Abstract
Candidate gene and genome-wide association studies (GWAS) have identified 11 independent susceptibility loci associated with bladder cancer risk. To discover additional risk variants, we conducted a new GWAS of 2422 bladder cancer cases and 5751 controls, followed by a meta-analysis with two independently published bladder cancer GWAS, resulting in a combined analysis of 6911 cases and 11 814 controls of European descent. TaqMan genotyping of 13 promising single nucleotide polymorphisms with P < 1 × 10−5 was pursued in a follow-up set of 801 cases and 1307 controls. Two new loci achieved genome-wide statistical significance: rs10936599 on 3q26.2 (P = 4.53 × 10−9) and rs907611 on 11p15.5 (P = 4.11 × 10−8). Two notable loci were also identified that approached genome-wide statistical significance: rs6104690 on 20p12.2 (P = 7.13 × 10−7) and rs4510656 on 6p22.3 (P = 6.98 × 10−7); these require further studies for confirmation. In conclusion, our study has identified new susceptibility alleles for bladder cancer risk that require fine-mapping and laboratory investigation, which could further understanding into the biological underpinnings of bladder carcinogenesis.
INTRODUCTION
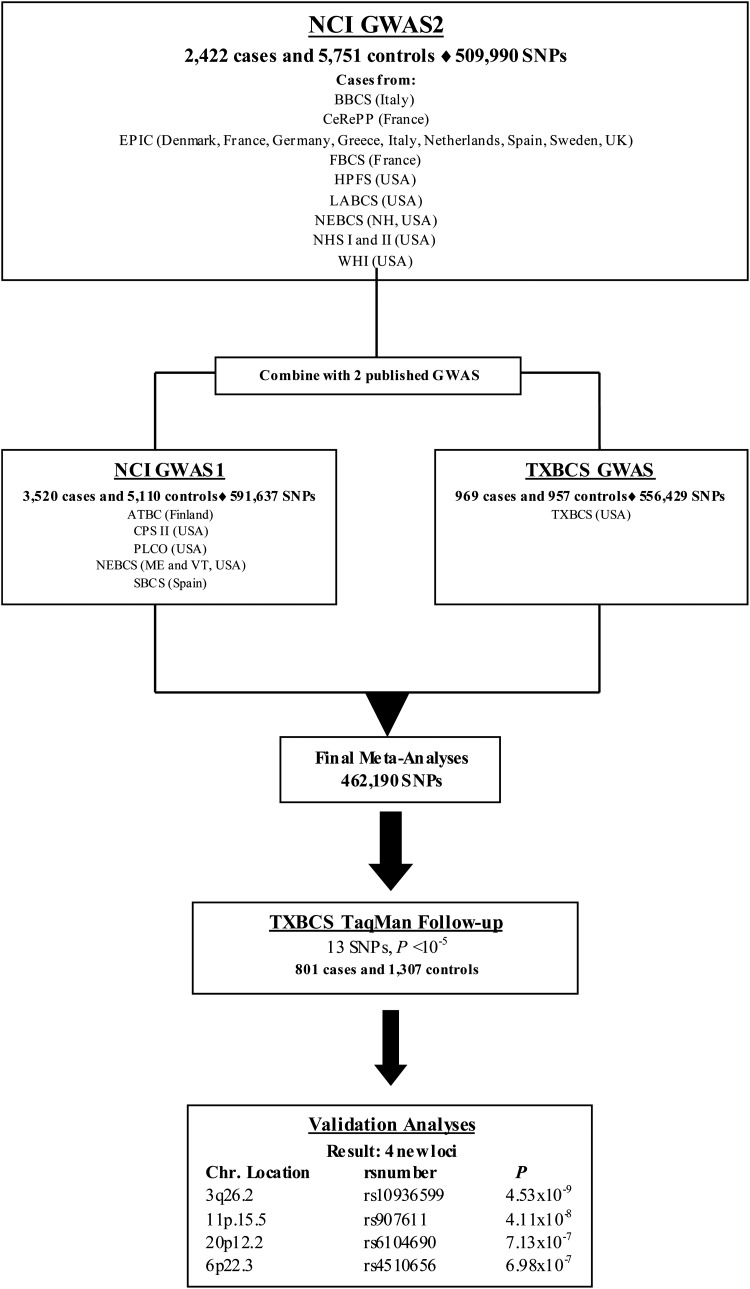
Each year ∼380 000 bladder cancers are diagnosed worldwide, with men being three to four times more likely than women to be diagnosed with the disease (1,2). Family history of bladder cancer is associated with ∼2-fold increased risk, suggesting shared genetic and potential environmental contribution to its etiology (3,4). Meta-analyses of established candidate genes and genome-wide association studies (GWAS) have identified 11 loci that harbor bladder cancer susceptibility alleles: 1p13.3 (GSTM1), 2q37.1 (UGT1A cluster), 3q28 (TP63), 4p16.3 (TMEM129 and TACC3-FGFR3), 5p15.33 (TERT-CLPTM1L), 8p22 (NAT2), 8q24.21, 8q24.3 (PSCA), 18q12.3 (SLC14A1), 19q12 (CCNE1) and 22q13.1 (CBX6, APOBEC3A) (5–14). Using an approach that estimates the number of susceptibility loci and the distribution of their effect sizes based on previously reported loci (15), many additional common single nucleotide polymorphisms (SNPs) for bladder cancer are yet to be discovered (9). To search for such loci, we performed a new scan of 2422 cases and 5751 controls (National Cancer Institute, NCI-GWAS2). We then conducted a meta-analysis with two previously independently published GWAS [NCI-GWAS1 and Texas Bladder Cancer Study (TXBCS)-GWAS] (9,10). Here, we present our GWAS analysis that included 6911 cases and 11814 cancer-free controls, and follow-up of promising loci with TaqMan genotyping in an independent set of 801 cases and 1307 controls from the TXBCS (TXBCS-TaqMan).
RESULTS
After applying standard quality control metrics, the new bladder cancer GWAS was comprised 2422 cases genotyped with the HumanHap 660w and HumanHap 610 (Illumina, San Diego, CA, USA) and 5751 cancer-free controls selected from previously scanned studies at the NCI (NCI-GWAS2) (Fig. 1, Supplementary Material, Table S1) (9,16–21). Meta-analysis of previously published data for 11 reported bladder cancer susceptibility loci (5–14), including data from NCI-GWAS2, confirmed bladder cancer risk associations close to or below genome-wide significance (Supplementary Material, Table S2). In addition, using a TaqMan-based deletion detection assay, we also confirmed an association with null versus present copy number variation in GSTM1 (1p13.3) (9) (Supplementary Material, Table S2).
Figure 1.
See Supplementary Material, Table S1 for details of study designs and sample sizes. The New England Bladder Cancer Study (NEBCS) represents a single study comprises Maine (ME) and Vermont (VT) components genotyped in NCI-GWAS1, and the New Hampshire (NH) component genotyped in NCI-GWAS2.
To identify new bladder cancer susceptibility loci, a meta-analysis was performed using the NCI-GWAS2 dataset with two previously published bladder cancer GWAS: NCI-GWAS1 (6,9), and TXBCS-GWAS (10) (Figure 1). The primary analytical approach employed logistic regression models for genotype trend effects (with 1 d.f.) adjusted for study group, age, sex and smoking status. NCI-GWAS1 and NCI-GWAS2 were further adjusted for six eigenvectors (see Materials and Methods, Supplementary Material, Fig. S1). After rigorous quality control metrics were applied across the sets of called genotypes from the three GWAS datasets (see Materials and Methods), 462 190 common genotyped SNPs were available for analysis (Figure 1). A manhattan plot shows the results based on the meta-analysis of the three GWAS (Supplementary Material, Fig. S2).
After excluding previously identified regions associated with bladder cancer in our primary analysis, we identified 13 SNPs with P < 1 × 10−5, for which we developed TaqMan assays for follow-up analysis in an independent set of 801 cases and 1307 controls not included in the GWAS analysis from the TXBCS (Figure 1). Combined meta-analysis for the 13 SNPs adjusted for smoking, gender, age, study group and six eigenvectors for NCI-GWAS2 and NCI-GWAS1, identified two new loci below genome-wide statistical significance (Table 1, Supplementary Material, Table S3): rs10936599 on 3q26.2 (P = 4.53 × 10−9) and rs907611on 11p15.5 (P = 4.11 × 10−8). Study-specific estimates are shown in Supplementary Material, Figure S3.
Table 1.
Associations for rs10936599 at 3q26.2, rs907611 at 11p15.5, rs6104690 at 20p12.2 and rs4510656 at 6p22.3 with bladder cancer risk
| SNP | Cytoband | Alleles | Minor allele | MAF | Cases | Controls | Group | ORadj | (95% CI) |
Ptrend | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10936599 | 3q26.2 | T/C | T | 0.24 | 2415 | 5751 | NCI-GWAS2 | 0.81 | 0.73 | 0.89 | |
| 0.24 | 3517 | 5107 | NCI-GWAS1 | 0.88 | 0.81 | 0.94 | |||||
| 0.24 | 969 | 957 | TXBCS-GWAS | 0.84 | 0.72 | 0.98 | |||||
| 0.25 | 790 | 1293 | TXBCS-TaqMan | 0.90 | 0.77 | 1.05 | |||||
| 7691 | 13 108 | Combined | 0.85 | 0.81 | 0.90 | 4.53 × 10−9 | |||||
| rs907611 | 11p15.5 | G/A | A | 0.31 | 2379 | 5720 | NCI-GWAS2 | 1.25 | 1.14 | 1.36 | |
| 0.33 | 2757 | 4314 | NCI-GWAS1 | 1.12 | 1.04 | 1.2 | |||||
| 0.32 | 968 | 955 | TXBCS-GWAS | 1.20 | 1.04 | 1.39 | |||||
| 0.31 | 797 | 1291 | TXBCS-TaqMan | 0.97 | 0.84 | 1.12 | |||||
| 6901 | 12 280 | Combined | 1.15 | 1.09 | 1.21 | 4.11 × 10−8 | |||||
| rs6104690 | 20p12.2 | G/A | G | 0.44 | 2409 | 5747 | NCI-GWAS2 | 0.89 | 0.82 | 0.96 | |
| 0.44 | 3510 | 5097 | NCI-GWAS1 | 0.89 | 0.84 | 0.95 | |||||
| 0.45 | 969 | 957 | TXBCS-GWAS | 0.91 | 0.79 | 1.04 | |||||
| 0.46 | 790 | 1287 | TXBCS-TaqMan | 0.89 | 0.78 | 1.02 | |||||
| 7678 | 13 088 | Combined | 0.89 | 0.85 | 0.93 | 7.13 × 10−7 | |||||
| rs4510656 | 6p22.3 | C/A | A | 0.45 | 2416 | 5747 | NCI-GWAS2 | 0.87 | 0.8 | 0.95 | |
| 0.43 | 3515 | 5106 | NCI-GWAS1 | 0.91 | 0.85 | 0.97 | |||||
| 0.45 | 968 | 957 | TXBCS-GWAS | 0.88 | 0.77 | 1.01 | |||||
| 0.46 | 798 | 1300 | TXBCS-TaqMan | 0.90 | 0.79 | 1.03 | |||||
| 7697 | 13 110 | Combined | 0.89 | 0.85 | 0.93 | 6.98 × 10−7 | |||||
OR adjusted for age, study group, smoking status and gender. Fixed-effects meta-analysis by stage was used to calculate the combined OR, 95% CI and P-trend.
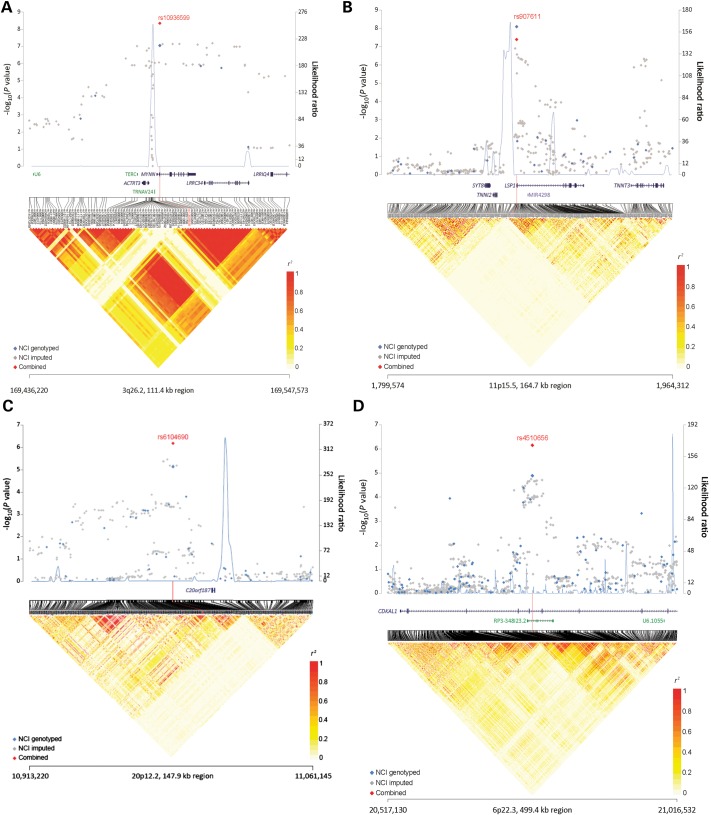
The most significant finding was for rs10936599 at 3q26.2 (ORadj per T allele = 0.85, 95% CI 0.81–0.90, P = 4.53 × 10−9; Table 1), which maps to a multigenic region that includes TERC, ACTRT3 (also known as ARPM1), MYNN and LRRC34 (Fig. 2a). Using ENCODE resources (22), including HaploReg (23) and RegulomeDB (24), there were 35 highly correlated SNPs with rs10936599 with r2 > 0.8 (based on 1000 Genomes CEU). We evaluated these 35 SNPs and found many of them to be located within the areas predicted to act as enhancers or promoter based on specific chromatin modification marks (Supplementary Material, Table S4). The 40 kb linkage disequilibrium (LD) block surrounding rs10936599, includes a coding synonymous variant rs10936599 (His6His) within MYNN, missense variants rs6793295 (Ser249Gly) and rs10936600 (Leu254Ile) within LRRC34 (Supplementary Material, Table S4). Imputation analysis did not yield any signals with a stronger association (Fig. 2a). RNA-sequencing analysis in tumor (n = 7) and adjacent normal (n = 5) bladder tissue showed expression of ACTRT3, MYNN and LRRC34 while expression of TERC was below the level of confident detection by RNA sequencing (Supplementary Material, Fig. S4a). Expression analysis of individual transcripts using more sensitive TaqMan assays in a larger set of bladder tissues (41 muscle-invasive tumors and 40 adjacent normal samples) showed higher expression of these transcripts in tumors compared with normal tissues: TERC (P = 3.2 × 10−21), MYNN (P = 2.4 × 10−4) and ACTRT3 (P = 4.8 × 10−4), but did not show significant differences in relation to rs10936599 genotypes (Supplementary Material, Fig. S5).
Figure 2.
(A–D) The −log10(P-value) (Y-axis) for NCI-GWAS1 and NCI-GWAS2 genotyped SNPs (blue) and imputed SNPs (gray) was plotted on the genomic coordinates (X-axis; NCBI genome build 37). The combined data for NCI-GWAS1, NCI-GWAS2, TXBCS-GWAS and TXBCS-TaqMan for four novel regions rs10936599 (3q16.2), rs907611 (11p15.5), rs6104690 (20p12.2) and rs4510656 (6p22.3) are shown in red.
The next most significant finding was rs907611 on 11p15.5, which maps upstream of the LSP1 gene (ORadj per A allele = 1.15, 95% CI 1.09–1.21, P = 4.11 × 10−8). Data for the rs907611 SNP were missing for one study. The American Cancer Society's Cancer Prevention Study II due to <90% completion rates, but there was no evidence of significant heterogeneity by study group among those studies with SNP data that passed quality control metrics (I2 = 9.1%, P = 0.36) (Supplementary Material, Fig. S3); TaqMan genotyping of 376 unique samples (145 cases and 231 controls) from seven studies showed 100% concordance with GWAS data for this SNP. The rs907611 SNP lies 130 bp upstream of the transcription start site for the longer transcript of the LSP1 gene (Fig. 2b). The 10 kb LD block includes 11 SNPs highly correlated (r2 > 0.8) with rs907611 (based on 1000 Genomes CEU), all located within the first exon and part of the intron of LSP1 gene, which also includes miRNA-4298 (Fig. 2b, Supplementary Material, Fig. S4b). Imputation analysis did not yield any signals with a stronger association (Fig. 2b). TaqMan expression analysis in the set of bladder tissues showed comparable expression of LSP1 and mir-4298 in relation to sample status (normal or tumor) and rs907611genotypes (Supplementary Material, Fig. S5). LSP1, mir-4298 expression levels did not show significant differences in relation to rs907611genotypes (Supplementary Material, Fig. S5).
In the analysis, there were also two notable regions that approached genome-wide significance. The first observed signal was marked by rs6104690 at 20p12.2, a non-genic region (ORadj per G allele = 0.89, 95% CI 0.85–0.93, P = 7.13 × 10−7) (Fig. 2c, Supplementary Material, Fig. S4c). Imputation analysis did not yield any signals with a stronger association (Fig. 2c). There was some suggestion of potential heterogeneity by study group for rs6104690 with an I2 value of 36.3 and P = 0.09 (Supplementary Material, Fig. S3). The study heterogeneity was driven by the Italy group composed of Environment and Genetics in Lung cancer Etiology Study (EAGLE) controls and BRESCIA cases, since when we removed this group the I2 = 0.0, P = 0.48. The 7 kb LD block includes seven SNPs with r2 > 0.8 with rs6104960 (based on 1000 Genomes CEU). HaploReg data for rs6104690 and seven linked SNPs shows that these variants are located within predicted enhancers, DNAse I hypersensitivity sites, and transcription factor binding sites (Supplementary Material, Table S4). The second observed signal was marked by rs4510656 at 6p22.3 (ORadj per A allele = 0.89, 95% CI 0.85–0.93, P = 6.98 × 10−7), which maps to the intronic region of CDKAL1 (Figure 2d). The 17 kb LD block at 6p22.3 includes 22 SNPs highly correlated (r2 > 0.8) with rs4510656 (based on 1000 Genomes CEU), all located within the CDKAL1 gene (Supplementary Material, Fig. S4d). Imputation analysis did not yield any signals with a stronger association (Fig. 2d). TaqMan expression analysis in the set of bladder tissues showed significantly higher levels of CDKAL1 expression in tumors compared with normal tissue (P = 6.98 × 10−4), but did not show significant differences by rs4510656 genotypes (Supplementary Material, Fig. S5).
Within the combined NCI-GWAS1 and GWAS2 datasets, we tested for multiplicative and additive interactions and smoking status (ever/never) for all bladder cancer susceptibility SNPs. We found evidence of significant interaction with smoking on the multiplicative scale for the SNP rs1495741 at NAT2, while 11 of the 14 bladder susceptibility SNPs showed evidence of additive interactions in NCI-GWAS2 and NCI-GWAS1 datasets, including rs10936599 and rs4510656, Supplementary Material, Table S5. Analyses of SNP–SNP interactions for the 14 identified bladder cancer susceptibility loci did not suggest evidence for interaction after adjustment for multiple comparisons (data not shown). Analysis of SNP associations by tumor stage and grade suggested a stronger association between rs907611 and rs10936599 and lower grade tumors, Supplementary Material, Table S6.
DISCUSSION
Using our new NCI-GWAS2 data combined with two published bladder cancer GWAS (9,10) and follow-up TaqMan data from for 13 SNPs, we report two new susceptibility regions on 3q26.2 and 11p15.5, plus two suggestive regions on 20p12.2 and 6q22.3 that need further confirmation. As suggested by the Park model, the larger sample size led to the discovery of additional susceptibility regions marked by common, low-penetrance variants (15). Further, using the largest dataset of bladder cancer cases to date, we have refined the risk estimates for previously published loci and provide further support for additive interactions with smoking as noted previously (25). Imputation analysis in all four regions did not yield any signals with a stronger association.
Our strongest signal was for the rs10936599 SNP within 3q26.2, which has been associated in a GWAS of colorectal cancer (26) and approached genome-wide significance in a GWAS of multiple sclerosis (27). The rs10936599 SNP is in complete LD (r2 = 1.0 in 1000 Genomes, CEU) with SNP rs2293607 located upstream of the TERC transcript, which has been shown to affect mRNA folding and telomere length in functional studies (28,29). The telomerase RNA component (TERC) gene located within this region is a strong candidate for the association with bladder cancer. TERC serves as a template for extension of telomeres by the telomerase reverse transcriptase (TERT) and the ability of cells to maintain long telomeres is proposed as a mechanism associated with cell proliferation and cancer (30). Interestingly, a genetic variant within the TERT region has been associated with increased risk of bladder cancer in two GWAS (9,14). We found significantly higher TERC mRNA expression in muscle-invasive bladder tumors compared with adjacent normal bladder tissues, and a previous report showed significantly higher TERC mRNA expression in muscle-invasive compared with superficial bladder tumors (31). Cumulatively, these data suggest that TERC may have functional relevance for bladder cancer susceptibility through its function in bladder tissue. However, the possible functional effects of other genes (MYNN and ACTRT3) in the associated LD block cannot be excluded as a molecular cause of this association.
The LD block rs907611 SNP at 11p15.5 and its associated LD block includes a part of the LSP1 gene and miRNA-4298. LSP1 is an intracellular F-actin binding protein, which in mouse models is important for regulation of leukocyte recruitment to inflamed sites (32). Chronic inflammation is an important factor in bladder cancer initiation and development, as evidenced by the role of infection with schistosomiasis, a urinary parasite associated with bladder cancer risk primarily in Northern Africa (33). The rs907611 SNP has also been associated with ulcerative colitis, one of the major types of inflammatory bowel disease (34), and an independent marker in this region has also been associated with breast cancer risk (35). miRNA-4298, found to be expressed both in normal and tumor bladder tissue, is also an interesting candidate for this association signal since miRNAs have documented importance for carcinogenesis (36). Genetic variants within miRNAs might affect the level of miRNA expression, secondary structure, stability and the efficiency of interaction with their targets (37).
The rs6104690 SNP lies within a region of chromosome 20p12.2 with no known genes and there is little data suggesting any functional mechanisms. The associated LD block at 6p22.3 maps within the CDKAL1 gene and we showed differences in expression for CDKAL1 with higher levels found in tumors compared with normal bladder tissues. CDKAL1 variants have been previously associated with insulin secretion and type 2 diabetes risks as well as Crohn's disease (38–41); however, the rs4510656 SNP associated with bladder cancer seems to be an independent signal from these variants based on low LD (r2 < 0.20).
In summary, we refined risk estimates for 11 previously identified bladder cancer susceptibility loci and report up to four new susceptibility loci. We present functional data for three of the new bladder cancer susceptibility regions (3q26.2 and 11p15.5, and 6q22.3), supporting their potential relevance to bladder carcinogenesis. Interestingly, the 3q26.2 and 11p15.5, and 6q22.3 regions associated with bladder cancer have been identified in GWAS studies of other diseases, suggesting pleiotropy for these genetic effects (42). These new observations raise questions and provide new avenues for investigation that could shed light on shared mechanisms between cancers and other diseases. At the same time, we recognize that each of the four susceptibility regions will need to be carefully mapped to determine the optimal variants for estimating risk. Identification of bladder cancer susceptibility variants as described here provide a foundation to improve risk prediction and explore the complex interplay of genes and environmental exposures involved in bladder carcinogenesis.
MATERIALS AND METHODS
Study participants
The samples and studies for the NCI-GWAS2 are listed in Supplementary Material, Table S1. Cases and controls were non-Hispanic Caucasians of European origin. Cases were defined as histologically confirmed primary carcinoma of the urinary bladder including carcinoma in situ (ICD-0-2 topography codes C67.0-C67.9 or ICD9 codes 188.1–188.9). Each participating study obtained informed consent from study participants and approval from the corresponding Institutional Review Boards. Participating studies obtained institutional certification permitting data sharing in accordance with the NIH policy for sharing of data obtained in NIH supported or conducted GWAS.
Genotyping and quality control
For NCI-GWAS2, we performed genotyping on cases and controls for the New Hampshire component of the New England Bladder Cancer Study (NEBCS-NH). Samples from the other two study centers (Maine and Vermont) in the NEBCS were genotyped in NCI-GWAS1 with a dense SNP array as reported previously ((9), Supplementary Material, Table S1). We genotyped cases and used existing control data for four cohort studies already subjected to rigorous quality control metrics: the European Prospective Investigation Into Cancer and Nutrition Study (EPIC), Women's Health Initiative (WHI), Health Professionals Follow-up Study (HPFS) and Nurses' Health Study I and II (NHS I and II), which have been a part of Cancer Genetic Markers of Susceptibility (CGEMS) (Supplementary Material, Table S1). We genotyped cases for four case–control studies, the Los Angeles Bladder Cancer Study (LABCS), the French Center for Research on Prostate Diseases (CeRePP), the French Bladder Study (FBCS) and the Brescia Bladder Cancer Study (BBCS). For LABCS, CeRePP, FBCS and BBCS studies where we genotyped cases only, we created in silico study groups based on comparable geographic/demographic parameters, which resulted in three new ‘study groups’, specifically, Europe (which comprises data from EPIC, CeRePP, and FBCS), Multiethnic Cohort (MEC)/LA (which comprises cases from LABCS and controls from the MEC) and Italy (which comprises cases from BBCS and controls from the EAGLE).
DNA samples for cases in NCI-GWAS2 were selected for genotyping based on pre-genotyping quality control measures performed for GWAS at the Cancer Genomics Research Laboratory of the NCI (CGR). 1504 samples were attempted at CGR on the Illumina HumanHap660 chips for NCI-GWAS2. The 1504 bladder cancer case samples, mapped to 1483 unique individuals from seven studies: HPFS, BBCS, FBCS, CeRePP, WHI, LABCS and NHS I and II. A total of 816 EPIC bladder cases were genotyped in UK and mapped to 772 unique individuals. In addition, 742 samples from NEBCS-NH were genotyped on Illumina HumanHap610 chip at the CGR, mapping to 720 unique individuals. Genotype clusters were estimated with samples by study with preliminary completion rates >98% per individual study (namely, EPIC, WHI, LABCS, NEBCS-NH, CeRePP, FBCS, BBCS, HPFS and NHS I and II). Genotypes for the analytical build were based on study-specific clustering. SNP assays with locus call rates <90% were excluded. The number of SNPs available for association analysis for NCI-GWAS2 was 509 990. After quality control metrics were applied to the full data set, 462 190 SNPs overlapped across the three GWAS datasets. Access to the NCI-GWAS1 and GWAS2 genotypes are available through dbGAP identifier, phs000346.v2, at http://www.ncbi.nlm.nih.gov/gap.
Additional participants were excluded based on: (i) completion rates <94–96% (n = 126 samples), (ii) heterozygosity of <27% or >33% (n = 13), (iii) gender discordant subjects (n = 26), (iv) one from each first-degree relative pairs including unexpected duplicates (n = 33). Seventy-five expected duplicates were evaluated and yielded an average concordance rate of 99.93%.
Assessment of the population substructure of study participants was performed with Genotyping Library and Utilities (GLU) struct.admix module by seeding the analysis with founder genotypes from three HapMap populations (build 26) (43). A set of 12 898 SNPs with extremely low pair-wise correlation (r2 < 0.004) was selected for this analysis (44–46). A total of 79 participants (15 cases and 64 controls) were estimated to have <80% HapMap CEU admixture (Supplementary Material, Fig. S6). Principal component analysis (PCA) of genotyped subjects (excluding one from each inferred closely related pairs) was performed with GLU struct.pca module (a similar procedure to EIGENSTRAT) (44,45). We ran a logistic regression model based on studies in NCI GWAS2 or studies in NCI GWAS1, respectively, and the eigenvectors that we adjusted for in the regression model were derived from principal components analysis, which were significantly associated with the case–control status based on each coefficient z statistic with P-value of <0.05, as has been shown previously (47–49). Based on this analysis, we adjusted for EV4 in NCI-GWAS1 and (EV1, 2, 3, 4, 6, 7) for NCI-GWAS2. We also evaluated PCs by each case–control set and adjustment by PCs by this alternate approach yielded similar association results (data not shown).
We estimated the inflation of the test statistic, λ, adjusted to a sample size of 1000 cases/1000 controls as per the method of de Bakker et al. λ (corrected) = 1 + (λ − 1) × [ncase−1 + ncont−1]/[2 × 10−3] (50). A quantile–quantile plot of NCI-GWAS2 data showed some enrichment of the test statistics at lower P-values compared with the expected uniform distribution (Supplementary Material, Fig. S7). The corrected estimated λ1000 is 1.004, whereas the uncorrected λ is 1.012. For the NCI-GWAS1 and TXBCS-GWAS, genotyping was conducted as previously described (9,10). A quantile–quantile plot of combined meta-analysis data from NCI-GWAS2, NCI-GWAS1 and TXBCS-GWAS showed a λ = 1.016 and after removal of SNPs 200 kb within the 14 bladder cancer susceptibility regions with tag SNPs λ = 1.015 (Supplementary Material, Fig. S8).
Custom TaqMan genotyping assays (ABI, Foster City, CA, USA) were designed and optimized for 13 SNPs. TaqMan genotyping for the 13 SNPs in TXBCS replication samples were conducted at MD Anderson Cancer Center. For the four SNPs with the most significant associations that achieved or approached genome-wide significance (rs10936599 at 3q26.2, rs907611 at 11p15.5, rs6104690 at 20p12.2 and rs4510656 at 6p22.3), a total of 425 samples were randomly chosen from NCI-GWAS scans for technical validation. One sample was excluded due to its low completion rate (<80%, i.e. no call for more than one assay), and the remaining 424 samples were merged into 376 unique individuals (145 cases and 231 controls). The overall concordance rate for 48 pairs of blind duplicates was 100%. A comparison of the genotypes from the GWAS scan and these TaqMan assays showed 100% concordance among the 376 unique individuals for these four SNPs. The Illumina Infinium cluster plots for the four novel associations, rs10936599 at 3q26.2, rs907611 at 11p15.5, rs6104690 at 20p12.2 and rs4510656 at 6p22.3 are shown in Supplementary Material, Figure S9.
Association analysis
Primary association analyses used logistic regression, adjusted for age (in 5-year categories), sex and study group. Based on the results of the PCA described above (Supplementary Material, Fig. S1), we further adjusted for eigenvectors in NCI GWAS datasets: EV4 for NCI-GWAS1 and (EV1, 2, 3, 4, 6, 7) for NCI-GWAS2. Each SNP genotype was coded as a count of minor alleles, with the exception of X-linked SNPs among men that were coded as 2 if the participant carried the minor allele and 0 if he carried the major allele (51). A score test with one degree of freedom was performed on all genetic parameters in each model to determine statistical significance. We assessed heterogeneity in genetic effects across study groups using the I2 statistic. Fixed-effects meta-analysis were conducted for the 13 SNPs assessed across the four datasets, using allelic odds ratios adjusted by age, sex, smoking status and study group, and eigenvectors for NCI-GWAS2 and NCI-GWAS1. Adjusted allelic odds ratios did not materially differ from unadjusted allelic odds ratios using genotype counts by case–control status (Table 1 and Supplementary Material, Fig. S3).
Assessment of gene–environment interactions for both additive and multiplicative models tests whether the observed joint effects OR for smoking and the genetic risk are significantly different than the expected joint effects OR. On a multiplicative scale, which evaluates whether the relative risk for smoking varies across levels of genetic risk, the expected joint effects are calculated as ORSNP*ORsmoking. On an additive scale, which evaluates whether the risk difference for smoking varies across levels of genetic risk, the expected joint effects are calculated as ORSNP + ORsmoking-1. Because the expected joint effects of the additive model are lower than the multiplicative, except when one of the two ORs for SNP or exposure are equal to 1.0, the observed joint effects were closer to the multiplicative than the additive expected effects, rejection of the null hypothesis for additive more than multiplicative interaction is more frequent.
To conduct a test for gene–environment interaction on the relative risk scale (multiplicative interaction), we used an Empirical Bayes (EB) model fitting procedure that can gain power by exploiting the assumption of gene–environment independence in the underlying population and yet is immune to bias when the independence assumption is violated (52,53). For gene–environment interaction testing on the additive scale (i.e. risk difference), we conducted a likelihood ratio test comparing an unconstrained and constrained model for joint effects using logistic regression models (54). Under the null hypothesis of an additive model, the OR for the joint effect of a given SNP and smoking status is constrained so that the risk difference associated with one exposure (e.g. smoking) is constant across levels of other exposure (e.g. SNP) and vice versa. All tests for gene–environment interactions were conducted using categorical variables (each SNP was coded as a dichotomous variable indicating the presence of any risk allele) to avoid complex numerical issues in the additive test and to make the additive and multiplicative tests comparable.
Polytomous logistic regression was used to obtain estimates of effect for different tumor subtypes. Case–case analyses with tumor type as an outcome were used to test for differences in effect size across subtypes. Polytomous logistic regression models were also used to test for trends and were calculated by constraining the effect size to increase linearly across levels of tumor grade and stage. SNP–SNP interactions were assessed using logistic regression models adjusted by study center, age sex and smoking status and including interaction terms.
Data analysis and management were performed with GLU (version 1.0), a suite of tools available as an open-source application for management, storage and analysis of GWAS data, and STATA.
Estimate of recombination hotspots
SequenceLDhot (55) that uses an approximate marginal likelihood method (56) was used to compute likelihood ratio statistics for a set of putative hotspots across the region of interest. We sequentially analyzed subsets of 100 controls of European background (by pooling five controls from each study). We used Phasev2.1 to infer the haplotypes as well as background recombination rates. The analysis was repeated with five non-overlapping sets of 100 pooled controls.
Imputation
Imputation was performed on 5942 cases and 10 861 control samples from NCI-GWAS2 and NCI-GWAS1 for the four newly identified bladder cancer susceptibility regions at 3q26.2, 11p15.5, 20p12.2 and 6p22.3. IMPUTE v2.2.2 (57) was used to impute 2-Mb intervals of Chr3: 168492101–170492101; Chr11: 874072–2874072; Chr20: 9988099–11988099 and Chr6: 19766697–21766697 (GRCh37/hg19) using a 1000 Genomes Phase 1 integrated variant set (release v3) updated on August 26, 2012.
Expression analysis
The full set of bladder tissue samples included 45 muscle-invasive tumors and 44 adjacent normal bladder samples. All tissue samples were purchased from Asterand (Detroit, MI, USA) after exemption #4715 by the National Institutes of Health (NIH) Office of Human Subjects Research. The samples were not included in GWAS and were genotyped separately for index GWAS markers with TaqMan assays used for GWAS validation studies. RNA-sequencing of tumor (n = 7) and adjacent normal (n = 5) bladder tissue samples was previously described (58). The high-quality RNA samples (RIN > 8.0) were sequenced with HiSeq 2000 to generate paired reads of 107 bp. Expression studies for individual transcripts were performed in total RNA samples extracted from tissues with MirVana to preserve miRNA fraction (Life Technologies). cDNA samples were synthesized with SuperScript III and random hexamers (Life Technologies). All the TaqMan expression assays were from Life Technologies: Hs00158885_m1 for LSP1, Hs00268536_g1 for TNNI2, Hs00610931_m1 for MYNN and Hs00214949_m1 for CDKAL1. Endogenous controls beta-2-microglobulin (B2M, assay Hs00187842_m1) and beta-actin (ACTB, assay 4352935) were used for normalization of expression.
Expression of mRNA and miRNA transcripts was analyzed according to relative quantification method and the differences were presented on log 2 scale as dCt = Ct (control) – Ct (target). dCt values can be converted to fold differences as fold = 2dCt. For all assays, reactions with water and 10 ng of genomic DNA were used as negative controls. The expression detection was performed on the ABI PRISM 7900HT SDS with cDNA prepared from ∼10 ng of total RNA, 0.25 µl of 20× TaqMan gene expression assays and 2.5 µl of 2× Gene Expression Master Mix in 5 µl reaction volume. The expression was measured in four technical replicates and average values were used for the analysis. An unpaired two-tailed t-test was applied when comparing expression of tumor and normal samples. Association between genotypes and expression of transcripts was analyzed by linear regression (P-trend) implemented in PLINK.
Data access
The CGEMS data portal provides access to individual level data for investigators from certified scientific institutions after approval of a submitted Data Access Request.
URLs
CGEMS portal, http://cgems.cancer.gov/; Cancer Genomics Research Laboratory, CGR, http://cgf.nci.nih.gov/; GLU, http://code.google.com/p/glu-genetics/; EIGENSTRAT, http://genepath.med.harvard.edu/~reich/EIGENSTRAT.htm; SNP500Cancer, http://snp500cancer.nci.nih.gov/; STRUCTURE, http://pritch.bsd.uchicago.edu/structure.html; STATA, http://www.stata.com/; http://www.ncbi.nlm.nih.gov/gap.
SUPPLEMENTARY MATERIAL
FUNDING
Funding support for individual studies that participated in the effort is as follows: ATBC (D.A.)—This research was supported in part by the Intramural Research Program of the NIH and the National Cancer Institute. Additionally, this research was supported by US Public Health Service contracts (N01-CN-45165, N01-RC-45035, N01-RC-37004 and HHSN261201000006C) from the National Cancer Institute, Department of Health and Human Services. EPIC (P.V.)—ICL—Europe Against Cancer Program of the European Commission (SANCO); IARC—International Agency for Research on Cancer; France—Ligue contre le Cancer Societe 3M, Mutuelle Generale de l'Education Nationale; Institut National de la Santé et de la Recherche Médicale (INSERM); Italy—Italian Association for Research on Cancer National Research Council; Spain—Health Research Fund (FIS) of the Spanish Ministry of Health; the CIBER en Epidemiología y Salud Pública (CIBERESP), Spain; ISCIII RETIC (RD06/0020); Spanish Regional Governments of Andalusia, Asturias, Basque Country, Murcia (N 6236) and Navarra and the Catalan Institute of Oncology; UK—Cancer Research UK Medical Research Council with additional support from the Stroke Association, British Heart Foundation, Department of Health, Food Standards Agency, the Wellcome Trust; The Netherlands—Dutch Ministry of Public Health Dutch Prevention Funds LK Research Funds Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF); Greece—Hellenic Ministry of Health, the Stavros Niarchos Foundation and the Hellenic Health Foundation; Germany—German Cancer Aid, German Cancer Research Center Federal Ministry of Education and Research (Grant 01-EA-9401); Sweden—Swedish Cancer Society, Swedish Scientific Council, Regional Government of Skane, Sweden and Denmark—Danish Cancer Society. FBCS (S.B.)—Ligue Contre le Cancer du Val-de-Marne; Fondation de France; Groupement d'Entreprises Françaises dans la Lutte contre le Cancer; Association pour la Recherche sur le Cancer, France (TXBCS—U01 CA 127615 (X.W.), R01 CA 74880 (X.W.) and P50 CA 91846 (X.W. and C.P.D.), UT MD Anderson Cancer Centre Research Trust (X.W.), Centre for Translational and Public Health Genomics at MD Anderson Cancer Centre. LABCS (M.P.)—NIH (grants R01CA65726, R01CA114665, 1R01CA114665 and 1P01CA86871). NEBCS (D.T.S.)—Intramural research program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics and intramural contract number NCI N02-CP-01037. NHS and HPFS (I.D.V.)—CA055075, P01 CA87969, R01 CA49449, UM1 CA176726, R01 CA67262 and UM1 CA167552. PLCO (M.P.P.)—The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 and RO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C), and study coordination at the GENEVA (N.C.) Coordination Center (U01HG004446) for the genotyping of the lung studies with controls from EAGLE study and part of the PLCO. Genotyping for the remaining part of PLCO and all ATBC and CPS-II samples were supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The PLCO is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health. SBCS (D.T.S.)—Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics and intramural contract number NCI N02-CP-11015. FIS/Spain 98/1274, FIS/Spain 00/0745, PI061614 and G03/174, Fundació Marató TV3, Red Temática Investigación Cooperativa en Cáncer (RTICC), Consolíder ONCOBIO, EU-FP7-201663; and RO1-CA089715 and CA34627. WHI (C.K.)—The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C and HHSN271201100004C.
Supplementary Material
Acknowledgments
Conflict of Interest statement. G.L.A. has a commercial research grant from Johnson & Johnson, Medivation and Wilex, has ownership interest (including patents) in Envisioneering Medical and is a consultant/advisory board member of Amgen, Augmenix, Steba Biotech, Viking Medical, Bayer, Bristol Myers Squibb, Cambridge Endo, Caris, GlaxoSmithKline, Janssen Biotech Inc., Myriad Genetics and Ortho-Clinical Diagnostics.
REFERENCES
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Silverman D.T., Devessa S.S., Moore L., Rothman N. In: Cancer Epidemiology and Prevention 3rd edn. Fraumeni J.F. Jr, Schottenfeld D., editors. New York, NY: Oxford University Press; 2006. pp. 1101–1127. [Google Scholar]
- 3.Kantor A.F., Hartge P., Hoover R.N., Fraumeni J.F., Jr Familial and environmental interactions in bladder cancer risk. Int. J. Cancer. 1985;35:703–706. doi: 10.1002/ijc.2910350602. [DOI] [PubMed] [Google Scholar]
- 4.Murta-Nascimento C., Silverman D.T., Kogevinas M., Garcia-Closas M., Rothman N., Tardon A., Garcia-Closas R., Serra C., Carrato A., Villanueva C., et al. Risk of bladder cancer associated with family history of cancer: do low-penetrance polymorphisms account for the increase in risk? Cancer Epidemiol. Biomarkers Prev. 2007;16:1595–1600. doi: 10.1158/1055-9965.EPI-06-0743. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Closas M., Malats N., Silverman D., Dosemeci M., Kogevinas M., Hein D.W., Tardon A., Serra C., Carrato A., Garcia-Closas R., et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Closas M., Ye Y., Rothman N., Figueroa J.D., Malats N., Dinney C.P., Chatterjee N., Prokunina-Olsson L., Wang Z., Lin J., et al. A genome-wide association study of bladder cancer identifies a new susceptibility locus within SLC14A1, a urea transporter gene on chromosome 18q12.3. Hum. Mol. Genet. 2011;20:4282–4289. doi: 10.1093/hmg/ddr342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiemeney L.A., Thorlacius S., Sulem P., Geller F., Aben K.K., Stacey S.N., Gudmundsson J., Jakobsdottir M., Bergthorsson J.T., Sigurdsson A., et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore L.E., Baris D.R., Figueroa J.D., Garcia-Closas M., Karagas M.R., Schwenn M.R., Johnson A.T., Lubin J.H., Hein D.W., Dagnall C.L., et al. GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis. 2011;32:182–189. doi: 10.1093/carcin/bgq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothman N., Garcia-Closas M., Chatterjee N., Malats N., Wu X., Figueroa J.D., Real F.X., Van Den Berg D., Matullo G., Baris D., et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 2010;42:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X., Ye Y., Kiemeney L.A., Sulem P., Rafnar T., Matullo G., Seminara D., Yoshida T., Saeki N., Andrew A.S., et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiemeney L.A., Sulem P., Besenbacher S., Vermeulen S.H., Sigurdsson A., Thorleifsson G., Gudbjartsson D.F., Stacey S.N., Gudmundsson J., Zanon C., et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat. Genet. 2010;42:415–419. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafnar T., Vermeulen S.H., Sulem P., Thorleifsson G., Aben K.K., Witjes J.A., Grotenhuis A.J., Verhaegh G.W., Hulsbergen-van de Kaa C.A., Besenbacher S., et al. European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum. Mol. Genet. 2011;20:4268–4281. doi: 10.1093/hmg/ddr303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang W., Fu Y.P., Figueroa J.D., Malats N., Garcia-Closas M., Chatterjee N., Kogevinas M., Baris D., Thun M., Hall J.L., et al. Mapping of the UGT1A locus identifies an uncommon coding variant that affects mRNA expression and protects from bladder cancer. Hum. Mol. Genet. 2012;21:1918–1930. doi: 10.1093/hmg/ddr619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J.H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landi M.T., Chatterjee N., Yu K., Goldin L.R., Goldstein A.M., Rotunno M., Mirabello L., Jacobs K., Wheeler W., Yeager M., et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landi M.T., Consonni D., Rotunno M., Bergen A.W., Goldstein A.M., Lubin J.H., Goldin L., Alavanja M., Morgan G., Subar A.F., et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michiels S., Laplanche A., Boulet T., Dessen P., Guillonneau B., Mejean A., Desgrandchamps F., Lathrop M., Sarasin A., Benhamou S. Genetic polymorphisms in 85 DNA repair genes and bladder cancer risk. Carcinogenesis. 2009;30:763–768. doi: 10.1093/carcin/bgp046. [DOI] [PubMed] [Google Scholar]
- 20.Siddiq A., Couch F.J., Chen G.K., Lindstrom S., Eccles D., Millikan R.C., Michailidou K., Stram D.O., Beckmann L., Rhie S.K., et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher F.R., Berndt S.I., Siddiq A., Jacobs K.B., Wang Z., Lindstrom S., Stevens V.L., Chen C., Mondul A.M., Travis R.C., et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum. Mol. Genet. 2011;20:3867–3875. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R., et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Closas M., Rothman N., Figueroa J.D., Prokunina-Olsson L., Han S.S., Baris D., Jacobs E.J., Malats N., De Vivo I., Albanes D., et al. Common genetic polymorphisms modify the effect of smoking on absolute risk of bladder cancer. Cancer Res. 2013;73:2211–2220. doi: 10.1158/0008-5472.CAN-12-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houlston R.S., Cheadle J., Dobbins S.E., Tenesa A., Jones A.M., Howarth K., Spain S.L., Broderick P., Domingo E., Farrington S., et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones A.M., Beggs A.D., Carvajal-Carmona L., Farrington S., Tenesa A., Walker M., Howarth K., Ballereau S., Hodgson S.V., Zauber A., et al. TERC polymorphisms are associated both with susceptibility to colorectal cancer and with longer telomeres. Gut. 2012;61:248–254. doi: 10.1136/gut.2011.239772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirabello L., Yu K., Kraft P., De Vivo I., Hunter D.J., Prescott J., Wong J.Y., Chatterjee N., Hayes R.B., Savage S.A. The association of telomere length and genetic variation in telomere biology genes. Hum. Mutat. 2010;31:1050–1058. doi: 10.1002/humu.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blasco M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- 31.Takihana Y., Tsuchida T., Fukasawa M., Araki I., Tanabe N., Takeda M. Real-time quantitative analysis for human telomerase reverse transcriptase mRNA and human telomerase RNA component mRNA expressions as markers for clinicopathologic parameters in urinary bladder cancer. Int. J. Urol. 2006;13:401–408. doi: 10.1111/j.1442-2042.2006.01300.x. [DOI] [PubMed] [Google Scholar]
- 32.Jongstra-Bilen J., Jongstra J. Leukocyte-specific protein 1 (LSP1): a regulator of leukocyte emigration in inflammation. Immunol. Res. 2006;35:65–74. doi: 10.1385/IR:35:1:65. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y.L., Amr S., Saleh D.A., Dash C., Ezzat S., Mikhail N.N., Gouda I., Loay I., Hifnawy T., Abdel-Hamid M., et al. Urinary bladder cancer risk factors in Egypt: a multicenter case-control study. Cancer Epidemiol. Biomarkers Prev. 2012;21:537–546. doi: 10.1158/1055-9965.EPI-11-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson C.A., Boucher G., Lees C.W., Franke A., D'Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A., et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calin G.A., Croce C.M. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 37.Kim H.K., Prokunina-Olsson L., Chanock S.J. Common genetic variants in miR-1206 (8q24.2) and miR-612 (11q13.3) affect biogenesis of mature miRNA forms. PLoS ONE. 2012;7:e47454. doi: 10.1371/journal.pone.0047454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S., et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 39.Pascoe L., Tura A., Patel S.K., Ibrahim I.M., Ferrannini E., Zeggini E., Weedon M.N., Mari A., Hattersley A.T., McCarthy M.I., et al. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56:3101–3104. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 40.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hancock D.B., Artigas M.S., Gharib S.A., Henry A., Manichaikul A., Ramasamy A., Loth D.W., Imboden M., Koch B., McArdle W.L., et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 46.Yu K., Wang Z., Li Q., Wacholder S., Hunter D.J., Hoover R.N., Chanock S., Thomas G. Population substructure and control selection in genome-wide association studies. PLoS ONE. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berndt S.I., Skibola C.F., Joseph V., Camp N.J., Nieters A., Wang Z., Cozen W., Monnereau A., Wang S.S., Kelly R.S., et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat. Genet. 2013;45:868–876. doi: 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan Q., Hsiung C.A., Matsuo K., Hong Y.C., Seow A., Wang Z., Hosgood H.D., III, Chen K., Wang J.C., Chatterjee N., et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet. 2012;44:1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savage S.A., Mirabello L., Wang Z., Gastier-Foster J.M., Gorlick R., Khanna C., Flanagan A.M., Tirabosco R., Andrulis I.L., Wunder J.S., et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat. Genet. 2013;45:799–803. doi: 10.1038/ng.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clayton D. Testing for association on the X chromosome. Biostatistics. 2008;9:593–600. doi: 10.1093/biostatistics/kxn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y.H., Chatterjee N., Carroll R.J. Shrinkage estimators for robust and efficient inference in haplotype-based case-control studies. J. Am. Stat. Assoc. 2009;104:220–233. doi: 10.1198/jasa.2009.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee B., Chatterjee N. Exploiting gene-environment independence for analysis of case-control studies: an empirical Bayes-type shrinkage estimator to trade-off between bias and efficiency. Biometrics. 2008;64:685–694. doi: 10.1111/j.1541-0420.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 54.Han S.S., Rosenberg P.S., Garcia-Closas M., Figueroa J.D., Silverman D., Chanock S.J., Rothman N., Chatterjee N. Likelihood ratio test for detecting gene (G)–environment (E) interactions under an additive risk model exploiting G–E independence for case-control data. Am. J. Epidemiol. 2012;176:1060–1067. doi: 10.1093/aje/kws166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics. 2006;22:3061–3066. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 56.Fearnhead P., Harding R.M., Schneider J.A., Myers S., Donnelly P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics. 2004;167:2067–2081. doi: 10.1534/genetics.103.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu Y.P., Kohaar I., Rothman N., Earl J., Figueroa J.D., Ye Y., Malats N., Tang W., Liu L., Garcia-Closas M., et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc. Natl. Acad. Sci. USA. 2012;109:4974–4979. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.