Abstract
Spinobulbar muscular atrophy (SBMA) is an inherited neuromuscular disorder caused by the expansion of a CAG repeat encoding a polyglutamine tract in exon 1 of the androgen receptor (AR) gene. SBMA demonstrates androgen-dependent toxicity due to unfolding and aggregation of the mutant protein. There are currently no disease-modifying therapies, but of increasing interest for therapeutic targeting is autophagy, a highly conserved cellular process mediating protein quality control. We have previously shown that genetic manipulations inhibiting autophagy diminish skeletal muscle atrophy and extend the lifespan of AR113Q knock-in mice. In contrast, manipulations inducing autophagy worsen muscle atrophy, suggesting that chronic, aberrant upregulation of autophagy contributes to pathogenesis. Since the degree to which autophagy is altered in SBMA and the mechanisms responsible for such alterations are incompletely defined, we sought to delineate autophagic status in SBMA using both cellular and mouse models. Here, we confirm that autophagy is induced in cellular and knock-in mouse models of SBMA and show that the transcription factors transcription factor EB (TFEB) and ZKSCAN3 operate in opposing roles to underlie these changes. We demonstrate upregulation of TFEB target genes in skeletal muscle from AR113Q male mice and SBMA patients. Furthermore, we observe a greater response in AR113Q mice to physiological stimulation of autophagy by both nutrient starvation and exercise. Taken together, our results indicate that transcriptional signaling contributes to autophagic dysregulation and provides a mechanistic framework for the pathologic increase of autophagic responsiveness in SBMA.
INTRODUCTION
Spinobulbar muscular atrophy (SBMA), one of the nine neurodegenerative diseases caused by CAG/glutamine tract expansions (1), is an X-linked, progressive disorder of adult onset primarily affecting lower motor neurons and skeletal muscle. The polyglutamine (polyQ) expansion critical for SBMA pathogenesis occurs in exon 1 of the androgen receptor (AR) gene. The mutant protein acquires a toxic gain of function by undergoing aberrant unfolding, oligomerization and metabolism dependent on binding the cognate ligands, such as testosterone and dihydrotestosterone (2,3). These steps are critical for mediating toxicity in target tissues, and the dependence of disease development on androgen underlies the exclusive incidence of SBMA in men (4). Additionally, the polyQ AR exhibits a partial loss of transactivation function (5,6), correlating with features of androgen insensitivity including gynecomastia and reduced fertility in SBMA patients. Despite progress over the past two decades in characterizing key aspects of neuromuscular toxicity and endocrine disruption in SBMA, mechanisms that are essential to pathogenesis remain incompletely understood and available therapies are of limited utility.
As the unfolded mutant protein is the proximal mediator of toxicity in SBMA, pathways that regulate cellular proteostasis have attracted considerable attention. Among these is macroautophagy (hereafter referred to as autophagy), a highly conserved catabolic process in which misfolded or dysfunctional proteins and organelles in the cytoplasm are enveloped in double-membraned structures known as autophagosomes (7–10). These autophagosomes are trafficked to and fuse with lysosomes, enabling degradation of their intraluminal contents. Regulation of autophagy is achieved through several processes, including the transcription factor EB (TFEB), which directs the expression of hundreds of autophagy- and lysosomal-related genes as part of the Coordinated Lysosomal Expression and Regulation (CLEAR) network (11–13). TFEB is a basic helix-loop-helix, leucine-zipper member of the MITF/TFE family and functions as a master regulator of autophagy and lysosomal biogenesis. Treatments that induce autophagy, including nutrient deprivation and inhibition of mammalian target of rapamycin (mTOR), also promote TFEB activity (11,12,14–16). Recent data indicate that this activity is antagonized by the transcription factor ZKSCAN3, a zinc-finger protein with Krüppel-associated box (KRAB) and SRE-ZBP/CTfin-51/AQ-1/Number 18 cDNA-homology (SCAN) domains that has been implicated in regulating TFEB target genes as a master repressor of autophagy (17). Despite evidence suggesting that changes in protein quality control occur in SBMA (18–21), the degree to which these regulatory transcription factors are affected in disease is unknown.
While the nuclear-localized polyQ AR is not an autophagic substrate, autophagy is able to degrade the mutant protein upon its sequestration within the cytoplasm (22). Furthermore, expression of the mutant AR itself is sufficient to induce autophagy in SBMA cell and animal models, although the mechanism underlying this observation has not been explored (18,19). Consistent with these findings, we have previously demonstrated that genetic manipulations that modulate autophagy have a significant influence on the phenotype of AR113Q knock-in mice. In these mice, gene targeting was used to exchange much of mouse Ar exon 1 with human sequence while inserting a glutamine tract encoded by 113 CAG repeats; the same strategy was used to generate control AR21Q knock-in mice (23–25). While AR21Q males are similar to wild-type littermates, AR113Q males exhibit hormone-dependent weight loss, deficits in muscle strength and early death (24,26). AR113Q males deficient for the unfolded protein response effector C/EBP-homologous protein show enhanced autophagy and exacerbated skeletal muscle atrophy (27). Conversely, genetic inhibition of autophagy through haploinsufficiency for Beclin-1, a component of the autophagic initiation complex, reduces levels of autophagy in AR113Q mice, mitigates skeletal muscle atrophy and prolongs survival (27). These data indicate that excessive, chronic activation of autophagy is detrimental to SBMA mice.
Here, we further explore the notion that autophagy is aberrantly activated in SBMA, and that autophagic dysregulation underlies deleterious contributions to disease pathogenesis. We test this hypothesis by analyzing aspects of autophagic induction, flux and regulation in both cellular and animal models of SBMA. We confirm that autophagy is induced in the context of the polyQ-expanded AR both in vitro and in vivo, implicate TFEB in mediating increased expression of autophagy-related genes in SBMA mice and patient tissue as well as establish that nutrient deprivation and exercise act as physiological stimuli to potently induce autophagy in SBMA mice. These results demonstrate the mechanism underlying aberrant upregulation of autophagy in SBMA and provide discrete targets in the autophagic pathway for further investigation and therapeutic design.
RESULTS
The polyQ AR promotes autophagy
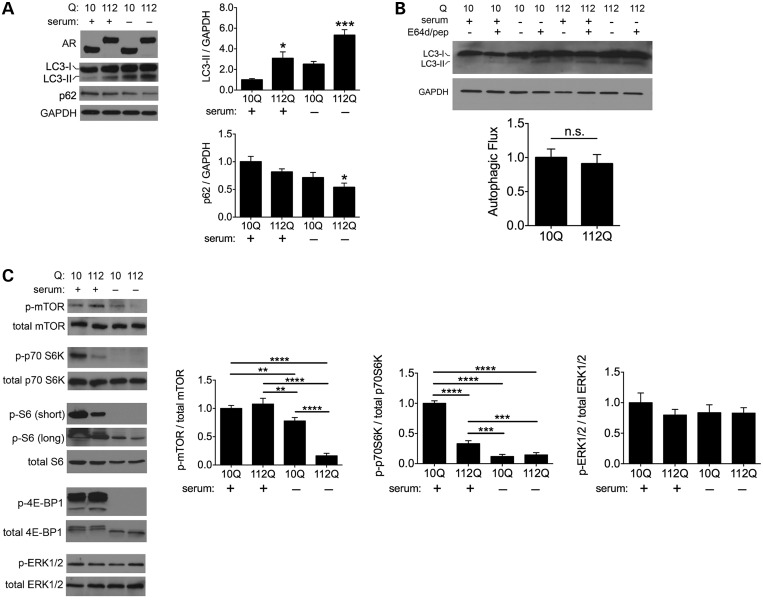
To investigate autophagy in SBMA, we initially studied PC12 cells that stably express tetracycline-inducible full-length human AR with 10 or 112 glutamines (28). We observed higher basal levels of the autophagosome marker LC3-II in cells expressing AR112Q (Fig. 1A). Serum starvation promoted robust conversion of LC3-I to LC3-II in both cell lines, but this process was significantly more pronounced in AR112Q cells (Fig. 1A). To determine whether these elevated LC3-II levels resulted from enhanced activation of autophagy or impaired flux, we examined levels of the autophagic substrate p62 and assessed levels of LC3-II with and without lysosomal inhibition. Both of these are standard assays for evaluating autophagic flux (29). AR112Q cells demonstrated significant clearance of p62 in response to serum starvation (Fig. 1A). Furthermore, treatment with the lysosomal protease inhibitors E64d and pepstatin A led to similarly enhanced accumulation of LC3-II in both AR10Q and AR112Q cells (Fig. 1B). These results indicate that autophagic markers accumulate to a greater extent with AR112Q expression primarily due to upregulation of autophagy rather than compromised autophagic flux.
Figure 1.
PolyQ AR promotes autophagy. (A) AR112Q cells demonstrate higher levels of autophagy. Tetracycline-regulated PC12 cells were induced to express AR10Q or AR112Q in the presence of 10 nM R1881 for 48 h. For the final 4 h, cells either were supplemented with serum or were deprived to induce autophagy. Lysates were analyzed for AR, LC3 and p62. Quantification of three independent experiments shown at right. Data are mean ± SEM. *P < 0.05, ***P < 0.001. (B) AR polyQ length does not affect autophagic flux. AR10Q and AR112Q cells were treated as in (A), with the final 4 h consisting of treatment with serum or serum starvation with or without E64d (10 µg/ml) and pepstatin A (10 µg/ml). Representative western blot is shown at top. Autophagic flux from three independent experiments was determined by quantifying normalized LC3-II band intensities and subtracting the difference of fed E64d/pepstatin A-treated and untreated cells from that of starved E64d/pepstatin A-treated and untreated cells (bottom). Data are mean ± SEM. n.s., not significant. (C) Expanded polyQ tract negatively correlates with mTOR pathway activity. Cells were treated as in (A) and lysates were analyzed for levels of phospho- and total mTOR, phosho- and total p70 S6 K, phospho- and total S6, phospho- and total 4E-BP1 as well as phosoph- and total ERK1/2. Quantifications of three independent experiments are shown at right. Data are mean ± SEM. **P < 0.01, ***P < 0.001 and ****P < 0.0001.
We then sought to define which signaling mechanisms were responsible for the increased activation of autophagy in SBMA. To do this, we first probed the mTOR pathway since this serine/threonine kinase serves as a principal regulator of autophagy (30–32). We found that phosphorylation of mTOR was decreased in both cell lines following serum starvation, but this reduction was significantly greater in AR112Q cells (Fig. 1C). Similarly, phosphorylation of p70 S6 kinase, a downstream effector of the mTOR pathway and other kinases, was lower in AR112Q cells at baseline (Fig. 1C). Serum starvation eliminated detectable phosphorylation of p70 S6 kinase and decreased phosphorylation of its substrate, the ribosomal protein S6, particularly in AR112Q cells (Fig. 1C). Similarly, phosphorylation of the mTOR target 4E-BP1 was diminished in both cell lines following serum starvation (Fig. 1C). Lower mTOR activity and phosphorylation are consistent with disinhibition of autophagy, since mTOR negatively regulates autophagic induction (30–32). In contrast, phosphorylation of extracellular-regulated kinases ERK1/2, which comprise another set of autophagy regulators that can function independently of mTOR (33–37), was unchanged regardless of serum supplementation (Fig. 1C), indicating that autophagy was selectively upregulated by serum deprivation due to inhibition of the mTOR pathway, and that this effect was accentuated in AR112Q cells.
Transcription factors that regulate autophagy partition in a glutamine length-dependent manner
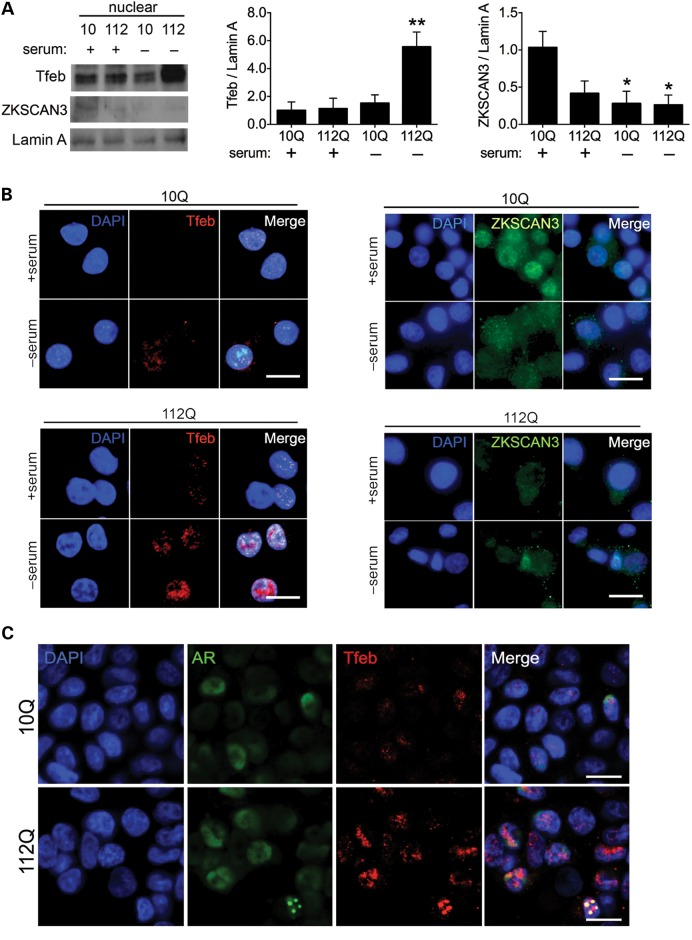
Recent studies have shown that TFEB controls the expression of autophagy-related genes, and that its activity and nuclear localization are negatively regulated by mTOR (14–16). Additionally, the zinc-finger protein, ZKSCAN3, has been characterized as a transcriptional antagonist to autophagy, functioning oppositely and in concert with TFEB (17). Given our observations of increased autophagy and enhanced responsiveness of mTOR activity in the context of AR112Q, we postulated that TFEB and ZKSCAN3 act as major determinants of autophagic upregulation in SBMA. To test this hypothesis, we prepared nuclear lysates of PC12 cells subjected to serum starvation and determined the relative localization of each transcription factor. Consistent with our findings of increased autophagic induction and reduced mTOR activity in AR112Q cells, we found that the transcriptional activator TFEB displayed significantly greater nuclear localization upon serum starvation, whereas the repressor ZKSCAN3 exhibited diminished nuclear localization even under fed conditions (Fig. 2A). Similarly, immunofluorescence demonstrated nuclear localization of TFEB in AR10Q cells with concomitant cytoplasmic translocation of ZKSCAN3 only after starvation. In contrast, nuclear TFEB and cytoplasmic ZKSCAN3 were detected in AR112Q cells even when serum supplemented (Fig. 2B). Following serum deprivation, we observed TFEB sequestration into AR112Q nuclear aggregates that occurred in a subset of cells (Fig. 2C). Taken together, our results suggest that autophagy is primed for activation with depressed mTOR activity in the presence of AR112Q, and that enhanced autophagic induction in SBMA corresponds to relocalization of the transcriptional regulators TFEB and ZKSCAN3.
Figure 2.
Transcription factors that regulate autophagy display glutamine length-dependent changes in intracellular localization. (A) AR112Q cells demonstrate increased TFEB nuclear localization and decreased ZKSCAN3 nuclear localization. Nuclear lysates were prepared from AR10Q and AR112Q cells after induction of AR expression for 48 h in the presence of 10 nM R1881, the final 4 h of which consisted of serum supplementation or deprivation. Lysates were analyzed for TFEB and ZKSCAN3, and quantifications from three independent experiments are shown at right. Data are mean ± SEM. *P < 0.05 and **P < 0.01. (B) AR10Q and AR112Q cells were treated as in (A) and stained with antibodies against TFEB (left panels) and ZKSCAN3 (right panels). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 2 µm. (C). AR10Q and AR112Q cells were induced to express AR for 48 h in the presence of 10 nM R1881, the final 4 h of which consisted of serum deprivation. Cells were stained for AR (green) and Tfeb (red). Nuclei were stained with DAPI. Scale bar, 2 µm.
polyQ AR increases autophagy in vivo
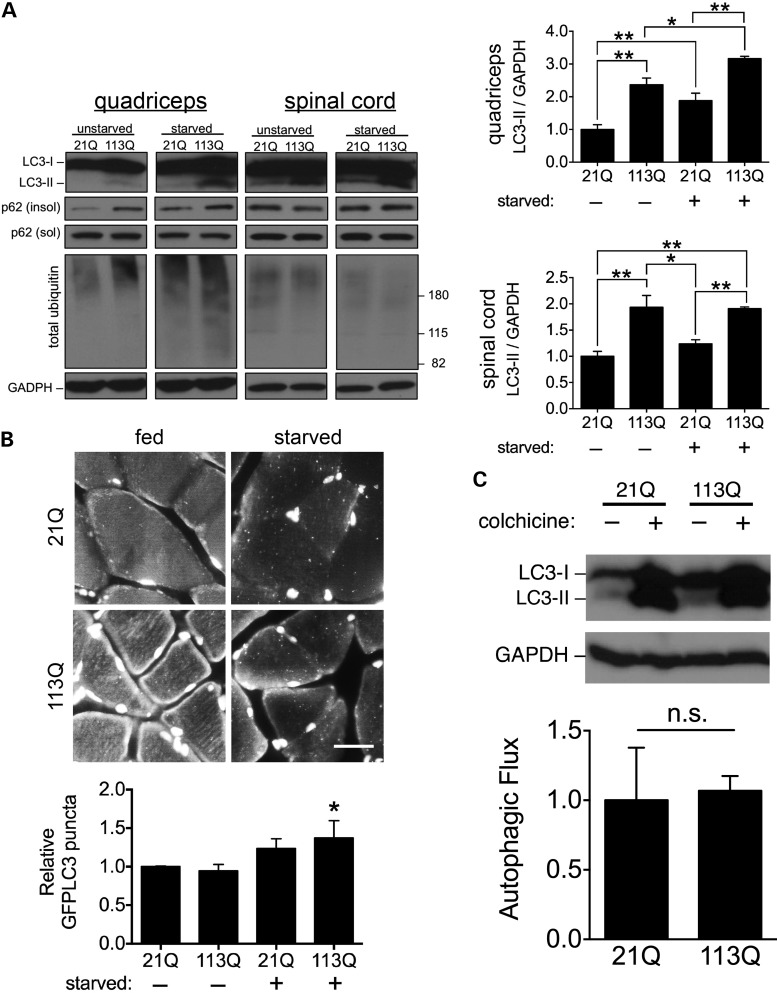
To corroborate the autophagic changes we observed in PC12 cells, we probed markers of autophagy in AR21Q and AR113Q knock-in male mice. Consistent with our previous observations, we found significantly higher LC3-I to LC3-II conversion in skeletal muscle and spinal cord in AR113Q males, and observed that this difference became more pronounced in muscle when autophagy was induced by starvation (Fig. 3A). Additionally, total ubiquitinated proteins and p62 from the insoluble fraction were also increased in AR113Q muscle (Fig. 3A). Autophagic puncta accumulated in skeletal muscle after starvation in AR21Q and AR113Q males, which we detected by co-expressing a GFP–LC3 transgene (38) in both knock-in lines. This accumulation of autophagic puncta occurred to a greater extent in starved AR113Q mice, in line with our biochemical data (Fig. 1B). To confirm that autophagic flux was intact in both knock-in lines, we inhibited autophagosome to lysosome fusion in vivo by administering colchicine (39). Starved, colchicine-treated mice of both genotypes showed similarly enhanced accumulation of LC3-II (Fig. 1C). We conclude that autophagic induction is increased in AR113Q knock-in males without accompanying loss of autophagic flux.
Figure 3.
Expression of the polyQ AR increases autophagy in vivo. (A) Autophagic markers are significantly increased in a glutamine length-dependent manner. Tissue lysates of quadriceps muscle and lumbar spinal cord were prepared from AR21Q and AR113Q knock-in male mice before and after starvation. Lysates were analyzed for LC3, p62 and total ubiquitin. Quantification (n = 3 mice per group) for relative LC3-II is shown at right. Data are mean ± SEM. *P < 0.05 and **P < 0.01. (B) GFP-positive autophagic puncta accumulate after starvation. AR21Q and AR113Q mice expressing GFP–LC3 were fed or starved for 48 h, quadriceps muscle harvested and sections stained for GFP. Nuclei were stained with DAPI. Scale bar, 10 µm. Quantification from three independent experiments are shown at bottom. Data are mean ± SEM. *P < 0.05 by ANOVA, compared with unstarved AR21Q mice. (C) Autophagic flux is intact in AR113Q mice. AR21Q and AR113Q knock-in mice were starved for 48 h and received once daily IP injections of either normal saline or 0.4 mg/kg colchicine. Lysates of quadriceps muscle were collected and analyzed for LC3. Quantification of autophagic flux (mean ± SEM) from three independent cohorts of mice (bottom panel) was determined by subtracting the level of LC3-II in colchicine-treated mice from that of LC3-II in saline-injected mice. Data are mean ± SEM. n.s., not significant.
AR113Q promotes TFEB activity in vivo
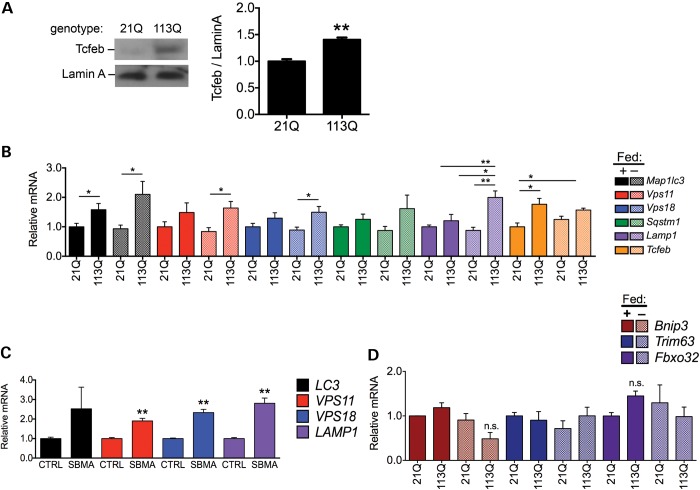
To test whether TFEB activity is increased in AR113Q mice, we prepared nuclear lysates of skeletal muscle from each knock-in line and assessed TFEB localization. We found that, as occurs in tetracycline-regulated PC12 cells, the mouse ortholog TCFEB displays greater nuclear localization in AR113Q compared with AR21Q male mice (Fig. 4A). To determine whether enhanced nuclear localization correlated with upregulation of autophagy-related genes, we assayed the expression profile of a select set of previously identified TFEB target genes (11–13). Real-time quantitative reverse transcription PCR (RT-PCR) confirmed that autophagy-related genes containing TFEB-binding sites in their promoter regions demonstrated increased expression levels in AR113Q mice, and that this difference was more pronounced for many genes after starvation (Fig. 4B). Similarly, we found that several TFEB target genes were also expressed at higher levels in skeletal muscle biopsies from SBMA patients than controls (Fig. 4C). These glutamine length- and starvation-dependent transcriptional changes in AR113Q mice were not observed in target genes of FOXO3a (Fig. 4D), another important regulator of autophagy in skeletal muscle (40,41). Taken together, these data indicate that SBMA results in increased activity specific to TFEB and, consequently, enhanced expression of autophagy-related genes.
Figure 4.
AR113Q promotes TFEB activity in vivo. (A) TFEB localizes to the nucleus to a greater extent in AR113Q mice. Nuclear lysates from quadriceps muscle were prepared from AR21Q and AR113Q knock-in mice and analyzed for TCFEB, the murine ortholog of TFEB. Relative TCFEB quantified at right from three independent experiments. Data are mean ± SEM. **P < 0.01. (B) TFEB target genes are upregulated in AR113Q mice. Total RNA was extracted from quadriceps muscle of fed and starved AR21Q and AR113Q knock-in mice (n = 6 per group), and transcripts of genes regulated by TCFEB were analyzed by quantitative RT-PCR. Data are mean ± SEM. *P < 0.05 and **P < 0.01. (C) TFEB targets are upregulated in SBMA patients. Transcripts of TFEB-regulated genes were analyzed by quantitative RT-PCR in muscle samples from SBMA and control patients (n = 3 patients per group). Color coding is same as in (B). Data are mean ± SEM. **P < 0.01. (D) FOXO3a targets are not upregulated in AR113Q mice. Transripts of FOXO3a target genes were analyzed by quantitative RT-PCR in the quadriceps muscle samples described in (B). Data are mean ± SEM. n.s., not significant.
Expanded polyQ AR enhances autophagic response to exercise
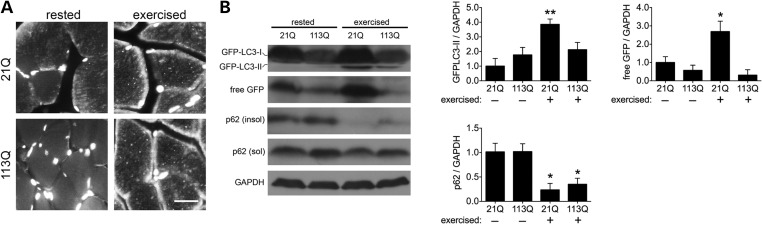
Finally, we sought to explore whether enhanced TFEB activity and increased expression of lysosomal and autophagy-related genes lead to an augemented autophagic response following an acute, physiologically relevant stimulus. Recently, He et al. established exercise as a potent inducer of autophagy in vivo (42). To corroborate the effect of exercise in AR knock-in mice, we ran AR21Q/GFP–LC3 and AR113Q/GFP–LC3 male mice on a treadmill and evaluated the formation of GFP-positive autophagic puncta in quadriceps muscle. After 80 min of exercise, both knock-in lines demonstrated accumulation of autophagic puncta akin to induction by starvation (Fig. 5A). We then asked if each AR line responded equivalently to autophagic induction by exercise. To accomplish this, we followed markers of autophagy biochemically and found that exercise stimulated a higher degree of GFP–LC3-II accumulation in AR21Q muscle (Fig. 5B). We hypothesized that this difference reflected enhanced clearance of autophagic substrates in AR113Q males after acute stimulation of autophagy. We tested this notion by evaluating levels of p62 and free GFP as indicators of autophagic flux. Free GFP is produced after GFP–LC3-II is delivered to lysosomes, and the LC3-II portion of the fusion protein is degraded (43–45). Western analysis of these autophagic markers after exercise revealed similar degradation of endogenous p62 in both knock-in lines, but a striking absence of free GFP in AR113Q mice; in contrast, free GFP robustly accumulated in skeletal muscle of exercised AR21Q mice (Fig. 5B). Our analysis suggests rapid clearance of the GFP–LC3 fusion protein in AR113Q males. We conclude that exercise promotes autophagy in both knock-in lines. The enhanced clearance of GFP–LC3 supports greater autophagic flux in skeletal muscle of AR113Q males, indicative of enhanced responsiveness to physiologic stimuli in these mice.
Figure 5.
Expanded polyQ AR enhances autophagic response to exercise. (A) Exercise induces autophagy in AR knock-in mice. AR21Q and AR113Q mice expressing GFP–LC3 were rested or subjected to 80 min of exercise. Quadriceps muscles were collected and stained for GFP. Nuclei were stained with DAPI. Scale bar, 10 µm. (B) AR113Q accentuates degradation of autophagic substrates in response to exercise. Mice were exercised for 80 min, and lysates of quadriceps muscle were collected and analyzed for GFP–LC3, free GFP and p62. Quantification from three independent experiments are shown at right. Data are mean ± SEM. *P < 0.05 and **P < 0.01.
DISCUSSION
Autophagy is a conserved catabolic process and an essential regulator of cellular proteostasis. Although enhancement of autophagy has attracted considerable attention as a means to remove cytoplamsic protein aggregates responsible for neurodegenerative diseases and thereby provide therapeutic benefit (46), the manner in which manipulation of the autophagic pathway affects SBMA is controversial. Previous work establishes that the principal site of toxic action for the expanded polyQ AR is the nucleus, and nuclear localization of the mutant protein is both essential for pathogenesis and prevents its efficient degradation by autophagy (22). Rather, clearance of nuclear-localized polyQ AR is primarily regulated by chaperone-dependent ubiquitination and proteasomal degradation (47,48). However, alternative evidence demonstrates a role for autophagic degradation of the polyQ AR in cell culture models that exogenously express the mutant protein (18,49). Interestingly, enhanced expression of the LC3- and ubiquitin-binding protein p62 ameliorates the disease phenotype of SBMA transgenic mice, in part by sequestering the polyQ AR into intranuclear aggregates (49). While these latter studies suggest that autophagy may contribute to clearance of the polyQ AR in some model systems, these beneficial effects in vivo may be relatively small compared with detrimental consequenes of chronically enhanced autophagy in tissues, such as skeletal muscle, that are affected by SBMA.
Supporting the notion that chronically enhanced autophagy contributes to the pathogenesis of SBMA, we previously demonstrated that levels of autophagy directly correlate with skeletal muscle atrophy in AR113Q knock-in mice (27). Here, we elaborate on these findings and demonstrate that the induction of autophagy by the polyQ AR is associated with a concomitant attenuation of the mTOR pathway. We show that this is associated with activation of TFEB, a master regulator of autophagy and lysosomal biogenesis. Skeletal muscle from AR113Q mice and SBMA patients demonstrate increased expression of TFEB target genes. Corresponding changes in autophagy are further revealed in enhanced responsiveness to physiological stimuli. AR113Q mice show an augmented response not only to the mTOR-regulated stimulus of chronic starvation, leading to a saturating rise in autophagic markers, but also to the BCL-2-regulated stimulus of acute exercise, leading to marked autophagic degradation of protein substrates (42,50). Our studies characterize a striking instance of neurodegenerative disease in which regulated control of autophagy is compromised, and we postulate that these changes effect deleterious contributions to pathogenesis including muscle atrophy.
Mechanisms of proteostasis are essential mediators of muscle integrity, and balancing the degradative and protective roles of protein quality control in muscle is pivotal for muscle maintenance and appropriately responding to nutrient deprivation, mechanical stress and disuse atrophy. The ubiqitin-proteasome system (UPS) serves a critical role in muscle protein catabolism, and enhanced or aberrant UPS activity is responsible for physiologic and pathologic forms of atrophy (51). However, it is unlikely that the UPS acts as the primary mediator of muscle wasting in SBMA, since the muscle-specific ubiquitin E3 ligases atrogin-1/MAFbx and MuRF1 that mediate muscle atrophy are not upregulated in AR113Q or AR97Q mice (27,52–54). In contrast, several groups have demonstrated that dysregulated autophagy is sufficient to promote atrophy in muscle. Loss of Runx1 models disuse and deneveration changes in skeletal muscle and causes atrophy through disinhibited autophagy (55). Similarly, denervation- and fasting-induced atrophy have been shown to occur through FOXO3-mediated induction of autophagy (40,41). Furthermore, increased autophagy is instrumental in mediating or enhancing myopathy in neuromuscular disorders (56–58). Our findings here are consistent with genetic and pharmacologic manipulations of isulin-like growth factor 1 that mitigate SBMA in a muscle-specific manner, since these ameliorative effects are dependent on enhanced activation of the autophagic repressor Akt (54,59,60). It will be important for future studies to weigh the therapeutic potential of chronically inducing autophagy to address proteinopathies against our observation of detrimentally elevated autophagy in SBMA.
We have delineated specific alterations in the autophagic pathway occuring in SBMA and particular effectors associated with these changes. It is likely that these alterations constitute a compensatory mechanism in response to pathogenic insults introduced by the polyQ AR. Although elucidating the involvement of TFEB and its regulatory pathways will be valuable for therapeutic targeting, the mechanism directly linking the mutant AR to changes in mTOR and TFEB remains to be defined. In addition, our identification of an enhanced autophagic response to exercise has important implications for determining the usefulness of exercise therapy in SBMA, not only in current (trial number 11-N-0171; NCT01369901) and future clinical trials of exercise, but also in accounting for the equivocal benefits of exercise seen thus far in patients (61,62). We note that that exercise has broad effects beyond activation of autophagy. Additionally, benefits of short-term induction of autophagy have been observed in motor neuron cell models of SBMA (22), raising the possibility that responses are dependent on cell type or induction duration. As such, it is possible that skeletal muscle and motor neurons exhibit differential responses to autophagic stimuli in vivo, and that beneficial effects of chronically increased autophagy could predominate in cell types other than skeletal muscle. Overall, our results broaden understanding of the contribution of autophagy to pathogenesis in SBMA, and these insights may further refine therapeutic strategies to confront the burden of neuromuscular disease.
MATERIALS AND METHODS
Materials
Lysosomal protease inhibitors E64d and pepstatin A, normal saline tablets and colchicine were from Sigma. For the preparation of nuclear lysates from mouse tissue, the following buffers were used: NB1 (10 mm Tris pH 8.0, 10 mm NaCl, 3 mm MgCl2, 0.5 mm dithiothreitol (DTT), 0.1% Triton X-100 and 0.1 M sucrose), NB2 (10 mm Tris pH 8.0, 10 mm NaCl, 3 mm MgCl2, 0.5 mm DTT, 0.1% Triton X-100 and 0.25 M sucrose), NB3 (10 mm Tris pH 8.0, 5 mm MgCl2, 0.5 mm DTT and 0.33 M sucrose), EB (25 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.8, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5 mm DTT and 25% glycerol) and Z (25 mm HEPES pH 7.8, 12.5 mm MgCl2, 1 mm DTT, 0.1 m KCl, 0.1% NP40 and 20% glycerol). Antibodies against indicated proteins and their respective vendors were as follows: AR (N-20) was from Santa Cruz; GAPDH (6C5) was from Abcam; GFP (A11122) was from Invitrogen; LC3 (NB600-1384) and p62 (2C11) were from Novus Biologicals; Lamin A (MAB3540), phospho-mTOR (09-213), phospho-ERK1/2 (12D4), total ERK1/2 (06-182) and total p70 S6K (07-402) were from Millipore; phospho-p70 S6K (9204), total mTOR (2972), S6 (2217), phospho-S6 (Ser235/236) (2211), 4E-BP1 (53H11) and phospho-4E-BP1 (Thr37/46) (2855) were from Cell Signaling; Tfeb (MBS120432) was from MyBioSource; ZKSCAN3 (AV33609) was from Sigma. Horseradish peroxidase-tagged secondary antibodies were from Bio-Rad. Alexa Fluor 488- and 594-conjugated secondary antibodies were from Invitrogen.
Cell culture
PC12 cells stably transfected with tetracycline-inducible forms of AR were described previously (28). AR expression was induced with 500 ng/ml of doxycycline (Clontech) in the presence of 10 nM R1881. Cells were maintained in phenol red-free Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 15% charcoal-stripped horse serum (Invitrogen), 10% charcoal-stripped fetal bovine serum (Thermo Scientific), 100 units/ml of penicillin/streptomycin (Invitrogen), 200 µg/ml of hygromycin B (Invitrogen) and 100 µg/ml of G418 (Invitrogen) in 15% CO2 at 37°C. Serum starvation was performed in Hank's Balanced Salt Solution (HBSS, Invitrogen) supplemented with doxycycline and R1881 for 4 h. To analyze autophagic flux, PC12 cells were induced to express AR for 44 h then fed or starved in HBSS with E64d/pepstatin A (10 µg/ml each) or DMSO vehicle for 4 h. Autophagic flux was then determined by subtracting the difference of LC3-II levels in serum-starved cells with and without E64d/pepstatin A treatment from that of LC3-II levels in serum-supplemented cells with and without E64d/pepstatin A.
Mouse studies
Generation of knock-in mice with targeted AR allele containing 21 or 113 CAG repeats has been previously described (23,24). GFP–LC3 transgenic mice were kindly provided by Jun-Lin Guan (University of Michigan) and have been described previously (38). Mice were group housed-in an SPF facility and provided with food and water ad libitum. Genotypes were confirmed by the polymerase chain reaction (PCR) of tail biopsy-derived DNA. AR knock-in mice were genotyped with forward primer 5′-CCAGAATCTGTTCCAGAGCGTG-3′ and reverse primers 5′-TGTTCCCCTGGACTCAGATG-3′ and 5′-GCACTCCAGGGCCGACTGCG-3′ in a 2 : 2 : 1 ratio, respectively. GFP–LC3 mice were genotyped with forward primer 5′-TCCTGCTGGAGTTCGTGACCG-3′ and reverse primer 5′-TTGCGAATTCTCAGCCGTCTTCATCTCTCTCGC-3′ to detect the GFP–LC3 transgene and in a separate reaction with forward primer 5′-TGAGCGAGCTCATCAAGATAATCAGGT-3′ and reverse primer 5′-GTTAGCATTGAGCTGCAAGCGCCGTCT-3′ to detect the endogenous LC3 allele.
Skeletal muscle and spinal cord were collected from adult male knock-in mice 13–15 weeks old and flash frozen for biochemical analysis or mounted in OCT for cryosectioning. For morphologic analysis of autophagy in skeletal muscle, AR knock-in mice were crossed with GFP–LC3 mice to generate bigenic 21Q/GFP–LC3 and 113Q/GFP–LC3 mice. To induce autophagy, mice were starved for 48 h and provided with water ad libitum. To analyze autophagic flux, mice were starved for 48 h and treated with intraperitoneal injections of normal saline or colchicine (0.4 mg/kg/day). Autophagic flux was then determined by subtracting the level of LC3-II in saline-treated mice from that of LC3-II in colchicine-treated mice.
For exercise studies, a treadmill protocol was implemented as previously described (42). Mice were trained on an Exer3/6 treadmill (Columbus Instruments) for 2days. On day 1, mice were run at 8 m/min for 5 min, and on day 2, those were run at 8 m/min for 5 min then 10 m/min for 5 min. After training, mice were run on day 3 for 40 min at 10 m/min, then for 30 min with increasing speed 1 m/min every 10 min, and finally for 10 min with increasing speed 1 m/min every 5 min for a total of 80 min of exercise. Mice were then immediately sacrificed and perfused with 4% paraformaldehyde and proximal hind limb muscles were collected. The University of Michigan Committee on Use and Care of Animals approved all procedures involving mice in compliance with the NIH Guidelines for the Care and Use of Experimental Animals.
Gene expression analysis
Total RNA was extracted from proximal hind limb muscle from AR knock-in mice after the indicated treatments in triplicate with Trizol (Sigma). Reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. Total RNA was similarly prepared from anonymized skeletal muscle biopsies from SBMA patients and controls as previously described (27). Quantitative PCR was performed with FastStart TaqMan Probe Master/Rox Master Mix (Roche) on a 7500 Real-Time PCR SDS System with the supplied software (Applied Biosystems) using gene-specific primers with carboxyfluorescein-labeled probes purchased from Applied Biosystems (Human: LAMP1, Hs00174766_m1; MAP1LC3, Hs01076567_g1; VPS11, Hs00222240_m1; VPS18, Hs00363585_m1. Mouse: bnip3, Mm01275601_g1; Fbxo32, Mm00499518_m1; Lamp1, Mm00495262_m1; Map1lc3, Mm00782868_sH; Sqstm1, Mm00448091_m1; Tcfeb, Mm00448968_m1; Trim63, Mm00185221_m1; Vps11, Mm01168594_m1; Vps18, Mm00552119_m1). Relative expression levels were calculated by comparing with the expression of 18S rRNA.
Analysis of protein expression
Cells were washed and collected in phosphate-buffered saline (PBS), then harvested and lysed in Radio Immunoprecipitation Assay (RIPA) buffer supplemented with phosphatase and protease inhibitors. Lysates were clarified at 15 000g for 15 min at 4°C. Nuclear and cytoplasmic lysates were prepared with the NE-PER Extraction Kit (Thermo Scientific) according to the manufacturer's instructions. For tissue analyses, proximal hind limb muscle and lumbar spinal cord from AR knock-in mice were minced, lysed and homogenized in RIPA buffer containing phosphatase and protease inhibitors. Lysates were clarified at 13 000g for 10 min at 4°C.
Nuclear lysates from proximal hind limb muscle were prepared as follows. Frozen tissue was pulverized, lysed in NB1 buffer, homogenized with a type A dounce homogenizer, mixed with NB2 buffer, layered underneath with NB3 buffer and nuclei were pelleted through the sucrose step gradient. After discarding the supernatant, nuclei were resuspended in EB buffer, homogenized with a type B dounce homogenizer and rotated for 30 min at 4°C. After pelleting the nuclear debris, the supernatant containing nuclear proteins was dialyzed overnight in Z buffer. Protein concentration for all lysates was determined by protein assay (Bio-Rad), and normalized protein samples were resolved in 10 or 15% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis gels after boiling at 100°C for 5 min in 6× SDS sample buffer. Proteins were then transferred onto nitrocellulose membranes in a semi-dry transfer apparatus (Bio-Rad). Proteins of interest were probed by indicated antibodies and detected by chemiluminescence. Densitometry measurements of protein bands were performed in ImageMeter (Flashscript).
Immunofluorescence
After induction of AR expression and serum starvation, PC12 cells were washed in PBS and fixed in 4% paraformaldehyde. Proximal hind limb muscles were collected for cryosectioning from mice after the indicated treatments and perfused with 4% paraformaldehyde. After perfusion, muscles were further fixed in 4% paraformaldehyde overnight, 15% sucrose for 4 h, 30% sucrose overnight, then flash frozen in liquid nitrogen and mounted in cryosection blocks with OCT Compound (Tissue-Tek). Sections were then cut at 10 µm using a Cryocut 1800 cryostat (Leica). Cells and muscle sections were stained with indicated antibodies and mounted with Vectashield medium with 4′,6-diamidino-2-phenylindole (Vector Labs). Fluorescence images were taken with a Zeiss Axio Imager microscope. GFP–LC3 puncta were quantified in ImageJ (NIH) using a GFP–LC3 quantification macro designed by Dagda and Chu (63,64).
Statistics
Statistical significance was determined by analysis of variance (ANOVA) with the Newman–Keuls multiple comparison test or unpaired two-tailed Student's t-test using Prism 6 (GraphPad). Differences were considered significant when P-values were <0.05.
FUNDING
This work was supported by grants from the National Institutes of Health (grant nos F31 NS076189 to J.P.C., R01 NS055746 to A.P.L., R01 CA144032 to D.M.R. and R01 NS032214 to D.E.M.).
ACKNOWLEDGEMENTS
We thank Dr Gregory Dressler for advice and expertise in isolating nuclei from mouse skeletal muscle, and Yuqing Sun and Chan Chung for experimental assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. doi:10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. doi:10.1016/S0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 3.Takeyama K., Ito S., Yamamoto A., Tanimoto H., Furutani T., Kanuka H., Miura M., Tabata T., Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–864. doi: 10.1016/s0896-6273(02)00875-9. doi:10.1016/S0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 4.Sobue G., Doyu M., Kachi T., Yasuda T., Mukai E., Kumagai T., Mitsuma T. Subclinical phenotypic expressions in heterozygous females of X-linked recessive bulbospinal neuronopathy. J. Neurol. Sci. 1993;117:74–78. doi: 10.1016/0022-510x(93)90157-t. doi:10.1016/0022-510X(93)90157-T. [DOI] [PubMed] [Google Scholar]
- 5.Mhatre A.N., Trifiro M.A., Kaufman M., Kazemi-Esfarjani P., Figlewicz D., Rouleau G., Pinsky L. Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nat. Genet. 1993;5:184–188. doi: 10.1038/ng1093-184. doi:10.1038/ng1093-184. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman A.P., Harmison G., Strand A.D., Olson J.M., Fischbeck K.H. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet. 2002;11:1967–1976. doi: 10.1093/hmg/11.17.1967. doi:10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. doi:10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. doi:10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J., Huang W.P., Stromhaug P.E., Klionsky D.J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. doi:10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. doi:10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 11.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. doi:10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Settembre C., Ballabio A. TFEB regulates autophagy: an integrated coordination of cellular degradation and recycling processes. Autophagy. 2011;7:1379–1381. doi: 10.4161/auto.7.11.17166. doi:10.4161/auto.7.11.17166. [DOI] [PubMed] [Google Scholar]
- 13.Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. doi:10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 14.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., Facchinetti V., et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. doi:10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. doi:10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pena-Llopis S., Vega-Rubin-de-Celis S., Schwartz J.C., Wolff N.C., Tran T.A., Zou L., Xie X.J., Corey D.R., Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. doi:10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chauhan S., Goodwin J.G., Manyam G., Wang J., Kamat A.M., Boyd D.D. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. doi:10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rusmini P., Bolzoni E., Crippa V., Onesto E., Sau D., Galbiati M., Piccolella M., Poletti A. Proteasomal and autophagic degradative activities in spinal and bulbar muscular atrophy. Neurobiol. Dis. 2010;40:361–369. doi: 10.1016/j.nbd.2010.06.016. doi:10.1016/j.nbd.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Pandey U.B., Nie Z., Batlevi Y., McCray B.A., Ritson G.P., Nedelsky N.B., Schwartz S.L., DiProspero N.A., Knight M.A., Schuldiner O., et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. doi:10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 20.Thomas M., Yu Z., Dadgar N., Varambally S., Yu J., Chinnaiyan A.M., Lieberman A.P. The unfolded protein response modulates toxicity of the expanded glutamine androgen receptor. J. Biol. Chem. 2005;280:21264–21271. doi: 10.1074/jbc.M500144200. doi:10.1074/jbc.M500144200. [DOI] [PubMed] [Google Scholar]
- 21.Mandrusiak L.M., Beitel L.K., Wang X., Scanlon T.C., Chevalier-Larsen E., Merry D.E., Trifiro M.A. Transglutaminase potentiates ligand-dependent proteasome dysfunction induced by polyglutamine-expanded androgen receptor. Hum. Mol. Genet. 2003;12:1497–1506. doi: 10.1093/hmg/ddg161. doi:10.1093/hmg/ddg161. [DOI] [PubMed] [Google Scholar]
- 22.Montie H.L., Cho M.S., Holder L., Liu Y., Tsvetkov A.S., Finkbeiner S., Merry D.E. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 2009;18:1937–1950. doi: 10.1093/hmg/ddp115. doi:10.1093/hmg/ddp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albertelli M.A., O'Mahony O.A., Brogley M., Tosoian J., Steinkamp M., Daignault S., Wojno K., Robins D.M. Glutamine tract length of human androgen receptors affects hormone-dependent and -independent prostate cancer in mice. Hum. Mol. Genet. 2008;17:98–110. doi: 10.1093/hmg/ddm287. doi:10.1093/hmg/ddm287. [DOI] [PubMed] [Google Scholar]
- 24.Yu Z., Dadgar N., Albertelli M., Scheller A., Albin R.L., Robins D.M., Lieberman A.P. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. Am. J. Pathol. 2006;168:195–204. doi: 10.2353/ajpath.2006.050619. doi:10.2353/ajpath.2006.050619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albertelli M.A., Scheller A., Brogley M., Robins D.M. Replacing the mouse androgen receptor with human alleles demonstrates glutamine tract length-dependent effects on physiology and tumorigenesis in mice. Mol. Endocrinol. 2006;20:1248–1260. doi: 10.1210/me.2006-0021. doi:10.1210/me.2006-0021. [DOI] [PubMed] [Google Scholar]
- 26.Yu Z., Dadgar N., Albertelli M., Gruis K., Jordan C., Robins D.M., Lieberman A.P. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J. Clin. Invest. 2006;116:2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Z., Wang A.M., Adachi H., Katsuno M., Sobue G., Yue Z., Robins D.M., Lieberman A.P. Macroautophagy is regulated by the UPR-mediator CHOP and accentuates the phenotype of SBMA mice. PLoS Genet. 2011;7:e1002321. doi: 10.1371/journal.pgen.1002321. doi:10.1371/journal.pgen.1002321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walcott J.L., Merry D.E. Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J. Biol. Chem. 2002;277:50855–50859. doi: 10.1074/jbc.M209466200. doi:10.1074/jbc.M209466200. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. doi:10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., Yonezawa K., Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. doi:10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. doi:10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. doi:10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 33.Petiot A., Ogier-Denis E., Blommaart E.F., Meijer A.J., Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. doi:10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 34.Pattingre S., Bauvy C., Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J. Biol. Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. doi:10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 35.Corcelle E., Nebout M., Bekri S., Gauthier N., Hofman P., Poujeol P., Fenichel P., Mograbi B. Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 2006;66:6861–6870. doi: 10.1158/0008-5472.CAN-05-3557. doi:10.1158/0008-5472.CAN-05-3557. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J.H., Horbinski C., Guo F., Watkins S., Uchiyama Y., Chu C.T. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. doi:10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., Whiteman M.W., Lian H., Wang G., Singh A., Huang D., Denmark T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol. Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. doi:10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. doi:10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju J.S., Varadhachary A.S., Miller S.E., Weihl C.C. Quantitation of ‘autophagic flux’ in mature skeletal muscle. Autophagy. 2010;6:929–935. doi: 10.4161/auto.6.7.12785. doi:10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., et al. Foxo3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. doi:10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. Foxo3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. doi:10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 42.He C., Bassik M.C., Moresi V., Sun K., Wei Y., Zou Z., An Z., Loh J., Fisher J., Sun Q., et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. doi:10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosokawa N., Hara Y., Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. doi:10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Gao W., Ding W.X., Stolz D.B., Yin X.M. Induction of macroautophagy by exogenously introduced calcium. Autophagy. 2008;4:754–761. doi: 10.4161/auto.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. doi:10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris H., Rubinsztein D.C. Control of autophagy as a therapy for neurodegenerative disease. Nat. Rev. Neurol. 2012;8:108–117. doi: 10.1038/nrneurol.2011.200. doi:10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- 47.Wang A.M., Miyata Y., Klinedinst S., Peng H.M., Chua J.P., Komiyama T., Li X., Morishima Y., Merry D.E., Pratt W.B., et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol. 2013;9:112–118. doi: 10.1038/nchembio.1140. doi:10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang A.M., Morishima Y., Clapp K.M., Peng H.M., Pratt W.B., Gestwicki J.E., Osawa Y., Lieberman A.P. Inhibition of hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J. Biol. Chem. 2010;285:15714–15723. doi: 10.1074/jbc.M109.098806. doi:10.1074/jbc.M109.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doi H., Adachi H., Katsuno M., Minamiyama M., Matsumoto S., Kondo N., Miyazaki Y., Iida M., Tohnai G., Qiang Q., et al. P62/SQSTM1 differentially removes the toxic mutant androgen receptor via autophagy and inclusion formation in a spinal and bulbar muscular atrophy mouse model. J. Neurosci. 2013;33:7710–7727. doi: 10.1523/JNEUROSCI.3021-12.2013. doi:10.1523/JNEUROSCI.3021-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alers S., Loffler A.S., Wesselborg S., Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. doi:10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonaldo P., Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 2013;6:25–39. doi: 10.1242/dmm.010389. doi:10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. doi:10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 53.Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., Goldberg A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. doi:10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palazzolo I., Stack C., Kong L., Musaro A., Adachi H., Katsuno M., Sobue G., Taylor J.P., Sumner C.J., Fischbeck K.H., et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63:316–328. doi: 10.1016/j.neuron.2009.07.019. doi:10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Blagden C., Fan J., Nowak S.J., Taniuchi I., Littman D.R., Burden S.J. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005;19:1715–1722. doi: 10.1101/gad.1318305. doi:10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ching J.K., Weihl C.C. Rapamycin-induced autophagy aggravates pathology and weakness in a mouse model of VCP-associated myopathy. Autophagy. 2013;9:799–800. doi: 10.4161/auto.23958. doi:10.4161/auto.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fanin M., Nascimbeni A., Angelini C. Muscle atrophy in limb girdle muscular dystrophy 2a: a morphometric and molecular study. Neuropathol. Appl. Neurobiol. 2013 doi: 10.1111/nan.12034. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 58.Dobrowolny G., Aucello M., Rizzuto E., Beccafico S., Mammucari C., Boncompagni S., Belia S., Wannenes F., Nicoletti C., Del Prete Z., et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. doi:10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 59.Palazzolo I., Burnett B.G., Young J.E., Brenne P.L., La Spada A.R., Fischbeck K.H., Howell B.W., Pennuto M. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum. Mol. Genet. 2007;16:1593–1603. doi: 10.1093/hmg/ddm109. doi:10.1093/hmg/ddm109. [DOI] [PubMed] [Google Scholar]
- 60.Rinaldi C., Bott L.C., Chen K.L., Harmison G.G., Katsuno M., Sobue G., Pennuto M., Fischbeck K.H. Insulin like growth factor (IGF)-1 administration ameliorates disease manifestations in a mouse model of spinal and bulbar muscular atrophy. Mol. Med. 2012;18:1261–1268. doi: 10.2119/molmed.2012.00271. doi:10.2119/molmed.2012.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalbello-Haas V., Florence J.M., Krivickas L.S. Therapeutic exercise for people with amyotrophic lateral sclerosis or motor neuron disease. Cochrane Database Syst. Rev. 2008;16:CD005229. doi: 10.1002/14651858.CD005229.pub2. [DOI] [PubMed] [Google Scholar]
- 62.Preisler N., Andersen G., Thogersen F., Crone C., Jeppesen T.D., Wibrand F., Vissing J. Effect of aerobic training in patients with spinal and bulbar muscular atrophy (Kennedy disease) Neurology. 2009;72:317–323. doi: 10.1212/01.wnl.0000341274.61236.02. doi:10.1212/01.wnl.0000341274.61236.02. [DOI] [PubMed] [Google Scholar]
- 63.Chu C.T., Plowey E.D., Dagda R.K., Hickey R.W., Cherra S.J., III, Clark R.S. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 2009;453:217–249. doi: 10.1016/S0076-6879(08)04011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dagda R.K., Zhu J., Kulich S.M., Chu C.T. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]