Abstract
Carney complex (CNC) is a hereditary disease associating cardiac myxoma, spotty skin pigmentation and endocrine overactivity. CNC is caused by inactivating mutations in the PRKAR1A gene encoding PKA type I alpha regulatory subunit (RIα). Although PKA activity is enhanced in CNC, the mechanisms linking PKA dysregulation to endocrine tumorigenesis are poorly understood. In this study, we used Förster resonance energy transfer (FRET)-based sensors for cAMP and PKA activity to define the role of RIα in the spatiotemporal organization of the cAMP/PKA pathway. RIα knockdown in HEK293 cells increased basal as well as forskolin or prostaglandin E1 (PGE1)-stimulated total cellular PKA activity as reported by western blots of endogenous PKA targets and the FRET-based global PKA activity reporter, AKAR3. Using variants of AKAR3 targeted to subcellular compartments, we identified similar increases in the response to PGE1 in the cytoplasm and at the outer mitochondrial membrane. In contrast, at the plasma membrane, the response to PGE1 was decreased along with an increase in basal FRET ratio. These results were confirmed by western blot analysis of basal and PGE1-induced phosphorylation of membrane-associated vasodilator-stimulated phosphoprotein. Similar differences were observed between the cytoplasm and the plasma membrane in human adrenal cells carrying a RIα inactivating mutation. RIα inactivation also increased cAMP in the cytoplasm, at the outer mitochondrial membrane and at the plasma membrane, as reported by targeted versions of the cAMP indicator Epac1-camps. These results show that RIα inactivation leads to multiple, compartment-specific alterations of the cAMP/PKA pathway revealing new aspects of signaling dysregulation in tumorigenesis.
INTRODUCTION
The cAMP pathway transduces the action of numerous hormones into a plethora of cellular functions ranging from metabolism and cellular excitability to cell growth and differentiation. These hormones bind to G-protein-coupled receptors (GPCRs) which in turn activate either Gs or Gi proteins to stimulate or inhibit cAMP synthesis by adenylyl cyclase, respectively. Although cAMP regulates some of these processes through direct binding to cyclic nucleotide gated channels or exchange factors for small G proteins Epac, a large majority of cAMP-dependent effects are mediated by the cAMP-dependent protein kinase (PKA) (1). In the absence of cAMP, PKA is a heterotetramer composed of two catalytic (C) and two regulatory (R) subunits. Binding of two cAMP molecules on each R subunit results in dissociation and activation of the C subunits. Two types of PKA, designated PKAI and PKAII, were originally identified based on the nature of their R subunit, termed RI and RII (2,3). Molecular cloning subsequently revealed the existence of two RI subunits (RIα and RIβ) and two RII subunits (RIIα and RIIβ) encoded by distinct genes, as well as up to four C subunits (Cα, Cβ, Cγ and PRKX) (1). Such molecular diversity gives rise to multiple PKA isozymes with distinct biochemical properties (4) and physiological functions (5). It also confers to the PKA system the capacity to self-regulate, a process in which the RIα subunit plays a unique role, acting as an universal buffer of unregulated PKA activity. Indeed, compensatory increase in RIα protein have been observed upon overexpression of C subunits in NIH 3T3 fibroblasts and pituitary AtT-20 cells (6), upon silencing of RIIβ subunits in Y1 adrenal cells (7) or upon invalidation of the genes encoding for RIβ, RIIα and RIIβ in mice (reviewed in (5)). Accordingly, RIα is the only PKA subunit which absence results in embryonic lethality in mice due to impaired development of the heart (8,9).
A crucial mechanism to achieve PKA signaling specificity is the subcellular targeting of the kinase via scaffold proteins called A-kinase anchoring proteins (AKAPs) (1,10). AKAPs cluster PKA to its downstream targets as well as signal termination enzymes such as phosphatases (11,12) and phosphodiesterases (PDEs) (13,14). For instance, in HEK293 cells, AKAP250 (gravin) binds PKAII and PDE4D to control cAMP levels beneath the plasma membrane (15). Although the majority of AKAPs described so far bind PKAII, it is now appreciated that PKAI is also specifically localized by AKAPs in various cell types (16–21). For instance, PAP7 specifically targets PKAI at the outer mitochondrial membrane to regulate the steroid acute response protein and cholesterol transport in steroid producing cells (22,23).
Mutations in various components of the cAMP pathway such as GPCRs, Gs, PKA and PDEs have been associated with endocrine diseases. Recently, gain-of-function mutations in the RIα gene PRKAR1A at 17q23-24 that decrease PKA sensitivity to cAMP and confer hormonal resistance were reported in patients with acrodysostosis, a rare form of skeletal dysplasia characterized by short stature, severe brachydactyly, facial dysostosis and nasal hypoplasia (24–26). In contrast, inactivating mutations of RIα have been found in patients with “Carney complex” (CNC) (27–29). CNC is an autosomal dominant multiple endocrine neoplasia syndrome defined by the association of “myxomas, spotty skin pigmentation and endocrine overactivity” (30,31). The most frequent endocrine manifestation is primary pigmented nodular adrenal disease (PPNAD), but acromegaly, thyroid adenomas or carcinomas, ovarian cysts or cancer and large cell calcifying Sertoli cell tumors are also associated with CNC. PPNAD is responsible for an ACTH-independent Cushing's syndrome linked to an autonomic secretion of cortisol by the two adrenals. Most often, medical treatment is insufficient and patients undergo bilateral adrenalectomy. A recent publication reported a total of 117 pathogenic PRKAR1A mutations identified to date (32). The majority of PRKAR1A inactivating mutations leads to a premature stop codon (97/117; 82.9%) and an unstable mRNA degraded by non-sense mediated mRNA decay (NMD) leading to a 50% reduction of cellular RIα level (32). Since a 17q22–24 allelic loss is often observed in tumors from CNC patients, these mutations may lead to a complete loss of PRKAR1A expression (28). Reduced levels of RIα are associated with an increase in total cellular or tissue PKA activity (8,28,33). Primary mouse embryonic fibroblasts lacking RIα exhibit constitutive PKA activation and are immortalized (34). Heterozygous mice with PRKAR1A inactivation develop tumors (35) and have fertility defects (36). Selective inactivation of PRKAR1A in the adrenal cortex of mice increases PKA activity and reproduces the essential features of PPNAD in humans with PRKAR1A mutations (37).
The mechanisms by which PRKAR1A inactivation increases PKA activity are not completely understood. Considering the major role of compartmentation for physiological cAMP/PKA signaling, it is crucial to understand the effects of PRKAR1A mutations at the subcellular level. The consequences of PRKAR1A inactivation might differ between subcellular compartments and/or the timing of PKA activation. RIα inactivation might also alter the cAMP buffering capacity of the cell, thus impacting subcellular cAMP levels (38–40). Over the last decade, new methods for monitoring intracellular PKA activity and cAMP levels in single living cells have been developed. A-kinase activity reporter (AKARs) and Epac-based cAMP sensors are genetically encoded probes based on Förster resonance energy transfer (FRET) that allow real-time monitoring of PKA activity and cAMP levels (41–44). Here, we used variants of these sensors targeted to various subcellular compartments to investigate the consequences of PRKAR1A inactivation on the subcellular dynamics of the cAMP/PKA pathway.
RESULTS
PRKAR1A silencing increases forskolin-stimulated global PKA activity in HEK293 cells
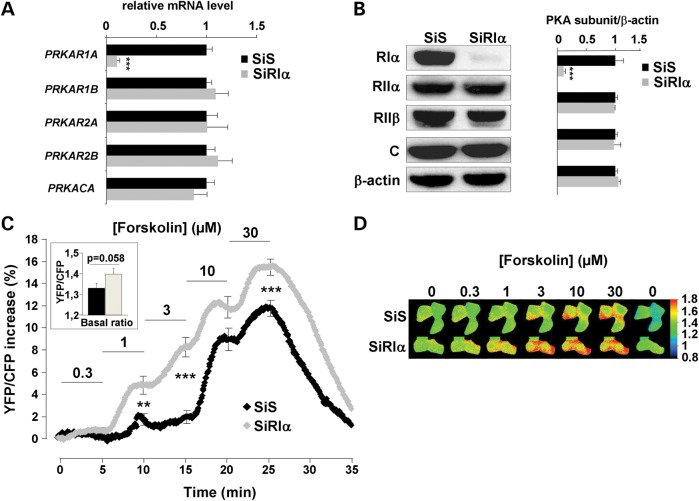
As stated above, the majority of PRKAR1A inactivating mutations introduce a premature stop codon and NMD (32). To mimic this situation, we used a small interfering RNA (SiRIα) to knockdown PRKAR1A expression in HEK293 cells. As shown in Figure 1A and B, PRKAR1A mRNA and protein levels were reduced by ∼80% in cells transfected with SiRIα for 48 h as compared with a scrambled siRNA (SiS) used as control. PRKAR1A silencing did not alter mRNA or protein levels for the other PKA subunits including RIIα, RIIβ and C.
Figure 1.
PRKAR1A silencing and real-time monitoring of global PKA activity with AKAR3 in HEK293 cells. (A) Quantitative PCR analysis and (B) western blot analysis of PKA regulatory subunits RIα, RIIα, RIIβ and catalytic C subunit in HEK293 cells 48 h after transfection with either scrambled (SiS) or RIα-specific (SiRIα) siRNA. PPIA (CYCLO) gene expression was used as reference in PCR. β-Actin was used as loading control in western blot. Data represent the average of four- and three-independent experiments for qPCR and western blot analysis, respectively. (C) HEK293 cells were cotransfected with AKAR3 and either SiS or SiRIα and FRET measurements were performed 48 h after. Average time course of the normalized YFP/CFP ratio upon challenge with increasing concentrations of forskolin (SiS, n = 61; SiRIα, n = 61). Each forskolin concentration was applied for 5 min, as indicated by the solid lines. Error bars at the end of each application indicate SEM. Bar graph in inset shows average basal YFP/CFP ratio value before forskolin application. Statistically significant differences between SiS and SiRIα are indicated as **P < 0.01; ***P < 0.001. (D) Pseudocolor images illustrate the variations of the YFP/CFP emission ratio elicited by forskolin in SiS- and SiRIα-transfected cells.
To investigate the consequences of PRKAR1A knockdown on total PKA activity in living cells, HEK293 cells were cotransfected with siRNA and the A-kinase activity reporter, AKAR3. AKAR3 includes a phosphothreonine-binding domain [forkhead associated domain 1 (FHA)] and a PKA consensus site inserted between optimized variants of the FRET pair CFP and YFP. The FHA domain binds the substrate domain when phosphorylated by PKA, thus allowing FRET between the two fluorophores (41). As shown in the inset of Figure 1C, the average basal YFP/CFP ratio tended to be higher in PRKAR1A-silenced cells (P = 0.058). As shown in the frames of Figure 1D and by the average time courses in Figure 1C, the activation started to be observed at 0.3 µm forskolin (P < 0.05 versus basal) in PRKAR1A-silenced cells, a concentration that had no effect in control cells. Moreover, for each forskolin concentration between 1 and 30 µm, PKA activity was higher in RIα-deficient cells compared with controls (Fig. 1C). Thus, RIα knockdown increased the potency and efficacy of global PKA activation in HEK293 cells.
PRKAR1A silencing increases global PKA activity in response to prostaglandin E1
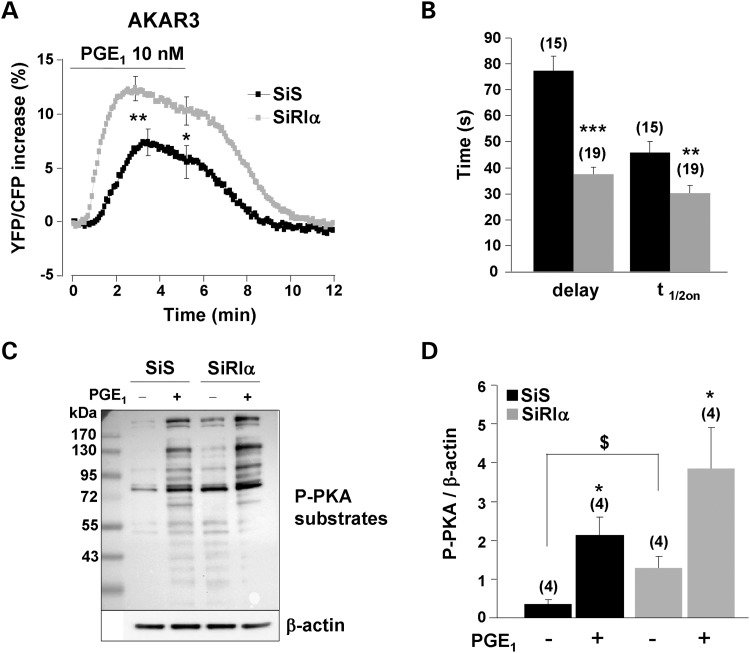
We next sought to determine whether a similar increase in PKA activity occurred upon physiological stimulation of the cAMP pathway. For this, HEK293 cells were challenged with prostaglandin E1 (PGE1) to stimulate endogenous prostanoid receptors positively coupled to adenylyl cyclase (AC) (45,46). The response to 10 nm PGE1 was nearly doubled in RIα-deficient cells, when measured at the peak or after 5 min (Fig. 2A). PKA activation upon PGE1 stimulation was not sustained, as indicated by a significant difference between peak and 5 min ratio values (P < 0.05 for both SiS and SiRIα). However, this time-dependent decrease was not different between control and RIα-deficient cells. Moreover, as depicted in Figure 2B, PRKAR1A knockdown reduced the initial delay between PGE1 application and AKAR3 phosphorylation and accelerated the on-rate kinetics of the response. In order to confirm these results by an independent approach, we used a PKA-phosphospecific antibody to analyze the phosphorylation state of endogenous PKA target proteins in total extracts from HEK293 cells (Fig. 2C and D). In control cells, the basal PKA phosphorylation level was low and could be strongly increased by PGE1 stimulation. In RIα-deficient cells, the phosphorylation of PKA substrates was augmented under basal condition, a result which is consistent with the increase in basal YFP/CFP ratio observed in Figure 1C, and phosphorylation of PKA substrates was further enhanced by PGE1 stimulation. Altogether, these results indicate that RIα knockdown increases both constitutive and stimulated PKA activity in HEK293 cells.
Figure 2.
Upregulation of global PKA activity in RIα-deficient HEK293 cells. (A) Average time course of the normalized YFP/CFP ratio upon PGE1 (10 nm) stimulation in control cells (SiS, n = 15) and RIα-deficient cells (SiRIα, n = 19) expressing AKAR3. PGE1 application started at time zero and lasted 5min before washout. SEM is indicated at the peak and at the end of the PGE1 application. Statistically significant differences between SiS and SiRIα are indicated as *P < 0.05, **P < 0.01. (B) Kinetic parameters of the PGE1 response in the experiments depicted in (A). The delay between the application of PGE1 and the initial response (Delay) and the time required to reach half-maximal response (t1/2on) are indicated. Statistically significant differences between groups are indicated as **P < 0.01, ***P < 0.001. (C) Phosphorylation of PKA substrates as detected with a Phospho-PKA substrate antibody in total proteins (50 µg/lane) from HEK293 cells transfected with SiS or SiRIα and stimulated or not with 10 nm PGE1 for 3 min. (D) Quantification of four-independent experiments as in (C). In each experimental condition, the entire lane was quantified and normalized to β-actin. Statistically significant differences are indicated as *P < 0.05 between control and PGE1 and as $P < 0.05 between SiS and SiRIα.
PRKAR1A silencing differentially alters PKA activity in distinct subcellular compartments
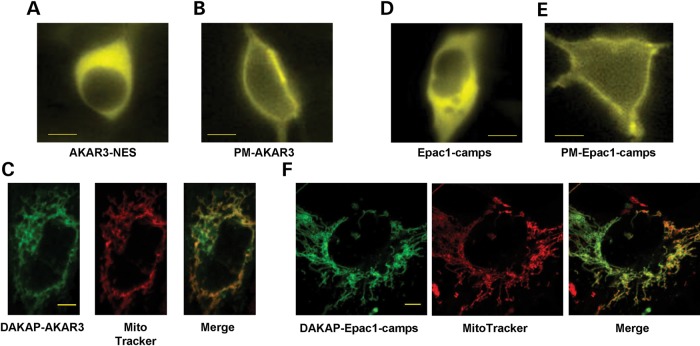
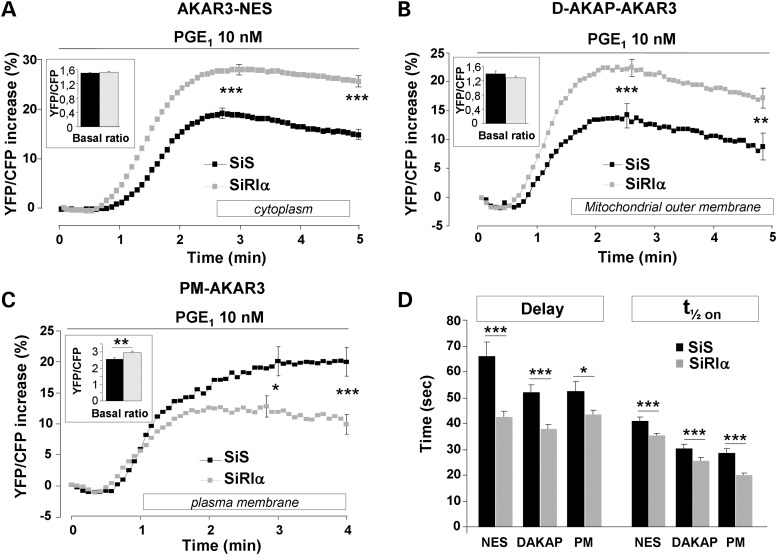
To determine whether RIα knockdown leads to specific alterations in subcellular PKA activity, we next used AKAR3 sensors targeted to specific compartments. A variant of AKAR3 containing a nuclear export signal (AKAR3-NES; Fig. 3A) was used to measure PKA activity in the cytoplasm (41). We also used an AKAR3 variant fused with a lipid anchor that allows targeting to the plasma membrane (PM-AKAR3; Fig. 3B) (41). Excessive steroid hormone production is a hallmark of endocrine tumors in CNC (30). The first step in steroid hormone biosynthesis is cholesterol transport into mitochondria (47), a process regulated by PKAI at the outer mitochondrial membrane (22). We evaluated how PRKAR1A inactivation affects PKA activity in this compartment using a variant of AKAR3 targeted to the mitochondrial outer membrane (DAKAP-AKAR3, which colocalized with the mitochondrial dye MitoTracker® Red, Supplementary Material, Fig. S3C) (41). As shown in the insets of Figure 4A and B, the average basal YFP/CFP ratios were similar in RIα-deficient cells and controls in the cytoplasm and at the outer membrane of mitochondria. Conversely, the basal YFP/CFP ratio was significantly higher in RIα-deficient cells than in controls at the plasma membrane (inset of Fig. 4C, P = 0.002).
Figure 3.
Subcellular localization of AKAR3 and Epac1-camps indicators in HEK293 cells. All images were taken 48 h after transfection of the various FRET sensors. YFP images of representative cells transfected with AKAR3-NES (A), PM-AKAR3 (B), Epac1-camps (D) and PM-Epac1-camps (E) obtained with epifluorescent microscopy. Scale bars: 10 µm. (C)Confocal YFP images of DAKAP-AKAR3, MitoTracker® Red staining and colocalization (Pearson's coefficient: 0.87). (F) Confocal YFP images of DAKAP-Epac1-camps, MitoTracker® Red staining and colocalization (Pearson's coefficient: 0.78). In (C) and (F) scale bars represent 5 µm.
Figure 4.
Comparative effects of PGE1 on PKA activity in distinct subcellular compartments of control and RIα-deficient HEK293 cells. Average time course of PKA activation by 10 nm PGE1 in (A) the cytoplasm, as monitored with AKAR3-NES (SiS, n = 105; SiRIα, n = 121); (B) the outer mitochondrial membrane, as reported by DAKAP-AKAR3 (SiS, n = 65; SiRIα, n = 87) and (C) the plasma membrane, as reported by PM-AKAR3 (SiS, n = 50; SiRIα, n = 97). (A–C) PGE1 application started at time zero and was maintained throughout the experiment as indicated by the solid line. SEM is indicated at the peak and at the end of the stimulation. The bar graphs in inset indicate the values of the average basal YFP/CFP ratio before PGE1 application. (D) Kinetic parameters (delay and t1/2on values) of PKA activation in the experiments as in (A–C). Statistically significant differences are indicated as *P < 0.05; **P < 0.01; ***P < 0.001.
Activation of PKA by PGE1 was elevated in the cytoplasm of RIα-deficient cells, both at the peak (P < 0.001) and after 5 min (P < 0.001). The response was transient in both groups with a similar significant decrease at 5 min (P < 0.001 for both SiS and SiRIα) (Fig. 4A). Similar results were found at the outer membrane of the mitochondria: in RIα-deficient cells, the response was higher than in the control group at the peak (P < 0.001) and after 5 min (P = 0.001). In both groups, the response was transient with a significant difference between the response at 5 min and the peak (P < 0.001 for both SiS and SiRIα) (Fig. 4B). In contrast to the cytoplasmic and the outer mitochondrial membrane compartments, PGE1 stimulation of plasma membrane PKA was decreased in RIα-deficient cells compared with controls, both at the peak (P = 0.013) and at 4 min (P < 0.001). The increase in PKA activity occurred with a significantly shorter delay and faster on-rate kinetics in RIα-deficient cells versus control cells (Fig. 4D) in all subcellular compartments, similarly to the pattern observed for global PKA activity. In order to confirm that basal PKA activity is increased at the plasma membrane, whereas the response to PGE1 is lower in SiRIα compared with SiS cells, we studied the phosphorylation of vasodilator-stimulated phosphoprotein (VASP), a plasma membrane-associated target of PKA (48). VASP is involved in focal adhesions, cell–cell contacts, microfilaments, and highly dynamic membrane regions (49) and was therefore used to probe cAMP-dependent PKA phosphorylation in this subcellular compartment. We transfected HEK293 cells with VASP and compared its phosphorylation by PKA at Ser157 in SiS and SiRIα cells. As shown in Supplementary Material, Figure S1, PGE1-induced VASP phosphorylation was markedly attenuated in SiRIα compared with SiS cells, an effect likely due to a significant increase in basal VASP phosphorylation (P < 0.01). Taken together, our measurements of VASP phosphorylation confirm the pattern of PKA activity observed with the membrane-targeted AKAR3-PM sensor.
Differences in PKA activity between plasma membrane and cytoplasmic compartments observed in HEK293 cells are conserved in human PPNAD cells
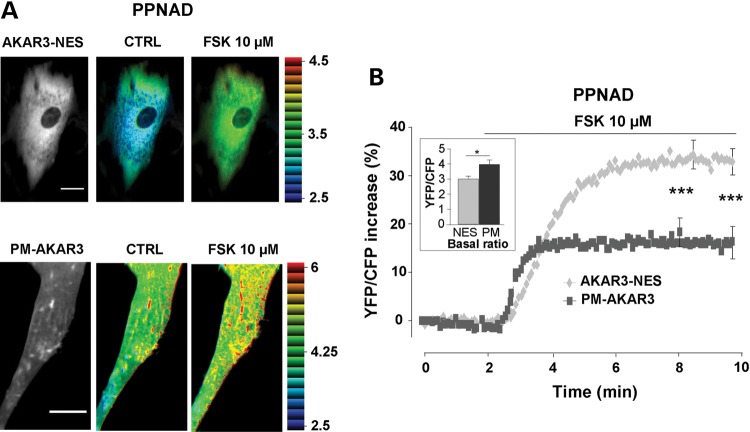
In Supplementary Material, Figure S2, the time course of PKA activation at the plasma membrane and in the cytoplasm upon PGE1 application are compared on the same graph in control and RIα-deficient cells (data are the same as those in Fig. 4A and C). As shown in the left panel of Supplementary Material, Figure S2, in control HEK293 cells, PKA activation occurred earlier at the plasma membrane than in the cytoplasm, but the amplitude of the response was similar. This indicates that the difference in the extent of PKA activation between the plasma membrane and cytoplasmic compartments observed in RIα-deficient HEK293 cells (Supplementary Material, Fig. S2, right panel) was not due to the different targeting of the sensors. This allowed comparing directly the subcellular PKA activity in human primary adrenal cells from two patients with PPNAD disease due to inactivating PRKAR1A mutation. Adenoviruses encoding AKAR3-NES and PM-AKAR3 allowed expression of both probes in human PPNAD cells (Fig. 5A). As shown in the inset of Figure 5B, the basal YFP/CFP ratio was higher in cells expressing PM-AKAR3 versus AKAR-NES, compatible with higher basal PKA activity at the plasma membrane. Activation of PKA by FSK (10 µm) was nearly doubled in the cytoplasm of PPNAD cells compared to that at the plasma membrane (P < 0.001) similarly to the observation in RIα-deficient HEK293 cells (Supplementary Material, Fig. S2, right panel).
Figure 5.
PKA activity gradients in human adrenal cells from patients with PPNAD disease. (A) Left: YFP images of PPNAD cells expressing AKAR3-NES and PM-AKAR3 (scale bar = 20 µm). Right: intensity modulated display pseudocolor images of the YFP/CFP emission ratio in control Ringer solution (CTRL) and upon forskolin application (FSK 10 µm) in the same cells. (B) Average PKA activation by forskolin (FSK 10 µm) in the cytoplasm (n = 23 cells from two patients) and at the plasma membrane (n = 16 cells from two patients). Bar graph in inset shows average basal YFP/CFP ratio values obtained with AKAR3-NES in the cytoplasm (NES) and with PM-AKAR3 at the plasma membrane (PM). Statistically significant differences are indicated as *P < 0.05; ***P < 0.001.
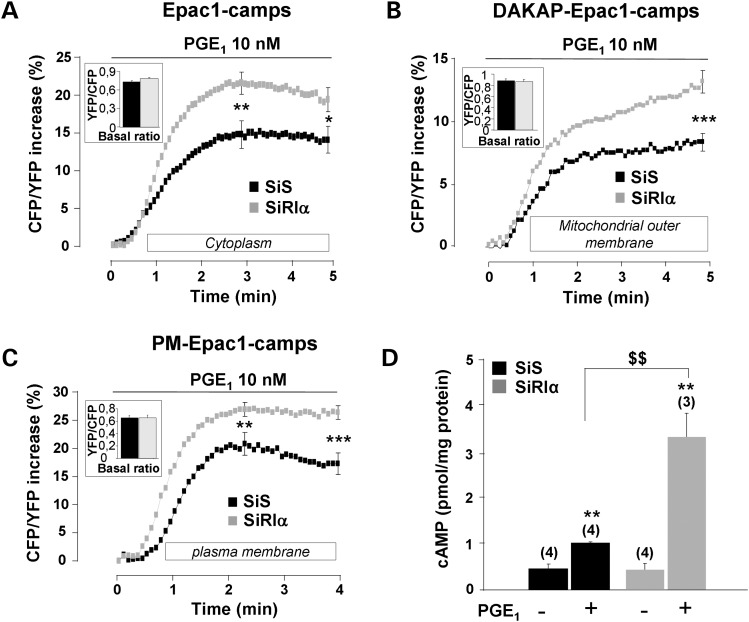
PRKAR1A silencing increases cAMP level in response to prostaglandin E1 in distinct subcellular compartments and in the whole cell
To determine whether altered PKA activities in RIα knockdown cells are due to changes in cAMP levels, subcellular pools of cAMP were measured using Epac-based cAMP sensors (42). The cytosolic Epac1-camps biosensor showed uniform distribution throughout the cytoplasm of transfected HEK293 cells (Fig. 3D), whereas fusion with a lipid anchor tag targets the sensor to the plasma membrane (PM-Epac1-camps, Fig. 3E) (50). We also generated a DAKAP-Epac1-camps sensor to investigate how PRKAR1A inactivation affects cAMP levels at the mitochondrial outer membrane. We verified that this sensor, similarly to the DAKAP-AKAR3 sensor, colocalized with MitoTracker® Red (Fig. 3F). In Epac1-camps, cAMP binding decreases FRET between CFP and YFP. Thus, cAMP elevation is reported by an increase of the CFP/YFP emission ratio. As shown in the insets of Figure 6A to C, the average basal CFP/YFP ratio measured with the different cAMP sensors did not differ between SiS and SiRIα cells. However, cAMP elevation in response to 10 nm PGE1 was significantly increased in cells with silenced RIα at the peak and after 5 min, in the cytoplasm (P < 0.05), at the plasma membrane (P < 0.01) and at 5 min at the outer membrane of mitochondria (P < 0.001). We next confirmed that global cAMP levels were increased after PGE1 stimulation in RIα-silenced cells using enzyme immunoassay. As shown in Figure 6D, total cAMP accumulation in response to PGE1 was increased by about 3-fold in RIα-silenced cells (P < 0.01). Altogether, these results indicate that upon RIα knockdown, the enhanced PKA responses to PGE1 stimulation are due at least in part to increased cAMP levels.
Figure 6.
Upregulation of cAMP levels after PGE1 stimulation in RIα-deficient HEK293 cells. Average time course of cAMP elevation in response to 10 nm PGE1 in (A) the cytoplasm, as reported by the cAMP sensor Epac1-camps (SiS, n = 61; SiRIα, n = 76); (B) the outer mitochondrial membrane, as reported by DAKAP-Epac1-camps (SiS, n = 65; SiRIα, n = 60) and (C) the plasma membrane, as reported by PM-Epac1-camps (SiS, n = 66; SiRIα, n = 88). (A–C) PGE1 application started at time zero and was maintained throughout the experiment as indicated by the solid line. In (A) and (C), SEM is indicated at the peak and at the end of the stimulation, and in (B) at 5 min (corresponding to maximal value). (D) Total cAMP levels measured by enzyme immunoassay in SiS and SiRIα treated HEK293 cells challenged with 10 nm PGE1 for 5 min. Values are mean ± SEM. Statistically significant differences are indicated as **P < 0.05 between control and PGE1 and as $$P < 0.01 between SiS and SiRIα.
DISCUSSION
The cellular consequences of PRKAR1A inactivation in CNC are not fully explained. We and others have previously demonstrated dysregulation of the cAMP signaling pathway in tumor tissues as well as in in vitro and in vivo models (28,29,51). However, until now these alterations have only been studied at the tissue or whole cell level. In this study, our main objective was to explore the cAMP/PKA signaling at the subcellular level to probe whether CNC is associated with a spatiotemporal dysregulation of this pathway. Using live cell cAMP and PKA sensors targeted to distinct subcellular compartments, we show for the first time that cAMP/PKA signaling dysregulation due to PRKAR1A inactivation differs between subcellular compartments.
In HEK293 cells, PRKAR1A silencing was not associated with a compensatory increase of other regulatory subunits of PKA (Fig. 1), as sometimes found in CNC tumors or animal models (33,52,53). Moreover, catalytic subunit levels were also unchanged. These results make it unlikely that the different responses observed in PRKAR1A-silenced cells are due to compensatory changes in the expression of the remaining PKA subunits.
Using AKAR3, we first tested whether PRKAR1A knockdown modified total PKA activation following AC activation by forskolin. It was observed that both the sensitivity and the extent of PKA stimulation were enhanced in cells with decreased RIα expression (Fig. 1). This result is consistent with in vitro measurements in human PPNAD tumors (28,54) and in CNC patient lymphocytes with a PRKAR1A inactivating mutation (55). A similar increase in global PKA activity was obtained upon PGE1 stimulation, showing that PRKAR1A inactivation also impacts hormonal stimulation subjected to physiological feedback regulatory mechanisms. In HEK293 cells, prostanoid receptors were used to demonstrate the compartmentalized nature of the cAMP/PKA pathway (15,43,45,46,56,57). Therefore, we investigated whether PRKAR1A inactivation modified PKA activation by PGE1 in subcellular compartments. Our results identify a loss of the spatiotemporal control of PKA activity upon PRKAR1A knockdown. Similarly to global PKA activity, PRKAR1A inactivation leads to higher stimulated PKA activity in the cytosolic compartment and at the outer membrane of the mitochondria. This elevated PKA activity at the outer mitochondrial membrane may provide an explanation for the increased cortisol production in PPNAD caused by PRKAR1A inactivation (30). Within the three compartments we studied, RIα depletion accelerated the kinetics of the response, with both the lag between PGE1 application and the onset of the response and the time to reach half-maximal stimulation being decreased (Fig. 4D). Interestingly, while the PGE1-induced PKA activation was delayed in the cytoplasm compared with the plasma membrane in control HEK293 cells, PRKAR1A inactivation abolished this difference (Supplementary Material, Fig. S2). These results are in good agreement with PKA regulatory subunits exerting a significant buffering of cAMP (38,39) which is predicted to shape cAMP signals and PKA-mediated phosphorylation gradients by mechanistic model studies (40,57). Such a buffering was recently shown to participate in the localized β2-adrenergic receptor cAMP signaling in cardiomyocytes (58). We tested this hypothesis using cAMP indicators based on Epac1 to monitor cAMP levels in the cytoplasm, at the outer membrane of mitochondria and at the plasma membrane. We found that indeed, PGE1 stimulation of cAMP was increased in all compartments in HEK293 cells with RIα inactivation (Fig. 6). These results are reminiscent of recent studies where an increase in cAMP levels in bone lesions from Prkar1a+/− mice and from children with neonatal-onset multisystem inflammatory disease along with an increase in PGE2 was also shown (59,60). From these results, we conclude that enhanced cAMP levels due to the loss of cAMP buffering in RIα-depleted cells is at least partly responsible for enhanced PKA activity observed in the whole cell, the cytoplasm and at the outer mitochondrial membrane upon PGE1 stimulation. Obviously, this conclusion cannot apply to the plasma membrane, where a paradoxical decrease in PGE1 stimulation of PKA activity was observed despite increased cAMP levels. However, this may simply reflect the prominent increase in basal PKA activity observed in this compartment upon inactivation of RIα, as indicated by the increased basal FRET ratio of PM-AKAR3 (Fig. 4C) and the hyperphosphorylation of VASP (Supplementary Material, Fig. S1), an established plasma membrane target of PKA that was used previously to probe PKA activity in this compartment (61,62). These results are consistent with the increased basal PKA activity previously reported in mouse embryos and adrenals with targeted inactivation of Prkar1a, presumably caused by free catalytic subunits (8,37).
The marked increase in basal PKA activity at the plasma membrane observed here could be related to the presence of a pool of PKAI at the plasma membrane in HEK293 cells. Indeed, RIα can be targeted to the plasma membrane by ezrin in T cells (16), by alpha4 integrins in Jurkat cells (17) and by smAKAP, a newly discovered RI-specific AKAP expressed in a large number of tissues (21). The catalytic subunits of PKA can be myristoylated at Gly1 and this was shown to increase its affinity to membranes (63). The membrane binding motif of the myristoylated C subunit steers the enzyme toward membranes independently of the regulatory subunits or an AKAP, which may provide yet another mechanism to confine the catalytic subunits in this compartment upon RIα invalidation (64). Importantly, similar findings were obtained when subcellular PKA activity was monitored in human adrenal cells isolated from patients with PPNAD disease caused by PRKAR1A mutations (Fig. 5): the basal FRET ratio was increased at the plasma membrane, and PKA activation by forskolin was reduced compared with the cytoplasm. This indicates that the differences identified upon PRKAR1A knockdown in HEK293 cells are relevant to the pathophysiology of CNC and tumor development.
In conclusion, the present study confirms that RIα is essential to maintain physiological regulation of PKA activity (5) and demonstrates that its inactivation differentially alters PKA activity in specific subcellular compartments of HEK293 cells. In addition, we demonstrate that RIα acts as a major buffer for cAMP, which provides a mechanism for the accelerated kinetics and overall exacerbated PKA activation observed in RIα-depleted cells. This shows for the first time that spatiotemporal dynamic alterations of the cAMP pathway are an important aspect of cAMP dysregulation in neoplasias harboring genetic alterations of key components of this pathway.
MATERIALS AND METHODS
Reagents
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum, l-glutamine, penicillin, streptomycin, opti-MEM, trypsin and PBS were obtained from Invitrogen (Invitrogen Life Technologies). Forskolin and prostaglandin E1 (PGE1) were obtained from Sigma-Aldrich. The following antibodies were used at concentrations advised by the manufacturers: antibodies against RIα, RIIα, RIIβ and C (BD Transduction Laboratories), anti phospho-PKA substrate (target sequence RRXS/T, Cell Signaling Technology), antibody against β-actin-HRP (Santa Cruz), antibodies against VASP and phospho VASP (Ser157) (Cell Signaling Technology).
Cell culture, transfection and infection
Human Embryonic Kidney 293 cells (HEK293) were grown in DMEM supplemented with 10% fetal bovine serum, 2 mm l-glutamine and antibiotics and maintained at 37°C in a humidified atmosphere containing 5% CO2. For FRET experiments, 2 × 105 cells were plated on 2 cm diameter glass coverslips. For western blots, cells were seeded on 6-well plates at a density of 8 × 105 cells per well. To achieve specific knockdown of RIα, a 21-mer RNA duplex targeting exon 7 was designed (SiRIα, UGAAUGGGCAACCAGUGUUdTdT). A scrambled sequence of the 21-mer RNA duplex was used as control (SiS, CAGUCGCGUUUGCGACUGGdTdT). The duplexes were obtained from Dharmacon and delivered into cells with Lipofectamine 2000 (Invitrogen Life Technologies) according to the manufacturer's instructions. For western blot experiments, cells were transfected with 100 pmol of either SiS or SiRIα. For FRET experiments, cells were cotransfected with 0.4 µg of FRET sensor DNA and 25 pmol of either SiS or SiRIα. Experiments were performed 48 h after transfection.
Human adrenals were obtained after informed consent from two CNC patients undergoing surgery for PPNAD. Adrenal tissue collection was approved by the ethics committee of Cochin Hospital. These two patients carried an heterozygous inactivating PRKAR1A mutation (c.109C > T and c.709(-7-2)del6) (32,65,66). Adrenals were immediately immersed in culture medium until cell dissociation. A small piece of tissue was rinsed once in phosphate-buffered isotonic saline (PBS), trimmed of fat and minced as finely as possible with small surgical scissors, in a sterilized petri dish. Cells in the mashed tumor tissue were dispersed by mechanical disaggregation and enzymatic digestion with 2 mg/ml of Collagenase type I in DMEM Ham's/F12 medium (Sigma) supplemented with 50 units/ml penicillin, 50 mg/ml streptomycin, 2% Ultroser G2 (Biosepra), and ITS (5 µg/ml insulin, 5 µg/ml transferrin and 5 ng/ml selenium) and incubated 40 min in the incubator at 37°C, with gentle shaking. Cells were further dispersed by pipetting and filtering them through a sterile 70 µm cell strainer (BD Falcon) and then centrifuged for 5 min at 107 g. The cell pellet was resuspended in the same medium, counted and dispersed in cell culture dishes. Cells were grown in DMEM/F-12 Ham's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine and antibiotics and maintained at 37°C in a humidified atmosphere containing 5% CO2. Dishes were examined daily for growth and the medium was changed every 3–4 days. When reaching confluency (7 days), cells were trypsinized and sub-cultured. For experimentation, cells were used at Passage 3. Cells were plated on 2 cm diameter glass coverslips at a density of 50 000 cells/dish. The next day, cells were infected with adenoviruses encoding the cytoplasmic and plasma membrane-targeted version of AKAR3 (AKAR3-NES and PM-AKAR3, respectively) at a multiplicity of infection of 500 pfu/cell. FRET experiments were performed 24–48 h after infection.
Analysis of RNA by quantitative PCR
Total RNA extracted from HEK293 cells was treated with DNase and further purified with the RNeasy Mini kit and RNase-free DNase Set (Qiagen) according to the manufacturer's instructions. Purified RNA was reverse transcribed with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems), and expression levels of target genes were analyzed by quantitative PCR using a LightCycler Fast Start SYBR Green kit (Roche Diagnostics) according to the manufacturer's instructions. The PCR conditions and primer sequences are indicated in Supplementary Material, Table S1. Relative quantification of target cDNA was determined by calculating the difference in cross-threshold (CT) values after normalization to the housekeeping gene, PPIA [peptidylprolyl isomerase A (cyclophilin A)] (ΔCT method).
Western blot
Cells were lysed in buffer containing (in mm): Tris–HCl 50 (pH 7.4), NaCl 150, EDTA 5, EGTA 1, Triton X-100 1%, protease and phosphatase inhibitor cocktails (Roche Diagnostics, Meylan, France) and centrifuged for 5 min at 800g. Proteins in the supernatant were quantified using a BCA assay (Sigma-Aldrich). Similar amounts of proteins were separated on a 10% SDS–PAGE, then electrotransferred to PVDF membrane, and analyzed by immunoblotting.
FRET-based reporters of cAMP and PKA activity
The cAMP sensor Epac1-camps was used to evaluate cytoplasmic cAMP levels (42). Plasma membrane and mitochondrial outer membrane-targeted versions of Epac1-camps were constructed by fusing the 10 N-terminal amino acids of the kinase Lyn (50) and the targeting motif from DAKAP1a (67) at the N-terminus of Epac1-camps. The A-kinase activity reporter 3 (AKAR3) used for live cell measuring of PKA activity was described previously (41). Differentially targeted versions of AKAR3 were used to monitor PKA activity in specific subcellular compartments: AKAR3-NES for the cytoplasm, PM-AKAR3 for the plasma membrane and DAKAP1-AKAR3 for the outer membrane of mitochondria (41). Adenoviruses encoding AKAR3-NES and PM-AKAR3 were generated using the ViraPower Adenoviral Expression System (Invitrogen) according to the manufacturer's protocol.
FRET imaging
Cells were maintained in a control Ringer solution containing (in mm): NaCl 121.6, KCl 5.4, MgCl2 1.8; CaCl2 1.8; NaHCO3 4, NaH2PO4 0.8, d-glucose 5, sodium pyruvate 5, HEPES 10, adjusted to pH 7.4. Control or drug-containing solutions were applied by placing the cell at the opening of a 250-µm (inner diameter) capillary tube. Images were captured every 5 s using the ×40 objective of a Nikon TE 300 inverted microscope connected to a software-controlled (Metafluor, Molecular Devices) cooled charge coupled (CCD) camera (Sensicam QE, PCO). CFP was excited during 300 ms by a Xenon lamp (Nikon) using a 440/20BP filter and a 455LP dichroic mirror. Dual emission imaging of CFP and YFP was performed using an Optosplit II emission splitter (Cairn Research) equipped with a 495LP dichroic mirror and BP filters 470/30 and 535/30, respectively. All experiments were performed at room temperature (21–25°C).
Confocal microscopy
HEK293 cells were transfected with the mitochondrial sensors DAKAP-AKAR3 and DAKAP-Epac1-camps. After 48 h, cells were incubated for 30 min with 100 nm MitoTracker® Red (Invitrogen) at 37°C and rinsed with the same Ringer solution as used for FRET experiments (see above). Images were acquired with a Leica SP5 confocal microscope using a Plan-Apochromat 63x/1.2 water-immersion objective. YFP was excited at 514 nm and emitted light was collected between 524 and 568 nm. MitoTracker® Red was excited at 578 nm and emitted light was collected between 588 and 800 nm.
cAMP assays
HEK293 cells were transfected with SiS or SiRIα for 48 h and then washed with Ringer solution and stimulated or not with 10 nm PGE1 for 5 min. After stimulation 250 µl ice-cold buffer containing 0.1 m HCl and 0.5% Triton-X 100 was added to the cells and they were incubated for 30 min on ice. The cell lysate was then centrifuged at 845g for 30 min at 4°C. The supernatant was removed and used for cAMP measurement using a direct cAMP antibody-based ELISA kit from New East Biosciences. Total protein concentration was determined by bicinchonic acid assay.
Data analysis
Western blot quantification was performed using quantity one software (Bio-Rad). For FRET measurements, average fluorescence intensity of the entire cell was measured. Background was subtracted and YFP intensity was corrected for CFP spillover before calculating the ratio. Ratio images were obtained with ImageJ software (National Institutes of Health). ImageJ software was also used to merge images of mitochondrial sensors with MitoTracker® Red. Pearson's coefficient was calculated using the ImageJ toolbox JACoP (68). Kinetic parameters (delay, the time between application of the drug and initial increase in ratio and t1/2on, the time required to reach half-maximal effect) were calculated using Microsoft Excel software. Average time course of the ratio represent the mean of all the cells measured in a given experimental condition. The data were normalized to the ratio measured before the stimulus and expressed as percent change of the ratio measured at zero time. Values were expressed as mean ± SEM. Paired Student's t-test was used for statistical evaluation within the same group. When two groups were compared, unpaired Student's t-test was used.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Agence Nationale de la Recherche (ANR-08-GENOPAT-007 to J.B., ANR-2010 BLAN 1139-01 to G.V.), the Fondation de France (FDF2006 005665 to G.V.) and the National Institutes of Health (R01 DK073368 to J.Z., HL107960 to W.R.). L.C. is recipient of a fellowship from the Association pour la Recherche sur le Cancer and B.R. of a fellowship from the Conny-Maeva foundation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Rodolphe Fischmeister and Charlene Depry for critical reading of the manuscript. We thank Drs Ana Maria Gomez and Ana Llach for help with confocal microscopy and Dr Cora Reiβ for providing the plasmid encoding VASP.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Tasken K., Aandahl E.M. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 2.Corbin J.D., Keely S.L., Park C.R. The distribution and dissociation of cyclic adenosine 3′:5′-monophosphate-dependent protein kinases in adipose, cardiac, and other tissues. J. Biol. Chem. 1975;250:218–225. [PubMed] [Google Scholar]
- 3.Reimann E.M., Walsh D.A., Krebs E.G. Purification and properties of rabbit skeletal muscle adenosine 3′,5′-monophosphate-dependent protein kinases. J. Biol. Chem. 1971;246:1986–1995. [PubMed] [Google Scholar]
- 4.Dostmann W.R., Taylor S.S., Genieser H.G., Jastorff B., Doskeland S.O., Ogreid D. Probing the cyclic nucleotide binding sites of cAMP-dependent protein kinases I and II with analogs of adenosine 3′,5′-cyclic phosphorothioates. J. Biol. Chem. 1990;265:10484–10491. [PubMed] [Google Scholar]
- 5.Amieux P.S., McKnight G.S. The essential role of RI alpha in the maintenance of regulated PKA activity. Ann. N. Y. Acad. Sci. 2002;968:75–95. doi: 10.1111/j.1749-6632.2002.tb04328.x. [DOI] [PubMed] [Google Scholar]
- 6.Uhler M.D., McKnight G.S. Expression of cDNAs for two isoforms of the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 1987;262:15202–15207. [PubMed] [Google Scholar]
- 7.Mantovani G., Lania A.G., Bondioni S., Peverelli E., Pedroni C., Ferrero S., Pellegrini C., Vicentini L., Arnaldi G., Bosari S., et al. Different expression of protein kinase A (PKA) regulatory subunits in cortisol-secreting adrenocortical tumors: relationship with cell proliferation. Exp. Cell. Res. 2008;314:123–130. doi: 10.1016/j.yexcr.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Amieux P.S., Howe D.G., Knickerbocker H., Lee D.C., Su T., Laszlo G.S., Idzerda R.L., McKnight G.S. Increased basal cAMP-dependent protein kinase activity inhibits the formation of mesoderm-derived structures in the developing mouse embryo. J. Biol. Chem. 2002;277:27294–27304. doi: 10.1074/jbc.M200302200. [DOI] [PubMed] [Google Scholar]
- 9.Yin Z., Jones G.N., Towns W.H., 2nd, Zhang X., Abel E.D., Binkley P.F., Jarjoura D., Kirschner L.S. Heart-specific ablation of prkar1a causes failure of heart development and myxomagenesis. Circulation. 2008;117:1414–1422. doi: 10.1161/CIRCULATIONAHA.107.759233. [DOI] [PubMed] [Google Scholar]
- 10.Scott J.D., Dessauer C.W., Tasken K. Creating order from chaos: cellular regulation by kinase anchoring. Annu. Rev. Pharmacol. Toxicol. 2013;53:187–210. doi: 10.1146/annurev-pharmtox-011112-140204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coghlan V.M., Perrino B.A., Howard M., Langeberg L.K., Hicks J.B., Gallatin W.M., Scott J.D. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 12.Marx S.O., Kurokawa J., Reiken S., Motoike H., D'Armiento J., Marks A.R., Kass R.S. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 13.Dodge K.L., Khouangsathiene S., Kapiloff M.S., Mouton R., Hill E.V., Houslay M.D., Langeberg L.K., Scott J.D. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 2001;20:1921–1930. doi: 10.1093/emboj/20.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasken K.A., Collas P., Kemmner W.A., Witczak O., Conti M., Tasken K. Phosphodiesterase 4D and protein kinase a type II constitute a signaling unit in the centrosomal area. J. Biol. Chem. 2001;276:21999–22002. doi: 10.1074/jbc.C000911200. [DOI] [PubMed] [Google Scholar]
- 15.Willoughby D., Wong W., Schaack J., Scott J.D., Cooper D.M. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J. 2006;25:2051–2061. doi: 10.1038/sj.emboj.7601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruppelt A., Mosenden R., Gronholm M., Aandahl E.M., Tobin D., Carlson C.R., Abrahamsen H., Herberg F.W., Carpen O., Tasken K. Inhibition of T cell activation by cyclic adenosine 5′-monophosphate requires lipid raft targeting of protein kinase A type I by the A-kinase anchoring protein ezrin. J. Immunol. 2007;179:5159–5168. doi: 10.4049/jimmunol.179.8.5159. [DOI] [PubMed] [Google Scholar]
- 17.Lim C.J., Han J., Yousefi N., Ma Y., Amieux P.S., McKnight G.S., Taylor S.S., Ginsberg M.H. Alpha4 integrins are Type I cAMP-dependent protein kinase-anchoring proteins. Nat. Cell. Biol. 2007;9:415–421. doi: 10.1038/ncb1561. [DOI] [PubMed] [Google Scholar]
- 18.Di Benedetto G., Zoccarato A., Lissandron V., Terrin A., Li X., Houslay M.D., Baillie G.S., Zaccolo M. Protein kinase A type I and type II define distinct intracellular signaling compartments. Circ. Res. 2008;103:836–844. doi: 10.1161/CIRCRESAHA.108.174813. [DOI] [PubMed] [Google Scholar]
- 19.Means C.K., Lygren B., Langeberg L.K., Jain A., Dixon R.E., Vega A.L., Gold M.G., Petrosyan S., Taylor S.S., Murphy A.N., et al. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc. Natl. Acad. Sci. USA. 2011;108:E1227–E1235. doi: 10.1073/pnas.1107182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pidoux G., Witczak O., Jarnaess E., Myrvold L., Urlaub H., Stokka A.J., Kuntziger T., Tasken K. Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J. 2011;30:4371–4386. doi: 10.1038/emboj.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgers P.P., Ma Y., Margarucci L., Mackey M., van der Heyden M.A., Ellisman M., Scholten A., Taylor S.S., Heck A.J. A small novel A-kinase anchoring protein (AKAP) that localizes specifically protein kinase A-regulatory subunit I (PKA-RI) to the plasma membrane. J. Biol. Chem. 2012;287:43789–43797. doi: 10.1074/jbc.M112.395970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Degenhardt B., Tobin D., Yao Z.X., Tasken K., Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol. Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- 23.Carlson C.R., Lygren B., Berge T., Hoshi N., Wong W., Tasken K., Scott J.D. Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J. Biol. Chem. 2006;281:21535–21545. doi: 10.1074/jbc.M603223200. [DOI] [PubMed] [Google Scholar]
- 24.Linglart A., Menguy C., Couvineau A., Auzan C., Gunes Y., Cancel M., Motte E., Pinto G., Chanson P., Bougneres P., et al. Recurrent PRKAR1A mutation in acrodysostosis with hormone resistance. N. Engl. J. Med. 2011;364:2218–2226. doi: 10.1056/NEJMoa1012717. [DOI] [PubMed] [Google Scholar]
- 25.Nagasaki K., Iida T., Sato H., Ogawa Y., Kikuchi T., Saitoh A., Ogata T., Fukami M. PRKAR1A mutation affecting cAMP-mediated G protein-coupled receptor signaling in a patient with acrodysostosis and hormone resistance. J. Clin. Endocrinol. Metab. 2012;97:E1808–E1813. doi: 10.1210/jc.2012-1369. [DOI] [PubMed] [Google Scholar]
- 26.Linglart A., Fryssira H., Hiort O., Holterhus P.M., Perez de Nanclares G., Argente J., Heinrichs C., Kuechler A., Mantovani G., Leheup B., et al. PRKAR1A and PDE4D mutations cause acrodysostosis but two distinct syndromes with or without GPCR-signaling hormone resistance. J. Clin. Endocrinol. Metab. 2012;97:E2328–E2338. doi: 10.1210/jc.2012-2326. [DOI] [PubMed] [Google Scholar]
- 27.Casey M., Vaughan C.J., He J., Hatcher C.J., Winter J.M., Weremowicz S., Montgomery K., Kucherlapati R., Morton C.C., Basson C.T. Mutations in the protein kinase A R1alpha regulatory subunit cause familial cardiac myxomas and Carney complex. J. Clin. Invest. 2000;106:R31–R38. doi: 10.1172/JCI10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirschner L.S., Carney J.A., Pack S.D., Taymans S.E., Giatzakis C., Cho Y.S., Cho-Chung Y.S., Stratakis C.A. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nat. Genet. 2000;26:89–92. doi: 10.1038/79238. [DOI] [PubMed] [Google Scholar]
- 29.Groussin L., Kirschner L.S., Vincent-Dejean C., Perlemoine K., Jullian E., Delemer B., Zacharieva S., Pignatelli D., Carney J.A., Luton J.P., et al. Molecular analysis of the cyclic AMP-dependent protein kinase A (PKA) regulatory subunit 1A (PRKAR1A) gene in patients with Carney complex and primary pigmented nodular adrenocortical disease (PPNAD) reveals novel mutations and clues for pathophysiology: augmented PKA signaling is associated with adrenal tumorigenesis in PPNAD. Am. J. Hum. Genet. 2002;71:1433–1442. doi: 10.1086/344579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertherat J. Carney complex (CNC) Orphanet J. Rare Dis. 2006;1:21. doi: 10.1186/1750-1172-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carney J.A., Gordon H., Carpenter P.C., Shenoy B.V., Go V.L. The complex of myxomas, spotty pigmentation, and endocrine overactivity. Medicine (Baltimore) 1985;64:270–283. doi: 10.1097/00005792-198507000-00007. [DOI] [PubMed] [Google Scholar]
- 32.Horvath A., Bertherat J., Groussin L., Guillaud-Bataille M., Tsang K., Cazabat L., Libe R., Remmers E., Rene-Corail F., Faucz F.R., et al. Mutations and polymorphisms in the gene encoding regulatory subunit type 1-alpha of protein kinase A (PRKAR1A): an update. Hum. Mutat. 2010;31:369–379. doi: 10.1002/humu.21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson-White A., Hundley T.R., Shiferaw M., Bertherat J., Sandrini F., Stratakis C.A. Protein kinase-A activity in PRKAR1A-mutant cells, and regulation of mitogen-activated protein kinases ERK1/2. Hum. Mol. Genet. 2003;12:1475–1484. doi: 10.1093/hmg/ddg160. [DOI] [PubMed] [Google Scholar]
- 34.Nadella K.S., Kirschner L.S. Disruption of protein kinase a regulation causes immortalization and dysregulation of D-type cyclins. Cancer Res. 2005;65:10307–10315. doi: 10.1158/0008-5472.CAN-05-3183. [DOI] [PubMed] [Google Scholar]
- 35.Veugelers M., Wilkes D., Burton K., McDermott D.A., Song Y., Goldstein M.M., La Perle K., Vaughan C.J., O'Hagan A., Bennett K.R., et al. Comparative PRKAR1A genotype–phenotype analyses in humans with Carney complex and prkar1a haploinsufficient mice. Proc. Natl. Acad. Sci. USA. 2004;101:14222–14227. doi: 10.1073/pnas.0405535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burton K.A., McDermott D.A., Wilkes D., Poulsen M.N., Nolan M.A., Goldstein M., Basson C.T., McKnight G.S. Haploinsufficiency at the protein kinase A RI alpha gene locus leads to fertility defects in male mice and men. Mol. Endocrinol. 2006;20:2504–2513. doi: 10.1210/me.2006-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahut-Barnola I., de Joussineau C., Val P., Lambert-Langlais S., Damon C., Lefrancois-Martinez A.M., Pointud J.C., Marceau G., Sapin V., Tissier F., et al. Cushing's syndrome and fetal features resurgence in adrenal cortex-specific Prkar1a knockout mice. PLoS Genet. 2010;6:e1000980. doi: 10.1371/journal.pgen.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beavo J.A., Bechtel P.J., Krebs E.G. Activation of protein kinase by physiological concentrations of cyclic AMP. Proc. Natl. Acad. Sci. USA. 1974;71:3580–3583. doi: 10.1073/pnas.71.9.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbin J.D., Sugden P.H., Lincoln T.M., Keely S.L. Compartmentalization of adenosine 3′:5′-monophosphate and adenosine 3′:5′-monophosphate-dependent protein kinase in heart tissue. J. Biol. Chem. 1977;252:3854–3861. [PubMed] [Google Scholar]
- 40.Saucerman J.J., Zhang J., Martin J.C., Peng L.X., Stenbit A.E., Tsien R.Y., McCulloch A.D. Systems analysis of PKA-mediated phosphorylation gradients in live cardiac myocytes. Proc. Natl. Acad. Sci. USA. 2006;103:12923–12928. doi: 10.1073/pnas.0600137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen M.D., Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem. Biophys. Res. Commun. 2006;348:716–721. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- 42.Nikolaev V.O., Bunemann M., Hein L., Hannawacker A., Lohse M.J. Novel single chain cAMP sensors for receptor-induced signal propagation. J. Biol. Chem. 2004;279:37215–37218. doi: 10.1074/jbc.C400302200. [DOI] [PubMed] [Google Scholar]
- 43.DiPilato L.M., Cheng X., Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc. Natl. Acad. Sci. USA. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponsioen B., Zhao J., Riedl J., Zwartkruis F., van der Krogt G., Zaccolo M., Moolenaar W.H., Bos J.L., Jalink K. Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep. 2004;5:1176–1180. doi: 10.1038/sj.embor.7400290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terrin A., Di Benedetto G., Pertegato V., Cheung Y.F., Baillie G., Lynch M.J., Elvassore N., Prinz A., Herberg F.W., Houslay M.D., et al. PGE(1) stimulation of HEK293 cells generates multiple contiguous domains with different cAMP: role of compartmentalized phosphodiesterases. J. Cell. Biol. 2006;175:441–451. doi: 10.1083/jcb.200605050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willoughby D., Baillie G.S., Lynch M.J., Ciruela A., Houslay M.D., Cooper D.M. Dynamic regulation, desensitization, and cross-talk in discrete subcellular microdomains during beta2-adrenoceptor and prostanoid receptor cAMP signaling. J. Biol. Chem. 2007;282:34235–34249. doi: 10.1074/jbc.M706765200. [DOI] [PubMed] [Google Scholar]
- 47.Jefcoate C. High-flux mitochondrial cholesterol trafficking, a specialized function of the adrenal cortex. J. Clin. Invest. 2002;110:881–890. doi: 10.1172/JCI16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smolenski A., Bachmann C., Reinhard K., Honig-Liedl P., Jarchau T., Hoschuetzky H., Walter U. Analysis and regulation of vasodilator-stimulated phosphoprotein serine 239 phosphorylation in vitro and in intact cells using a phosphospecific monoclonal antibody. J. Biol. Chem. 1998;273:20029–20035. doi: 10.1074/jbc.273.32.20029. [DOI] [PubMed] [Google Scholar]
- 49.Reinhard M., Halbrugge M., Scheer U., Wiegand C., Jockusch B.M., Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 51.Ragazzon B., Cazabat L., Rizk-Rabin M., Assie G., Groussin L., Fierrard H., Perlemoine K., Martinez A., Bertherat J. Inactivation of the Carney complex gene 1 (protein kinase A regulatory subunit 1A) inhibits SMAD3 expression and TGF beta-stimulated apoptosis in adrenocortical cells. Cancer Res. 2009;69:7278–7284. doi: 10.1158/0008-5472.CAN-09-1601. [DOI] [PubMed] [Google Scholar]
- 52.Griffin K.J., Kirschner L.S., Matyakhina L., Stergiopoulos S., Robinson-White A., Lenherr S., Weinberg F.D., Claflin E., Meoli E., Cho-Chung Y.S., et al. Down-regulation of regulatory subunit type 1A of protein kinase A leads to endocrine and other tumors. Cancer Res. 2004;64:8811–8815. doi: 10.1158/0008-5472.CAN-04-3620. [DOI] [PubMed] [Google Scholar]
- 53.Griffin K.J., Kirschner L.S., Matyakhina L., Stergiopoulos S.G., Robinson-White A., Lenherr S.M., Weinberg F.D., Claflin E.S., Batista D., Bourdeau I., et al. A transgenic mouse bearing an antisense construct of regulatory subunit type 1A of protein kinase A develops endocrine and other tumours: comparison with Carney complex and other PRKAR1A induced lesions. J. Med. Genet. 2004;41:923–931. doi: 10.1136/jmg.2004.028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson-White A., Meoli E., Stergiopoulos S., Horvath A., Boikos S., Bossis I., Stratakis C.A. PRKAR1A Mutations and protein kinase A interactions with other signaling pathways in the adrenal cortex. J. Clin. Endocrinol. Metab. 2006;91:2380–2388. doi: 10.1210/jc.2006-0188. [DOI] [PubMed] [Google Scholar]
- 55.Robinson-White A.J., Leitner W.W., Aleem E., Kaldis P., Bossis I., Stratakis C.A. PRKAR1A inactivation leads to increased proliferation and decreased apoptosis in human B lymphocytes. Cancer Res. 2006;66:10603–10612. doi: 10.1158/0008-5472.CAN-06-2200. [DOI] [PubMed] [Google Scholar]
- 56.Rich T.C., Fagan K.A., Tse T.E., Schaack J., Cooper D.M., Karpen J.W. A uniform extracellular stimulus triggers distinct cAMP signals in different compartments of a simple cell. Proc. Natl. Acad. Sci. USA. 2001;98:13049–13054. doi: 10.1073/pnas.221381398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rich T.C., Xin W., Mehats C., Hassell K.A., Piggott L.A., Le X., Karpen J.W., Conti M. Cellular mechanisms underlying prostaglandin-induced transient cAMP signals near the plasma membrane of HEK-293 cells. Am. J. Physiol. Cell. Physiol. 2007;292:C319–C331. doi: 10.1152/ajpcell.00121.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nikolaev V.O., Moshkov A., Lyon A.R., Miragoli M., Novak P., Paur H., Lohse M.J., Korchev Y.E., Harding S.E., Gorelik J. Beta2-adrenergic receptor redistribution in heart failure changes cAMP compartmentation. Science. 2010;327:1653–1657. doi: 10.1126/science.1185988. [DOI] [PubMed] [Google Scholar]
- 59.Tsang K.M., Starost M.F., Nesterova M., Boikos S.A., Watkins T., Almeida M.Q., Harran M., Li A., Collins M.T., Cheadle C., et al. Alternate protein kinase A activity identifies a unique population of stromal cells in adult bone. Proc. Natl. Acad. Sci. USA. 2010;107:8683–8688. doi: 10.1073/pnas.1003680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almeida M.Q., Tsang K.M., Cheadle C., Watkins T., Grivel J.C., Nesterova M., Goldbach-Mansky R., Stratakis C.A. Protein kinase A regulates caspase-1 via Ets-1 in bone stromal cell-derived lesions: a link between cyclic AMP and pro-inflammatory pathways in osteoblast progenitors. Hum. Mol. Genet. 2011;20:165–175. doi: 10.1093/hmg/ddq455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blackman B.E., Horner K., Heidmann J., Wang D., Richter W., Rich T.C., Conti M. PDE4D and PDE4B function in distinct subcellular compartments in mouse embryonic fibroblasts. J. Biol. Chem. 2011;286:12590–12601. doi: 10.1074/jbc.M110.203604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie M., Rich T.C., Scheitrum C., Conti M., Richter W. Inactivation of multidrug resistance proteins disrupts both cellular extrusion and intracellular degradation of cAMP. Mol. Pharmacol. 2011;80:281–293. doi: 10.1124/mol.111.071134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gangal M., Clifford T., Deich J., Cheng X., Taylor S.S., Johnson D.A. Mobilization of the A-kinase N-myristate through an isoform-specific intermolecular switch. Proc. Natl. Acad. Sci. USA. 1999;96:12394–12399. doi: 10.1073/pnas.96.22.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaffarogullari E.C., Masterson L.R., Metcalfe E.E., Traaseth N.J., Balatri E., Musa M.M., Mullen D., Distefano M.D., Veglia G. A myristoyl/phosphoserine switch controls cAMP-dependent protein kinase association to membranes. J. Mol. Biol. 2011;411:823–836. doi: 10.1016/j.jmb.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Groussin L., Jullian E., Perlemoine K., Louvel A., Leheup B., Luton J.P., Bertagna X., Bertherat J. Mutations of the PRKAR1A gene in Cushing's syndrome due to sporadic primary pigmented nodular adrenocortical disease. J. Clin. Endocrinol. Metab. 2002;87:4324–4329. doi: 10.1210/jc.2002-020592. [DOI] [PubMed] [Google Scholar]
- 66.Groussin L., Horvath A., Jullian E., Boikos S., Rene-Corail F., Lefebvre H., Cephise-Velayoudom F.L., Vantyghem M.C., Chanson P., Conte-Devolx B., et al. A PRKAR1A mutation associated with primary pigmented nodular adrenocortical disease in 12 kindreds. J. Clin. Endocrinol. Metab. 2006;91:1943–1949. doi: 10.1210/jc.2005-2708. [DOI] [PubMed] [Google Scholar]
- 67.Ma Y., Taylor S.S. A molecular switch for targeting between endoplasmic reticulum (ER) and mitochondria: conversion of a mitochondria-targeting element into an ER-targeting signal in DAKAP1. J. Biol. Chem. 2008;283:11743–11751. doi: 10.1074/jbc.M710494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolte S., Cordelieres F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.