on the RTs of the iRT-peptides in the same run (empirical iRT). While the measured RT in subsequent runs can vary based on the loading and other factors the empirical iRT remains almost stable. The average CV of the RT across 9 runs was 2.5% while the CV of the empirical iRT was 0.6%. In principle an iRT based on-the-fly calibration using each peptide as a reference could be implemented into the instrument software to further improve scheduling.
Discussion
Scheduled MRM [33] has been shown to largely solve the limitations of MRM in the number of targets that can be measured in a single run. It is based on the knowledge when a specific peptide will elute from the column in a LC-MRM experiment. The acquisition happens only in a defined window around this anticipated elution time. In general, the smaller the window the more different peptides can be targeted without compromising data quality. However, if the windows are chosen too small peptides may elute outside of the window, resulting in inconclusive data.
The minimum window size that can be used for practical reasons depends therefore strongly on the accuracy of the RT prediction. Currently one has to determine the RTs manually by prior measurements before each experiment and then use these measured values as predictions for the following analytical runs. With this simple approach one can in principle predict with high accuracy the RTs of specific peptides for a specific set-up. However, the prior measurements need to be performed with much lower multiplexing. In the case of our test-set with 1232 transitions, which we measure in a single run, in order to achieve similar dwell-times in un-scheduled mode one would need to perform multiple runs only to define the RTs. If the set-up would be changed, e.g. after a clogged column, the whole procedure would need to be repeated. In practice this makes high multiplexing unfeasible and can be a reason that the full potential of MRM workflows is not exploited.
We have shown that iRT, a normalized RT derived from an empirical measurement and a set of standard peptides, combines the advantages of both, the empirical and the in-silico method, and can be used for highly precise prediction of RTs for large-scale scheduled LC-MRM measurements with a single calibration run before the analytical runs.
RT prediction used on prior measurement of standard peptides has been described before [34]. However, such normalization runs are always relative to the standard set and cannot be disconnected from the underlying data set. This is a major obstacle for transferring the RT across labs and different setups. We have defined the iRT-C18 scale to relate an experimental RT to a fixed gold standard of defined landmark peptides for C18 based resins. This reduces the required information to a single number that is a fixed dimensionless attribute of a peptide sequence, which serves as a parameter to accurately predict RT without any context to the original measurement for any system with compatible LC conditions and where the RT of the reference peptides can be measured.
We have shown that the linearity assumption is valid across different LC-systems. While the use of other mobile phases can lead to changes in the elution order, which would require the definition of a new scale [31] the linearity is largely conserved for different resin types typically used in proteomics experiments [35]. The iRT concept shows some similarity to the SSRCalc approach [36]. Both systems are based on a determination of the RTs of some reference peptides in order to derive the linear equation that is necessary to describe the current chromatographic setup (n and m in equations 2–3). SSRCalc predicts the relative RT of a peptide from physicochemical properties using a model based on empirical measurements that is applicable to all sequences [20, 36, 37]. The iRT of a peptide is derived from a direct empirical measurement of a specific peptide. It is therefore subject to less variance and predicts the RT more accurately. In fact iRT can almost capture all sources of variance: The peptide-intrinsic retention behavior is described by a fully empirical score, and variance introduced by the specific LC-setup is covered by a prior calibration run with the iRT-peptides (as it is the case for in silico methods). The in silico approach also predicts RTs with high correlation (0.95) between measured and predicted RTs. However, the variance is much higher. For accurate non-empirical relative RT prediction of peptides that are potentially modified, a very detailed model would have to be used that takes into account the dynamic molecular structure of the peptide, dynamic behavior of the stationary phase, and how the solvation shell changes depending on the percentage acetonitrile and ions in the mobile phase[38], which is beyond the capabilities of most mass spectrometric labs.
iRT is an open concept that could be used with any combination of reference peptides given that they elute over a broad enough range. Nonetheless we provide here a fixed set of iRT-peptides with a scale that is applicable to all reversed-phase C18-resin based chromatography systems. We recommend using this scale for future publications of MRM-assay and generally spectral libraries. The sequences of the underlying iRT-peptides have been carefully chosen. They are disclosed and can be synthesized or are commercially available as a ready-to-use kit. iRTs in the newly defined scale could also be derived independently of the 11 iRT-peptides using publicly available peptide iRTs such as our published list of 94 peptides in Supplementary Table 2. The iRT concept and the specific iRT-C18 scale have been implemented in Skyline [24], the most widely used MRM software with support for instruments from all major triple quadrupole vendors. The openness of the iRT concept allows for the translation of any standard peptide based scale into iRT-C18 or vice versa.
Transferability of data and methods between laboratories is required in large-scale biomarker studies as well as in the preclinical and clinical validation phase, where multisite robustness is a prerequisite [4]. The consistent use of an iRT scale will enable the rapid and simple transfer of MRM assays from machine to machine as well as from laboratory to laboratory. This opens the possibility to assemble any given combination of target peptides with iRT to pathway- or disease-specific panels of scheduled MRM assays with a single calibration run.
In many biomarker studies, observed fold-changes for candidate or verified markers are in the range of 2–5 [39–41]. Accurate quantification of peptides in MRM experiments is therefore required in targeted proteomics-based pipelines for biomarker verification [6, 39] or in the characterization of protein interaction networks in signaling pathways [42]. When measuring our standard set of 150 precursors at a defined ratio of 5:1, the coefficient of variance was markedly reduced with smaller RT window size.
We showed that the accuracy of RT prediction can be further increased by correcting for the residual variance in the LC-setup by combining it with on-the-fly calibration [23]. In our set of 148 peptides, only one peptide was missed using on-the-fly RT calibration with small windows over nine runs using three different amounts of total protein while without this feature up to 80% would have been missed under the same conditions. The missed peptide was the first eluting peptide after the first reference peptide which indicates that on-the-fly calibration using only one peptide as offset may not be very stable for all LC setups. It is therefore recommended to increase window size for early eluting peptides before the on-the-fly calibration is effective. iRT combined with on-the-fly calibration of RT shifts provide a valuable and unique setup for the robust and accurate measurement of large sets of target peptides in a single scheduled LC-MRM run.
We have applied the iRT concept here in the context of targeted proteomics but it could be equally useful outside LC-MRM applications. The iRT-C18 value is a stable peptide property for common LC-systems. The value of large scale spectral libraries [43, 44] would be considerably increased if each peptide in these libraries would be assigned its iRT value to accurately predict RTs across laboratories and chromatographic set-ups.
Supplementary Material
Supplementary Figure 1: Screenshot Skyline of the iRT user interface showing the same data as Figure 2.
Supplementary Figure 2: Comparison of linearity of iRT-peptides across LC systems, mass spectrometers and chromatographic gradients. Upper left: Eksigent nanoLC ultra 2D plus, ABI/Sciex QTRAP5500, 30min gradient, R2=0.999; Upper right: Eksigent nanoLC ultra 2D plus/Thermo Scientific LTQ Orbitrap XL, 60min gradient, R2=0.996; Lower left: Proxeon easy-nLC/Thermo Scientific FT Ultra, 40min gradient, R2=0.995; Lower right: Proxeon easy-nLC/Thermo Scientific TSQ Vantage, 30min gradient R2=0.999.
Supplementary Figure 3: Variation in quantification accuracy increases strongly with acquisition window size. The expected log2 intensity ratio was 2.32 (5-fold regulation) based on sample preparation.
Supplementary Figure 4: Average absolute retention time deviation (measured – predicted) for 148 peptides as a function of the number of reference peptides used to determine the linear transformation (bootstrap). Prediction accuracy increases with the number of reference peptides.
Supplementary Table 1: Transition list, sequences and iRTs for iRT peptides
Supplementary Table 2: iRTs of selected potential human housekeeping proteins
Acknowledgments
LR, CE, RO, and OR thank Alexander Schmidt for help in selecting the iRT-peptides and acknowledge Alexander Leitner for support in mass spectrometry. BM, JC, and MM were supported by a sponsored research agreement from ThermoFisher and NIH grant P41 RR011823.
Abbreviations
- RT
Retention time
Footnotes
Competing Interests
Authors LR, CE, RO, and OR are employees of Biognosys AG, Switzerland. This work was partly funded by Biognosys AG. Biognosys sells the iRT-peptides as a product (RT-Kit).
References
- 1.Domon B, Aebersold R. Nature biotechnology. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 2.Beck M, Schmidt A, Malmstroem J, Claassen M, et al. Molecular systems biology. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagaraj N, Wisniewski JR, Geiger T, Cox J, et al. Molecular systems biology. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addona TA, Abbatiello SE, Schilling B, Skates SJ, et al. Nat Biotech. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jovanovic M, Reiter L, Picotti P, Lange V, et al. Nature methods. 2010;7:837–842. doi: 10.1038/nmeth.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whiteaker JR, Lin C, Kennedy J, Hou L, et al. Nat Biotech. 2011;29:625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuster B, Schirle M, Mallick P, Aebersold R. Nature reviews Molecular cell biology. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 8.Lange V, Picotti P, Domon B, Aebersold R. Molecular systems biology. 2008;4 doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson L, Hunter CL. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 12.Petritis K, Kangas LJ, Ferguson PL, Anderson GA, et al. Analytical chemistry. 2003;75:1039–1048. doi: 10.1021/ac0205154. [DOI] [PubMed] [Google Scholar]
- 13.Shinoda K, Tomita M, Ishihama Y. Bioinformatics (Oxford, England) 2008;24:1590–1595. doi: 10.1093/bioinformatics/btn240. [DOI] [PubMed] [Google Scholar]
- 14.Palmblad M, Ramstrom M, Markides KE, Hakansson P, Bergquist J. Analytical chemistry. 2002;74:5826–5830. doi: 10.1021/ac0256890. [DOI] [PubMed] [Google Scholar]
- 15.Strittmatter EF, Ferguson PL, Tang K, Smith RD. Journal of the American Society for Mass Spectrometry. 2003;14:980–991. doi: 10.1016/S1044-0305(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 16.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, et al. Molecular & cellular proteomics: MCP. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Mant CT, Hodges RS. J Chromatogr A. 2003;1010:45–61. doi: 10.1016/s0021-9673(03)00877-x. [DOI] [PubMed] [Google Scholar]
- 18.Shibue M, Mant CT, Hodges RS. J Chromatogr A. 2005;1080:58–67. doi: 10.1016/j.chroma.2005.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi T. Analytical and Bioanalytical Chemistry. 2007;389:1659–1660. [Google Scholar]
- 20.Guo D, Mant CT, Taneja AK, Parker RJM, Rodges RS. Journal of Chromatography A. 1986;359:499–518. [Google Scholar]
- 21.Gorshkov AV, Tarasova IA, Evreinov VV, Savitski MM, et al. Analytical chemistry. 2006;78:7770–7777. doi: 10.1021/ac060913x. [DOI] [PubMed] [Google Scholar]
- 22.Krokhin OV, Craig R, Spicer V, Ens W, et al. Mol Cell Proteomics. 2004;3:908–919. doi: 10.1074/mcp.M400031-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Kiyonami R, Schoen A, Zabrouskov V. Thermo Fisher Scientific. 2010 Application Note 503. [Google Scholar]
- 24.MacLean B, Tomazela DM, Shulman N, Chambers M, et al. Bioinformatics (Oxford, England) 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Maclean B, Tomazela DM, Abbatiello SE, Zhang S, et al. Analytical chemistry. 2010;82:10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiter L, Rinner O, Picotti P, Hüttenhain R, et al. Nat Meth. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 28.R development core team. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- 29.Krokhin OV, Spicer V. Analytical chemistry. 2009;81:9522–9530. doi: 10.1021/ac9016693. [DOI] [PubMed] [Google Scholar]
- 30.Gilar M, Xie H, Jaworski A. Analytical chemistry. 2010;82:265–275. doi: 10.1021/ac901931c. [DOI] [PubMed] [Google Scholar]
- 31.Riddle L, Guiochon G. Chromatographia. 2006;64:1–7. [Google Scholar]
- 32.Picotti P, Rinner O, Stallmach R, Dautel F, et al. Nat Meth. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 33.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, et al. Molecular & Cellular Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Mirzaei H, Brusniak MY, Mueller LN, Letarte S, et al. Molecular & Cellular Proteomics. 2009;8:1934–1946. doi: 10.1074/mcp.M800569-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarasova IA, Guryca V, Pridatchenko ML, Gorshkov AV, et al. Journal of chromatography. 2009;877:433–440. doi: 10.1016/j.jchromb.2008.12.047. [DOI] [PubMed] [Google Scholar]
- 36.Krokhin OV, Craig R, Spicer V, Ens W, et al. Mol Cell Proteomics. 2004;3:908–919. doi: 10.1074/mcp.M400031-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Browne CA, Bennett HP, Solomon S. Analytical biochemistry. 1982;124:201–208. doi: 10.1016/0003-2697(82)90238-x. [DOI] [PubMed] [Google Scholar]
- 38.Oda A, Kobayashi K, Takahashi O. Journal of chromatography. 2011;879:3337–3343. doi: 10.1016/j.jchromb.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Addona TA, Shi X, Keshishian H, Mani DR, et al. Nat Biotech. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kälin M, Cima I, Schiess R, Fankhauser N, et al. European Urology. 2011 doi: 10.1016/j.eururo.2011.06.038. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 41.Hyung S-W, Lee MY, Yu J-H, Shin B, et al. Molecular & Cellular Proteomics. 2011 [Google Scholar]
- 42.Bisson N, James DA, Ivosev G, Tate SA, et al. Nat Biotech. 2011;29:653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- 43.Deutsch EW, Lam H, Aebersold R. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picotti P, Lam H, Campbell D, Deutsch EW, et al. Nat Methods. 2008;5:913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Screenshot Skyline of the iRT user interface showing the same data as Figure 2.
Supplementary Figure 2: Comparison of linearity of iRT-peptides across LC systems, mass spectrometers and chromatographic gradients. Upper left: Eksigent nanoLC ultra 2D plus, ABI/Sciex QTRAP5500, 30min gradient, R2=0.999; Upper right: Eksigent nanoLC ultra 2D plus/Thermo Scientific LTQ Orbitrap XL, 60min gradient, R2=0.996; Lower left: Proxeon easy-nLC/Thermo Scientific FT Ultra, 40min gradient, R2=0.995; Lower right: Proxeon easy-nLC/Thermo Scientific TSQ Vantage, 30min gradient R2=0.999.
Supplementary Figure 3: Variation in quantification accuracy increases strongly with acquisition window size. The expected log2 intensity ratio was 2.32 (5-fold regulation) based on sample preparation.
Supplementary Figure 4: Average absolute retention time deviation (measured – predicted) for 148 peptides as a function of the number of reference peptides used to determine the linear transformation (bootstrap). Prediction accuracy increases with the number of reference peptides.
Supplementary Table 1: Transition list, sequences and iRTs for iRT peptides
Supplementary Table 2: iRTs of selected potential human housekeeping proteins