Abstract
Objectives
An additive genetic risk score (GRS) for coronary heart disease (CHD) has previously been associated with incident CHD in the population-based Greek European Prospective Investigation into Cancer and nutrition (EPIC) cohort. In this study, we explore GRS-‘environment’ joint actions on CHD for several conventional cardiovascular risk factors (ConvRFs), including smoking, hypertension, type-2 diabetes mellitus (T2DM), body mass index (BMI), physical activity and adherence to the Mediterranean diet.
Design
A case–control study.
Setting
The general Greek population of the EPIC study.
Participants and outcome measures
477 patients with medically confirmed incident CHD and 1271 controls participated in this study. We estimated the ORs for CHD by dividing participants at higher or lower GRS and, alternatively, at higher or lower ConvRF, and calculated the relative excess risk due to interaction (RERI) as a measure of deviation from additivity.
Results
The joint presence of higher GRS and higher risk ConvRF was in all instances associated with an increased risk of CHD, compared with the joint presence of lower GRS and lower risk ConvRF. The OR (95% CI) was 1.7 (1.2 to 2.4) for smoking, 2.7 (1.9 to 3.8) for hypertension, 4.1 (2.8 to 6.1) for T2DM, 1.9 (1.4 to 2.5) for lower physical activity, 2.0 (1.3 to 3.2) for high BMI and 1.5 (1.1 to 2.1) for poor adherence to the Mediterranean diet. In all instances, RERI values were fairly small and not statistically significant, suggesting that the GRS and the ConvRFs do not have effects beyond additivity.
Conclusions
Genetic predisposition to CHD, operationalised through a multilocus GRS, and ConvRFs have essentially additive effects on CHD risk.
Keywords: Epidemiology, Genetics
Strengths and limitations of this study.
Strengths of the study are the population-based prospective cohort design of the underlying study and the minimal concern for population stratification.
Another strength is the use of a multilocus genetic risk score using SNPs with documented association with CHD.
The main limitation of this study stems from the modest numbers of incident CHD cases, not withstanding the fact that the underlying cohort was large and was followed for approximately 10 years.
Introduction
Coronary heart disease (CHD) is a leading cause of death and disability worldwide.1 Lifestyle and environmental factors, such as cigarette smoking, physical inactivity, chronodisruption and unhealthy diets, play a significant role in its development and are largely responsible for increased risk of this disease.2 3 In addition, compelling evidence from the literature suggest a genetic basis for CHD4 so that genetic data may identify individuals who have an inherited predisposition to develop CHD.
During the past few years, genome-wide association studies (GWAS) have successfully identified a large number of chromosomal loci and genetic variants that are robustly associated with CHD,5–11 although their effects on risk are generally fairly small. To combine the relatively small effects of individual genes and to better capture the complex relationship between genetics and CHD, genotypes at multiple genetic variants have previously been combined into scores calculated according to the number of risk alleles carried.12 13 To date, several studies have examined the utility of different genetic risk scores (GRSs) to identify participants at increased CHD risk.14–18 Ripatti et al16 reported that a GRS based on a series of genetic variants from GWAS for myocardial infarction or CHD was associated with risk of CHD, and that the upper quintile of individuals of European ancestry who carried the most risk alleles had a roughly 1.7 times increased risk of CHD when compared with those in the lowest quintile of GRS. Using a similar approach, we have shown that a GRS based on nine documented genetic variants from GWAS is associated with incident CHD in the population-based Greek European Prospective Investigation into Cancer and nutrition (EPIC) cohort.19
Despite the success of GWAS in identifying novel genetic contributors to CHD, the heritability of common disorders cannot be adequately explained by the genes that have been discovered; moreover, for the most part, we do not know how these recently discovered loci interact with the environment and what role such interactions play in the development of the disease.20 21 Testing such interactions is thus a new frontier for large-scale GWAS of CHD,22 and some initial findings support the important role of environmental exposures in influencing the magnitude of the genetic associations with cardiovascular disease23 or other common diseases and traits.24 25
The aim of the current study was to explore potential GRS-‘environment’ interaction effects on CHD for several important conventional cardiovascular risk factors (ConvRFs), including smoking, hypertension, type-2 diabetes mellitus (T2DM), body mass index (BMI), physical activity and adherence to the Mediterranean diet (MedDiet). We have used resources generated in the Greek-EPIC cohort in which medically documented incident cases of CHD26 are recorded during an extended follow-up of this population-based cohort.
Methods
Study population
The EPIC is a longitudinal study aimed at investigating the role of biological, nutritional, lifestyle and environmental factors in the aetiology of cancer and other chronic diseases. The study has been described in detail elsewhere.27 28 The recruitment of Greek-EPIC participants was from 1994 to 1999. The active follow-up of study participants is repeated every 2–4 years. In each round, the focus of follow-up is on the update of information related to health status of the participants. For this analysis, exposure data at enrolment and follow-up data until the end of 2009 for outcomes are considered.
By December 2009, 788 participants were diagnosed with an incident, medically confirmed, CHD or stroke event and were considered eligible for a study also evaluating genetic predisposition.19 For each case, an attempt was made to choose two control participants matched for sex, age (±2 years) and date of recruitment (±6 months). Both cases and controls were free of CHD and stroke at baseline; the final study sample consisted of 788 cases (494 CHD, 320 stroke, 26 both diseases) and 1345 controls. For each study participant, a buffy coat sample was drawn from the Greek-EPIC bio-repository and genomic DNA was extracted. CHD events included myocardial infarction, angina and other ischaemic heart disease (cardiac arrest, presence of cardiac and vascular implants and grafts), with several cases following in more than one categories.26 28 All procedures were in accordance with the Helsinki Declaration and all participants provided written informed consent.
Selection of genetic variants, genotyping and GRS calculation
We constructed a multilocus GRS by using nine previously reported genetic variants associated with myocardial infarction or CHD from GWAS, with convincing replication evidence in populations with European ancestry,6 10 16 29 30 as previously described.19 The variants used were: rs11206510 at 1p32 near PCSK9, rs646776 at 1p13 near CELSR2-PSRC1-SORT1, rs17465637 at 1q41 in MIA3, rs6725887 at 2q33 in WDR12, rs9349379 at 6p24 in PHACTR1, rs1746048 at 10q11 near CXCL12, rs1122608 at 19p13 near LDLR, rs9982601 at 21q22 near SLC5A3-MRPS6-KCNE2 and the lead variant (rs1333049) at locus 9p21 near CDKN2A/2B identified by the Wellcome Trust Case Control Consortium.7
Genotyping was performed blindly as to case–control status with the TaqMan allelic discrimination system on the ABI 7900HT platform using custom genotyping assays and probes designed by Applied Biosystems, Inc (Foster City, California, USA). Replicate quality control samples yielded 100% concordance and call rates exceeded 98%. All genotypes were analysed in the Nutrition and Genomics Laboratory, Jean Mayer US Department of Agriculture, Human Nutrition Research Center on Aging at Tufts University, Boston, Massachusetts, USA.
A GRS was computed for each individual as the sum of the number of risk alleles across all nine variants, after weighting each one by its estimated effect size in the discovery samples5 10 as generally used16–18 and previously described.19 In this study, the minimum and maximum weighted GRS values were, respectively, 4.6 and 17.7 in control participants and 5.7 and 18.8 in CHD cases.
Conventional risk factors for CHD
We evaluated GRS-‘environment’ interaction effects on CHD for several important conventional ConvRFs for which information was collected at enrolment. These factors were: smoking status, hypertension, T2DM, BMI, waist-to-hip ratio, physical activity, energy intake and adherence to the MedDiet. Participants were characterised as current, former or never smokers and were considered as hypertensive if they met one of the following criteria: (1) their measured arterial blood pressure was 140 mm Hg or higher systolic, or 90 mm Hg or higher diastolic and (2) self-reported intake of an antihypertensive treatment. T2DM was identified through self-reported T2DM-spesific medication use or self-reported medical diagnosis of T2DM. Weight, height, waist and hip circumference were measured using standard procedures, and BMI was calculated in kg/m2. With respect to physical activity, we used a metabolic equivalent index (MET-value) that expresses the amount of energy per kilogram of body weight expended during an average day.31 Dietary information of the participants was measured at baseline using a validated interviewer-administered food frequency questionnaire (FFQ).32 The frequency of consumption of about 200 foods and recipes that are common in Greece was reflected at the FFQ. The daily energy intake was assessed by recording participants’ energy intake (in kcal). Adherence to the MedDiet was assessed with a MedDiet score that incorporates the salient characteristics of this diet, that is, high intake of plant foods and olive oil, low intake of meat and dairy products and moderate intake of alcohol. This score, with values from 0 to 9 (higher scores indicate greater adherence to the MedDiet), is associated with death from CHD, with lower values predicting higher incidence of death from CHD.28 33
Statistical analysis
For this study, we have used all incident CHD cases and all available control participants and we have proceeded through unconditional logistic regression.
Mean values of quantitative characteristics, as well as percentages for qualitative ones, by sex and case–control status, were calculated for descriptive purposes. We evaluated whether CHD incidence is related to the aforementioned ConvRFs using logistic regression, adjusting for age, sex and GRS. We evaluated ORs for CHD, as estimates of the incidence rate ratios, in relation to age, sex and higher or lower risk with respect to GRS (above or equal to vs below the sex-specific median score in controls) and, alternatively, on the basis of smoking status (current vs never/former smoker), hypertension (yes vs no), T2DM (yes vs no), physical activity (below vs above or equal to the sex-specific median), energy intake (below vs above or equal to the sex-specific median), MedDiet score (below vs above or equal to the median score of 4.0), BMI (above or equal vs below 25 kg/m2) or waist-to-hip ratio (above or equal to vs below the sex-specific median).
In order to access the nature of the joint effects of GRS and ConvRFs, we calculated the relative excess risk due to interaction (RERI), as defined by Rothman.34 RERI is an estimate of excess or deficit risk that is attributable to the interaction between two exposures, in this case GRS and each one of the ConvRFs; it measures deviation from additivity of effects independently of the risk scale of the outcome. From the ORs of the logistic regression, we computed the RERIs between GRS and ConRFs as follows35; let X+ and Y+ denote the presence of the risk factors X (GRS in our analysis) and Y (conventional factor) and X– and Y– denote the absence of these risk factors. Then, by considering that the OR estimates the relative risk (RR) we have that
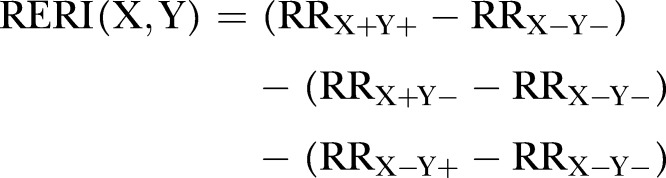 |
that is, RERI(X,Y)=(ORX+Y+−1)−(ORX+Y–−1)−(ORX–Y+−1)
The necessary variance estimators of RERI for the construction of 95% CIs were derived using the standard δ method.35 All statistical analyses were conducted using the Stata Statistical Software, release V.11 (StataCorp 2009, StataCorp LP).
Results
Of the 1839 study participants with genotype data (494 patients with incident CHD only and 1345 controls), 91 participants had missing data for one or more of the ConvRFs; thus, our analyses were restricted to 477 CHD cases and 1271 controls with complete datasets. The characteristics of the study participants at enrolment according to sex and case–control status are given in table 1.
Table 1.
Characteristics of conventional cardiovascular risk factors and genetic risk score for incident CHD cases and controls in the Greek-EPIC cohort
| Cases (n=477) |
Controls (n=1271) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Men (n=331) |
Women (n=146) |
Men (n=784) |
Women (n=487) |
|||||
| Age (years) | 60.1 | (11.4) | 66.2 | (6.9) | 60.6 | (10.9) | 65.6 | (7.3) |
| Body mass index (kg/m2) | 28.7 | (3.8) | 31.1 | (5.5) | 28.0 | (3.9) | 29.8 | (4.9) |
| Waist-to-hip ratio | 0.97 | (0.06) | 0.87 | (0.07) | 0.96 | (0.07) | 0.85 | (0.09) |
| Physical activity (MET-h/day) | 33.8 | (5.6) | 33.6 | (3.7) | 34.7 | (6.0) | 34.5 | (4.5) |
| Energy intake (kJ) | 9250.8 | (3000.8) | 6733.7 | (2021.7) | 9370.9 | (2700.4) | 7028.7 | (2330.5) |
| MedDiet score * | 4.4 | (1.7) | 4.1 | (1.6) | 4.4 | (1.7) | 4.2 | (1.6) |
| Hypertensive, n (%)† | 224 | (67.7) | 131 | (89.7) | 452 | (57.7) | 318 | (65.3) |
| Type 2 diabetes, n (%)‡ | 68 | (20.5) | 51 | (34.9) | 66 | (8.4) | 58 | (11.9) |
| Current smokers, n (%) | 138 | (41.7) | 13 | (8.9) | 269 | (34.3) | 34 | (7.0) |
| Weighted GRS§ | 12.6 | (2.0) | 12.9 | (2.1) | 12.3 | (2.1) | 12.3 | (2.1) |
Data are expressed as mean (SD) unless otherwise indicated.
*The range of the MedDiet score is from 0 to 9, with higher values indicating greater adherence to the MedDiet.33
†Defined as a systolic blood pressure of 140 mm Hg or higher or a diastolic blood pressure of 90 mm Hg or higher, or self-reported receipt of an antihypertensive treatment.
‡ Identified through self-reported T2DM-specific medication use or self-reported medical diagnosis of T2DM.
§The minimum and maximum weighted GRS values were 4.6 and 18.8.
CHD, coronary heart disease; EPIC, European prospective investigation into cancer and nutrition; GRS, genetic risk score; MedDiet, Mediterranean diet; MET, metabolic equivalent.
The association of ConvRFs with CHD incidence in this prospective cohort study is illustrated in table 2. As expected, smoking, hypertension, T2DM and an increased BMI and waist-to-hip ratio were all associated with a substantial increase in the risk of CHD, whereas higher levels of physical activity and energy expenditure (as reflected in an increased energy intake)36 were associated with a decrease in risk. Greater adherence to the MedDiet was also associated with an 11% decreased risk of CHD, although this association was not statistically significant.
Table 2.
ORs for CHD incidence by conventional risk factors in the Greek-EPIC cohort*
| OR (95% CI) | p Value | |
|---|---|---|
| Smoking status (current vs never/former smokers) | 1.39 (1.08 to 1.80) | 0.012 |
| Hypertension (yes vs no) | 2.16 (1.68 to 2.78) | <0.001 |
| Type 2 diabetes mellitus (yes vs no) | 3.36 (2.52 to 4.47) | <0.001 |
| Physical activity (≥sex-specific median vs <sex-specific median) | 0.70 (0.56 to 0.87) | 0.002 |
| Energy intake (≥sex-specific median vs <sex-specific median) | 0.75 (0.60 to 0.93) | 0.011 |
| MedDiet score (≥sex-specific median vs<sex-specific median) | 0.89 (0.71 to 1.11) | 0.299 |
| Body mass index (≥25 vs <25 kg/m2) | 1.45 (1.08 to 1.96) | 0.015 |
| Waist-to-hip ratio (≥sex-specific median vs <sex-specific median) | 1.46 (1.17 to 1.81) | 0.001 |
*Association tested with unconditional logistic regression adjusted for age, sex and genetic risk score; median values according to the overall sample (cases and controls combined).
CHD, coronary heart disease; EPIC, European prospective investigation into cancer and nutrition; MedDiet, Mediterranean diet.
We then examined the impact on CHD risk of the joint presence of genetic predisposition and ConvRFs by modelling the data through unconditional logistic regression, adjusting for age and sex. Specifically, we estimated ORs for CHD incidence depending on participants having a higher or lower GRS and simultaneously as being at higher or lower risk on the basis of a conventional risk factor. Table 3 gives the distribution of CHD cases and controls by GRS and each ConvRF (lower vs higher risk for CHD) in men and women. As shown in table 4, in all instances the joint presence of higher GRS and higher-risk ConvRF is associated with a substantial increase in the risk of CHD, compared with the joint presence of lower GRS and lower-risk ConvRF. In addition, participants with higher GRS values (high-risk genetic predisposition) and simultaneously at higher risk because of a ConvRF are characterised by an OR for CHD that is higher than the OR among individuals with high-risk genetic predisposition who belong to the lower risk category of the respective ConvRF (smoking status, OR 1.70 vs 1.49; hypertension, OR 2.72 vs 1.21; T2DM, OR 4.13 vs 1.34; physical activity, OR 1.86 vs 1.25; energy intake, OR 1.75 vs 1.43; MedDiet score, OR 1.51 vs 1.24; BMI, OR 2.01 vs 1.47; waist-to-hip ratio, OR 1.88 vs 1.25).
Table 3.
Distribution of CHD cases and controls by genetic risk score and conventional cardiovascular risk factors (lower/higher risk for CHD), in men and women
| Men (n=1115) |
Women (n=663) |
|||
|---|---|---|---|---|
| Cases (n=331) | Controls (n=784) | Cases (n=146) | Controls (n=487) | |
| Lower/higher risk | Lower/higher risk | Lower/higher risk | Lower/higher risk | |
| GRS (lower risk: <sex-specific median of controls; higher risk: ≥sex-specific median of controls) | 150/181 (45/55) | 400/384 (51/49) | 60/86 (41/59) | 243/244 (50/50) |
| Smoking status (lower risk: never/former smokers; higher risk: current smokers) | 193/138 (58/42) | 515/269 (66/34) | 133/13 (91/9) | 453/34 (93/7) |
| Hypertension (lower risk: no; higher risk: yes) | 107/224 (32/68) | 332/452 (42/58) | 15/131 (10/90) | 169/318 (35/65) |
| Type 2 diabetes mellitus (lower risk: no; higher risk: yes) | 263/68 (79/21) | 718/66 (92/8) | 95/51 (65/35) | 429/58 (88/12) |
| Physical activity (lower risk: ≥sex-specific median; higher risk: <sex-specific median) | 148/183 (45/55) | 410/374 (52/48) | 64/82 (44/56) | 254/233 (52/48) |
| Energy intake (lower risk: ≥sex-specific median; higher risk: <sex-specific median) | 153/178 (46/54) | 405/379 (52/48) | 63/83 (43/57) | 254/233 (52/48) |
| MedDiet score (lower risk: ≥4; higher risk: <4) | 224/107 (68/32) | 545/239 (69/31) | 93/53 (64/36) | 328/159 (67/33) |
| Body mass index (lower risk: <25 kg/m2; higher risk: ≥ 25 kg/m2) | 51/280 (15/85) | 160/624 (20/80) | 14/132 (10/90) | 70/417 (14/86) |
| Waist-to-hip ratio (lower risk: <sex-specific median; higher risk: ≥sex-specific median) | 147/184 (44/56) | 410/374 (52/48) | 60/86 (41/59) | 255/232 (52/48) |
Data are numbers (% in parenthesis). Median values for GRS are based on controls only,19 whereas for conventional risk factors median values are based on cases and controls combined.
CHD, coronary heart disease; GRS, genetic risk score; MedDiet, Mediterranean diet.
Table 4.
ORs for CHD occurrence by genetic risk score and, alternatively, the indicated conventional cardiovascular risk factors in the Greek-EPIC cohort (CHD cases: n=477; controls: n=1271)*
| 1st (reference) GRS: lower risk ConvRF: lower risk |
2nd GRS: lower risk ConvRF: higher risk |
3rd GRS: higher risk ConvRF: lower risk |
4th GRS: higher risk ConvRF: higher risk |
Relative Excess Risk due to Interaction (RERI) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| N | OR (95% CI) | N | OR (95% CI) | N | OR (95% CI) | N | Estimate (95% CI) | p Value | |
| Smoking status (lower risk: never/former smokers higher risk: current smokers) | 630 | 1.75 (1.22 to 2.49) | 223 | 1.49 (1.15 to 1.92) | 664 | 1.70 (1.19 to 2.41) | 231 | –0.54 (–1.31 to 0.24) | 0.18 |
| Hypertension (lower risk: no; higher risk: yes) | 318 | 2.07 (1.45 to 2.94) | 535 | 1.21 (0.81 to 1.80) | 305 | 2.72 (1.92 to 3.83) | 590 | 0.44 (–0.27 to 1.16) | 0.22 |
| Type 2 diabetes mellitus (lower risk: no; higher risk: yes) | 740 | 3.72 (2.45 to 5.63) | 113 | 1.34 (1.05 to 1.71) | 765 | 4.13 (2.79 to 6.12) | 130 | 0.07 (–1.94 to 2.07) | 0.95 |
| Physical activity (lower risk: ≥sex-specific median; higher risk: <sex-specific median) | 425 | 1.36 (0.99 to 1.88) | 428 | 1.25 (0.92 to 1.71) | 451 | 1.86 (1.36 to 2.54) | 444 | 0.25 (–0.32 to 0.81) | 0.39 |
| Energy intake (lower risk: ≥sex-specific median; higher risk: <sex-specific median) | 439 | 1.47 (1.07 to 2.03) | 414 | 1.43 (1.05 to 1.94) | 436 | 1.75 (1.29 to 2.39) | 459 | –0.14 (–0.76 to 0.47) | 0.65 |
| MedDiet score (lower risk: ≥4.0; higher risk: <4.0) | 574 | 1.03 (0.73 to 1.43) | 279 | 1.24 (0.95 to 1.60) | 616 | 1.51 (1.10 to 2.08) | 279 | 0.25 (–0.29 to 0.79) | 0.36 |
| Body mass index (lower risk: <25 kg/m2; higher risk: ≥25 kg/m2) | 143 | 1.56 (0.99 to 2.46) | 710 | 1.47 (0.84 to 2.56) | 152 | 2.01 (1.28 to 3.15) | 743 | –0.02 (–0.82 to 0.78) | 0.96 |
| Waist-to-hip ratio (lower risk: <sex-specific median; higher risk: ≥sex-specific median) | 433 | 1.40 (1.02 to 1.93) | 420 | 1.25 (0.92 to 1.71) | 439 | 1.88 (1.39 to 2.55) | 456 | 0.23 (–0.35 to 0.80) | 0.44 |
*Association tested with unconditional logistic regression adjusted for age and sex. Statistically significant results (p≤0.05) are in italic fonts.
CHD, coronary heart disease; ConvRF, conventional cardiovascular risk factor; EPIC, European prospective investigation into cancer and nutrition; GRS, genetic risk score; MedDiet, Mediterranean diet.
Relative excess risks due to interaction (RERIs) between the GRS and each one of the ConvRFs are presented in the last column of table 4. There is some evidence for superadditivity with respect to hypertension, and, on the contrary, some evidence for subadditivity with respect to smoking. Nevertheless, in all instances, RERI values are fairly small and the 95% CIs cover the null values of RERI, suggesting that the GRS and the conventional risk factors do not have effects beyond additivity.
Discussion
In a sizable case–control study nested in the population-based Greek-EPIC cohort, we have found that genetic predisposition to CHD, operationalised through a multilocus GRS (the sum of high-risk alleles in 9 genetic variants) and ConvRFs have essentially additive influence on CHD risk. In other words, people at high risk for CHD because of genetic susceptibility tend to have additively increased RR when also exposed to any of the investigated conventional risk factors. This is highlighted by the fact that, while among people with low genetic risk, only five of the eight investigated ConvRFs were documentable as ‘statistically significant’, all eight were documentable as such among people at high genetic risk.
Evaluation of joint effects in a multiplicative scale through interaction terms in logistic regression and other models that rely on similar principles are very valuable on account of their flexibility and provision of insights on causal pathways. Additive models (and deviations from additivity), however, as evaluated in this paper, convey straightforward answers to questions of preventive and clinical importance by pointing to individual change of risk in relation to values of conventional risk factors and specified genetic risk background.34 37 The results of the present study indicate that persons at high genetic risk for CHD increase this risk when they move into a high-risk category of a conventional cardiovascular risk factor no more than persons at low genetic risk, although they end up with a higher overall risk on account of the joint presence of high-risk genetic predisposition and ConvRF. Our results are not incompatible with those of previous investigations focusing on joint effects of genetic predisposition, assessed in variable ways and selected ConvRFs for CHD.38 In this respect, Tavani et al39 have previously examined the joint effect of a family history of heart disease, taken as a proxy for genetically determined predisposition to the disease, and selected adult life risk factors on the risk of the disease and have shown that a substantial increase in heart disease is evident when a family history and the environmental risk factors are present.
In the present investigation, we found no evidence of superadditive or subadditive effect of the GRS in conjunction with several ConvRFs. This does not preclude that such interactions does not exist between ConvRFs that were not studied in the present investigation and genetic variants not included in the GRS, over and beyond issues related to statistical power.21 40 41 It does appear, however, that the joint effects of genetic and non-genetic risk factors tend, generally, to be additive.
Strengths of the present nested case–control investigation are the population based prospective cohort design of the underlying study, the minimal concern for population stratification and the use of single-nucleotide polymorphisms with documented association with CHD. In this investigation, the effect estimates for the ConvRFs used (smoking, hypertension, etc), as well as the genetic factors which were components of the GRS were comparable with those reported in the literature that argues for the validity of the database used.10 16 Nevertheless, the use of single baseline measurements of ConvRFs can lead to underestimation of associations with CHD risk (through regression dilution bias).42 For example, the association between smoking and cardiovascular disease is intrinsically underestimated in cohort studies, since a proportion of smokers stop after data collection, and the RR falls rapidly after stopping. Correcting for within-person variation in lifestyle factors over time may result in more informative estimates of CHD risk associated with these factors, particularly for the risks associated with continued smoking and the benefits of regular physical activity,43 and, therefore, future studies should take these influences into account. The main limitation of this study stems from the modest numbers of incident CHD cases, not withstanding the fact that the underlying cohort was large and was followed for approximately 10 years. In addition, due to lack of available data on certain conventional risk factors of CHD, such as blood cholesterol levels, we were not able to examine in this study their joint relations with the GRS used.
In conclusion, this study provides evidence that genetic and conventional cardiovascular risk factors tend to have additive consequences on CHD, an issue that may be of preventive importance when genetic predisposition could not be assessed through an ad hoc genetic risk score but simply through a positive family history.
Supplementary Material
Footnotes
Contributors: NY, AT, JMO and DT participated in study concept and design. NY, AT and DT participated in acquisition of data. NY, MK, AT, JMO and DT participated in analysis and interpretation of the data. NY, MK, AT, JMO and DT participated in drafting and critical revision of the manuscript for important intellectual content. MK, NY and DT participated in statistical analysis. AT and JMO obtained funding. NY, AT and JMO provided administrative, technical and material support. AT and NY participated in study supervision.
Funding: This study was supported by the Hellenic Health Foundation and the Stavros Niarchros Foundation; and by contracts 53-K06-5-10 and 58-1950-9-001 from the US Department of Agriculture Research.
Competing interests: None.
Patient consent: Obtained.
Ethics approval: All procedures were in accordance with the Helsinki Declaration. The study protocol was approved by the ethics committees of the International Agency for Research on Cancer and the Medical School of the University of Athens.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2095–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yusuf S, Hawken S, Ounpuu S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet 2004;364:937–52 [DOI] [PubMed] [Google Scholar]
- 3.Hu FB. Diet and lifestyle influences on risk of coronary heart disease. Curr Atheroscler Rep 2009;11:257–63 [DOI] [PubMed] [Google Scholar]
- 4.Vaidya D, Yanek LR, Moy TF, et al. Incidence of coronary artery disease in siblings of patients with premature coronary artery disease: 10 years of follow-up. Am J Cardiol 2007;100:1410–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samani NJ, Deloukas P, Erdmann J, et al. Large scale association analysis of novel genetic loci for coronary artery disease. Arterioscler Thromb Vasc Biol 2009;29:774–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007;316:1488–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009;41:334–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007;316:1491–3 [DOI] [PubMed] [Google Scholar]
- 12.Yiannakouris N, Trichopoulou A, Benetou V, et al. A direct assessment of genetic contribution to the incidence of coronary infarct in the general population Greek EPIC cohort. Eur J Epidemiol 2006;21:859–67 [DOI] [PubMed] [Google Scholar]
- 13.Humphries SE, Drenos F, Ken-Dror G, et al. Coronary heart disease risk prediction in the era of genome-wide association studies: current status and what the future holds. Circulation 2010;121:2235–48 [DOI] [PubMed] [Google Scholar]
- 14.Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med 2008;358:1240–9 [DOI] [PubMed] [Google Scholar]
- 15.Paynter NP, Chasman DI, Pare G, et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA 2010;303:631–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet 2010;376:1393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies RW, Dandona S, Stewart AF, et al. Improved prediction of cardiovascular disease based on a panel of single nucleotide polymorphisms identified through genome-wide association studies. Circ Cardiovasc Genet 2010;3:468–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thanassoulis G, Peloso GM, Pencina MJ, et al. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet 2012;5:113–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yiannakouris N, Katsoulis M, Dilis V, et al. Genetic predisposition to coronary heart disease and stroke using an additive genetic risk score: a population-based study in Greece. Atherosclerosis 2012;222:175–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manolio TA. Cohort studies and the genetics of complex disease. Nat Genet 2009;41:5–6 [DOI] [PubMed] [Google Scholar]
- 21.Ordovas JM, Tai ES. Why study gene-environment interactions? Curr Opin Lipidol 2008;19:158–67 [DOI] [PubMed] [Google Scholar]
- 22.Lanktree MB, Hegele RA. Gene-gene and gene-environment interactions: new insights into the prevention, detection and management of coronary artery disease. Genome Med 2009;1:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Do R, Xie C, Zhang X, et al. The effect of chromosome 9p21 variants on cardiovascular disease may be modified by dietary intake: evidence from a case/control and a prospective study. PLoS Med 2011;8:e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamza TH, Chen H, Hill-Burns EM, et al. Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson's disease modifier gene via interaction with coffee. PLoS Genet 2011;7:e1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surakka I, Isaacs A, Karssen LC, et al. A genome-wide screen for interactions reveals a new locus on 4p15 modifying the effect of waist-to-hip ratio on total cholesterol. PLoS Genet 2011;7:e1002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misirli G, Bamia C, Dilis V, et al. Validation of self-reported incident cardiovascular disease events in the Greek EPIC cohort study. Italian J Public Health 2012;9:e7538 [Google Scholar]
- 27.Riboli E, Hunt KJ, Slimani N, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr 2002;5:1113–24 [DOI] [PubMed] [Google Scholar]
- 28.Dilis V, Katsoulis M, Lagiou P, et al. Mediterranean diet and CHD: the Greek European prospective investigation into cancer and nutrition cohort. Br J Nutr 2012;108:699–709 [DOI] [PubMed] [Google Scholar]
- 29.Schunkert H, Gotz A, Braund P, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation 2008;117:1675–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preuss M, Konig IR, Thompson JR, et al. A Genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet 2010;3:475–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trichopoulou A, Gnardellis C, Lagiou A, et al. Body mass index in relation to energy intake and expenditure among adults in Greece. Epidemiology 2000;11:333–6 [DOI] [PubMed] [Google Scholar]
- 32.Gnardellis C, Trichopoulou A, Katsouyanni K, et al. Reproducibility and validity of an extensive semiquantitative food frequency questionnaire among Greek school teachers. Epidemiology 1995;6:74–7 [DOI] [PubMed] [Google Scholar]
- 33.Trichopoulou A, Costacou T, Bamia C, et al. Adherence to a Mediterranean diet and survival in a Greek population. N Engl J Med 2003;348:2599–608 [DOI] [PubMed] [Google Scholar]
- 34.Rothman KJ. Modern epidemiology. Boston, Toronto: Little Brown and Co, 1986 [Google Scholar]
- 35.Hosmer DW, Lemeshow S. Confidence interval estimation of interaction. Epidemiology 1992;3:452–6 [DOI] [PubMed] [Google Scholar]
- 36.Morris JN, Marr JW, Clayton DG. Diet and heart: a postscript. BMJ 1977;2:1307–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Mutsert R, de Jager DJ, Jager KJ, et al. Interaction on an additive scale. Nephron Clin Pract 2011;119:c154–7 [DOI] [PubMed] [Google Scholar]
- 38.Lee YC, Lai CQ, Ordovas JM, et al. A Database of gene-environment interactions pertaining to blood lipid traits, cardiovascular disease and type 2 diabetes. J Data Mining Genomics Proteomics 2011;2:pii:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tavani A, Augustin L, Bosetti C, et al. Influence of selected lifestyle factors on risk of acute myocardial infarction in subjects with familial predisposition for the disease. Prev Med 2004;38:468–72 [DOI] [PubMed] [Google Scholar]
- 40.Talmud PJ. Gene-environment interaction and its impact on coronary heart disease risk. Nutr Metab Cardiovasc Dis 2007;17:148–52 [DOI] [PubMed] [Google Scholar]
- 41.Carty CL, Heagerty P, Heckbert SR, et al. Interaction between fibrinogen and IL-6 genetic variants and associations with cardiovascular disease risk in the Cardiovascular Health Study. Ann Hum Genet 2010;74:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emberson JR, Whincup PH, Morris RW, et al. Extent of regression dilution for established and novel coronary risk factors: results from the British Regional Heart Study. Eur J Cardiovasc Prev Rehabil 2004;11:125–34 [DOI] [PubMed] [Google Scholar]
- 43.Emberson JR, Whincup PH, Morris RW, et al. Lifestyle and cardiovascular disease in middle-aged British men: the effect of adjusting for within-person variation. Eur Heart J 2005;26:1774–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


