Abstract
Resveratrol (trans-3,4′,5-trihydroxystilbene) is a natural antioxidant with cardiovascular and cancer preventive properties, that is currently at the stage of pre-clinical studies for human cancer prevention. Beside its known effects on protein coding genes, one possible mechanism for resveratrol protective activities is by modulating the levels of non-coding RNAs. Here, we analyzed the effects of resveratrol on microRNA populations in human SW480 colon cancer cells. We establish that resveratrol treatment decreases the levels of several oncogenic microRNAs targeting genes encoding Dicer1, a cytoplasmic RNase III producing mature microRNAs from their immediate precursors, tumor suppressor factors such as PDCD4 or PTEN, as well as key effectors of the TGFβ signaling pathway, while increasing the levels of miR-663, a tumor-suppressor microRNA targeting TGFβ1 transcripts. We also show that, while upregulating several components of the TGFβ signaling pathway such as TGFβ receptors type I (TGFβR1) and type II (TGFβR2), resveratrol decreases the transcriptional activity of SMADs, the main effectors of the canonical TGFβ pathway. Our results establish that protective properties of resveratrol may arise at least in part from its capability to modify the composition of microRNA populations in cells, and suggest that the manipulation of the levels of key microRNAs, such as miR-663, may help to potentiate the anti-cancer and anti-metastatic effects of resveratrol.
Keywords: Colon cancer, microRNAs, miR-663, resveratrol, SW480 cells, TGFβ
1. Introduction
Colorectal cancer (CRC) is the third most common malignancy and the fourth biggest cause of cancer mortality worldwide [1, 2]. Despite the increased use of screening strategies such as fetal occult blood testing, sigmoidoscopy and colonoscopy, more than one third of patients with colorectal cancer will ultimately develop metastatic disease [2]. It is now widely accepted that genetic factors play key roles in the predisposition to colorectal cancer development and progression. Thus, more than 11% of patients with colorectal cancer have at least one first degree relative with the same disease [3], however twin studies suggest that roughly 35% of CRCs are inherited [4]. Understanding the genes and pathways that cause CRC are thus of greatest interest, for they will allow to improve both the diagnoses and prognoses of the disease as well as to optimize its prevention and treatment. The genetic basis of familial CRC has been actively analyzed in the last decades, and the various CRC sub-types linked to several gene mutations responsible for initiating them [5]. For example, Lynch syndrome, formerly known as hereditary non-polyposis CRC, is characterized by rapid progression of colorectal tumors, related to germline mutation of one of the DNA mismatch repair (MMR) genes: MLH1, MSH2, MSH6 or PMS2 [6].
In addition, germline mutations in APC, a gene encoding a gatekeeper that acts as a negative regulator of the WNT signaling pathway, were initially found in Familial Adenomatous Polyposis (FAP), a syndrome that accounts for less than 1% of CRC in the United States, but then somatic mutations of APC have been found in the great majority of sporadic CRCs [6]. Indeed, activating mutations of the canonical WNT signaling pathway, which signals through the nuclear relocalization of β-CATENIN, have been found in more than 90% of CRCs [7]. Once in the nucleus, β-CATENIN functions as a cofactor for transcription factors of the T-cell factor/lymphoid enhancing factor (TCF/LEF) family, which regulate the transcription of genes mainly determining cell fate and regulating cell proliferation [7].
On the other hand, the TGFβ signaling pathway is involved in a number of biological processes, including cell proliferation, differentiation, migration and apoptosis [8]. It is one of the most commonly altered cellular signaling pathways in human cancers [9]. TGFβ signaling is initiated by the binding of TGFβ ligands to type II receptors (TGFβR2). Three TGFβ isoforms (TGFβ1, TGFβ2 and TGFβ3) are expressed in mammalian epitheliums, each being encoded by a unique gene and expressed in both a tissue-specific and developmentally regulated manner, with TGFβ1 being the most abundant and ubiquitously expressed isoform. Once bound by TGFβ, TGFβR2 recruits, phosphorylates and thus activates of the type I TGFβ receptor (TGFβR1). TGFβR1 then phosphorylates two transcriptional regulators, namely SMAD2 and SMAD3, which subsequently bind to SMAD4. This results in the nuclear translocation of SMAD complexes, allowing SMADs to interact with transcription factors controlling the expression of a multitude of TGFβ responsive genes [10]. TGFβ1 is usually considered a tumor suppressor, due to its cytostatic activity in epitheliums. However, on advanced stages of tumors, TGFβ1 behaves as a tumor promoter, due to its capability to enhance angiogenesis, epithelial-to-mesenchymal transition, cell motility and metastasis [11-13]. Not only the expression of TGFβ1 in both tumor and plasma was found to be significantly higher in patients with metastasic colorectal cancer, but increasing colorectal tumor stage was correlated with higher TGFβ1 expression in tumor tissues [14].
MicroRNAs (miRNAs) are short non-coding RNAs which regulate the translation and/or degradation of target messenger RNAs, and whose molecular malfunctions are associated with cancers [15, 16]. Depending on the effects of their downregulation or over-expression, miRNAs have been described either as oncogenic (onco-miRs) or tumor suppressors. For example, genomic amplification and over-expression of miR-17-92 miRNAs is found in B-cell lymphomas as well as in breast and lung cancers [15, 16]. MiR-21 is over-expressed in several cancers, including CRCs, gliomas, as well as breast, gastric, prostate, pancreas, lung, thyroid and cervical cancers [17]. On the other hand, let-7 miRNAs, frequently downregulated in cancers like lung, colon or other solid tumors, are therefore considered as tumor suppressor miRNAs in these types of cancers [15]. Also, miR-15 and miR-16-1 suppress tumorigenicity by inhibiting cell proliferation and promoting apoptosis of cancer cells [18]. Beside miR-21, several miRNAs are overexpressed in CRCs, such as for example miR-17, miR-25, miR-26a or miR-181a [19, 20]. Furthermore, several miRNAs, including miR-21, have been shown to activate metastasis by acting on multiple signaling pathways and targeting various proteins that are key players in this process [19, 21].
Resveratrol (trans-3,4′,5-trihydroxystilbene) is a dietary polyphenolic, non-flavonoid antioxidant derived from grapes, berries, peanuts, and other plant sources. Resveratrol has various health benefits, such as cardiovascular and cancer preventive properties [22-24], and it is currently at the stage of pre-clinical studies for human cancer prevention [24, 25]. Resveratrol induces apoptosis by up-regulating pro-apoptotic genes while simultaneously down-regulating anti-apoptotic genes. Resveratrol induces the redistribution of CD95 and other death receptors in lipid rafts, thus contributing to their sensitization to death receptor agonists [26]. In addition, resveratrol causes growth arrest at G1 and G1/S phases by inducing the expression of CDK inhibitors p21/CDKN1A and p27/CDKN1B [23]. However, the molecular bases for resveratrol effects remain unclear.
Here, we address the effects of resveratrol treatment on miRNA populations from human SW480 cells. We show that resveratrol modulates the levels of miRNAs targeting both oncogenes and tumor suppressor genes, which provides a rational for manipulating the levels of miRNAs depending on the nature and the stage of tumors to optimize the anti-tumors effects of resveratrol.
2 Materials and methods
2.1 Micro-array analyses
RNAs extracted with TRIzol (Invitrogen, Carlsbad, California, USA) were subsequently subjected to DNase digestion (Turbo-DNase from Ambion, Invitrogen, Carlsbad, California, USA). MiRNA micro-array analyses were done at the Ohio State University micro-array facility. Data were submitted to MIAME database with the accession numbers to be received after confirmation.
2.2 Cell culture, transfection and treatments
SW480 cells were maintained in culture following standard procedures. Transfections were done using Lipofectamine 2000 (Invitrogen). Cells were treated for 14 hours with either the vehicle (ethanol) or resveratrol 50 μM (Sigma-Aldrich, Saint Louis, Missouri, USA). Whenever needed, cells were either mock-treated or stimulated for 14 hours with 100 ng/ml TGFβ1 (Invitrogen).
2.3 Preparation of clones and subclones
The 307 nt-long 3′-UTR (untranslated region) of the TGFβ1 gene was cloned by PCR from genomic DNA extracted from HEK-293 cells. Luc-TGFβ constructs were prepared by inserting the TGFβ1 3′-UTR downstream of the luciferase gene in the XbaI site of the pGL3-Control vector (Promega, Madison, Wisconsin, USA). The sequences of the oligonucleotides used for cloning are available upon request.
2.4 Mutagenesis
The two sets of overlapping miR-663 target sites in TGFβ1 3′-UTRs, respectively containing three and two miR-663 sites, (site 663-1: CCCCGCCCCGCCCCGCC, starting at nt 8, and site 663-2: CCCCGCCCCGCC, starting at nt 43) were respectively mutated to M663-1 (CGGCGCGCGGCCGGGCC) and M663-2 (GCCGCGCCGCGG) using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, Agilent Technologies, Inc., Santa Clara, California, USA). M663-1,2 mutant carry both mutations simultaneously.
2.5 Luciferase assays
SW480 (1 × 106) in 6-well plates were transfected with 0.4 μg of DNA (Promega pGL3-Control vector or derived constructs), or with RNA 50 nM final, i.e., pre-miR Precursor Molecule-Negative Control #1 (Ambion) or either pre-miR-663 precursor RNA (Ambion) or a miR-663 antisense inhibitory RNA (663-I, Ambion), along with 20 ng of Renilla luciferase control vector (pRL-TK from Promega). Assays were performed 48 hours after transfection using the Dual Luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. TGFβ signaling was tested in sixplicates in 12-wells plates using the Cignal SMAD Reporter Assay Kit from Super Array Bioscience Corporation (SABioscience, Qiagen, Valencia, California, USA). This kit is designed to monitor the activity of the canonical TGFβ-induced signal transduction pathway, i.e., to give a measure of the rate of SMAD2 and/or SMAD3 nuclear transcriptional activity in conjunction with SMAD4 on a reporter construct containing a functional SMAD binding site.
2.6 RNase-protection assays
Assays were performed using the mirVana miRNA Probe Construction Kit and the mirVana miRNA Detection Kit from Ambion. RNAs were incubated overnight at 46 °C in the presence of 8 × 104 cpm of [32P]-labelled antisense RNA probe and then digested for 40 minutes at 37 °C using a 1/50 dilution of the provided RNAse A/RNase T1 solution. The relative intensities of the bands were determined using the Adobe Photoshop software (Adobe Systems Incorporated, San Jose, California, USA).
2.7 Western blots
Anti- TGFβ1 and anti-SMAD3 antibodies were from Cell Signaling Technology (Danvers, Massachusetts, USA). Anti- TGFβR1, anti- TGFβR2, anti-SMAD7, anti-PDCD4 and anti-E-CADHERIN antibodies were from Abcam (Abcam Inc., Cambridge, Massachusetts, USA). Anti-PTEN and anti-GAPDH antibodies were from Santa Cruz Biotechnology (Santa Cruz, California, USA). The relative intensities of the bands on Western blots were determined using the Adobe Photoshop software.
2.8 Statistical analysis
Results are expressed as mean ± S.D. The statistical significance of the results was determined by Student t tests using the Microsoft Excel software (Microsoft Campus, Redmond, Washington, USA).
3. Results
3.1 Resveratrol modulates the levels of miRNAs targeting known tumor suppressors as well as components of the TGFβ signaling pathway
MiRNA micro-arrays (MIAME accession number) showed that a 14 hours resveratrol treatment of SW480 cells significantly (P < 0.05) increased the levels of 22 miRNAs while decreasing those of 26 other miRNAs (Table 1). The changes in miRNA levels were generally limited, with a few exceptions such as a 17 and 11 fold upregulation of miR-146b-5p and miR-1, respectively. This suggests that resveratrol effects on miRNA populations are not global, but rather miRNA-specific. Several miRNAs downregulated by resveratrol, as for example miR-17, miR-21, miR-25, or miR-92a-2, are generally considered as onco-miRs [15, 16]. and are known to be overexpressed in CRCs [19, 20]. Surprisingly, miR-16-1 was also downregulated by resveratrol, while usually working along miR-15 to suppress tumorigenicity by inhibiting cell proliferation and promoting apoptosis of cancer cells [18].
Table 1.
Human microRNAs whose levels changed significantly (P < 0,05) following resveratrol treatment of SW480 cells, as determined on miRNA micro-arrays.
| *hsa-miR- | Resveratrol / Control | Parametric P values | *hsa-miR- | Resveratrol / Control | Parametric P values |
|---|---|---|---|---|---|
|
|
|
||||
| 206 | 2.748 | 0.0004106 | 26a | 0.391 | 0.0153595 |
| 560 | 2.924 | 0.0005005 | 594 | 0,622 | 0,01711 |
| 194-2 | 2.435 | 0.0005049 | 30a-3p | 0.374 | 0.0175082 |
| 181a2 | 0.32 | 0.0012911 | 615 | 3.113 | 0.0175787 |
| 639 | 2.467 | 0.0016422 | 663 | 1.934 | 0.0183065 |
| 1 | 11.106 | 0.0024567 | 17 | 0.432 | 0.0208079 |
| 801 | 3.117 | 0.0027415 | 565 | 0.505 | 0.0234496 |
| 21 | 0.437 | 0.0040256 | 103-1 | 0.481 | 0.0263589 |
| 424 | 0.492 | 0.004825 | 565 | 1.762 | 0.0269349 |
| 196a1 | 0.237 | 0.0063464 | 103-2 | 0.608 | 0.028218 |
| 659 | 5.35 | 0.0063984 | 340 | 2.975 | 0.0282464 |
| 323 | 1.926 | 0.0066204 | 363*-5p | 2.568 | 0.028538 |
| 572 | 1.972 | 0.0072194 | 631 | 0.47 | 0.0293983 |
| 560 | 2.084 | 0.0082416 | 638 | 1.712 | 0.0320884 |
| 657 | 0.473 | 0. 0096236 | 494 | 2.038 | 0.032807 |
| 92a-2 | 0.504 | 0.0101895 | 30d | 0.587 | 0.0339924 |
| 23a | 0.236 | 0.0108057 | 622 | 2.724 | 0.0363927 |
| 16-1 | 0.588 | 0.0110056 | 23b | 0.484 | 0.0366713 |
| 25 | 0.586 | 0.0117457 | 102 | 0.547 | 0.0414935 |
| 146a | 0.591 | 0.0121974 | 574 | 1,501 | 0.0441683 |
| 497 | 1.691 | 0.0126286 | 205 | 0.595 | 0.0472807 |
| 29c | 0.58 | 0.0127887 | 629 | 0.53 | 0.0485583 |
| 30c-1 | 2.204 | 0.0138886 | 146b-5p | 17.667 | 0.0490768 |
| 100- 1/2 | 0.423 | 0.0147436 | 30e-5p | 0.496 | 0.0494118 |
Human microRNAs were arranged based upon increasing P values.
In silico analysis using TargetScan (http://www.targetscan.org/) showed that several of miRNAs under resveratrol control potentially target known tumor suppressor genes, such as components of the mismatch repair machinery (MLH3, MSH2 and MSH3), PDCD4 and PTEN, as well as several effectors and regulators of the TGFβ signaling pathway (Table 2). Of note, the same miRNAs may also potentially target DICER1 transcripts, with encode the RNase III producing mature miRNas from their immediate precursors in the cytoplasm. A majority of the miRNAs potentially targeting the above targets were downregulated by resveratrol (Table 1). Altogether, these results indicate that some of resveratrol antitumor properties may arise from its effects on the levels of miRNAs targeting transcripts encoding key regulators of cell homeostasis.
Table 2. Putative mRNA targets of human microRNAs whose levels were affected by resveratrol treatment of SW480 cells.
| Putative target mRNA | Main functions of the encoded factors | Role in oncogenesis | hsa-miR- downregulated by resveratrol | hsa-miR- upregulated by resveratrol |
|---|---|---|---|---|
| TGFβ1 | Binds TGFβR1, TGFβR2 and TGFβR3 / Cytostatic / Induces EMT / Anti-inflammatory / Activation of hsa-miR-21 maturation / Reduces the immune response | Tumor-suppressor at pre-malignant stage / Pro-metastasis at malignant stage | 663 | |
| TGFβ3 | Binds TGFβR1, TGFβR2 and TGFβR3 | Not clear / Possibly tumor suppressor in skin / Inhibits colon metastasis in TGF m 1-/-Rag2- mice | 29c / 631 | |
| TGFβR1 / ALK5 | SMAD2,3 phosphorylation / TGFβ1-3 signal transduction | Mutated in CRCs | 21 / 181a2 | 1 / 206 |
| TGFβR2 | TGFβR1 phosphorylation and activation / TGFβ1-3 signal transduction | Mutated in CRCs | 17 / 23a / 23b / 103-1 / 103-2 / 181a2 | |
| TGFβR3 / BETAGLY CAN | Sequestration of TGFβ1-3 (secreted form) / Presentation of TGFβ2 to TGFβ receptors (trans-membrane form) / Loss facilitates EMT in neoplastic cells / TGFβ-independent effects? | Downregulation correlated with tumor progression or metastasis | 16-1 / 23a / 23b / 103-1 / 103-2 / 424 | 1 / 206 /497 |
| BAMBI | TGFβ decoy receptor impairing the formation of TGFβR1- TGFβR2 complexes / Related to TGFβR1, but lacks the intracellular kinase domain / Overexpressed in metastasis | Increases cellular growth / Enhances WNT signaling and inhibit of TGFβ-SMAD signaling | 17 / 146a / 205 | 146b-5p |
| SMAD2 | Regulatory SMAD phosphorylated and relocated in the nucleus in association with SMAD4 under TGFβ signaling / Transcriptional regulation | Mutated in CRCs | 16-1 / 25 / 30d / 30e-5p / 92a-2 / 146a / 205 / 181a2 / 424 | 30c-1 / 146b-5p / 206 / 340 |
| SMAD3 | same as SMAD2 | Mutated in CRCs / Overexpressed in late stage sporadic CRCs / pSMAD3L mediates Mesenchymal cell invasion | 16-1 / 23a / 23b /103-1 / 103-2 / 146a / 424 | 1 / 206 / 146b-5p / 497 / 663 |
| SMAD4 | Interacts with phosphorated regulatory SMADs and relocates them the nucleus / Mediates TGFβ canonical signaling / Transcriptional regulation / Lost or decreased in colon cancer | Tumor suppressor | 16-1 / 23a / 23b / 25 / 26a / 92a-2 / 146a / 205 / 424 | 1 / 206 / 146b-5p / 206 / 340 / 497 |
| SMAD7 | Inhibitory SMAD / Interference with regulatory SMADs activation / Negative regulator of BMP and TGFβ signaling pathways / Decreases WNT signaling by recruiting SMURF2 to βCATENIN / Inhibition of NF-κB signaling | Oncogenic by inhibition of TGFβ-mediated growth inhibition / Tumor-supressor by inhibition of TGFβ-induced metastasis and WNT signaling | 16-1 / 17 / 21 / 25 / 92a-2 / 181a2 / 424 | 340 / 497 |
| SMURF1 | E3 ubiquitin ligase / Ubiquitinates SMAD1 and SMAD5 / Degradation of type I receptors, regulatory SMADS, SMAD4 and SMAD7 | Not clear | 16-1 / 17 / 25 / 29c / 92a-2 103-2 / 340 / 424 / 659 | 1 / 206 / 194-2 / 103-1 /497 |
| SMURF2 | E3 ubiquitin ligase / Ubiquitinates SMAD1 and SMAD2 / Degradation of type I receptors, regulatory SMADS, SMAD4 and SMAD7 / SMAD2-SMURF2 complexes mediate the degradation of the transcriptional corepressor SnoN | Highly expressed in lymph node metastases / Decreases WNT signaling by ubiquitinating βCATENIN | 16-1 / 21 / 424 | |
| DICER1 | RNase III / Pre-miRNA processing in cytoplasm / miR-103 validated target | Inhibition drifts epithelial cancer toward mesenchymal fate | 16-1 / 17 / 21 / 23a / 23b / 25 / 29c / 92a-2 / 103-1 / 103-2 /181a2 | 1 / 206 |
| PTEN | PI3K phosphatase / Inhibition of cell proliferation | Tumor suppressor | 17 / 23a / 23b / 25 / 26a / 29c / 92a-2 / 103-1 /103-2 / 181a2 / 205 | 494 |
| PDCD4 | Upregulated during apoptosis / Inhibits cell proliferation and tumor angiogenesis pathways / miR-21 validated target / Downregulated in CRCs | Tumor suppressor | 16-1 / 17 / 21 / 23a / 23b /160 / 181a2 / 424 | 340 / 497 |
| MLH3 | Mut-L homolog 3 / DNA Mismatch repair | Tumor-suppressor | 16-1 / 23a / 23b / 29c / 30d / 30e-/ 5p / 103-1 / 103-2/ 424 | 1 / 206 / 30c-1/ 497 |
| MSH2 | Mut-S homolog 2 / DNA Mismatch repair | Tumor-suppressor | 21 | |
| MSH3 | Mut-S homolog 3 / DNA Mismatch repair | Tumor-suppressor | 17 | |
| E-CADHERI N / CDH1 | Adherens Junctions / Maintenance of epithelial phenotype / Lost in EMT and metastasis | Inhibitor of invasion | 17 / 23a / 23b / 25 / 92a-2 / 103-1 / 103-2 |
3.2 The levels of tumor suppressor factors encoded by putative target transcripts of oncogenic miRNAs downregulated by resveratrol increase following resveratrol treatment
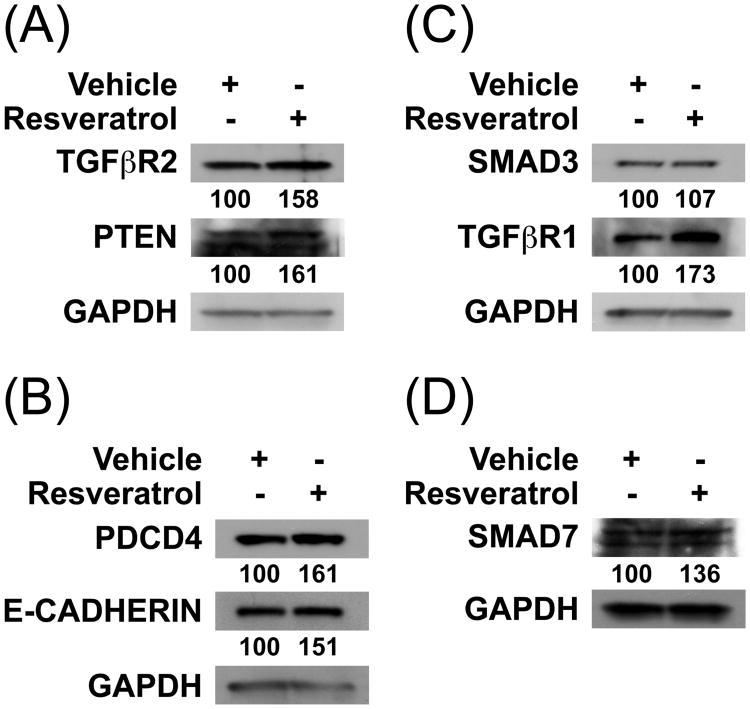
Although the absolute levels of miRNAs upregulated or downregulated by resveratrol as well as their relative activities on their target transcripts in SW480 cells cannot be definitely ascertained, the above results suggested that resveratrol treatment may change the levels of several of the factors encoded by transcripts targeted by these miRNAs (Table 2). We thus investigated the effects of resveratrol on several factors known to behave as tumor suppressors, such as PTEN and PDCD4, as well as on critical effectors of the TGFβ signaling pathway. Indeed, the levels of TGFβ receptors type II and type I (TGFβR2 and TGFβR1, respectively) increased by roughly 60% and 70%, respectively (Figures 1A and 1C, respectively). The levels of both PTEN, a phosphatase targeting PI3K (Phosphoinositide 3-kinase), and PDCD4, a factor upregulated during apoptosis, also increased by roughly 60% (Figures 1A and 1B, respectively). The levels of E-CADHERIN, a component of adherens junctions implicated in the maintenance of epithelial phenotype, and of SMAD7, a negative regulator of TGFβ signaling, respectively increased by nearly 50% and 35% (Figures 1B and 1D, respectively). In contrast, the upregulation of SMAD3 following resveratrol treatment was almost neglectible (Figure 1C), which may possibly arise from the fact that SMAD3 transcripts are also potentially targeted by five miRNAs upregulated by resveratrol, including miR-1 and miR-146b-5p, the two miRNAs which showed the greatest upregulation by resveratrol (Table 1). Nevertheless, resveratrol treatment of SW480 cells globally lead to the increase of the levels of effectors of TGFβ signaling pathway as well as of several tumor suppressor factors.
Figure 1. The levels of factors encoded by putative target transcripts of miRNAs donwregulated by resveratrol increase correspondingly.
The levels in SW480 cells of tumor suppressors as well as key effectors of TGFβ signaling pathway were assessed by Western blotting using the indicated antibodies. The relative levels following resveratrol treatment are given in percent of the corresponding control vehicle sample. Panels A to D were from different gels.
3.3 Resveratrol upregulates miR-663, a miRNA targeting TGFβ1 transcripts
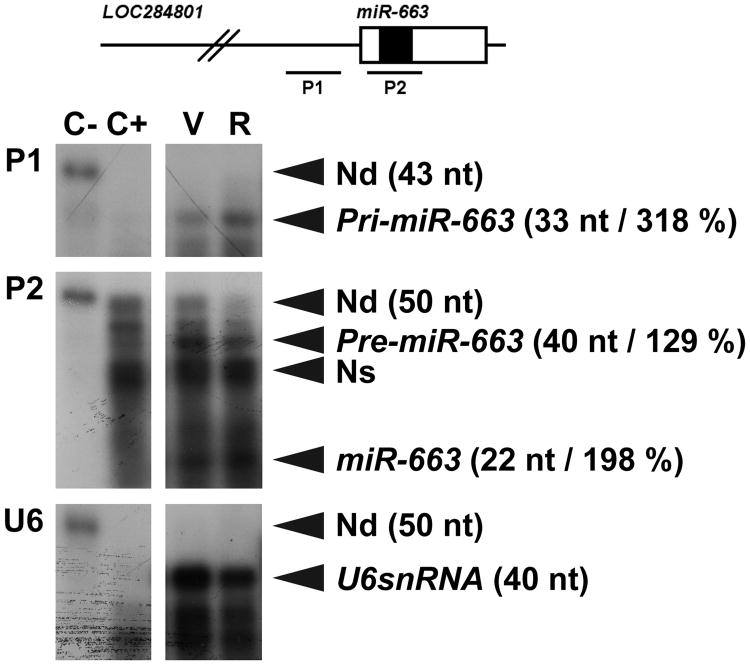
The 3′-UTR of TGFβ1 is short (307 nt in length) and is potentially targeted by no more than 28 miRNAs, which probably results from the existence of multiple levels of regulation of the rate of TGFβ1 signaling. Remarkably, however, resveratrol upregulated one and only one of these miRNAs in SW480 cells, namely miR-663, a miRNA previously shown to be downregulated in hormone refractory prostate cancer cells [27]. (Table 1). Of note, TGFβ1 behaves as a tumor promoter on advanced stages of tumorigenesis, due to its capability to enhance angiogenesis, epithelial-to-mesenchymal transition, cell motility and metastasis [11-13]. Especially, it has been shown that increasing colorectal tumor stage correlates with higher TGFβ1 expression in tumor tissues [14]. We therefore double-checked the upregulation of miR-663 by resveratrol using RNase-protection assays. This experiment suggested that the levels of mature miR-663 in SW480 cells may increase by roughly 100% following resveratrol treatment (Figure 2), a result in good agreement with those of miRNA micro-array experiments (Table 1). Of note, the levels of miR-663 primary transcripts, namely pri-miR-663, increased 50% further, suggesting that resveratrol may also regulate the processing of miR-663 from its primary transcripts (Figure 2).
Figure 2. Resveratrol increases the levels of miR-663 as well as those of pri-miR-663 and pre-miR-663 differentially.
Total RNAs (0.6 μg) extracted from SW480 cells treated for 14 hours with either the vehicle (V) or resveratrol (R) were hybridized with a radiolabeled RNA antisense probe (P1, P2 or U6) in the presence of 4.4 μg of yeast tRNA. The schematic structure of the LO284801 locus, whose transcripts represent miR-663 primary transcripts, is presented on the unscaled top drawing. Open and filled boxes represent miR-663 precursor (pre-miR-663) and mature miR-663, respectively. P1 protects a 33 nt-long fragment starting 184 nt upstream of pre-miR-663. Pre-miR-663 and miR-663 respectively generate 40 and 22 nt-long protected fragments from P2, which corresponds to one of the miRNA micro-array probes. The relative intensity of the signal following resveratrol treatment is given between parentheses in percent of the vehicle sample, calculated using U6snRNA as an internal control. The efficiency of RNase digestion was assessed in parallel (two left lanes). Samples and controls were from the same gels. C-, no RNase (15 fold dilution); C+, RNase; Nd, non digested; Ns, non specific.
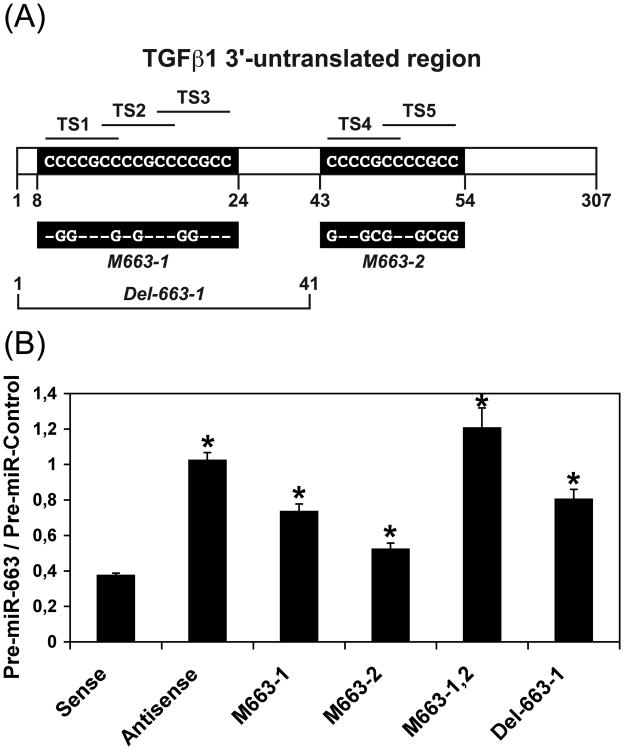
The effects of miR-663 on TGFβ1 transcripts were subsequently analyzed in SW480 cells transfected with either a control RNA (pre-miR-Control) or pre-miR-663 along with a construct containing the whole 307 nt-long 3′-UTR of TGFβ1 inserted downstream the luciferase gene in the pGL3-Control vector in sense or in antisense orientation. TGFβ1 3′-UTR namely contains five miR-663 target sites (TS) (Figure 3A) arranged in two sets of three and two overlapping sites, respectively (sites TS1 to TS3, starting at nt 8, and sites TS4 and TS5, starting at nt 43). Pre-miR-663 reduced the luciferase activity produced from sense Luc- TGFβ1 construct by about 60%, while remaining without effects on antisense construct (Figure 3B). The lack of the first 41 nt of TGFβ1 3′-UTR, which contain the three first miR-663 sites (i.e., TS1 to TS3), in Luc- TGFβ-Del-663-1, or the mutation of the three same sites by replacing a few nt by the complementary nt in Luc-TGFβ-M663-1 (Figure 3A), still allowed a residual down-regulation of the luciferase activity of roughly 20%, while mutating the two last miR-663 sites (TS4 and TS5) in Luc- TGFβ-M663-2 still lead to about 50% decrease of the luciferase activity (Figure 3B). However, the luciferase activity produced from a construct bearing both sets of mutations (Luc- TGFβ-M663-1,2) remained essentially unaffected by resveratrol. Collectively, these results suggest that miR-663 may target TGFβ1 transcripts in vivo.
Figure 3. MiR-663 targets TGFβ1 transcripts.
(A) Schematic representation (not to scale) of the 307 nt-long 3′-untranslated region of TGFβ1 transcripts. The positions of the two sets of overlapping miR-663 target sites (TS1 to TS5) are given in nucleotides. Their sequences (CCCCGCC) are given in black boxes. Nucleotides modified in the two single mutant constructs (Luc- TGFβ-M663-1 and Luc- TGFβ-M663-2) are given below the figure (dashes correspond to conserved nucleotides). Luc-TGFβ-M663-1,2 contains both sets of mutated nucleotides. The first 41 nucleotides of TGFβ1 3′-untranslated region is lacking in Luc- TGFβ-Del-663-1.
(B) Effects of pre-miR-663 on Luc- TGFβ constructs. SW480 cells were transfected with wild type Luc- TGFβ (WT) constructs both in sense and antisense orientation, with mutated Luc- TGFβ constructs, (M663-1, M663-2, and M663-1,2), or with a Luc- TGFβ construct bearing a deletion of the first 41 nt of TGFβ1 3′-UTR (Del-663-1), along with either a Control RNA (Pre-miR-Control) or pre-miR-663. The sequences of the two mutated series of overlapping miR-663 target sites are given in panel A. Values represent the mean ± standard deviation (n = 4). *, Significantly different from Pre-miR-Control, P < 0.001 (df = 6).
3.4 Resveratrol decreases TGFβ1 expression by both miR-663-dependent and miR-663-independent mechanisms
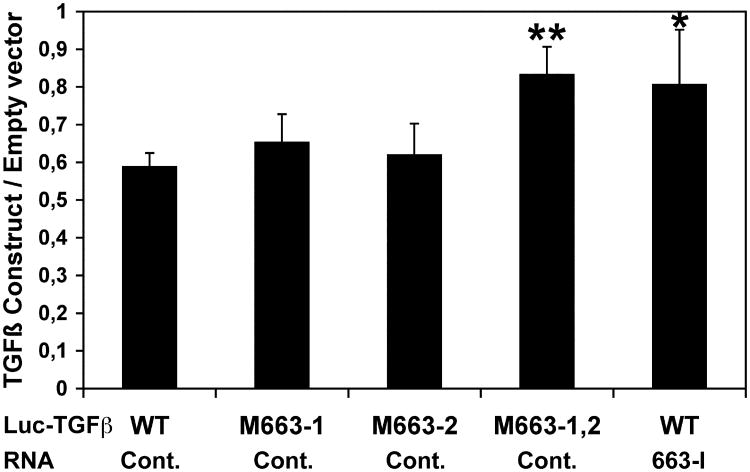
We then checked if the downregulation of TGFβ1 by resveratrol in SW480 cells was miR-663-dependent. As shown by luciferase assays, resveratrol as expected decreased the expression of wild type Luc- TGFβ-WT as well as of both Luc- TGFβ-M663-1 and Luc- TGFβ-M663-2 mutant constructs, an effect impaired by the mutation of the two sets of overlapping miR-663 target sites in Luc-TGFβ-M663-1,2 (Figure 4). On the other hand, resveratrol effects on wild-type Luc- TGFβ-WT construct were blocked by cotransfecting the cells with a miR-663 antisense inhibitory oligonucleotide (663-I). These results provide a further evidence that resveratrol should target the 3′-UTR of TGFβ1 transcripts through the upregulation of miR-663 (Figure 4).
Figure 4. Resveratrol downregulates TGFβ1 transcripts through miR-663.
SW480 cells were transfected with wild type (WT) or mutated (M663-1, M663-2, and M663-1,2) Luc- TGFβ constructs, along with either a Control RNA (Cont.) or a miR-663 antisense inhibitory RNA (663-I) before resveratrol treatment. Bars show the ratios of the relative levels of expression of the different constructs to those of the empty pGL3-Control vector. Values represent the mean ± standard deviation (n = 4). * and **, Significantly different from the corresponding WT / Control, *, P = 0.05, **, P = 0.017 (df = 6).
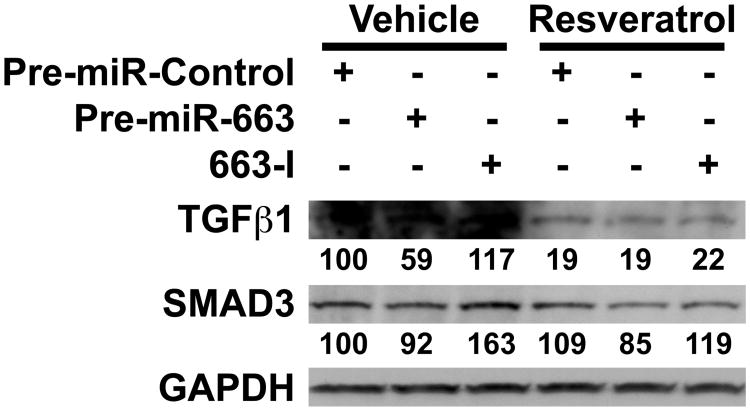
Taking into account that SMAD3 3′-UTR also contains three putative miR-663 target sites, we then investigated the combined effects of resveratrol, pre-miR-663 and 663-I on both endogenous TGFβ1 and SMAD3. Without resveratrol treatment, both pre-miR-663 and 663-I worked as expected on TGFβ1, although with various efficiencies, i.e., pre-miR-663 and 663-I respectively decreased and increased the levels of TGFβ1 (Figure 5). Of note, in the absence of resveratrol, TGFβ1 levels were more affected by pre-miR-663 than by 663-I, suggesting that miR-663 may normally not exert too tight a control on TGFβ1 levels in SW480 cells. In agreement with the above results, resveratrol downregulated sharply TGFβ1 levels. This was probably due to its upregulation of miR-663, as pre-miR-663 proved unable to lower further TGFβ1 levels (Figure 5). Still, 663-I effects on TGFβ1 surprisingly remained neglectible in the presence of resveratrol. By comparison, in the absence of resveratrol, SMAD3 levels, while being only marginally affected by pre-miR-663, increased sharply following transfection with 663-I, indicating that endogenous miR-663 usually works to keep SMAD3 levels below a certain threshold (Figure 5). In contrast, resveratrol effects on SMAD3 were rather limited, as previously established in section 3.2 (compare Figure 5 and Figure 1C). Strikingly, however, pre-miR-663 and 663-I kept working as expected on SMAD3 in the presence of resveratrol. This suggests that, at least in SW480 cells, resveratrol can also control the expression of TGFβ1, but not of SMAD3, through genetic circuitries independent of miR-663, possibly by decreasing the levels of TGFβ1 positive regulators targeted by some of the resveratrol-upregulated miRNAs.
Figure 5. Resveratrol decreases the levels of TGFβ1 mainly in a miR-663-independent manner.
SW480 cells were transfected with either a control RNA (pre-miR-Control), pre-miR-663 or an antisense miR-663 inhibitory RNA (663-I) before resveratrol treatment. The levels of TGFβ1 and SMAD3 were then assessed on Western blots. The relative levels of TGFβ1and SMAD3 are given in percent of the corresponding Pre-miR-Control Vehicle-treated samples.
3.5 Resveratrol decreases the activation of SMADs through the canonical TGFβ1 signaling pathway
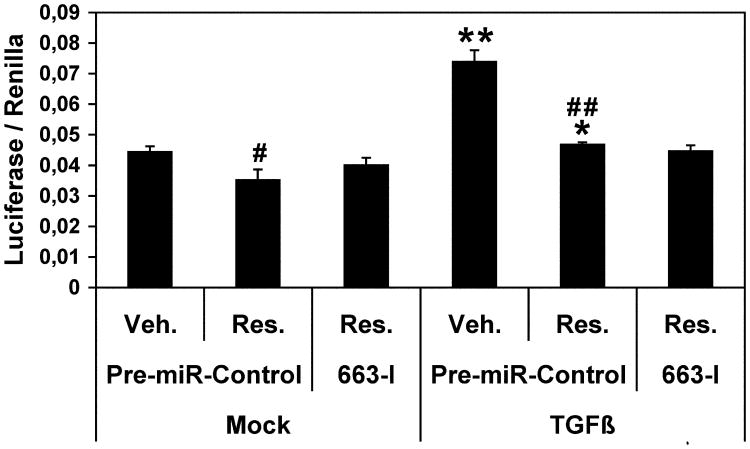
The activation of the TGFβ1 canonical pathway results in the C-terminal phosphorylation and activation of SMAD2 and/or SMAD3, which then interact with SMAD4, that leads to their nuclear import. In the nucleus, SMADs collaborate with transcriptional activators and repressors to control the expression of a number of TGFβ target genes. Given the above results, we then checked the effects of resveratrol on the expression of a SMAD2/SMAD3/SMAD4 luciferase reporter construct in order to measure the degree of activation of the above SMADS (Figure 6). Without TGFβ1 treatment, resveratrol slightly but significantly decreased the basal level of expression of the reporter construct, which was possibly due to its effects on endogenous TGFβ1 (see section 3.4). As expected, a treatment with TGFβ1 robustly increased the expression of the reporter construct in presence of the vehicle. Quite surprisingly, the activation of the reporter construct was nearly completely abolished by resveratrol treatment (Figure 6). Of note, transfecting SW480 cells with 663-I before resveratrol treatment remained without measurable consequence. This suggests that resveratrol, while restoring the levels of key components of the TGFβ signaling pathway, such as TGFβR1 and TGFβR2 (see section 3.2), nevertheless worked to put a cap to the overall activity of this SMAD-dependent pathway in a miR-663 independent manner, in agreement with the results of Figure 5.
Figure 6. Resveratrol decreases the expression of a SMAD2/SMAD3/SMAD4 luciferase reporter contruct.
After cotransfection with the SMAD2/SMAD3/SMAD4 luciferase reporter plasmid along with either a Control RNA (pre-miR-Control) or a miR-663 antisense inhibitory RNA (663-I), SW480 cells were mock-treated or treated with TGFβ for 14 hours, and subsequently treated with the vehicle (Veh.) or resveratrol (Res.) for another 14 hours. The Firefly luciferase activity was measured 48 hours after transfection and then normalized to the Renilla luciferase activity. Values represent the mean ± standard deviation (n = 6). * and **, TGFβ1-treated significantly different from the corresponding Mock-treated, *, P < 0.03, **, P < 0.002; # and ##, Resveratrol-treated significantly different from the corresponding Vehicle-treated, #, P < 0.03, ##, P < 0.005 (df = 10).
4. Discussion
Our study points to the importance of miRNA regulation in the modulation of the levels of several tumor suppressors and TGFβ effectors by resveratrol in SW480 colon cancer cells, and provides the very first data concerning the targeting of TGFβ1 transcripts by miR-663. It provides strong support that: (a) some of the protective effects of resveratrol come from its downregulation of miRNAs over-expressed in CRCs, such as miR-17, miR-21, miR-25, miR-26a, miR-92a-2, miR-103-1 and -103-2, or miR-181a2; (b) the increase of the levels of proteins encoded by putative target transcripts of these miRNAs roughly correlates with the downregulation of these miRNAs by resveratrol; (c) resveratrol upregulates miR-663, one of the few miRNAs targeting TGFβ1 transcripts; (d) resveratrol also controls TGFβ1 levels and the rate of TGFβ signaling through SMAD2/SMAD3/SMAD4 transcription factors using miR-663-independent mechanisms.
MiRNA micro-arrays showed that resveratrol downregulated several miRNAs known for their strong oncogenic potentials, including miR-21 or miRNAs from the miR-17-92 cluster. This is an important result, for miR-21 for example is over-expressed in several cancers, including CRCs, gliomas, as well as breast, gastric, prostate, pancreas, lung, thyroid and cervical cancers [17]. MiR-21 has been shown to function as an onco-miR, due to its targeting of transcripts encoding key regulators of cell proliferation and apoptosis such as PTEN and PDCD4 [28-30]. Especially, PDCD4 is downregulated in a number of cancers, and its suppression in lung and colorectal cancers is associated with poor patient prognosis [31, 32]. Indeed, the levels of these two anti-tumor factors increased following resveratrol treatment of SW480 cells. Also, the downregulation of both miR-103-1 and miR-103-2 by resveratrol may possibly be critical to its anti-metastatic effects. Namely, these miRNAs have been recently shown to induce epithelial-to-mesenchymal transition by targeting Dicer1 transcripts [33], in relation with the fact that a global reduction of miRNA abundance appears a general trait of human cancers, playing a causal role in the transformed phenotype [34-36]. Interestingly, the lower metastatic propensity in SW480 cells as compared with SW620 human colon cancer cells, both derived from the primary tumor and a metastasis of the same patient, respectively [37], was associated with a lower level of expression of miR-103 [33]. Finally, we also show that the levels of several effectors of TGFβ pathway signaling, and especially TGFβR1 and TGFβR2, increase following resveratrol treatment in correlation with the downregulation of miRNAs whose consensus target sequences are present in their 3′-UTRs. This may be the reason why resveratrol favors the tumor suppressor, i.e., the cytostatic activity of TGFβ1 in epitheliums [11, 13] at the early stage of cancers. Altogether, the modification of the levels of no less than 48 miRNAs following resveratrol treatment of SW480 colon cancer cells, some well known to be oncogenic but several of them remaining essentially un-characterized at the present time, is likely to be critical for resveratrol protective effects.
Furthermore, epidemiological studies suggest that as many as 25% of all cancers may be due to chronic inflammation [38, 39]. As miR-21 in particular is induced by inflammation [40], and resveratrol is known as an anti-inflammatory agent, the downregulation of miR-21 by resveratrol may be an important step in controlling cell proliferation and further tumorigenesis. In this respect, it is important to note that TGFβ also functions as a purveyor of immune privilege. While beneficial in initiating and controlling immune responses and maintaining immune homeostasis, immunosuppressive pathways mediated by TGFβ may obscure immune surveillance mechanisms, resulting in failure to recognize or respond adequately to self, foreign, or tumor-associated antigens [41]. It will thus be important to investigate the possible links between the targeting of TGFβR1 or SMAD3 and the development of local inflammation, especially considering that the maturation of miR-21 is increased upon TGFβ signaling in a SMAD3-dependent but SMAD4-independent pathway, with SMAD3 binding to miR-21 primary-transcript and interacting with the microprocessor complex [42].
Among miRNAs upregulated by resveratrol, we identified miR-663, a miRNA previously shown to be down-regulated in hormone refractory prostate cancer cells [27]. We present evidence that miR-663 targets constructs containing TGFβ1-3′-UTR, and that the levels of TGFβ1 in SW480 cells are inversely correlated with the levels of miR-663. We also show that resveratrol decreases the expression of Luc- TGFβ1 constructs in a miR-663-dependent manner. However, while resveratrol expectedly sharply decreased endogenous TGFβ1 levels in SW480 cells, this effect paradoxically remained essentially unaffected by transfecting the cells with either pre-miR-663 or 663-I. Of note, we previously observed the same kind of counter-intuitive result with JunB, whose 3′-UTR contains two putative target sites for miR-663. Namely, while miR-663 overexpression reduced both the expression of Luc-JunB constructs and the accumulation of endogenous JunB in THP-1 monocytic cells, resveratrol showed to decrease the expression of endogenous JunB essentially in a miR-663-independent manner (Tili et al, unpublished results). Together, these results suggest that one of the effects of resveratrol may be to relocate endogenous miR-663 along with its target transcripts in particular regions of the cell where they may become inaccessible to both ectopically expressed miR663 and miR-663 antisense inhibitory RNAs. In this respect, the hairpin precursor of miR-663 has been recently shown to be specifically bound by the nuclear factor TDP-43, a Drosha-associated protein which also represents the major component of the inclusions found in the brain of patients with a variety of neurodegenerative diseases [43]. It is thus possible that TDP-43 or other similar factors may bind miR-663 following resveratrol treatment. Further experiments should shed more light about this hypothesis. It is nevertheless also possible that resveratrol may control the expression of TGFβ1 and/or JunB through regulators possibly targeted by other miRNAs sensitive to this compound.
Of note, miR-663 has just been described as a tumor suppressor miRNA that induces mitotic catastrophe growth arrest in human gastric cancer cells [44]. While this paper do not provide the reader with a mechanistic explanation of this effect, it is tempting to speculate that the anti-tumor effects of miR-663 may be due, at least in part, to its effects on TGFβ1 and SMAD3 accumulation, given that TGFβ1 has been shown to promote invasion and metastasis of gastric cancer through its activation of TGFβR1-ALK5/SMAD3 pathway [45]. The effects of miR-663 described here may thus be of primary importance, for TGFβ1 has been shown to enhance angiogenesis, epithelial-to-mesenchymal transition, cell motility and metastasis at later stages of cancers [11-13]. Especially, the expression of TGFβ1 in both tumor and plasma was found to be significantly higher in patients with metastasic colorectal cancer, and increasing colorectal tumor stage was correlated with higher TGFβ1 expression in tumor tissue [14]. In this respect, it should be emphasized that, in relation with the multiple levels of possible regulation of the rate of TGFβ signaling, the 307-nt long 3′-UTR of TGFβ1 transcripts represents a potential target for 28 miRNAs only. While two of these miRNAs have two putative target sites in TGFβ1 3′UTR, this UTR contains only one target site for all the other miRNAs, but miR-663. Therefore, the fact that miR-663 may potentially target 5 different sites in TGFβ1 3′-UTR suggests that this miRNA could represent a critical TGFβ1 regulator which may possibly be called upon action in emergency situations such as those when cells begin to proliferate anarchically. The multiplicity of miR-663 targets sites in TGFβ1 3′-UTR further suggests that the effects of this miRNA may be dose-dependent, which would also help to explain the intriguing fact that the expression of both TGFβ1 and JunB expression seemed to become miR-663-independent following resveratrol treatment of SW480 and THP-1 cells, respectively (this paper, and Tili et al, unpublished results). Of note, previous publications suggested that striking phenotypes may be driven for the most part through small changes in the cellular concentration of key factors. For instance, in the B cell compartment, miR-150 curtails the activity of the c-Myb transcription factor in a dose-dependent fashion over a narrow range of miRNA and c-Myb concentrations [46].
Furthermore, it has been proposed that an increase in the absolute number of mRNA targets may impact the balance and/or half-life of “free” miRNA, miRNA-RISC, and MiRNA-RISC-mRNA complexes [47]. Thus, given the dual role of TGFβ1 as both a tumor-suppressor at pre-malignant stage and promoter of metastasis at malignant stage [8, 11-13], it would be interesting to determine if the impact of miR-663 on TGFβ1 transcripts may vary accordingly to the stage of tumor. Finally, as SMAD2 and SMAD3 phosphorylated at both linker and C-terminal regions transmit malignant TGFβ signaling in later stages of human CRC [48], and as SMAD7 can act either as a tumor-promoter, by inhibiting the canonical TGFβ pathway, or a as tumor suppressor, especially by promoting the ubiquitination and thus the proteasomal degradation of β-CATENIN, a key transducer of canonical WNT signaling [7, 49], it is probable that the interactions between TGFβ signaling and miRNAs sensible to resveratrol, and especially miR-663, may be generally both dose- and context-dependent.
As a last remark, the fact that miR-663, miR-21 and TGFβ1 have been implicated in the regulation of cell proliferation, tumor apparition and development, metastasis formation and innate immunity, strongly suggests that the capability of resveratrol to behave at the same time as an anti-tumor, anti-metastatic, anti-proliferation and anti-inflammatory agent most probably arises from its effects on the expression of a small set of critical endogenous miRNAs having the abilities to impact the cell proteome globally.
Finally, miRNAs have the promise to become biomarkers for different stages of cancer, both for diagnosis and prognosis. Our analyses of microRNAs that change following resveratrol treatment of SW480 colon cancer cells showed that many of their putative target transcripts encode effectors of the TGFβ signaling pathway. The discovery that resveratrol may potentially allow to control the behavior of the TGFβ signaling pathway opens the possibility of using resveratrol in colon cancers where this pathway is impaired or work to favor metastasis. Furthermore, using microRNA analyses turns out to be a powerful technique not only to classify tumors but also to identify the spectrum of genes where a certain drug/chemical might act. Thus, from our experiments, it starts to become clear that the use of resveratrol would be especially beneficial in the type of cancers where the TGFβ pathway is implicated. Of note, resveratrol use would have to be carefully correlated with the stages of cancers, knowing that TGFβ can have two faces, i.e. anti-and pro-metastatic.
In conclusion, further investigation of the effects of resveratrol on miRNA populations in tumors, and especially in CRCs, may allow to improve the diagnostic and prognostic of several types of cancers, as well as to optimize the use of this molecule as an anti-tumor agent. Importantly, a critical attention will have to be given to the model organism. Namely, while resveratrol anticarcinogenic potential has been linked with data primarily from human cell culture systems, evidence that resveratrol can inhibit carcinogenesis in several organ sites emerged from results of cancer prevention and therapy studies in laboratory animal models [24]. As miR-663 is found only in primates, our results come as a warning that studies in animal may not always allow to predict accurately the molecular effects of resveratrol in human, especially when it comes to miRNAs.
Acknowledgments
This work was founded by the Ohio State University and the French Cancer League.
Footnotes
Conflict of Interest: The authors declare no conflict of interests.
References
- 1.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:3538. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 3.Olsson L, Lindblom A. Family hitory of colorectal cancer in a Sweden county. Fam Cancer. 2003;2:87–93. doi: 10.1023/a:1025734200635. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Cheah PY. Recent advances in colorectal cancer genetics and diagnostics. Crit Rev Oncol Hematol. 2009;69:45–55. doi: 10.1016/j.critrevonc.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 7.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 8.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 9.Akhurst RJ. TGF beta signaling in health and disease. Nat Genet. 2004;36:790–2. doi: 10.1038/ng0804-790. [DOI] [PubMed] [Google Scholar]
- 10.Elliott RL, Blobe GC. Role of transforming growth factor beta in human cancer. J Clin Oncol. 2005;23:2078–93. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 11.Massagué J. TGFbeta in cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padua D, Massagué J. Role of TGFbeta in metastasis. Cell Res. 2009;1:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 13.Tian M, Schiemann WP. The TGF-beta paradox in human cancer: an update. Future Oncol. 2009;5:259–71. doi: 10.2217/14796694.5.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langenskiöld M, Holmdahl L, Falk P, Angenete E, Ivarsson ML. Increased TGF-beta 1 protein expression in patients with advanced colorectal cancer. J Surg Oncol. 2008;97:409–15. doi: 10.1002/jso.20961. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM. MiRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 16.Tili E, Michaille JJ, Gandhi V, Plunkett W, Sampath D, Calin GA. 2007. miRNAs and their potential for use against cancer and other diseases. Future Oncol. 2007;5:521–7. doi: 10.2217/14796694.3.5.521. [DOI] [PubMed] [Google Scholar]
- 17.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–20. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 19.Rossi S, Kopetz S, Davuluri R, Hamilton SR, Calin GA. MicroRNAs, ultraconserved genes and colorectal cancers. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Belaguli N, Berger DH. MicroRNA and Colorectal cancer. World J Surg. 2009;33:638–4. doi: 10.1007/s00268-008-9865-5. [DOI] [PubMed] [Google Scholar]
- 21.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs – the micro steering wheel of tumor metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 22.Csiszar A, Smith K, Labinskyy N, Orosz Z, Rivera A, Ungvari Z. Resveratrol attenuates TNFa-induced activation of coronary arterial endothelial cells: role of NF-kB inhibition. Am J Physiol Heart Circ Physiol. 2006;291:H1694–9. doi: 10.1152/ajpheart.00340.2006. [DOI] [PubMed] [Google Scholar]
- 23.Shankar S, Singh G, Srivastava RK. Chemoprevention by resveratrol: molecular mechanisms and therapeutic potential. Front Biosci. 2007;12:4839–54. doi: 10.2741/2432. [DOI] [PubMed] [Google Scholar]
- 24.Bishayee A. Cancer prevention and treatment with resveratrol: from rodent studies to clinical trials. Cancer Prev Res. 2009;2:409–18. doi: 10.1158/1940-6207.CAPR-08-0160. [DOI] [PubMed] [Google Scholar]
- 25.Athar M, Back JH, Tang X, Kim KH, Kopelovich L, Bickers DR, et al. Resveratrol: a review of preclinical studies for human cancer prevention. Toxicol Appl Pharmacol. 2007;224:274–83. doi: 10.1016/j.taap.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delmas D, Rébé C, Micheau O, Athias A, Gambert P, Grazide S, et al. Redistribution of CD95, DR4 and DR5 in rafts accounts for the synergistic toxicity of resveratrol and death receptor ligands in colon carcinoma cells. Oncogene. 2004;23:8979–86. doi: 10.1038/sj.onc.1208086. [DOI] [PubMed] [Google Scholar]
- 27.Lin SL, Chiang A, Chang D, Ying SY. Loss of miR-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–24. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 30.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–25. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Knösel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 200;200:640–6. doi: 10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- 32.Mudduluru G, Medved F, Grobholz R, Jost C, Gruber A, Leupold JH, et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 33.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A microRNA targeting Dicer for metastasis control. Cell. 2010;141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 34.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 35.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 36.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–9. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 37.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–69. [PubMed] [Google Scholar]
- 38.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–80. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 39.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 40.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahl SM, Wen J, Moutsopoulos N. TGF-beta: a mobile purveyor of immune privilege. Immunol Rev. 2006 Oct;:213, 213–27. doi: 10.1111/j.1600-065X.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- 42.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buratti E, De Conti L, Stuani C, Romano M, Baralle M, Baralle F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–81. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 44.Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L, et al. Tumor-suppressive miR-663 gene induces mitotic catastrophe growth arrest in human gastric cancer cells. Oncol Rep. 2010;24:105–12. doi: 10.3892/or_00000834. [DOI] [PubMed] [Google Scholar]
- 45.Yoo YA, Kang MH, Kim JS, Oh SC. Sonic hedgehog signaling promotes motility and invasiveness of gastric cancer cells through TGF-beta-mediated activation of the ALK5-Smad3 pathway. Carcinogenesis. 2008;29:480–90. doi: 10.1093/carcin/bgm281. [DOI] [PubMed] [Google Scholar]
- 46.Xiao C, Colado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 47.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–39. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuzaki K, Kitano C, Murata M, Sekimoto G, Yoshida K, Uemura Y, et al. Smad2 and Smad3 phosphorylated at both linker and COOH-terminal regions transmit malignant TGF-beta signal in later stages of human colorectal cancer. Cancer Res. 2009;69:5321–30. doi: 10.1158/0008-5472.CAN-08-4203. [DOI] [PubMed] [Google Scholar]
- 49.Yan X, Liu Z, Chen Y. Regulation of TGF-β by Smad7. Acta Biochim Biophys Sin. 2009;41:263–72. doi: 10.1093/abbs/gmp018. [DOI] [PMC free article] [PubMed] [Google Scholar]