Summary
Several gene disruption strategies have been described in Candida albicans to create homozygous mutants. We describe here a recyclable mini-blaster cassette containing C. albicans URA3 gene and 200 bp flanking repeats that is useful for disruption of C. albicans genes. The cassette can be used to create unmarked homozygous mutants which can be complemented at the HIS1 gene locus. This strategy of creating gene disruptions and subsequent complementation can be used to study gene function.
Keywords: genetics, mutants, complementation
1. Introduction
Candida albicans is a major fungal systemic pathogen in humans. The ability of this fungus to cause lethal infections has fueled the need to study its gene function to better understand aspects like host-pathogen interactions and virulence. The genetic architecture of this pathogen poses two significant obstacles [1]. First, C. albicans is a diploid so both alleles of a gene must be altered. Second, it is an asexual organism, so gene disruption and complementation depends upon successive manipulations of a single strain.
Loss-of-function or null mutations provide a simple avenue toward interpretation of gene function. In this chapter, we will discuss a detailed protocol to introduce a null mutation in a C. albicans gene of interest. We describe use of the mini-blaster strategy to delete both alleles of a gene by alternate transformation and recombination using a recyclable cassette containing the C. albicans URA3 marker [2]. This strategy is based on the original Ura-blaster design of Alani and Kleckner [3] that was modified for C. albicans by Fonzi and Irwin [4]. The mini-blaster, or “Ura-blister,” has shorter direct repeats than the Ura-blaster, thus facilitating PCR amplification. Specifically, the mini-blaster cassette carries the URA3-dpl200 marker (figure 1), which constitutes the URA3 gene and 200 bp flanking repeats. The flanking repeats permit homologous excision and re-utilization of the URA3 marker. The cassette is amplified and targeted using primers bearing 100 bp of homology to the gene locus. These same primers may be used for a more conventional dual-marker gene disruption procedure [5]. Transformation with the PCR product confers a Ura+ phenotype, creating a heterozygous mutant with one allele of the targeted gene replaced by the cassette. Subsequent growth of the Ura+ heterozygote under non-selective conditions, followed by selection on 5-FOA (5-Fluoroorotic acid) plates, yields a Ura− heterozygous strain. 5-FOA is used for identification and selection of strains that are Ura−, as it is toxic to cells that have the URA3 gene. These cells can synthesize the enzyme orotidine-5′-phosphate decarboxylase converting 5-FOA into a toxic compound and are therefore unable to grow on plates containing 5-FOA.The transformation and 5-FOA selection are repeated with the Ura− heterozygote to delete the second allele of the gene, thus generating an unmarked homozygous mutant. Additionally, we describe how use of a distinct primer set for deletion of the second allele can minimize same-allele integration of the cassette in the heterozygous mutant.
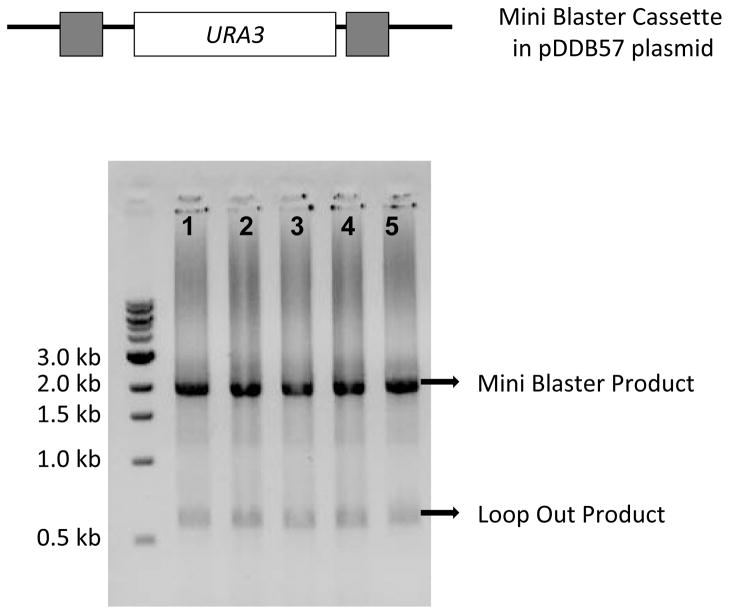
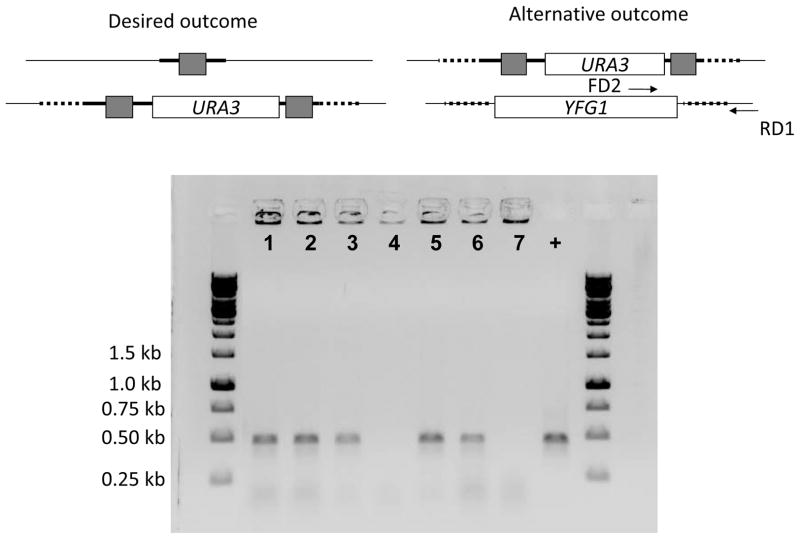
Figure 1.
PCR amplification of the mini blaster cassette from the pDDB57 plasmid. Grey boxes in the diagram indicate the 200 bp repeats flanking the URA3 ORF. In the gel picture, lanes 1 to 5 show the ~2000 bp long cassette and a ~750 bp long loop-out product.
The mini-blaster strategy enables one to generate unmarked deletion mutants, thus permitting creation of multiply mutant strains. This strategy is particularly important for deleting genes with similar sequences or overlapping function where mutations for two or more similar genes in the same background are required to assess gene function. Complementation with a wild type gene copy in these strains can be carried out at the HIS1 gene locus, using vector pDDB78 [6]. We note that pDDB78-based complementation can also be used for homozygous mutants created with ARG4 and URA3 cassettes in the BWP17 strain background [5]. We will discuss the complementation procedure as well. Phenotypic analysis following complementation of each of the genes individually in the deletion background can provide assessment of individual gene function.
2. Materials
2.1 Primer Design
-
Primer set 1 for disrupting first allele of a gene:
F1 (forward primer 1): 100 bp upstream of start codon + adapter sequence (T TTC CCA GTC ACG ACG TT)
R1 (reverse primer 1): 100 bp downstream of stop codon (reverse complement) + adapter sequence (G TGG AAT TGT GAG CGG ATA)
-
Primer set 2 for disrupting second allele (optional):
F2 (forward primer 2): 100 bp downstream of start codon + adapter sequence (T TTC CCA GTC ACG ACG TT)
R2 (reverse primer 2): 100 bp upstream of stop codon (reverse complement) + adapter sequence (G TGG AAT TGT GAG CGG ATA)
Detect primers to confirm cassette integration at the targeted locus.
Complementation primers for complementing the homozygous mutant with a wild type copy of the deleted gene.
2.2 E. coli Plasmid Extraction/Purification
LB+Ampicillin (100 μg/mL) liquid medium: 0.5 % yeast extract, 1 % NaCl, 1 % tryptone, 100 μg/mL Ampicillin.
Plasmid DNA Miniprep kit (Fermentas GeneJET™ Plasmid Miniprep Kit)
2.3 PCR Reaction to amplify the mini-blaster cassette
2.4 Transformation of the mini-blaster cassette into C. albicans
C. albicans strain BWP17 [5]
YPD+Uri (80 μg/μL) plates: 2% glucose, 2% bacto-peptone, 1% bacto-yeast extract, 2% bacto-agar, 80 μg/μL of uridine
YPD+Uri (80 μg/μL) liquid medium: 2% glucose, 2% bacto-peptone, 1% bacto-yeast extract, 80 μg/μL of uridine
Ura-blister cassette PCR product
LATE (0.1M LiOAc in 1X TE buffer): 0.372g/L 1mM EDTA disodium salt, 1.21g/L Tris, 10.2g/L Lithium acetate, pH 7.5
Calf thymus DNA (~10 mg/mL) (Sigma D8661-1ML)
PLATE (8 mL 50% w/v PEG3350 (Sigma) + 1 mL 10X TE + 1 mL 1 M LiOAc): For 50% w/v PEG3350 dissolve 50g PEG3350 in 100 mL (final volume) of distilled water and filter sterilize after mixing; For 10x TE 3.72g/L 1mM EDTA disodium salt, 12.1g 10mM TRIS, pH 7.5; For 1M LiOAc 102g/L Lithium acetate pH 7.5.
CSM-URA plates: 2% glucose, 0.67% yeast nitrogen base (without amino acids), 0.077% of CSM-URA dropout medium (MP Biomedicals, LLC), 2% bacto-agar
2.5 Marker recycling step
YPD+Uri (80 μg/μL) liquid medium (see item 3, section 2.4)
CSM-URA + 5FOA (1g/litre) plates: 2% glucose, 0.67% yeast nitrogen base (without amino acids), 0.077% of CSM-URA dropout medium (MP Biomedicals, LLC), 2% bacto-agar, 5-FOA (1g/litre)
YPD+Uri (80 μg/μL) plates (see item 2, section 2.4)
2.6 Confirmation of Heterozygous mutants
10 μM forward detect primer (FD1)
10 μM reverse detect primer (RD1)
10X PCR buffer with Mg2+: contains 100 mM Tris-HCl pH 9, 15mM MgSO4, 100 mM KCl, 80mM (NH4)2SO4, 0.5% np-40; final MgCl2 concentration of 1.5mM)
dNTPs
Taq DNA polymerase (Denville Scientific Inc.)
2.7 PCR Reaction to amplify the mini-blaster cassette for disruption of second allele
10 μM forward primer (F1 or F2, if desired)
10 μM reverse primer (R1 or R2, if desired)
10X Ex Taq Buffer PCR buffer (contains 20 mM Mg2+)
dNTPs (2.5mM each)
TaKaRa Ex Taq™ TaKaRa Ex Taq™ DNA polymerase (Takara Bio Inc)
Template Plasmid: pDDB57 [2]
2.8 Transformation of the mini-blaster cassette into heterozygous C. albicans recipient
Use reagents in section 2.4, but use the heterozygous yfg1::dpl200/YFG1 strain as the transformation recipient instead of YFG1/YFG1 strain BWP17.
2.9 Preparation of C. albicans Genomic DNA
YPD+Uri (80 μg/μL) liquid medium (see item 3, section 2.4)
TENTS: 1% SDS, 2% Triton X-100, 0.1 M NaCl in 1XTE, filter sterilized.
Acid washed beads (425–600μm), sterilized by autoclaving.
Phenol/chloroform/isoamyl alcohol (25:24:1, v/v).
Ethanol.
1x Tris-EDTA (1xTE), pH 7.4
10 mg/mL RNase.
10 M NH4OAc.
2.10 PCR amplification of gene of interest using complementation primers
10 μM Comp forward primer (CF)
10 μM comp reverse primer (CR)
10X Ex Taq Buffer PCR buffer (contains 20 mM Mg2+)
dNTPs (2.5mM each)
TaKaRa Ex Taq™ DNA polymerase (Takara Bio Inc)
Genomic DNA: Reference strain (see Note 1)
2.11 Construction of complementation plasmid by homologous recombination in S. cerevisiae and plasmid DNA recovery by mechanical disruption
S. cerevisiae BY4741Δ trp strain [7]
YPD plates (see item 2, section 2.4)
YPD liquid medium (see item 3, section 2.4)
pDDB78 plasmid, Pubmed accession number pending [6]
PCR product: wild type gene amplified using complementation primers
Restriction Enzymes NotI and EcoRI (New England Biolabs)
10X NE Buffer 3 (New England Biolabs)
100X BSA (New England Biolabs)
Calf thymus DNA (~10 mg/mL) (Sigma)
PLATE (8 mL 50% PEG3350 (Sigma) + 1 mL 10X TE + 1 mL 1 M LiOAc)
CSM-TRP plates: 2% glucose, 0.67% yeast nitrogen base (without amino acids), 0.074% of CSM-TRP dropout medium (MP Biomedicals, LLC), 2% bacto-agar
CSM-TRP liquid medium: 2% glucose, 0.67% yeast nitrogen base (without amino acids), 0.074% of CSM-URA dropout medium (MP Biomedicals, LLC)
Plasmid DNA Miniprep kit (Fermentas GeneJET™ Plasmid Miniprep Kit)
Acid washed glass beads (size:425–600μm)
2.12 E. coli Transformation
Chemically competent E. coli
LB+Ampicillin (100 μg/mL) plates (see item 1, section 2.1)
Plasmid DNA Miniprep kit (Fermentas GeneJET™ Plasmid Miniprep Kit)
2.13 Transformation of complementation plasmid in C. albicans
C. albicans homozygous mutants
YPD+Uri (80 μg/μL) plates (see item 2, section 2.4)
YPD+Uri (80 μg/μL) liquid medium (see item 3, section 2.4)
Restriction Enzyme NruI (New England Biolabs)
10 X NE Buffer 3 (New England Biolabs)
LATE (0.1M LiOAc in 1X TE buffer)
Calf thymus DNA (~10 mg/mL) (Sigma)
PLATE (8 mL 50% PEG3350 (Sigma) + 1 mL 10X TE + 1 mL 1 M LiOAc)
CSM-HIS plates: 2% glucose, 0.67% yeast nitrogen base (without amino acids), 0.077% of CSM-URA dropout medium (MP Biomedicals, LLC), 2% bacto-agar, 80 μg/μL of uridine
3. Methods
The methods that follow describe a gene disruption procedure in C. albicans in a step-by-step format. We begin with a description of the primer design procedure to be used to amplify the recyclable mini-blaster cassette. This cassette will be transformed into a C. albicans reference strain to integrate by homologous recombination and consequently replace both of the alleles of a specific gene of interest in sequential steps (Figure 2). We will follow up with a detailed protocol to describe the cassette recycling and subsequent detection strategy to confirm the deletion of both alleles in an unmarked homozygous deletion mutant.
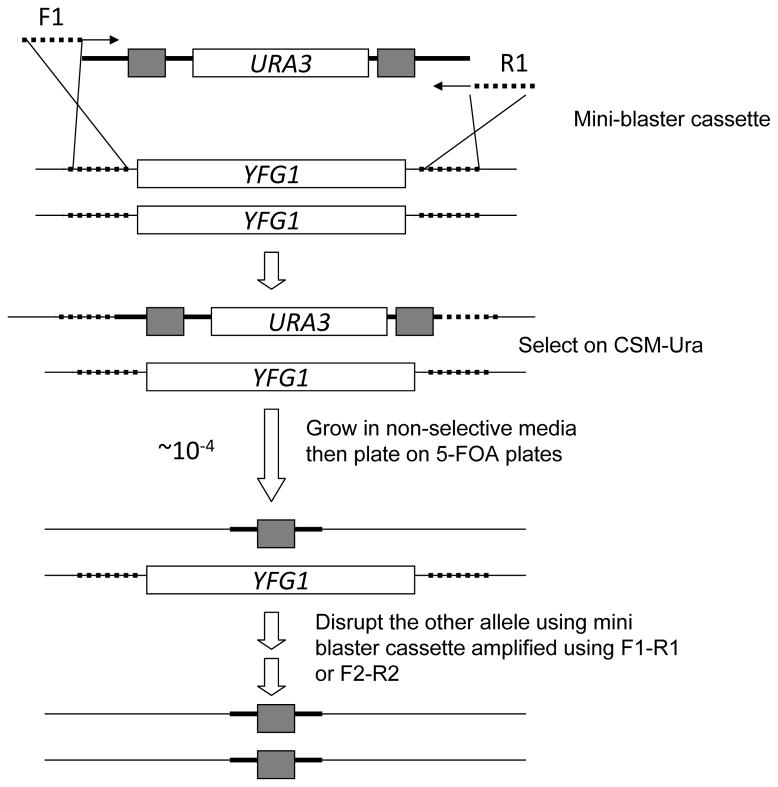
Figure 2.
Schematic showing the mini blaster cassette from the plasmid pDDB57. Outcomes of subsequent transformation events are indicated step-wise in the figure. Dotted lines indicate the 100 bp long regions in the primer sequences bearing homology to the gene locus of interest (YFG1 = Your Favourite Gene 1)
3.1 Primer Design
Refer to the CGD website (http://www.candidagenome.org/) to retrieve the coding sequence of your gene of interest plus 250 base pairs upstream of the start codon and 250 base pairs downstream of the stop codon.
For generating primer set 1; use 100 base pairs of non-coding sequence upstream of the start codon and add the adapter sequence at the 3′ end to generate the forward primer of set 1 (F1; see Materials section 2.1). For the corresponding reverse primer, use 100 base pairs of non-coding sequence downstream of the stop codon (remember to reverse complement) and add the adapter sequence at the 3′ end to generate the reverse primer of set1 (R1; see Materials section 2.1). These primers will be used to delete one copy of the gene of interest to generate a heterozygote.
For generating primer set 2; use 100 base pairs of coding sequence downstream from the start codon and add the adapter sequence at the 3′ end to generate the forward primer of set 2 (F2). For the corresponding reverse primer, use 100 base pairs of coding sequence upstream of the stop codon (remember to reverse complement) and add the adapter sequence at the 3′ end to generate the reverse primer of set 2 (R2) (see Note 2).
For generation of detect primers: Design a forward detect primer 150–300 bp upstream of the start codon (FD1). A corresponding reverse detect should be designed 150–300 bp downstream of the stop codon (RD1). Design an internal gene specific forward primer (FD2) in the region 50–300 bp upstream of the stop codon.
-
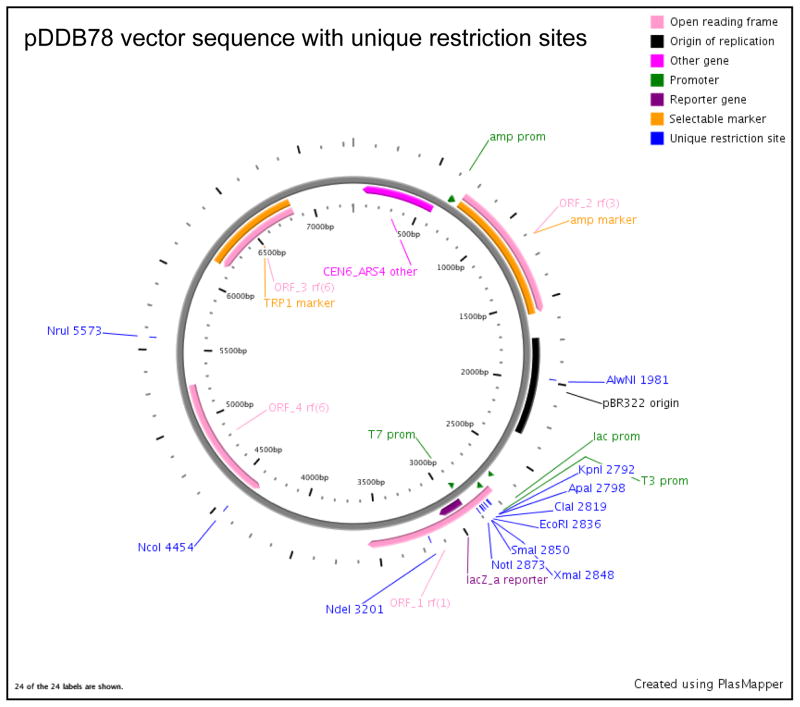
For generation of complementation primers, refer to the CGD website to retrieve the full coding sequence of the gene of interest plus 5′ and 3′ non-coding regions. Typically, ~1500 bp of 5′ non-coding sequence and ~300 bp of 3′ non-coding sequence is sufficient to cover the promoter and terminator sequence, respectively. This may vary depending on the intergenic distance and the orientation of the neighboring genes. Design primers covering the entire above retrieved sequence using the Primer 3 software (http://frodo.wi.mit.edu/primer3/). To the forward and reverse primers designed using the software, add the following adapter sequences (these 40-mer sequences flank the NotI and EcoRI restriction site in the pDDB78 vector, figure 6) at the 5′ end to give the following primers:
Comp forward (CF): TTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCT+ primer 3 forward sequence
Comp reverse (CR): TCGACCATATGGGAGAGCTCCCAACGCGTTGGATGCATAG+ primer 3 reverse sequence
Figure 6.
A restriction map of pDDB78 plasmid generated using the plasmapper program (http://wishart.biology.ualberta.ca/PlasMapper/) indicating the location of unique restriction sites.
3.2 E. coli Transformation, Plasmid Extraction/Purification
Streak out E. coli strain AMB900 (contains plasmid pDDB57) and AMB906 (contains the plasmid pDDB78) on LB+Ampicillin (100 μg/mL) plates to give single colonies. Incubate overnight at 37°C. Pick and grow a single colony of each in 5 ml of LB+Ampicillin (100 μg/mL) liquid medium overnight.
The next day, spin down the cultures and extract plasmid DNA using the Fermentas GeneJET™ Plasmid Miniprep Kit (see Note 3). Store both plasmid preps at −20°C until use. The plasmid pDDB57 can be diluted 1:50 to serve as template for subsequent PCR amplification steps.
3.3 PCR amplification of the mini-blaster cassette
-
A typical PCR reaction composition to amplify the cassette from the plasmid pDDB57 is listed below (see Note 4):
10X PCR buffer 5 μL 5 mM dNTPs 4 μL 10 μM forward primer (F1) 1 μL 10 μM reverse primer (R1) 1 μL Takara Ex Taq™ Polymerase 0.5 μL pDDB57 template (1:50) 1 μL dH20 37.5 μL
Total Volume 50 μL -
The PCR program is listed below (see Note 5):
Step 1 94°C for 5 min Step 2 94°C for 1 min Step 3 56°C for 2 min Step 4 72°C for 3 min Step 5 30 times to Step 2 Step 6 72°C for 8 min Step 7 4°C/End Check 5 μL of the PCR product on a 0.8% DNA agarose gel containing ethidium bromide to confirm a size of ~2000 bp (Figure 1). Typically a smaller amplicon of ~750 bp is also detected, representing loss of the URA3 insert due to cross-priming on the direct repeats. Store the PCR reaction at −20°C until use.
3.4 Transformation of the mini blaster cassette into C. albicans
Streak out C. albicans strain BWP17 [5] for single colonies on a YPD+Uri plate (see Note 6). Grow at 30°C for 2 days.
Culture a single colony in 5 mL YPD+Uri liquid media. Incubate overnight at 30°C with agitation until the culture is saturated.
The following day, dilute the cells to 1:200 in 50 mL YPD+Uri liquid media. Typically, this corresponds to an OD600 = 0.2 on our spectrophotometer.
Incubate the diluted culture at 30°C for 4–5 hou rs for the cells to undergo two doublings. This would correspond to an OD600 = 0.8 if the starting OD600 was 0.2 (see Note 7).
When the C. albicans culture has reached OD600 = 0.8, pour it into a 50 mL conical tube, and spin at low speed (~1000 g) for 5 min.
Discard the supernatant, and wash the cell pellet by gently resuspending it in 5 mL sterile dH2O (do not vortex).
Spin at low speed for 5 min, and discard the supernatant.
Resuspend the cell pellet in 500 μL of LATE.
-
To set up C. albicans transformation reactions, add the following:
LATE cell suspension 100 μL Calf thymus DNA 10 μL PCR reaction from step 3.2 25 μL Mix gently.
Incubate for 30 min at 30°C.
Add 700 μL of freshly made PLATE, and incubate overnight at 30°C (see Note 8).
Heat shock the cell mixture at 44°C for 15 min.
Spin cells down for 30 s at low speed, and aspirate the supernatant.
Wash the cell pellet by resuspending in 1 mL YPD+Uri.
Spin cells down for 30 s at low speed, and decant the supernatant.
Resuspend the cells gently in 100 μL of YPD+Uri, and spread on CSM-Ura plates.
Grow for 2 days at 30°C.
Pick 12 colonies from each transformation plate, streak for singles on CSM-Ura plates, and incubate for 2 days at 30°C. These tra nsformants should be heterozygous mutants.
3.5 URA3 marker excision by recombination
Pick a single colony of each of the 12 transformants (item 19, section 3.4) and inoculate in 2 mL YPD+Uri liquid media.
Grow at ~20–24 hours at 30°C with shaking.
Take 50 μL of the overnight culture solution and plate onto CSM-URA+5FOA (see Note 9) plates.
Incubate for 2 days at 30°C.
Pick one colony from each plate and streak for singles on YPD+Uri plates.
Grow for 2 days at 30°C. These strains should include heterozygous mutants (yfg1::dpl200/YFG1) as well as mitotic recombinants that have become homozygous for the non-disrupted allele of the gene (YFG1/YFG1).
3.6 Confirmation of Heterozygous mutants
-
Colony PCR from single colonies using 2 primers. An example of a typical colony PCR reaction is listed below:
10X PCR buffer 5 μL 5 mM dNTPs 4 μL 10 μM forward detect primer (FD1) 1 μL 10 μM reverse detect primer (RD1) 1 μL dH20 38.5 μL
Total Volume 49.5 μL Add a single colony to the PCR Mixture. Retain some cells from each of the colony to be tested so that it can be further processed if the desired PCR results are obtained.
Before adding the tubes to the PCR block, start the program and pause it at the first step at 94°C. When the block has reached 94 °C, place the tubes containing the PCR mixture and the colony over the block. Allow the thermocycler to boil the colony at 94 °C for 5 min.
Pause the reaction again and add 0.5μL of Taq DNA Polymerase to each tube.
-
Unpause the program again and let it run to completion (see Note 10). A typical PCR programis as follows:
Step 1 94°C for 2 min Step 2 94°C for 45 s Step 3 50°C for 45 s Step 4 72°C for 3 min Step 5 35 times to Step 2 Step 6 72°C for 12 min Step 7 4°C/End Run 10 μL of the above reaction on 0.8% agarose gel containing ethidium bromide. Figure 3 shows an example of a colony PCR from disruption of one allele of a gene of size ~1500 bp. The heterozygous mutant contains the ~1500 bp band corresponding to the wild type allele size (along with some the flanking upstream and downstream regions) as well as the disrupted allele band containing the cassette fragment of ~750 bp (refer to lanes labeled 7, 9 and 12).
Figure 3.
Colony PCR based detection strategy for heterozygous mutants indicating the positions of detect primers (FD1 and RD1). Shown in the gel picture are colony PCRs from 14 independent transformants (lane 1 to 14). Plus indicates positive control band in the reference strains encompassing an ORF of size ~1500 bp. Presence of the wild type ORF band (~1500 bp) and a gene disruption band (~750bp) indicates a heterozygous mutant; as observed in lanes 7, 9 and 12.
3.7 Creation of Homozygous mutants
Use primer set 1 (F1 and R1) or primer set 2 (F2 and R2) to amplify and check the cassette as per step 3.3. We use primer set 2 when we have been unable to create homozygous mutants through repeated use of primer set 1. Primer set 2 creates a PCR product that is preferentially targeted to the non-disrupted YFG1 allele in the yfg1::dpl200/YFG1 heterozygote.
Transform the cassette from the above step into the heterozygous strains (obtained after step 3.5) in the same way as the protocol described in step 3.4. Streak the transformants on CSM-URA plates for single colonies.
Perform a colony PCR check to ascertain integration of the cassette at the desired (previously undisrupted locus). Colony PCR using a single colony, this time, using FD2 and RD1 primer sets. The absence of a wild type band would indicate a putative homozygous mutant.
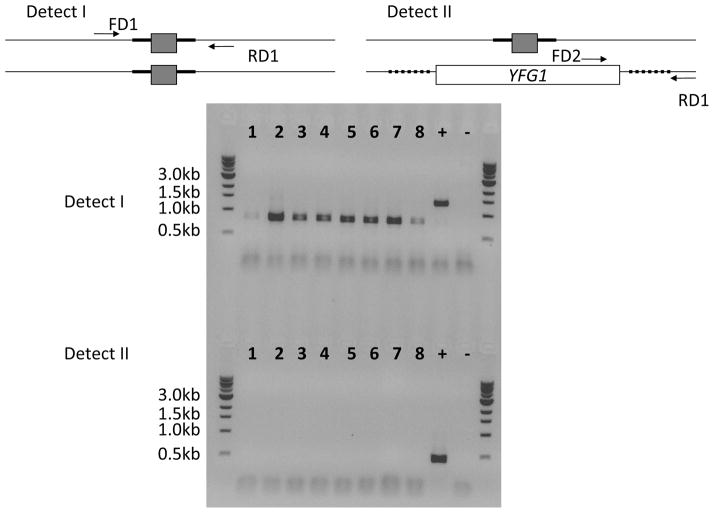
Run 10 μL of the above reaction on 0.8% agarose gel containing ethidium bromide. Figure 4 shows an example of a colony PCR indicating integration of the cassette into a previously undisrupted locus. Loss of the ~500 bp wild type band indicates a putative homozygote (refer to lanes labeled 4 and 7). Use the same colony for the next marker recycling step. Although we are relying on a negative result at this step, we will confirm our observation of a putative homozygous mutant in the next step by performing genomic DNA extractions and subsequent PCR test.
Follow the same instructions for marker recycling (refer step 3.5). The strains at this stage are predicted to be homozygous Ura− mutants. Streak for singles on YPD + Uri plates to prepare frozen stocks and to perform genomic DNA extractions for genotype verification.
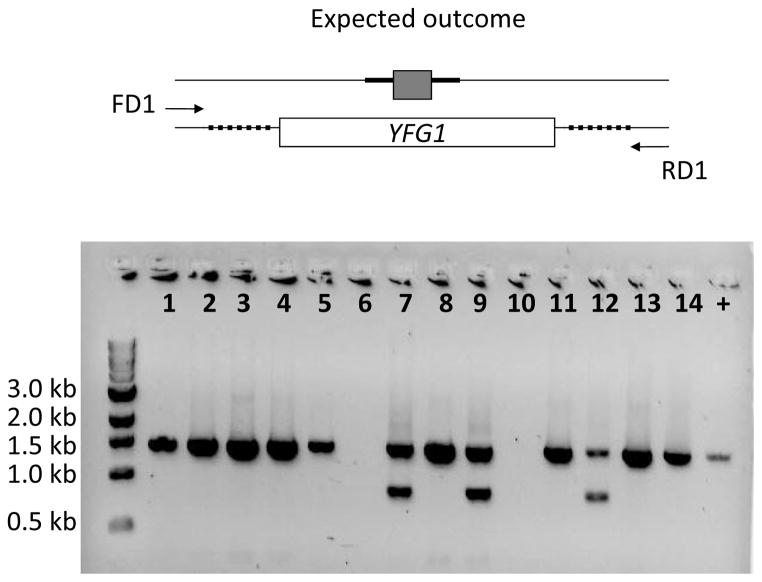
Figure 4.
Colony PCR based detection for putative homozygous mutants indicating the positions of detect primers (FD2 and RD1). Lanes 1 to 7 show PCR analysis on independent colonies. Two possible situations indicated by desired and alternative outcome are shown in the diagram. In the gel picture, lanes marked 4 and 7 indicate an expected outcome situation where the previously undisrupted allele is replaced by the cassette leading to loss of a wild type detect band (refer to the positive control lane).
3.8 C. albicans genomic DNA preparation to confirm homozygous mutants
Inoculate a 5 mL culture of YPD or YPD+Uri liquid media with C. albicans the positive homozygous strains from step 5 above and a reference strain (see Note 1). Incubate the culture overnight with agitation at 30°C.
Spin down the 5 mL culture at low speed, and aspirate supernatant.
Add 500 μL TENTS, and resuspend.
Transfer the resuspended mixture to a fresh microfuge tube containing 200 μL of sterile acid washed beads.
Add 500 μL phenol/choloroform/isoamylalcohol.
Vortex for 2 min.
Spin down tubes at 5000 gin a benchtop microfuge at 4°C for 10 min.
Transfer the aqueous (top) phase to a new microfuge tube.
Add 1 mL 100% ethanol.
Place at −20°C for at least 1 h.
Spin down tubes at 14,000 rpm at 4°C for 15 min.
Aspirate supernatant, and resuspend in 200 μL 1xTE.
Add 1 μL 10 mg/mL RNase.
Incubate at room temperature for 30 min.
Add 40 μL 10 M NH4OAc.
Add 500 μL 100% ethanol, and mix by inversion.
Place at −20°C for 30 min.
Spin down tubes at 14,000 rpm for 5 min. Decant supernatant.
Add 1 mL 70% Ethanol to the pellet, and immediately decant.
Place tubes open in a speed vacuum for 5–10 min or until dry.
Resuspend pellet gently in 50 μL 1xTE (see Note 11). Dilute 1:100 (1μL in 100 μL distilled water) to be used as template for PCR in the subsequent step.
-
Set up PCRs using the following PCR protocol. Use both forward detect primers FD1 and FD2 with reverse detect RD1, in separate reactions, to confirm a homozygous strain.
10X PCR buffer 5 μL 5 mM dNTPs 3 μL 10 μM forward detect primer (FD1) 1 μL 10 μM reverse detect primer (RD1) 1 μL Taq DNA Polymerase 0.5 μL Genomic DNA template (1:100) 1 μL dH20 37.5 μL
Total Volume 50 μL A typical PCR program is as follows:
Step 1 94°C for 2 min Step 2 94°C for 45 s Step 3 50°C for 45 s Step 4 72°C for 3 min Step 5 35 times to Step 2 Step 6 72°C for 12 min Step 7 4°C/End Run 10μL of the reaction on a 0.8% agarose gel containing ethidium bromide. The homozygous mutant contains the ~750 bp gene disruption band (labeled “Detect I” in Figure 5) and lacks the control band using a forward primer (FD2) intrinsic to the gene of interest (labeled “Detect II” in Figure 5). It should be noted that for Detect I strategy, we observe two closely spaced gene disruption bands as a second primer set F2-R2 was used to disrupt the second allele. Size differences between the double bands should be 200 bp.
Figure 5.
Genomic DNA confirmation for putative homozygous mutants indicating the positions of detect primers (FD1, FD2 and RD1) in the diagram. Lanes marked 1 to 8 represent transformants that yielded a negative result in the previous step. Detection strategy I using primers FD1 and RD2 confirms loss of the ~1500 bp wild type ORF band and presence of a gene disruption band (~750 bp). Note that we observe two closely spaced gene disruption bands as a second primer set, F2-R2, was used to disrupt the second allele. Size differences between the double bands should be 200 bp. Detection strategy II using primers FD2 and RD1 additionally confirms loss of the wild type band (~500 bp). Plus sign indicates corresponding positive controls using reference strain genomic DNA. Minus sign indicates no template controls.
3.9 PCR amplification of the gene of interest using complementation primers
-
1
A typical PCR composition to amplify the coding sequence of the gene of interest with the 5′ and 3′ non-coding sequences is as follows (Use genomic DNA of the reference strain diluted 1:100 from step 3.8 as template, see Note 4):
10X PCR buffer 5 μL 5 mM dNTPs 4 μL 10 μM forward primer (F1) 1 μL 10 μM reverse primer (R1) 1 μL Takara Ex Taq™ Polymerase 0.5 μL Genomic DNA template (1:100) 1 μL dH20 37.5 μL
Total Volume 50 μL -
2
The PCR program is listed below (see Note 10):
Step 1 94°C for 5 min Step 2 94°C for 1 min Step 3 56°C for 2 min Step 4 72°C for 3 min Step 5 30 times to Step 2 Step 6 72°C for 8 min Step 7 4°C/End -
4
Check 10 μL of the PCR product on a 0.8% DNA agarose gel containing ethidium bromide to confirm a band of wild type gene size. Store the PCR reaction at −20°C until use.
3.10 Construction of complementation plasmid by homologous recombination in S. cerevisiae and plasmid DNA recovery by mechanical disruption
Streak out S. cerevisiae strain BY4741Δ trp [7] for single colonies on a YPD plate. Incubate at 30°C for 2 days.
Culture a single colony in 5 mL YPD liquid media overnight at 30°C with agitation.
-
The following day, digest 3 μL of the pDDB78 DNA prep using the following restriction enzymes (see Note 12):
Extracted plasmid DNA 3 μL 10X NE Buffer 3 2 μL 100X BSA 0.2 μL NotI 0.2 μL EcoRI 0.2 μL dH2O 14.4 μL
Total Volume 20 μL Mix gently.
Allow the reaction to digest for 2 h at 37°C.
Run 10 μL of digestion reaction on an agarose gel containing ethidium bromide. Both NotI and EcoRI sites lie in the polylinker region close (Figure 6) to each other, so a linearized vector sequence of ~7300 bp should be apparent on the gel. Inactivate the restriction enzyme by incubating the digest at 65°C for 20 minutes.
-
Place 500 μL of the overnight saturated culture of strain BY4741Δ trp in each of 3 separate microfuge tubes. Centrifuge at 1450 g for 2 min and remove most of the supernatant (leave ~100 μL). Add the following in order to three tubes separately:
No DNA: 10 μL of calf thymus DNA
Cut vector: add 1–3 μL of digested vector + 10 μL of calf thymus DNA
Cut vector + PCR insert: add 1–3 μL of digested vector + 10 μL of calf thymus DNA + 5 μL of PCR from step 3.9.
Mix gently.
Add 500 μL of freshly made PLATE, and incubate overnight at 30°C.
The next day, centrifuge the tubes at 4000 rpm for 2 min, wash pellet with distilled water and plate onto CSM-TRP plates. Incubate plates at 30°C for 3 days.
Pick 10 colonies (more if desired) from the vector + insert plate and culture each in 5ml CSM-TRP liquid medium at 30°C with agitation (see Note 13). The ‘cut vector’ plate serves as a negative control in the experiment to quantitate the number of transformants obtained by transforming with the vector alone. It should have no colonies or very few colonies compared to the ‘cut vector plus PCR insert’ plate.
The next day, spin down the cells, discard the supernatant, and add 250 μL resuspension solution from the GeneJET™ Plasmid Miniprep Kit (Fermentas) kit (see Note 14). Add the resuspended solution to a screw-cap microfuge tube containing 500 μL glass beads.
Vortex the screw cap tubes at top speed in a bench microfuge for 5 min to disrupt cells.
Add 250 μL lysis buffer, mix 4–6 times and incubate for 5 min.
Add 350 μL of neutralizing buffer, mix immediately and centrifuge at top speed (>4500 g) for 10 min.
Transfer the supernatant to the filter unit provided with the kit. Centrifuge at top speed for ~ 30–60 s and discard the eluate in the collection unit.
Add 700 μL of wash solution (containing ethanol, refer to kit instructions) and centrifuge again at top speed for ~30–60 s. Discard the eluate in the collection unit.
Centrifuge again for 60 s at to remove any residual solution on the filter.
Transfer the filter unit onto a fresh microfuge tube and add 30–50 μL of elution buffer.
Centrifuge at top speed for 1 min and store the 30–50 μL eluate in the microfuge tube at −20°C for use in subsequent step of E. coli transformation.
3.11 E. coli Transformation
Thaw chemically competent E. coli on ice.
Add 25 μL of undiluted complementation plasmid from yeast preps to appropriate volume (~50 μL) of thawed chemically competent E. coli.
Incubate on ice for 10 min.
Heat shock mixture at 42°C for 45 s.
Place on ice for 5 min.
Add 1 mL LB, and incubate for 1 h at 37°C with a gitation.
Spin down cells at low speed for 1 min, decant supernatant, and resuspend cell pellet in 100 μL dH2O.
Spread on LB+Amp plates. Incubate overnight at 37°C.
Select 8 white colonies, and inoculate separately into 2 mL LB+Amp liquid media.
Incubate overnight at 37°C with agitation.
Spin down cultures and extract plasmid DNA using the GeneJET™ Plasmid Miniprep Kit (Fermentas) kit.
Digest the plasmids using appropriate restriction enzymes to confirm the sequence (see Note 15).
3.12 Transformation of complementation plasmid into C. albicans
-
1
Streak out the C. albicans homozygous mutant strain for single colonies on a YPD+Uri plate (see Note 6). Incubate at 30°C for 2 days.
-
2
Culture a single colony in 5 mL YPD+Uri liquid media overnight at 30°C with agitation until saturated.
-
3
The following day, dilute the sample to 1:200 in 50 mL YPD+Uri liquid media. Typically, this corresponds to an OD600 = 0.2 on our spectrophotometer.
-
5
Incubate the diluted culture at 30°C for 4–5 hours for the cells to undergo two doublings. This would correspond to an OD600 = 0.8 if the starting OD600 was 0.2 (see Note 7).
-
6
Digest 10μL of the pDDB78 DNA and the complementation plasmid separately using the following restriction enzymes (see Note 16):
Extracted plasmid DNA 10 μL 10X NE Buffer 3 2 μL NruI 0.5 μL dH2O 7.5μL
Total Volume 20 μL -
7
Mix gently.
-
8
Incubate at 37°C for 2–4 hours and confirm for linearized plasmid on an agarose gel containing ethidium bromide (optional, to confirm digestion).
-
9
When the C. albicans culture has reached OD600 = 0.8, pour it into a 50 mL conical tube, and spin at low speed (~1000 g) for 5 min.
-
10
Discard the supernatant, and wash the cell pellet by gently resuspending it in 5 mL sterile dH2O (do not vortex).
-
11
Spin at low speed for 5 min, and discard the supernatant.
-
12
Resuspend the cell pellet in 500 μL of LATE.
-
13
To set up C. albicans transformation reactions, add the following:
LATE cell suspension 100 μL Calf thymus DNA 10 μL PCR reaction from step 3.2 25 μL -
14
Mix gently.
-
15
Incubate for 30 min at 30°C.
-
16
Add 700 μL of freshly made PLATE, and incubate overnight at 30°C ( see Note 8).
-
17
Heat shock the cell mixture at 44°C for 15 min.
-
18
Spin cells down for 30 s at low speed, and aspirate the supernatant.
-
19
Wash the cell pellet by resuspending in 1 mL YPD+Uri.
-
20
Spin cells down for 30 s at low speed, and decant the supernatant.
-
21
Resuspend the cells gently in 100 μL of YPD+Uri, and plate on CSM-HIS plates.
-
22
Incubate for 2 days at 30°C.
-
23
Pick 12 colonies from each transformation plate, streak on CSM-HIS plates, and incubate for 2 days at 30°C. These transformants should be His+ mutants and complemented for the deleted gene. Check for the absence of wild type gene sequence (in the PDDB78-only transformants) and the presence of the wild type gene sequence (in the complementation plasmid transformants); using colony PCR procedure described in step 3.6. The detect primer sets FD2 and RD1 can be used for this purpose. Follow up with phenotypic analyses to confirm the genotype.
Acknowledgments
This work was supported by NIH grant R01 AI067703 to APM. We thank Dr. Carol A. Woolford for her comments on this chapter.
Footnotes
We typically extract genomic DNA from reference strains BWP17 (Arg− Ura− His−) [5], DAY185 (Arg+ Ura+ His+) [8], or DAY286 (Arg+ Ura+ His−) [9].
These primer sequences provide unique regions of homology to the undisrupted allele in a heterozygote; where one allele has been disrupted using the primer set 1. Thus we designate the primers F2 and R2 as optional above (section 2.1), because homozygous deletions for many genes can be accomplished simply through repeated use of F1-R1 amplified PCR products.
Plasmid DNA can be extracted and purified using several methods or several commercially available kits depending on the number of samples. For our purpose, the GeneJET™ Plasmid Miniprep Kit (Fermentas) worked well.
We have used the high fidelity Takara Ex Taq™ DNA polymerase (with proofreading activity) in two instances for amplification of the mini-blaster cassette and the disrupted gene of interest (for complementation purposes). Any other high fidelity DNA polymerase of choice could be used as well. For all other detect PCR reaction described in the paper we recommend use of Taq DNA polymerase due to cost considerations. For our detection PCRs on colony PCR and genomic DNA preps, the Taq DNA Polymerase from Denville Scientific was used.
The following PCR reaction is optimal to amplify the mini-blaster cassette. The PCR reaction produce ~2000 bp long cassette with a background loop-out product of ~750 bp size. We do not perform PCR extraction/purification as long as the ~2000 bp long band is present.
C. albicans strains that are Ura− (with the essential URA3 gene disrupted) require supplementation with uridine at 80 mg/l because disruption of URA3 blocks the ability to synthesize uridine. Supplementation with uracil, which is typical for S. cerevisiae media recipes, is not adequate for C. albicans Ura− cell growth.
The doubling time of C. albicans is ~1.5 h.
Incubating transformations in PLATE for greater than 16 h significantly reduces the transformation efficiency.
5-FOA plates should be preferably prepared fresh and kept in dark at 4°C until use. It is advisable to pick up and streak colonies to YPD plates after a maximum of 2–3 days of incubation on 5-FOA at 30°C.
This is a generalized PCR program that works for most C. albicans ORFs less than 4 kb in length. Larger ORFs may require longer extension times.
We try to avoid shearing genomic DNA by limiting vortexing.
The restriction sites for NotI and EcoRI lie in the polylinker sequence of the pDDB78 plasmid; simultaneous digestion with these enzymes linearizes the plasmid which promotes in vivo homologous recombination with the PCR product in the S. cerevisiae strain.
S. cerevisiae strains bearing episomal plasmids need to grown in selective conditions (CSM-TRP) in order to retain the plasmid.
We have used the modified GeneJET™ Plasmid Miniprep Kit (Fermentas) protocol to recover plasmids from S. cerevisiae cells. However, other commercially available kits or user developed protocols with other plasmid DNA prep kits may be used.
Refer to the pDDB78 plasmid map to select restriction enzymes. For other vectors, use appropriate enzymes keeping in mind the gene sequence and the vector sequence. If your gene of interest has an NruI site in the sequence, use vector pRYS2 digested with SrfI for complementation [10].
The purpose of digesting both the complementation vector and the backbone vector (pDDB78) is to make both the complemented and the mutant strain His+ for downstream analysis. 100X BSA is not required for NruI activity.
References
- 1.Berman J, Sudbery PE. Candida Albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- 2.Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spreghini E, Davis DA, Subaran R, Kim M, Mitchell AP. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot Cell. 2003;2:746–755. doi: 10.1128/EC.2.4.746-755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis DA, Bruno VM, Loza L, Filler SG, Mitchell AP. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics. 2002;162:1573–1581. doi: 10.1093/genetics/162.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault JS, et al. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell. 2008;19:2741–2751. doi: 10.1091/mbc.E08-02-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]