Abstract
The arrest of replication forks due to collisions with transcription complexes leads to genomic instability and cell death. Mechanisms that promote the progression of replication forks past transcription complexes are therefore essential for propagation and preservation of the genome. Recent studies of E. coli directly investigate the consequences of collisions of the replisome with RNAP polymerase (RNAP) in vitro and provide novel mechanisms by which these encounters may be resolved. Additionally, recent in vivo and in vitro studies support the longstanding hypothesis that auxiliary DNA helicases promote replication through roadblocks such as transcription complexes. Here we review past and recent advances that formulate our current understanding of how the bacterial replisome deals with transcription complexes along the path of chromosome duplication.
Keywords: replication, transcription, replisome, RNA polymerase, collision, helicase, recombinational repair, transcription-coupled repair, genomic instability
Introduction
How replication forks contend with transcription complexes during the course of DNA replication has been an outstanding question in biology for over 20 y. Replication fork arrest due to encounters with transcription complexes can lead to DNA damage response, mutagenesis and chromosomal deletions.1,2 This suggests replication forks may occasionally collapse after colliding with a RNA polymerase (RNAP). Mechanisms that prevent collapse of the fork following replisome-RNAP collisions are therefore necessary for propagation and preservation of the genome. In bacteria, replisome-RNAP collisions occur frequently since the rate of replication is 12–30 times greater than the rate of transcription and there is no separation of these processes.3 Bacteria must therefore employ mechanisms that efficiently deal with regular conflicts between replication and transcription in a manner that does not compromise genomic integrity.
In Vivo Studies of Replisome-RNA Polymerase Collisions
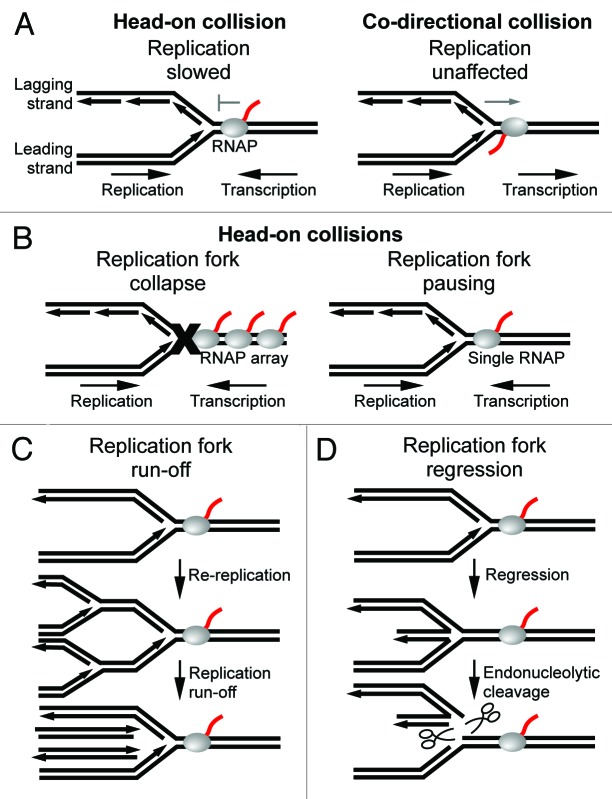
Most studies of replisome-RNAP collisions have been performed in E. coli and B. subtillus cells where the replication origin (oriC) and highly transcribed regions such as ribosomal (rRNA) operons are well defined. For example, a seminal study used electron microscopy to examine the directional effect of ribosomal transcription on fork progression by inverting ribosomal operons, which are normally co-directionally aligned with replication fork movement.4 Later studies similarly investigated the directional effect of transcription on replication of plasmid DNA in E. coli by two-dimensional gel electrophoresis of replication fork intermediates.5 More recently, replication fork progression through genome-wide transcription units has been investigated in B. subtillus using DNA microarrays.1,6 Together, these studies find replication to be slower through head-on transcription units, which oppose the fork, as compared with co-directional transcription units, which face the same direction as the fork (Fig. 1A). A similar bias of replication being inhibited by head-on transcription has been observed in yeast, which suggests that the outcome of replisome-RNAP collisions may be conserved.7
Figure 1. Effects of transcription complexes on the progression of replication forks. (A) Replication is slowed upon collision with head-on transcription complexes (left), but is unaffected by co-directional transcription complexes (right). (B) Replication forks are arrested and collapse upon encountering a head-on RNAP array (left). Replication forks pause, but remain intact upon encountering a single head-on RNAP (right). (C) Replication fork pausing due to a head-on collision may lead to double-strands breaks as a result of replication run-off of upstream forks. (D) Replication forks may regress following a head-on collision which leads to endonucleolytic cleavage of the DNA by RuvABC.
Interestingly, the severity of replication fork arrest due to head-on transcription seems to be correlated with the level of gene expression. For example, a recent study in B. subtillus demonstrated that only heavily transcribed genes such as ribosomal operons significantly impeded replication when they were inverted to oppose the fork.1 This explains why the organization of chromosomes was selected to minimize replication through highly expressed head-on transcription units. For example, highly expressed and essential genes such as ribosomal operons are co-directional with replication in all known bacteria.8 In eukaryotes, head-on collisions within ribosomal operons are prevented by protein-nucleic acid complexes called replication fork barriers which block replisomes from entering the 3′ end of these genes.9 Similar to bacteria, highly expressed genes in higher eukaryotes are in close proximity to replication origins and are directed away from origins which minimizes the frequency of head-on collisions in the cell.10 Taken together, having replication oppose transcription within highly expressed genes is probably deleterious to cells, presumably due to genomic instability caused by frequent replication fork arrest. This idea is supported by the aforementioned study in B. subtillus which demonstrated that reversing the orientation of ribosomal genes resulted in induction of the SOS response, genomic instability and cell death.1
Inhibition of replication within head-on ribosomal genes may be attributed to specific transcription regulatory mechanisms that promote the abundant synthesis of rRNA. For example, Nus antitermination factors promote transcription of ribosomal operons by increasing the rate of transcription elongation and preventing premature termination by the essential transcription termination factor Rho.11,12 Furthermore, transcription from ribosomal promoters is enhanced 200–300 fold due to the combined action of cis-acting upstream sequences (UP elements) and the transcription factor Fis.13 Transcription of ribosomal operons is also modulated in response to nutrient availability by the stringent response regulator (p)ppGpp along with transcription factor DksA.13 In this way, ribosomal transcription may be rapidly upregulated when growth conditions are optimal. Indeed, transcription of rRNA can account for 50% of total RNA synthesis in the cell.13 Such high transcription activity leads to the formation of RNAP arrays as revealed by electron microscopy.14 RNAP arrays are likely to facilitate cooperation among RNAPs, which has been shown to promote transcription through pause sites and high affinity protein roadblocks within highly expressed genes.15 The combined force of multiple RNAP motors directed against replisome movement provides a plausible explanation of why replication forks are arrested and probably collapse within inverted ribosomal operons (Fig. 1B, left). Head-on transcription inhibition of replication, however, is not exclusive to inverted ribosomal genes. Thus, a relatively high level of gene expression, such as from the strong T7A1 promoter, may be the only requirement for impeding replication within head-on genes.16
Although it is clear that replication is arrested within highly expressed head-on transcription units, the fate of the replication fork following collisions with head-on transcription complexes remains elusive. For example, a recent in vivo study of E. coli indicated that replication through inverted (head-on) ribosomal genes did not require RecA. This suggests that replication forks remain intact following head-on collisions since RecA is necessary for recombinational repair and reassembly of collapsed forks.17 In contrast, more recent reports suggest that replication forks collapse within inverted ribosomal genes in E. coli and B. subtillus cells.1,18 The recent finding that replication is only significantly impeded within highly expressed head-on transcription units suggests that replication forks pause after colliding with a single head-on RNAP, but collapse after colliding with a head-on RNAP array—which presumably only forms within highly expressed genes (Fig. 1B).1 However, since highly transcribed regions are exclusively co-directional with replication in wild-type cells, replication forks probably remain mostly intact as they traverse the genome. This idea is supported by the fact that only 18% of cells require reassembly during the course of replication in E. coli.19
Importantly, genomic integrity may be compromised as a result of head-on collisions even within transcription units that are not highly expressed.1,2 For example, a recent in vivo study of B. subtilus demonstrated that inverting the rpoB gene, which encodes the large β subunit of RNAP, resulted in mutations within the gene.1 The rpoB gene is among the longest transcription units in bacteria and therefore probably contains several RNAPs which increases the possibility of fork arrest. Consequently, longer genes were selected to be co-directional with replication which minimizes gene mutations that may occur due to head-on collisions.20 DNA deletions have also been observed at the site of a head-on collision along plasmid DNA in E. coli.2 Similarly, recombination of DNA due to head-on collisions has been demonstrated in yeast, which is referred to as transcription associated recombination.7 Replication forks may infrequently dissociate or stall when encountering a single head-on RNAP which may lead to DNA recombination. For example, since replication is repeatedly initiated from the origin in bacteria, prolonged stalling of the replisome may result in double-strand breaks due to replication run-off of an upstream fork (Fig. 1C). Furthermore, a stalled replication fork may regress which can also lead to double-strand breaks due to endonucleolytic cleavage of the DNA by the Holiday junction resolvase RuvABC (Fig. 1D). The observed mutations and deletions near collision sites may be due to erroneous recombinational repair following fork collapse. However, it is also possible that mutagenesis may result from error-prone DNA polymerase activity since translesion DNA polymerases have been proposed to switch with the replicative DNA polymerase after it becomes arrested.21 Alternatively, error-prone DNA polymerases may promote mutagenesis during recombinational repair of the fork since translesion DNA polymerases may act at recombination intermediates.22 Further studies are needed to determine the mechanisms by which DNA is altered at head-on collision sites.
Most in vivo studies have investigated the effects of entire transcription units on the progression of replication. The characteristics of transcription complexes, however, vary according to their particular phase of transcription and thus may have different effects on replication depending on their location along genes. For example, during transcription initiation RNAP forms an open promoter complex then repeatedly synthesizes short abortive RNA products while remaining bound to the promoter. Transcription initiation complexes are highly unstable. Thus, they are not likely to have a significant effect on replication. As RNAP leaves the promoter it undergoes a major conformational change which facilitates the transition from initiation phase to elongation phase. In contrast to initiation complexes, RNAP elongation complexes are highly stable and processive and are therefore likely to have the greatest effect on replication. RNAP undergoes a final structural transition during termination which can occur by a factor-dependent or factor-independent mechanism that leads to displacement of the RNAP and transcript from DNA. A recent in vivo study of E. coli investigated the effects of transcription initiation and termination complexes on the progression of the replication fork.16 Surprisingly, replication was impeded at the 3′ end of a co-directional transcription unit, which suggests the replisome has difficulty passing a co-directional transcription termination complex. Replisome pausing at the terminator was proposed to be due to RNAP backtracking since deleting GreA and GreB, which inhibit backtracking, exacerbated fork arrest. It was also found that replication was impeded after a head-on collision with an initiation complex, which is highly unstable and therefore not expected to affect fork movement. These studies indicate that replisome activity may be affected by different structural conformations of RNAP along DNA (i.e., initiation, elongation, termination complexes).
In Vitro Studies of Replisome-RNA Polymerase Collisions
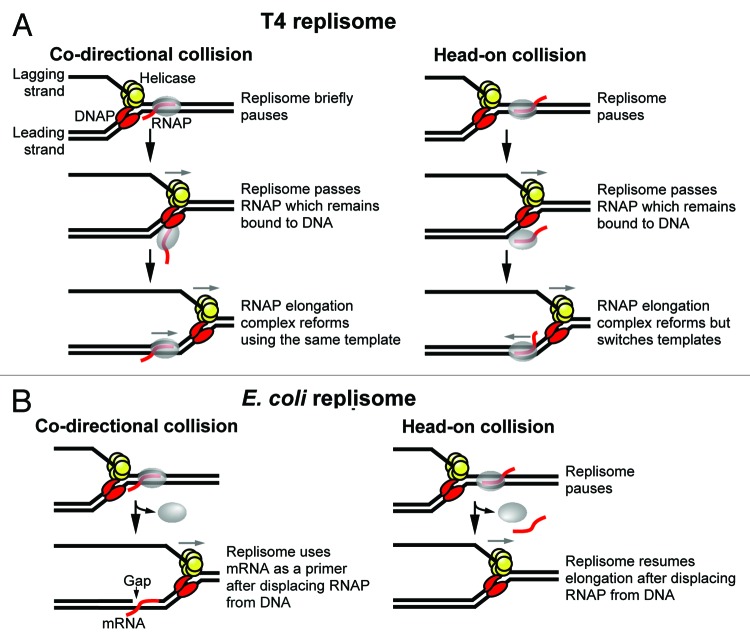
Insight into the outcome of replisome-RNAP collisions at the molecular level has been provided by in vitro studies. Bruce Alberts’ laboratory was the first to reconstitute collisions of a replication apparatus with RNAP in vitro using T4 as a model system.23,24 These studies found that the replisome paused upon collison with RNAP in either orientation, however, the duration of the pause was longer during a head-on collision which supports the general consensus that replication is slower through head-on vs. co-directional transcription (Fig. 1A). Strikingly, these studies suggested that the RNAP and transcript remained bound to the DNA as the replisome passed by (Fig. 2A). For example, in the case of a co-directional collision the transcription complex remained active as indicated by the ability of RNAP to resume transcription elongation following the collision (Fig. 2A, left).24 A head-on transcription complex also retained the ability to extend its transcript following a collision, however, the RNAP and transcript switched strands; the lagging strand was used as a template prior to the collision, whereas the newly synthesized leading strand was used as a template after the collision (Fig. 2A, right).23 These results are difficult to reconcile with the fact that the replisome occupies both strands during replication elongation—the leading strand polymerase binds to the leading strand and the replicative helicase binds to the lagging strand (Fig. 2A). Therefore, in either orientation, the transcript and RNAP must dissociate from the DNA in order to allow passage of the replisome. It was therefore proposed that the RNAP and transcript temporarily disengaged from the template but remained tethered to the DNA, and that the transcription complex reformed after the replisome passed by (Fig. 2A). It is now widely accepted that displacement of the RNA destabilizes a transcription elongation complex and results in release of the RNAP from DNA. Therefore, it remains unclear how the RNAP and transcript might disengage from the template then re-associate into an active elongation complex. Nevertheless, the ability of a bona fide transcription elongation complex to assemble from individual components of an elongation complex (i.e., RNAP core, transcript, template) has been well documented.25 Thus, further investigation of whether an RNAP elongation complex may temporarily disengage from the DNA to allow passage of a replication fork may be warranted.
Figure 2. Models of replisome-RNAP collisions performed in vitro. (A) (left) The T4 replisome briefly pauses upon encountering a co-directional RNAP. The RNAP and transcript remain bound to the DNA as the replisome passes then an active RNAP elongation complex reforms using the same template. (right) The T4 replisome pauses after colliding with a head-on transcription complex then passes the RNAP which remains bound to the DNA. An active RNAP elongation complex reforms using the newly synthesized leading strand as a template. (B) (left) The E. coli replisome uses mRNA as a primer after colliding with a co-directional RNAP that is displaced from the DNA. (right) The E. coli replisome pauses after colliding with a head-on RNAP then resumes elongation after displacing the RNAP from DNA.
The consequences of a collision of the replisome with a head-on RNAP has been recently investigated in vitro using E. coli as a model system.26 Similar to the findings of the T4 study, the E. coli replisome paused during a head-on collision with RNAP (Fig. 2B, right). However, in contrast to the T4 study, RNAP was displaced from the DNA. Importantly, the replisome remained intact following a head-on collision as demonstrated by the lack of a requirement for reloading the replicative DnaB helicase. These findings support previous in vivo studies which suggest that replication forks are able to pause for extended periods without collapsing.9 However, recall that stalled replisomes eventually dissociate due to replication run-off (Fig. 1C).9
Recent in vitro studies have also investigated a collision of the E. coli replisome with a co-directional RNAP.27 Surprisingly, leading strand synthesis was terminated upon collision with the RNAP, but was then reinitiated by using the transcript as a primer (Fig. 2B, left). This process therefore resulted in a gap in the leading strand. It was further found that the replisome remained intact and bound to the DNA after the collision, whereas the RNAP was displaced. Thus, in contrast to the results of a head-on collision, the replisome was not impeded by a co-directional RNAP, in agreement with in vivo studies. Importantly, the finding that a gap was left in the leading strand following a co-directional collision supports the discontinuous model of replication in which both the leading and lagging strands are synthesized in a discontinuous fashion.28 These data therefore suggest the possibility that discontinuities detected in the leading strand in vivo may be due, at least in part, to co-directional replisome-RNAP collisions. Future studies are needed to determine whether transcription complexes contribute to the formation of gaps in the leading strand in vivo.
Role of RNA Polymerase Modulators in Resolving Collisions
The in vitro studies described above focused on the effects of halted RNAP elongation complexes on replication. These data may therefore more accurately reflect the outcome of replication fork encounters with RNAPs stalled at lesions in the DNA. Several mechanisms of DNA repair involving recombination factors are available to deal with collapsed forks at lesions which ultimately promote replication restart.29 However, since RNAP is highly abundant in the cell, transcription complexes probably become arrested at lesions prior to replication forks.30 The arrest of a single RNAP due to DNA damage has been shown to lead to a backed-up array of RNAPs which probably impedes replication, especially when opposing the direction of the fork (Fig. 3A, left).31
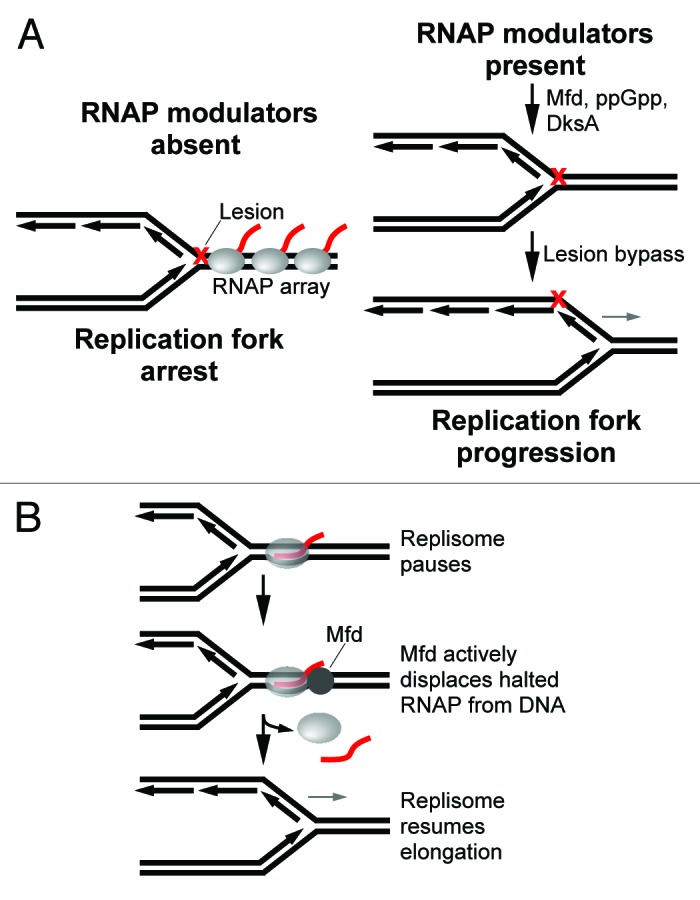
Figure 3. Models of RNAP modulator activity in reducing conflicts between replication and transcription. (A) (left) Replication fork blocking RNAP arrays may form at lesions in the DNA when RNAP modulators are absent. (right) RNAP modulators indirectly promote replication by preventing the formation of RNAP arrays. DNA repair pathways may then promote replication past DNA damage. (B) Mfd promotes fork progression after a head-on collision by facilitating displacement of the RNAP block ahead of the stalled fork.
Genetic studies have found that RNAP modulators such as Mfd, ppGpp, DksA and GreA increased cell viability in DNA repair deficient strains following exposure to UV (UV) light31,32 (reviewed in ref. 30). Mutations in RNAP that decrease the stability of an elongation complex had similar effects of suppressing growth defects of DNA repair deficient strains under the same conditions.30-32 These data suggest that RNAP modulators reduce conflicts between replication and transcription by dislodging or reviving stalled transcription complexes that might otherwise lead to replication blocking RNAP arrays (Fig. 3A).31 Indeed, ppGpp which destabilizes RNAP open promoter complexes in collaboration with DksA has been shown to prevent the formation of an RNAP array upstream from a DNA lesion in vitro.31 The transcription-repair coupling factor Mfd is also likely to prevent RNAP arrays due to DNA damage. Mfd is an ATP dependent DNA translocase that displaces a halted RNAP from DNA and recruits the nucleotide excision repair machinery to the site.33 Mfd therefore facilitates repair of the transcribed strand, a process referred to as transcription-coupled repair (TCR) (reviewed in refs. 33 and 34). Importantly, Mfd also displaces RNAP arrested by other types of impediments such as protein blocks and secondary structure of DNA.35-37 Thus, TCR may act as a general mechanism to dissociate highly stable halted transcription complexes which may interfere with other DNA transactions such as replication. TCR has therefore been hypothesized to promote replication by removing RNAP blocks from the chromosome.38 Indeed, a recent report demonstrated that Mfd promotes direct restart of the replication fork after a head-on collision by facilitating displacement of the blocking RNAP ahead of the stalled fork (Fig. 3B).26 However, since mfd cells grow normally, other helicases are likely to play a more dominant role in promoting replication through head-on transcription complexes (discussed further below). Nevertheless, cells lacking Mfd exhibit a greater lapse in replication following exposure to UV light as compared with wild-type cells.39 These data suggest that Mfd promotes replication through transcription complexes stalled by lesions in the DNA (Fig. 3A).39 Alternatively, Mfd may play a role in upregulating transcription during the SOS response and thus indirectly promote replication following the onset of DNA damage. Future studies are required to determine whether Mfd directly resolves conflicts between replication and transcription in vivo. GreA/B, which promote transcription elongation by stimulating endonuclease activity of a backtracked RNAP, may also prevent the formation of replication blocking RNAP arrays by reducing RNAP pausing due to backtracking.31 Finally, recombination factors may be involved in promoting replication past formidable RNAP blocks, perhaps as a last resort when the fork regresses or collapses.31,32 It remains uncertain, however, whether RNAP modulators and recombination factors are necessary for resolving collisions between replication and transcription in the absence of DNA damage.
Role of Auxiliary Helicases in Resolving Collisions
Helicases UvrD and Rep have long been implicated in replication and thought to promote replication through protein blocks such as transcription complexes.40 However, until recently, convincing evidence indicating a role for UvrD and Rep in assisting replication through roadblocks has been lacking. UvrD and Rep are SF1 superfamily helicases that translocate along single-strand DNA (ssDNA) with 3′-5′ polarity. UvrD and Rep are 40% homologous and uvrD rep cells are inviable which suggests they share a common function that is essential for cell survival. rep cells replicate DNA at a slow rate and exhibit a greater number of replication forks along the chromosome, which is probably due to frequent fork stalling.41,42 Inhibition of UvrD results in reduced replication and filamented cells which indicates an important role in replication.43 UvrD and Rep are also involved in various replication repair processes such as replication restart and recombinational repair.44,45 Importantly, UvrD, Rep and Dda—a SF1 superfamily homolog in T4—displace high affinity protein-nucleic acid complexes such as the lac repressor during DNA unwinding which supports a role in dislodging roadblocks ahead of the fork.46
Recent in vivo data provide strong evidence that auxiliary helicases such as UvrD and Rep promote replication through transcription complexes and repressors in E. coli.47,48 For example, UvrD and Rep along with the SF2 family helicase DinG were shown to be required for cell growth when ribosomal genes were inverted.47 Further, two-dimensional gel electrophoresis revealed that replication forks accumulate within inverted ribosomal operons only when one or more of these helicases were absent.47 Significantly, Guy, et al. demonstrated that growth defects of rep uvrD cells were suppressed by a mutation in RNAP which reduced the stability of an elongation complex.48 Increased levels of ppGpp, which inhibit rRNA transcription and possibly destabilize elongation complexes also suppressed the growth defects of uvrD rep cells.48 Interestingly, these data suggest that transcription complexes represent the greatest barrier to replication in the cell. Recent in vivo studies of budding yeast have also demonstrated a requirement for auxiliary helicase activity in promoting replication through transcription complexes.49 Thus, the function of auxiliary helicases in facilitating replication through protein barriers such as RNAP appears to be conserved.
The mechanisms by which DNA helicases assist the replication fork through transcription complexes remains poorly understood. For example, Dda has been shown to promote replication by T4 DNA polymerase through RNAP (Fig. 4A).50 Since Dda translocates along ssDNA with 5′-3′ polarity it probably occupies the lagging strand where the replicative gp41 helicase normally functions. Thus, it remains uncertain whether gp41 and Dda cooperate at the T4 replication fork. Cooperativity between UvrD and DNA polymerase III was suggested by a genetic study which demonstrated a requirement for UvrD in promoting replication through an ectopic Tus-Ter termination complex (Fig. 4B).51 RecA was also required for this activity which suggests that UvrD is recruited to the fork following fork collapse. Similarly, Boubarkri H, et al. proposed that UvrD and Rep act on the leading strand after the fork has collapsed following a collision with RNAP.47 DinG, which translocates along ssDNA with 5′-3′ polarity, was proposed to act on the lagging strand and cooperate with either UvrD or Rep on the leading strand (Fig. 4C).47 DinG was also proposed to reduce conflicts between replication and head-on transcription by removing R-loops formed due to negative supercoils behind RNAP.47 A similar model of R-loops contributing to conflicts between replication and head-on transcription has been recently proposed in higher eukaryotes.52 Importantly, Guy et al. showed that Rep binds to the replicative DnaB helicase and that the two helicases cooperate in unwinding duplex DNA.48 Thus, Rep may be a component of the replication fork and act ahead of DNA polymerase on the leading strand (Fig. 4D). Guy, et al. further demonstrated that Rep or UvrD act on the leading strand to promote replication through an EcoRI mutant protein block in vitro (Fig. 4D).48 PcrA helicase—a SF1 superfamily homolog in B. subtilllus—was shown to have a similar effect. PcrA has been previously shown to substitute for UvrD in other cellular activities such as promoting replication through a Tus-Ter termination complex.51 Together, these observations suggest that SF1 helicases may not necessarily have to interact with replisome components to dislodge protein blocks from ahead of the fork. Thus, SF1 helicases may simply require naked ssDNA onto which they can load. Clearly, future studies are required to elucidate the specific activities of auxiliary helicases at the replication fork.
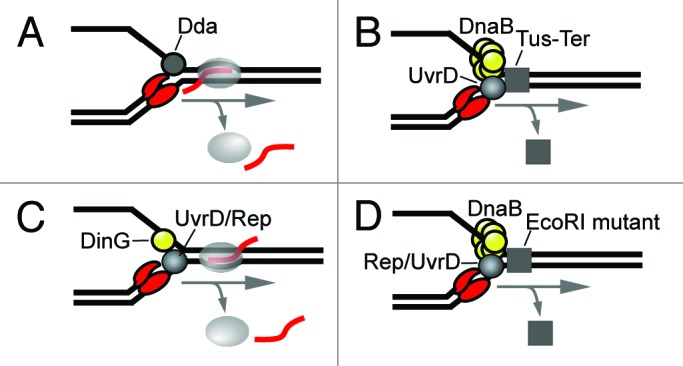
Figure 4. Models of auxiliary helicase activity in promoting replication through transcription complexes. (A) Dda helicase acts on the lagging strand to promote progression of the core T4 replisome through a co-directional RNAP in vitro. (B) UvrD helicase acts on the leading strand to promote replication through a Tus-Ter termination complex. (C) DinG acts on the lagging strand in collaboration with either UvrD or Rep on the lagging strand to promote replication through head-on transcription complexes. (D) Rep or UvrD act on the leading strand opposite DnaB to promote replication through a mutant EcoRI protein block.
Concluding Remarks
In conclusion, recent studies of E. coli and B. subtillus provide significant insight into the consequences of replisome-RNAP collisions and reveal factors and mechanisms that reduce or resolve conflicts that may arise following such encounters. However, as with most breakthroughs, several new questions have come to surface. For example, do replisome-RNAP collisions contribute to gaps observed in the leading strand in vivo? Are RNA transcripts utilized as primers during chromosomal replication? Do RNAP modulators facilitate fork progression following collisions in vivo? What causes DNA mutations at head-on collision sites? Does UvrD or Rep move with the fork? These and other intriguing questions regarding replisome-RNAP collisions represent important issues to be addressed in future studies.
Acknowledgments
We thank Richard Gourse and Isabel Kurth for critical reading of the manuscript.
Glossary
Abbreviations:
- RNAP
RNA polymerase
- TCR
transcription-coupled repair
- DNAP
DNA polymerase
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12122
References
- 1.Srivatsan A, et al. PLoS Genet. 2010;6:1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vilette D, Ehrlich SD, Michel B. Transcription-induced deletions in plasmid vectors: M13 DNA replication as a source of instability. Mol Gen Genet. 1996;252:398–403. doi: 10.1007/BF02173004. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg A, et al. DNA Replication. DNA Replication New York 1992; 2nd Ed 931. [Google Scholar]
- 4.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–5. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 5.Mirkin EV, Mirkin SM. Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol. 2005;25:888–95. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JD, Berkmen MB, Grossman AD. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A. 2007;104:5608–13. doi: 10.1073/pnas.0608999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–76. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy L, Roten CA. Genometric analyses of the organization of circular chromosomes: a universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene. 2004;340:45–52. doi: 10.1016/j.gene.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 9.Labib K, Hodgson B. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 2007;8:346–53. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huvet M, Nicolay S, Touchon M, Audit B, d’Aubenton-Carafa Y, Arneodo A, Thermes C. Human gene organization driven by the coordination of replication and transcription. Genome Res. 2007;17:1278–85. doi: 10.1101/gr.6533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–45. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–68. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 13.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–70. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 14.Quan S, Zhang N, French S, Squires CL. Transcriptional polarity in rRNA operons of Escherichia coli nusA and nusB mutant strains. J Bacteriol. 2005;187:1632–8. doi: 10.1128/JB.187.5.1632-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–27. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirkin EV, Castro Roa D, Nudler E, Mirkin SM. Transcription regulatory elements are punctuation marks for DNA replication. Proc Natl Acad Sci U S A. 2006;103:7276–81. doi: 10.1073/pnas.0601127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esnault E, et al. PLoS Genet. 2007;3:226. doi: 10.1371/journal.pgen.0030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–57. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maisnier-Patin S, Nordström K, Dasgupta S. Replication arrests during a single round of replication of the Escherichia coli chromosome in the absence of DnaC activity. Mol Microbiol. 2001;42:1371–82. doi: 10.1046/j.1365-2958.2001.02718.x. [DOI] [PubMed] [Google Scholar]
- 20.Omont N, Képès F. Transcription/replication collisions cause bacterial transcription units to be longer on the leading strand of replication. Bioinformatics. 2004;20:2719–25. doi: 10.1093/bioinformatics/bth317. [DOI] [PubMed] [Google Scholar]
- 21.Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–15. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 22.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–92. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Liu B, Alberts BM. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science. 1995;267:1131–7. doi: 10.1126/science.7855590. [DOI] [PubMed] [Google Scholar]
- 24.Liu B, Wong ML, Tinker RL, Geiduschek EP, Alberts BM. The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature. 1993;366:33–9. doi: 10.1038/366033a0. [DOI] [PubMed] [Google Scholar]
- 25.Daube SS, von Hippel PH. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992;258:1320–4. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- 26.Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327:590–2. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pomerantz RT, O’Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–6. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang TC. Discontinuous or semi-discontinuous DNA replication in Escherichia coli? Bioessays. 2005;27:633–6. doi: 10.1002/bies.20233. [DOI] [PubMed] [Google Scholar]
- 29.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–43. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 30.Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair (Amst) 2007;6:981–93. doi: 10.1016/j.dnarep.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–58. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 32.McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 33.Roberts J, Park JS. Mfd, the bacterial transcription repair coupling factor: translocation, repair and termination. Curr Opin Microbiol. 2004;7:120–5. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Deaconescu AM, Savery N, Darst SA. The bacterial transcription repair coupling factor. Curr Opin Struct Biol. 2007;17:96–102. doi: 10.1016/j.sbi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JS, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–67. doi: 10.1016/S0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 36.Chambers AL, et al. Mfd. Nucleic Acids Res. 2003;31:6409–18. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 38.Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–8. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 39.George DL, Witkin EM. Slow excision repair in an mfd mutant of Escherichia coli B/r. Mol Gen Genet. 1974;133:283–91. doi: 10.1007/BF00332704. [DOI] [PubMed] [Google Scholar]
- 40.Heller RC, Marians KJ. Non-replicative helicases at the replication fork. DNA Repair (Amst) 2007;6:945–52. doi: 10.1016/j.dnarep.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 41.Lane HE, Denhardt DT. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J Mol Biol. 1975;97:99–112. doi: 10.1016/s0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- 42.Colasanti J, Denhardt DT. The Escherichia coli rep mutation. X. Consequences of increased and decreased Rep protein levels. Mol Gen Genet. 1987;209:382–90. doi: 10.1007/BF00329669. [DOI] [PubMed] [Google Scholar]
- 43.Klinkert MQ, Klein A, Abdel-Monem M. Studies on the functions of DNA helicase I and DNA helicase II of Escherichia coli. J Biol Chem. 1980;255:9746–52. [PubMed] [Google Scholar]
- 44.Lestini R, Michel B. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 2007;26:3804–14. doi: 10.1038/sj.emboj.7601804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heller RC, Marians KJ. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J Biol Chem. 2005;280:34143–51. doi: 10.1074/jbc.M507224200. [DOI] [PubMed] [Google Scholar]
- 46.Yancey-Wrona JE, Matson SW. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992;20:6713–21. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–57. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS, et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell. 2009;36:654–66. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–34. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedinger P, Hochstrasser M, Jongeneel CV, Alberts BM. Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983;34:115–23. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- 51.Bidnenko V, Lestini R, Michel B. The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol Microbiol. 2006;62:382–96. doi: 10.1111/j.1365-2958.2006.05382.x. [DOI] [PubMed] [Google Scholar]
- 52.Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–24. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]