Abstract
Brain function and cognitive performance differ between men and women in some measures. The phenotypic variation may be partially due to sex differences in epigenomes and transcriptomes in specific brain regions [e.g. the prefrontal cortex (PFC)]. Genome-wide DNA methylation and gene expression were examined in postmortem PFC of 32 males and 14 females (all were Caucasians) using Illumina's 450K Methylation and HT-12 v4 Gene Expression BeadChips, respectively. Multiple linear regression, Pearson correlation and DAVID functional annotation analyses were applied to investigate sex-biased DNA methylation and gene expression, DNA methylation–gene expression correlation and gene ontology (GO) annotations overrepresented by differentially methylated and expressed genes. A total of 22 124 CpGs showed differential methylation between males and females (2.6 × 10−38 ≤ Pnominal ≤ 0.05), and the P-values of 8357 CpGs withstood multiple-testing correction (q < 0.05). A total of 1489 genes showed differential expression between males and females (4.1 × 10−36 ≤ Pnominal ≤ 0.05), and the P-values of 35 genes survived multiple-testing correction (q < 0.05). A significant correlation (Pcorrelation < 0.05) was observed between methylation levels of 585 differentially methylated CpGs (Pnominal ≤ 0.05) and expression levels of 188 differentially expressed host genes (Pnominal < 0.05). The GO terms enriched by these 188 genes (134 on autosomes and 54 on sex chromosomes) were assigned to 24 clusters, and 33 genes involved in the top cluster (enrichment score: 4.7) mainly participate in ribosome structure and function, RNA binding and protein translation. This study demonstrated sex-specific methylomic and transcriptomic profiles in the human PFC. Our findings suggest that sex-biased DNA methylation and gene expression could be either the cause or consequence of differential brain development between males and females.
INTRODUCTION
There are apparent sex differences in brain structure and function as well as susceptibility to neurological disorders. For example, cognitive function, such as working memory and attention, appears different between men and women (1). It is already known that the neuroanatomy of cognition includes a network of brain regions including dorsolateral and inferior prefrontal cortices (PFC) (2). However, the molecular basis of how men and women perform differently in perception, cognition, memory and neural function is poorly understood. It has been postulated that sex differences in brain function may be attributed to various genetic variants (3,4) and environmental factors such as prenatal stress (5), childhood abuse (6) and hormones (7,8).
There is substantial evidence supporting sex-biased gene expression. Studies using model organisms have shown sex-biased gene expression, and the number of sex-biased genes is large. For example, more than half of the genes in the Drosophila melanogaster genome show sex-biased expression (9), and most of the differences are attributable to genes that are expressed in reproductive tissues (10). By analyzing gene expression in multiple mouse somatic tissues, Yang et al. (11) found that more than 10 000 genes showed sex-biased expression, and these genes were enriched not only on sex chromosomes but also on several autosomes. Moreover, microarray studies of gene expression in postmortem human brains have also demonstrated sex-biased gene expression in both developing and adult human brains. Vawter et al. (12) reported sex-specific expression of genes located on sex chromosomes (one gene on X chromosome and five genes on Y chromosome) in three regions (dorsolateral PFC, anterior cingulated cortex and cerebellum) of postmortem brains. Weickert et al. (13) examined the transcriptome in postmortem PFC of subjects ranging in age from 1 month to 50 years, and found that the expression of 83 genes (25 on sex chromosomes and 58 on autosomes) differed between males and females. Although differential gene expression between males and females has been observed in numerous studies, the mechanism for the sex differences in gene expression is far from being understood.
Accumulating evidence suggests that, besides the genetic differences between males (with X and Y chromosomes) and females (with two X chromosomes), epigenetic modifications such as DNA methylation and histone modifications are also potential sources of sex differences in gene expression. A good example is X chromosome inactivation, a process by which one of the two copies of the X chromosome in females is inactivated. The silence of one of the two X chromosomes in females is largely due to a combination of DNA methylation and histone modifications (14). Gene imprinting, which appears in both sex and autosomal chromosomes, is also a type of epigenetic control of sex differences in gene expression. In imprinted genes, either the paternal or maternal allele is hypermethylated, thus leading to monoallelic expression (15). Additionally, environmental factors may affect brain activity and behavior as well as disease vulnerability differently between men and women (16,17). Since the effect of environmental factors on gene expression is potentially mediated by epigenetic events (e.g. DNA methylation) (18), environmental factors could be another source of epigenetic differences between males and females.
Based on the above findings, sex is believed to be an important predictor of DNA methylation and gene expression. Studies of the influence of sex on DNA methylation have provided strong evidence that sex affects genome-wide DNA methylation in both salvia and blood samples (19,20). Nevertheless, no study is known to have analyzed genome-wide DNA methylation and gene expression differences between males and females in human brain tissue samples simultaneously. The aim of the present study was to use the microarray approach to investigate postmortem PFC DNA methylation and gene expression differences between 32 males and 14 females (all with European ancestry) (Table 1). Functional annotations for those genes showing differential methylation and expression between males and females were analyzed by bioinformatics programs.
Table 1.
Demographic information of postmortem PFC tissues
| Males (n = 32) | Females (n = 14) | P-value (t-test) | |
|---|---|---|---|
| Ethnicity | Caucasian | Caucasian | |
| Age (years) (mean ± SD) | 56 ± 10 | 57 ± 6 | 0.896 |
| Postmortem interval (h) (mean ± SD) | 37.5 ± 14.8 | 33.1 ± 13.9 | 0.338 |
| Brain weight (g) (mean ± SD) | 1457.9 ± 96.0 | 1254.3 ± 107.1 | 3.3 × 10−6 |
| Brain pH (mean ± SD) | 6.6 ± 0.2 | 6.5 ± 0.3 | 0.293 |
| Alcohol use disorders [n (%)] | 16 (50%) | 7 (50%) |
RESULTS
Differentially methylated CpGs between males and females
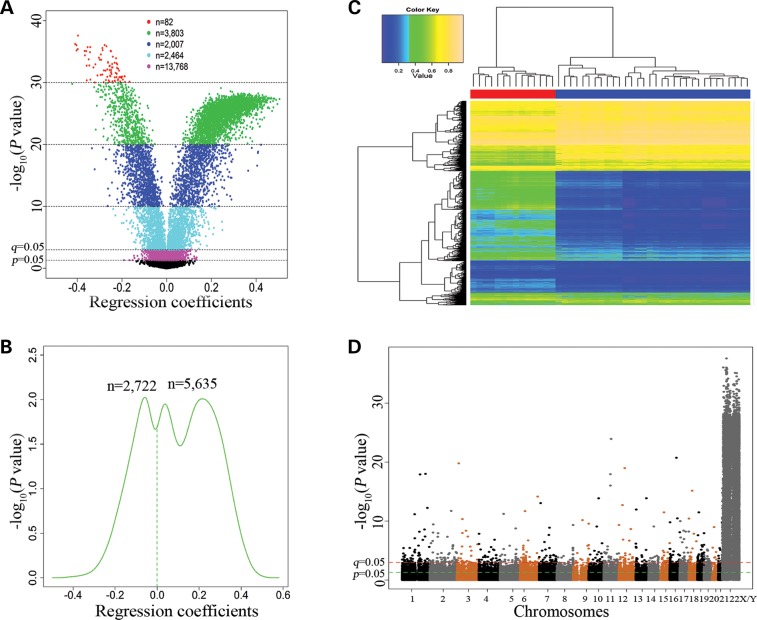
Genome-wide DNA methylation levels in postmortem PFC were examined using the Illumina Infinium HumanMethylation450 BeadChip and compared between males and females using multiple linear regress analysis (see Materials and Methods). Among the 430 011 CpGs (remained after data quality control), 420 419 (97.8%) were located on autosomes and 9592 (2.2%) were located on sex (X or Y) chromosomes. As displayed in the volcano plot (Fig. 1A), 22 124 CpGs (5.1%) showed differential methylation between males and females (Pnominal = 2.6 × 10−38–0.05, see Supplementary Material, Table S1). The P-values (Pnominal = 2.6 × 10−38–1.0 × 10−3) of 8357 CpGs (5587 CpGs in promoter regions, 1256 CpGs in gene bodies, 307 CpGs in 3′ UTRs and 1207 CpGs in intergenic regions) survived multiple-testing correction (1.3 × 10−32 ≤ q ≤ 0.05). The results of top 20 sex chromosomal CpGs (P ≤ 9.7 × 10−35) and top 20 autosomal CpGs (P ≤ 3.3 × 10−12) are presented in Table 2. The asymmetric density pattern (as shown in Fig. 1B) indicated that a greater proportion [67.4% or 5635 CpGs (including 476 on autosomes, 5152 on X chromosomes and 7 on Y chromosomes)] of the 8357 CpGs (q ≤ 0.05) were hypermethylated in females in comparison to males. Hierarchical clustering analysis was performed based on methylation levels (β-values) of 8357 CpGs (q ≤ 0.05). The heatmap showed that the 46 subjects were clustered into two distinct subgroups that were highly consistent with their sex status (Fig. 1C). The Manhattan plot (Fig. 1D) displayed the distribution of the P-values generated by multiple linear regression analysis of the sex effect on DNA methylation across all chromosomes. Among the 8357 CpGs that showed differential methylation between males and females (Pnominal <thinsp;1.0 × 10−3 or q ≤ 0.05), 7743 CpGs (92.7%) were located on sex chromosomes (7721 CpG on X chromosome and 22 on Y chromosome), whereas only 614 CpGs (7.3%) were located on autosomes.
Figure 1.
Genome-wide DNA methylation differences between 32 male and 14 female subjects. (A) Volcano plotting of regression coefficients of 430 011 CpGs against -log10(P-values). Regression coefficients were obtained from multiple linear regression analysis. (B) Kernel density estimation of the distributions of regression coefficients of 8357 CpGs (Pnominal < 0.001 or q ≤ 0.05). (C) Hierarchical clustering heatmap was constructed using methylation levels of 8357CpGs (Pnominal < 0.001 or q ≤ 0.05) across the 46 subjects (columns). Colors in the heatmap indicate methylation levels (blue to yellow: low to high methylation levels). The horizontal bars underneath the cluster tree indicate the sex status of the 46 subjects (red color: 14 females; blue color: 32 males). (D) A Manhattan plot showing sex differences in DNA methylation across the genome.
Table 2.
Top 20 sex chromosomal and top 20 autosonal CpGs with sex-biased methylation
| CpGs | Genes | Chromosome | Locationa | β (males)b | β (females)b | Effect sizec | Pnominald | qe |
|---|---|---|---|---|---|---|---|---|
| Top 20 sex chromosomal CpGs with sex-biased methylation | ||||||||
| cg11143827 | BCOR | X | Promoter | 0.94 | 0.54 | −0.40 | 2.6E−38 | 1.3E−32 |
| cg11516614 | X | Intergenic | 0.83 | 0.56 | −0.28 | 8.9E−37 | 1.2E−31 | |
| cg21010298 | BCOR | X | Promoter | 0.86 | 0.46 | −0.40 | 9.5E−37 | 1.2E−31 |
| cg01086462 | Y | Intergenic | 0.58 | 0.17 | −0.41 | 6.0E−37 | 1.2E−31 | |
| cg22223709 | PPP1R2P9 | X | Body | 0.94 | 0.67 | −0.27 | 1.4E−36 | 1.2E−31 |
| cg22655232 | PPP1R2P9 | X | Promoter | 0.88 | 0.54 | −0.34 | 1.9E−36 | 1.2E−31 |
| cg00455876 | Y | Intergenic | 0.76 | 0.40 | −0.35 | 2.0E−36 | 1.2E−31 | |
| cg03959079 | X | Intergenic | 0.84 | 0.55 | −0.29 | 2.6E−36 | 1.4E−31 | |
| cg11049634 | BCOR | X | Promoter | 0.92 | 0.52 | −0.40 | 4.0E−36 | 1.9E−31 |
| cg26327984 | PPP1R2P9 | X | Promoter | 0.93 | 0.65 | −0.29 | 6.0E−36 | 2.6E−31 |
| cg24183173 | BCOR | X | Promoter | 0.89 | 0.49 | −0.40 | 6.7E−36 | 2.7E−31 |
| cg27551771 | KIAA1210 | X | 3′UTR | 0.83 | 0.58 | −0.25 | 7.6E−36 | 2.7E−31 |
| cg05130312 | LOC286467 | X | Body | 0.83 | 0.46 | −0.37 | 8.0E−36 | 2.7E−31 |
| cg03161453 | BCOR | X | Promoter | 0.85 | 0.49 | −0.36 | 8.5E−36 | 2.7E−31 |
| cg22067189 | PPP1R2P9 | X | Promoter | 0.90 | 0.65 | −0.25 | 9.0E−36 | 2.7E−31 |
| cg20348344 | BCOR | X | Promoter | 0.94 | 0.59 | −0.34 | 1.3E−35 | 3.6E−31 |
| cg21797452 | LOC286467 | X | Body | 0.86 | 0.57 | −0.29 | 2.9E−35 | 7.8E−31 |
| cg04493908 | Y | Intergenic | 0.73 | 0.37 | −0.37 | 4.1E−35 | 1.0E−30 | |
| cg04690567 | PHF8 | X | 3′UTR | 0.81 | 0.57 | −0.24 | 6.8E−35 | 1.6E−30 |
| cg16440909 | MAMLD1 | X | Promoter | 0.84 | 0.54 | −0.30 | 9.7E−35 | 2.2E−30 |
| Top 20 autosomal CpGs with sex-biased methylation | ||||||||
| cg25294185 | RNASEH2C | 11 | Body | 0.17 | 0.07 | −0.10 | 1.2E−24 | 2.1E−22 |
| cg04946709 | LOC644649 | 16 | Body | 0.79 | 0.71 | −0.08 | 1.8E−21 | 2.2E−19 |
| cg11643285 | RFTN1 | 3 | Body | 0.73 | 0.85 | 0.12 | 1.6E−20 | 1.8E−18 |
| cg03691818 | KRT77 | 12 | Body | 0.05 | 0.11 | 0.06 | 1.1E−19 | 1.1E−17 |
| cg15817705 | 1 | Intergenic | 0.79 | 0.69 | −0.10 | 9.9E−19 | 1.0E−16 | |
| cg04858776 | 11 | Intergenic | 0.05 | 0.11 | 0.05 | 1.1E−18 | 1.2E−16 | |
| cg03618918 | 1 | Intergenic | 0.77 | 0.68 | −0.09 | 1.2E−18 | 1.3E−16 | |
| cg17232883 | 11 | Intergenic | 0.09 | 0.14 | 0.05 | 9.5E−17 | 8.9E−15 | |
| cg25304146 | WBP11P1 | 18 | Body | 0.65 | 0.57 | −0.07 | 7.0E−16 | 6.4E−14 |
| cg25568337 | ARID1B | 6 | Promoter | 0.15 | 0.21 | 0.06 | 7.1E−15 | 6.2E−13 |
| cg26355737 | TFDP1 | 13 | Body | 0.85 | 0.78 | −0.07 | 1.4E−14 | 1.2E−12 |
| cg22227586 | FAM35A | 10 | Promoter | 0.03 | 0.05 | 0.02 | 1.4E−14 | 1.2E−12 |
| cg26516287 | SCIN | 7 | Promoter | 0.77 | 0.81 | 0.04 | 8.9E−14 | 7.5E−12 |
| cg01225095 | YARS2 | 12 | Promoter | 0.11 | 0.15 | 0.04 | 1.9E−13 | 1.6E−11 |
| cg16727519 | H3F3A | 1 | Body | 0.57 | 0.51 | −0.05 | 5.8E−13 | 4.7E−11 |
| cg06710937 | 13 | Intergenic | 0.06 | 0.10 | 0.04 | 1.1E−12 | 8.9E−11 | |
| cg06759085 | NAB1 | 2 | 3'UTR | 0.78 | 0.71 | −0.07 | 1.9E−12 | 1.5E−10 |
| cg16169375 | C6orf108 | 6 | Promoter | 0.08 | 0.11 | 0.03 | 2.0E−12 | 1.6E−10 |
| cg20808136 | 15 | Intergenic | 0.84 | 0.78 | −0.06 | 3.3E−12 | 2.6E−10 | |
| cg03608000 | ZNF69 | 19 | Promoter | 0.06 | 0.08 | 0.02 | 3.3E−12 | 2.6E−10 |
aLocation of CpGs in promoter regions, gene bodies, 3’ UTRs or intergenic regions.
bMean CpG methylation levels in males (or females) measured by Illumina's 450 K Methylation BeadChip.
cRegression coefficients by multiple linear regression analysis.
dObserved P-values by multiple linear regression analysis.
eAdjusted P-values by R package qvalue, controlling the FDR at 0.05.
Differentially expressed genes between males and females
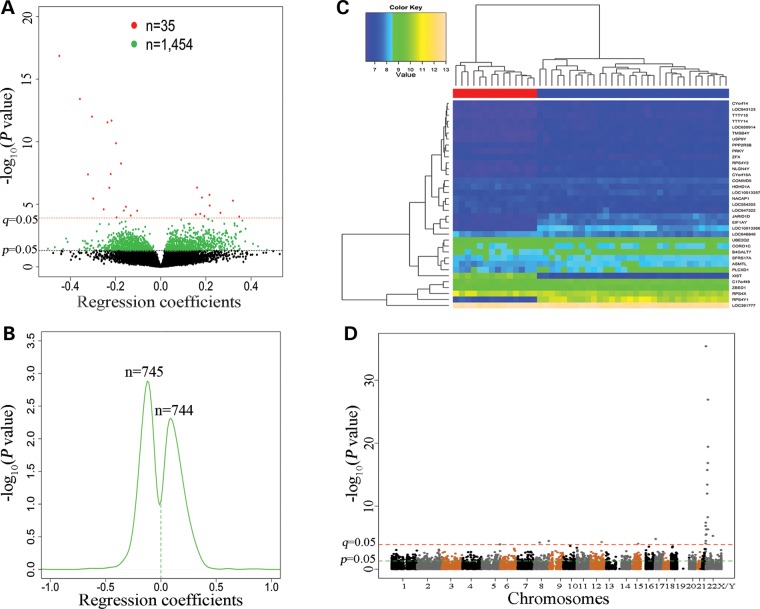
Genome-wide expression levels of genes were quantified using the Illumina HumanHT-12 v4 Expression BeadChip and compared between males and females using multiple linear regression analysis (see Materials and Methods). The regression analysis results of the 14 851 genes (remained after filtering out genes with low quality expression data) were summarized by volcano plotting (Fig. 2A). One thousand four hundred and eighty-nine genes were differentially expressed between males and females (Pnominal = 4.1 × 10−36–0.05, see Supplementary Material, Table S2). The P-values (4.1 × 10−36 ≤ Pnominal ≤ 1.1 × 10−4) of 35 genes (2.3%) survived multiple-testing correction (4.8 × 10−32 ≤ q ≤ 0.05) (Table 3). Density plotting of the regression coefficients (obtained from multiple linear regression analysis) of these 1489 genes showed that 744 genes (712 on autosomes and 32 on sex chromosome) were up-regulated and 745 genes (685 on autosomes and 60 on sex chromosomes) were down-regulated in females in comparison to males (Fig. 2B). Hierarchical clustering analysis was performed based on expression levels of the 35 genes (q ≤ 0.05). The heatmap showed that the 46 subjects were clustered into two distinct subgroups that were highly consistent with their sex status (Fig. 2C). The distribution of the P-values generated by multiple linear regression analysis of the sex effect on gene expression across all chromosomes was displayed by a Manhattan plot (Fig. 2D).
Figure 2.
Genome-wide gene expression differences between 32 male and 14 female subjects. (A) Volcano plotting of regression coefficients (obtained from multiple linear regression analysis) of 14 851 genes against −log10 (P-values). Red dots represent 35 genes with 4.07 × 10−36 ≤ Pnominal ≤ 1.13 × 10−4 (or q ≤ 0.05). Green dots represent 1464 genes with 1.57 × 10−4 ≤ Pnominal ≤ 0.05. (B) Kernel density estimation of the distributions of regression coefficients of the above 1489 (35 + 1464) genes (Pnominal ≤ 0.05). (C) Hierarchical clustering heatmap was constructed using methylation levels of 35 genes (rows) (q ≤ 0.05) across the 46 subjects (columns). Colors in the heatmap indicate expression levels (blue to yellow: low to high expression levels). The horizontal bars underneath the cluster tree indicate the sex status of the 46 subjects (red color: 14 females; blue color: 32 males). (D) A Manhattan plot showing sex differences in gene expression across the genome.
Table 3.
Top 35 genes with differential expression between males and females (q < 0.05)
| Genes | Chromosome | Expression (males)a | Expression (females)a | Effect sizeb | Pnominalc | qd |
|---|---|---|---|---|---|---|
| LOC647322 | 2 | 6.86 | 7.20 | 0.35 | 9.8E−05 | 3.4E−02 |
| LOC646849 | 3 | 7.76 | 7.95 | 0.22 | 1.3E−05 | 6.2E−03 |
| LOC391777 | 5 | 12.70 | 12.84 | 0.16 | 6.7E−05 | 2.5E−02 |
| B4GALT7 | 5 | 8.69 | 8.49 | −0.20 | 1.1E−04 | 3.8E−02 |
| COMMD5 | 8 | 7.49 | 7.39 | −0.10 | 3.3E−05 | 1.4E−02 |
| NACAP1 | 8 | 7.06 | 7.22 | 0.18 | 5.9E−05 | 2.3E−02 |
| CORO1C | 12 | 8.46 | 8.69 | 0.26 | 4.9E−05 | 2.0E−02 |
| UBE2Q2 | 15 | 8.70 | 8.88 | 0.19 | 8.8E−05 | 3.1E−02 |
| C17orf49 | 17 | 9.46 | 9.31 | −0.15 | 1.6E−05 | 7.4E−03 |
| XIST | X | 6.88 | 9.69 | 2.84 | 1.2E−34 | 7.3E−31 |
| LOC100133662 | X | 8.16 | 6.49 | −1.67 | 2.1E−26 | 4.8E−23 |
| ZFX | X | 6.57 | 6.74 | 0.16 | 4.7E−07 | 3.0E−04 |
| LOC554203 | X | 6.97 | 7.19 | 0.22 | 1.8E−06 | 1.0E−03 |
| HDHD1A | X | 7.21 | 7.40 | 0.18 | 2.9E−06 | 1.6E−03 |
| RPS4X | X | 10.14 | 10.44 | 0.32 | 5.2E−06 | 2.6E−03 |
| LOC100133578 | X | 7.30 | 7.03 | −0.25 | 2.6E−05 | 1.2E−02 |
| ASMTL | X/Ye | 8.32 | 7.99 | −0.32 | 4.1E−08 | 3.0E−05 |
| PLCXD1 | X/Ye | 8.68 | 8.07 | −0.63 | 1.4E−07 | 9.5E−05 |
| ZBED1 | X/Ye | 9.26 | 8.98 | −0.30 | 3.5E−06 | 1.9E−03 |
| SFRS17A | X/Ye | 8.38 | 8.23 | −0.16 | 3.2E−05 | 1.4E−02 |
| PPP2R3B | X/Ye | 6.77 | 6.64 | −0.13 | 8.0E−05 | 2.9E−02 |
| RPS4Y1 | Y | 10.43 | 7.19 | −3.24 | 4.1E−36 | 4.8E−32 |
| JARID1D | Y | 7.68 | 6.47 | −1.22 | 9.3E−31 | 3.6E−27 |
| EIF1AY | Y | 7.60 | 6.49 | −1.11 | 1.2E−27 | 3.6E−24 |
| RPS4Y2 | Y | 6.99 | 6.43 | −0.55 | 3.7E−20 | 7.3E−17 |
| CYorf15A | Y | 6.91 | 6.46 | −0.45 | 1.4E−17 | 2.4E−14 |
| NLGN4Y | Y | 6.98 | 6.37 | −0.61 | 1.8E−16 | 2.7E−13 |
| TMSB4Y | Y | 6.69 | 6.33 | −0.36 | 3.8E−14 | 5.0E−11 |
| USP9Y | Y | 6.64 | 6.35 | −0.30 | 1.0E−12 | 1.2E−09 |
| CYorf14 | Y | 6.59 | 6.37 | −0.22 | 2.1E−12 | 2.2E−09 |
| LOC643123 | Y | 6.60 | 6.36 | −0.24 | 2.8E−12 | 2.8E−09 |
| TTTY15 | Y | 6.53 | 6.33 | −0.20 | 1.3E−10 | 1.2E−07 |
| TTTY14 | Y | 6.63 | 6.45 | −0.18 | 5.5E−09 | 4.6E−06 |
| LOC650914 | Y | 6.62 | 6.39 | −0.22 | 3.9E−08 | 3.0E−05 |
| PRKY | Y | 6.72 | 6.48 | −0.23 | 4.8E−07 | 3.0E−04 |
aMean gene expression levels in males (or females) measured by Illumina's HT-12 v4 Gene Expression BeadChip.
bRegression coefficients by multiple linear regression analysis.
cObserved P-values by multiple linear regression analysis.
dAdjusted P-values by R package qvalue, controlling the FDR at 0.05.
eGenes are located on both X and Y chromosomes.
Correlation of DNA methylation and gene expression
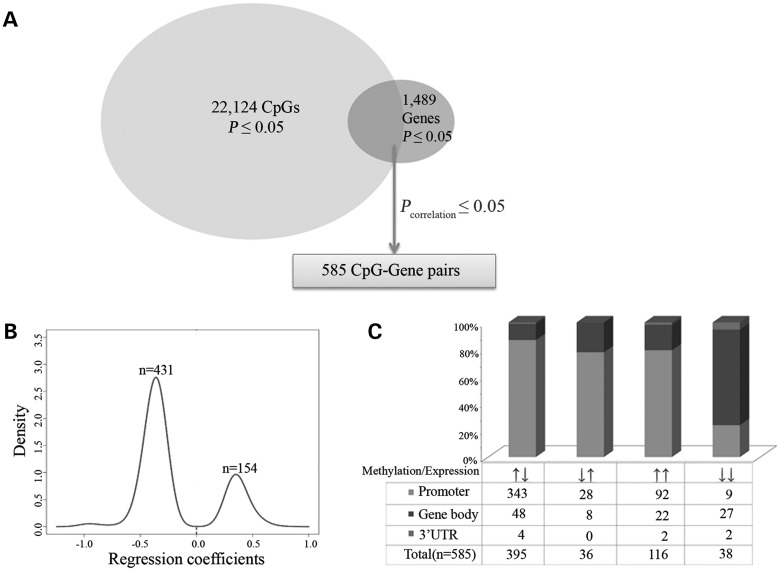
One thousand four hundred and three of the above 22 124 differentially methylated CpGs (Pnominal ≤ 0.05) were mapped to 547 of the above 1489 differentially expressed genes (Pnominal ≤ 0.05), thus generating 1436 CpG (methylation)–gene (expression) pairs. Based on the Illumina annotation file (see Materials and Methods), CpGs were classified into four groups based on their locations in promoter regions [from 1500 bp upstream to 200 bp downstream of the transcription start site (TSS), including all or part of the 5′ untranslated region (5′ UTR) and/or the first exon], gene bodies, 3′ UTRs or intergenic regions. Significant correlation was observed in 585 pairs (2.3 × 10−35 ≤ Pcorrelation ≤ 0.05, 1.2 × 10−32 < q < 0.044, see Supplementary Material, Table S3). The 585 pairs were classified into four subgroups in terms of the relative CpG methylation and gene expression alterations (up or down) in females in comparison to males as well as CpG locations in gene regions (promoter regions, gene bodies or 3′ UTRs) (Fig. 3). A negative correlation was observed in 431 (395 ↑↓ + 36↓↑) pairs (2.3 × 10−35 ≤ Pcorrelation ≤ 0.05, 1.2 × 10−32 < q < 0.044), whereas a positive correlation was observed in 154 (116↑↑ + 38↓↓) pairs (5.0 × 10−9 ≤ Pcorrelation ≤ 0.05, 2.6 × 10−7 < q < 0.043). The top 10 negatively (Pcorrelation ≤ 7.1 × 10−7, q < 3.3 × 10−5) and top 10 positively (Pcorrelation ≤ 1.2 × 10−4, q < 2.3 × 10−3) correlated CpG (methylation)–gene (expression) pairs are listed in Table 4.
Figure 3.
Sex-associated CpG (methylation)–gene (expression) pairs. (A) One thousand four hundred and three of the 22 124 differentially methylated CpGs (Pnominal ≤ 0.05) were mapped to 547 of the 1489 differentially expressed genes (Pnominal ≤ 0.05), thus generating 1436 CpG (methylation)–gene (expression) pairs [including 585 significant pairs (Pcorrelation ≤ 0.05)]. (B) Density plotting of correlation coefficients (obtained from the Pearson correlation analysis) of the 585 CpG (methylation)–gene (expression) pairs by the Kernel method. (C) Histogram of the 585 sex-specific CpG (methylation)–gene (expression) pairs (Pcorrelation ≤ 0.05). Arrows under the histogram indicate the direction of DNA methylation and gene expression changes in females (e.g. ↑↓ means increased DNA methylation levels but decreased gene expression levels in females) in comparison to males.
Table 4.
Top 10 negatively and top 10 positively correlated CpG (methylation)–gene (expression) pairs
| CpGs | Genes | Chr. | Location | Methylation analysis |
Expression analysis |
Correlation analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| βa | Pnominalb | qc | βa | Pnominalb | qc | r | Pcorrelationd | qcorrelationc | ||||
| Top 10 negatively correlated pairs | ||||||||||||
| cg12653510 | XIST | X | Body | −0.25 | 2.0E−32 | 1.8E−28 | 2.84 | 1.2E−34 | 7.3E−31 | −0.99 | 2.3E−35 | 1.2E−32 |
| cg11717280 | XIST | X | Promoter | −0.25 | 1.3E−27 | 8.1E−25 | 2.84 | 1.2E−34 | 7.3E−31 | −0.98 | 1.1E−32 | 2.7E−30 |
| cg26983535 | EIF1AY | Y | Promoter | 0.32 | 1.9E−24 | 3.2E−22 | −1.11 | 1.2E−27 | 3.6E−24 | −0.97 | 1.8E−27 | 3.1E−25 |
| cg05533223 | XIST | X | Body | −0.26 | 4.9E−26 | 1.2E−23 | 2.84 | 1.2E−34 | 7.3E−31 | −0.97 | 3.2E−27 | 4.1E−25 |
| cg01375382 | RPS4Y1 | Y | Promoter | 0.28 | 7.3E−22 | 9.2E−20 | −3.24 | 4.1E−36 | 4.8E−32 | −0.96 | 1.0E−26 | 1.1E−24 |
| cg03554089 | XIST | X | Body | −0.20 | 7.5E−19 | 7.8E−17 | 2.84 | 1.2E−34 | 7.3E−31 | −0.92 | 1.8E−19 | 1.5E−17 |
| cg20698282 | XIST | X | Promoter | −0.15 | 2.6E−18 | 2.6E−16 | 2.84 | 1.2E−34 | 7.3E−31 | −0.92 | 3.4E−19 | 2.5E−17 |
| cg02233183 | NLGN4Y | Y | Promoter | 0.02 | 4.1E−10 | 2.9E−08 | −0.61 | 1.8E−16 | 2.7E−13 | −0.78 | 2.3E−10 | 1.4E−08 |
| cg25032547 | TTTY15 | Y | Promoter | 0.15 | 1.5E−15 | 1.3E−13 | −0.20 | 1.3E−10 | 1.2E−07 | −0.75 | 1.9E−09 | 1.1E−07 |
| cg19917656 | GSTO2 | 10 | Promoter | 0.02 | 2.0E−02 | 6.0E−01 | −0.14 | 4.4E−02 | 3.8E−01 | −0.66 | 7.1E−07 | 3.3E−05 |
| Top 10 positively correlated pairs | ||||||||||||
| cg26251715 | TTTY14 | Y | Body | −0.15 | 4.6E−22 | 5.8E−20 | −0.18 | 5.5E−09 | 4.6E−06 | 0.74 | 5.0E−09 | 2.6E−07 |
| cg07900766 | SPAG16 | 2 | Promoter | 0.01 | 2.8E−02 | 7.2E−01 | 0.15 | 7.8E−04 | 1.5E−01 | 0.62 | 4.3E−06 | 1.8E−04 |
| cg02546818 | RPS4X | X | Promoter | 0.05 | 8.7E−12 | 6.7E−10 | 0.32 | 5.2E−06 | 2.6E−03 | 0.61 | 7.5E−06 | 2.8E−04 |
| cg01714671 | RPS4X | X | Body | 0.08 | 6.1E−18 | 6.0E−16 | 0.32 | 5.2E−06 | 2.6E−03 | 0.58 | 2.1E−05 | 6.6E−04 |
| cg17513789 | XIST | X | Promoter | 0.05 | 5.2E−05 | 3.0E−03 | 2.84 | 1.2E−34 | 7.3E−31 | 0.58 | 2.6E−05 | 7.9E−04 |
| cg00481499 | SPAG16 | 2 | Promoter | 0.01 | 2.7E−02 | 7.0E−01 | 0.15 | 7.8E−04 | 1.5E−01 | 0.55 | 6.9E−05 | 1.9E−03 |
| cg17279685 | XIST | X | Promoter | 0.03 | 1.6E−04 | 8.8E−03 | 2.84 | 1.2E−34 | 7.3E−31 | 0.54 | 9.8E−05 | 2.2E−03 |
| cg08859156 | RPS4X | X | Body | 0.03 | 1.5E−08 | 1.0E−06 | 0.32 | 5.2E−06 | 2.6E−03 | 0.54 | 1.1E−04 | 2.2E−03 |
| cg15319295 | XIST | X | Body | 0.02 | 1.9E−04 | 1.0E−02 | 2.84 | 1.2E−34 | 7.3E−31 | 0.54 | 1.2E−04 | 2.3E−03 |
| cg26944949 | LOC554203 | X | Body | 0.08 | 3.0E−07 | 1.9E−05 | 0.22 | 1.8E−06 | 1.0E−03 | 0.54 | 1.2E−04 | 2.3E−03 |
aEffect size (or regression coefficients) by multiple linear regression analysis.
bObserved P-values by multiple linear regression analysis.
cAdjusted P-values by R package qvalue, controlling the FDR at 0.05.
dCorrelation P-values by the Pearson correlation analysis.
Biological themes enriched by differentially methylated and expressed genes
We further analyzed the biological function of the 188 genes involved in the above 585 CpG methylation–genes expression pairs using the DAVID functional classification tool. The GO terms enriched by these 188 genes (134 on autosomes and 54 on sex chromosomes) were assigned to 24 clusters ranked by the biological significance of groups of genes based on all enriched annotation terms. Thirty-three genes included in the top cluster (enrichment score: 4.7) belong to 12 GO terms that are mainly involved in ribosome structure and function, RNA binding and protein translation (Table 5). Additionally, functional annotation analysis was performed using 54 genes (of the 188 genes) located on sex chromosomes. Eleven GO terms were found in the top cluster, and a majority of these GO terms are relevant to protein translation (see Supplementary Material, Table S4).
Table 5.
DAVID functional enrichment analysis results (Annotation Cluster 1)
| GO terms | Count | Fold enrichment | P-value | Genes |
|---|---|---|---|---|
| Translation | 17 | 4.9 | 3.8E−07 | MRPL4, VARS2, EIF2S3, RPS15A, RPL39, RPS4X, RPS8, RPS27, MRPL12, RPS16, RPL7, TSFM, EIF1AY, RPLP1, GSPT2, RPS4Y1, RPL36AL |
| Structural constituent of ribosome | 12 | 7.1 | 9.4E−07 | RPS27, MRPL4, MRPL12, RPS16, RPL7, RPLP1, RPS4Y1, RPS15A, RPS4X, RPL39, RPS8, RPL36AL |
| Translational elongation | 10 | 9.4 | 1.1E−06 | RPS27, RPS16, RPL7, TSFM, RPLP1, RPS4Y1, RPS15A, RPS4X, RPL39, RPS8 |
| Cytosolic ribosome | 9 | 10.6 | 1.9E−06 | RPS27, RPS16, RPL7, RPLP1, RPS4Y1, RPS15A, RPS4X, RPL39, RPS8 |
| Ribonucleoprotein complex | 19 | 3.5 | 6.3E−06 | MRPL4, RBM5, RPS15A, SF3B5, RPL39, RPS4X, PRPF4, BRCA1, RPS8, RPS27, MRPL12, RPS16, RPL7, DHX38, RPLP1, RPS4Y1, PPIL3, PABPC1, RPL36AL |
| Ribosomal subunit | 10 | 7.5 | 7.3E−06 | RPS27, MRPL12, RPS16, RPL7, RPLP1, RPS4Y1, RPS15A, RPS4X, RPL39, RPS8 |
| Ribosome | 12 | 5.3 | 1.5E−05 | RPS27, MRPL4, MRPL12, RPS16, RPL7, RPLP1, RPS4Y1, RPS15A, RPS4X, RPL39, RPS8, RPL36AL |
| Cytosolic part | 10 | 6.3 | 2.9E−05 | RPS27, RPS16, RPL7, RPLP1, IKBKG, RPS4Y1, RPS15A, RPS4X, RPL39, RPS8 |
| Cytosolic small ribosomal subunit | 6 | 14.3 | 5.6E−05 | RPS27, RPS16, RPS4Y1, RPS15A, RPS4X, RPS8 |
| Small ribosomal subunit | 6 | 9.1 | 4.9E−04 | RPS27, RPS16, RPS4Y1, RPS15A, RPS4X, RPS8 |
| Structural molecule activity | 15 | 2.3 | 4.4E−03 | MRPL4, RPS15A, RPL39, RPS4X, RPS8, RPS27, MRPL12, RPL7, RPS16, RPLP1, RPS4Y1, TUBA1B, DEDD2, RPL36AL, NPHP1 |
| RNA binding | 16 | 2.2 | 5.4E−03 | FUS, RBM41, RPUSD3, EXOSC5, RBM5, RPS15A, RPS4X, RPL39, MRPL12, RPL7, RPS16, EIF1AY, RPLP1, RPS4Y1, PABPC1, LRPPRC |
DISCUSSION
This study demonstrated differential methylomic and transcriptomic patterns in the PFC between men and women. It also provided evidence that sex-biased DNA methylation and sex-biased gene expression in a number of genes were significantly (either positively or negatively) correlated. Bioinformatics analysis suggests that genes with sex-biased DNA methylation and correlated sex-biased expression might be involved in cellular functions relevant to protein synthesis. The major findings from this study were discussed below.
First, sex differences in DNA methylation were dispersed across the genome but predominantly on the X chromosome. As shown in Figure 1D, among the 8357 CpGs that showed differential methylation between males and females (at the q ≤ 0.05 level), 7721 CpGs (92.4%) were located on the X chromosome. Moreover, 5152 (or 66.7%) of the 7721 CpGs were hypermethylated in females in comparison to males. These findings may have important implications for understanding the mechanism of X chromosome inactivation. It has long been known that X chromosome inactivation is required for generating an equivalent expression of X chromosome genes in males and females. However, the mechanism of the dosage compensation is understudied. Accumulating evidence suggests that one of the two X chromosomes is largely silenced by a combination of histone modifications and DNA methylation (14). The findings of the present study support that X chromosome inactivation is at least partially caused by hypermethylation of X chromosomal CpGs in females. Nevertheless, a smaller proportion (2569 or 33.3%) of the 7721 X chromosome CpGs showed significantly lower methylation levels in females than in males. The implication of hypomethylation of X chromosome CpGs in females is unknown and merits further investigation. Moreover, sex-biased DNA methylation has also been reported in non-brain samples such as placenta, peripheral blood or saliva (19,21). Thus, it is likely that sex-biased methylation of sex chromosome CpGs occurs widely in different organs or tissues in mammals.
Additionally, the present study demonstrated that only a small number of autosomal CpGs showed sex-biased methylation. Among the 8357 CpGs with differential methylation between males and females (at the q ≤ 0.05 level), only 614 CpGs (or 7.3%) were located on autosomes (Fig. 1D). This is consistent with our expectation that autosomal genes should not have a striking difference in DNA methylation between males and females. The differential methylation of the 614 autosomal CpGs between males and females is unlikely due to non-specificity of some of the Illumina 450 K microarray probes. Before performing statistical analysis, we excluded all those autosomal CpGs showing significant sex methylation differences resulting from the cross-reactive (or non-specific) probes that co-hybridize to both autosomal and sex chromosomes, as reported in a previous study (22). In the present study, 420 419 autosomal CpGs were detected by Illumina's 450 K Methylation BeadChip. The 614 differentially methylated CpGs only accounts for a very small proportion (0.1%) of the 420 419 autosomal CpGs. These results suggest that the majority of autosomal genes have similar methylation levels (and thus similar expression levels) in males and females. The small number of genes with sex-biased methylation may be involved in sexual dimorphism or phenotypic differences between males and females.
Secondly, we did observe sex-enriched expression of genes located on either autosomes or sex chromosomes. Nevertheless, the sex difference in gene expression was small. The small-scale differences in a number of genes are expected to have a large combined impact on cellular differences between males and females. As shown in Table 3, the expression of only 35 genes (9 on autosomes, 7 exclusively on the X chromosome, 14 exclusively on the Y chromosome and 5 on both X and Y chromosomes) was significantly different between males and females (at the q ≤ 0.05 level). Nineteen genes on the Y chromosome (including the 5 genes located on both X and Y chromosomes) were all up-regulated in males. The finding supports the mechanism of dosage compensation for expression of single-copy genes located on the Y chromosome. Several of these 19 genes (e.g. CYorf14, TTTY15 and TTTY14) are expressed mainly in the testis, suggesting a male-specific function in spermatogensis. Moreover, four (XIST, ZFX, HDHD1A and RPS4X) of the seven X chromosome genes (excluding the three unknown genes LOC554203, LOC100133578 and LOC100133662) were all up-regulated in females. There is evidence that about 15% of X-linked genes escape silencing by X chromosome inactivation and are expressed from both X chromosomes in female mammalians (23). The above four X chromosome genes (XIST, ZFX, HDHD1A and RPS4X) were among them; hence, their expression levels in females were higher than that in males because two alleles of the X chromosome genes were expressed in females. Among the nine autosomal genes showing differential expression between males and females, three were up-regulated but six were down-regulated in males in comparison to females. There are a number of possible causes for sex differences in autosomal gene expression. Wijchers and Festenstein (24) proposed several non-hormonal mechanisms that could explain the autosomal gene expression differences between males and females in somatic tissues. Apart from that, genetic variations and epigenetic modifications (e.g. DNA methylation-associated maternal or paternal allele imprinting) may also contribute to sex-biased autosomal gene expression. Additionally, in the present study, fewer sexual differentially expressed genes were identified in human brains compared with fruit fly or mouse somatic tissues. This may be due to (i) the sample size of human brain tissues (often not easily assessable) for the present study was moderate, thus genes with minor expression differences between men and women were not detected; and (ii) gene expression is usually tissue-specific, and the patterns of gene regulation and expression in the brain are complex.
Thirdly, the present study indicates that the correlation of DNA methylation and gene expression is complex. DNA methylation not only inhibits but also promotes gene expression. As shown in Figure 3, we obtained 585 CpG methylation–gene expression pairs (431 negatively and 154 positively correlated), and 472 (80.7%) of the 585 CpGs involved in the 585 pairs were located in gene promoter regions. The 585 CpGs were queried against the SNPnexus database (http://www.snp-nexus.org/) to see whether any of them were potentially located in transcription factor binding sites (TFBS). Nineteen (12.3%) CpGs involved in the 154 positively correlated pairs were potentially located in TFBS, while 50 (11.6%) CpGs of 431 negatively correlated pairs were potentially located in TFBS. The χ2 test did not show significant difference between the numbers of the positively and negatively correlated CpGs that are potentially located in TFBS (χ2 = 0.06, P = 0.808). Of interest, the proportion of negatively correlated CpGs in promoter regions was 86.1% (n = 371), which was significantly higher than that (65.6%) of the positively correlated CpGs (n = 101) in promoter regions (χ2 = 30.58, P = 3.2 × 10−8). This is consistent with previous findings that promoter DNA methylation tended to repress gene transcription. Several previous studies have also demonstrated a negative correlation of promoter CpG methylation and gene expression levels (25,26). These findings including ours support the hypothesis that methylation of promoter CpGs may prevent binding of transcription factors to promoter regions, thus leading to transcription inhibition. Nevertheless, the mechanism regarding the positive correlation of CpG methylation and gene expression is not clear. The influence of DNA methylation on gene transcription has been suggested to be dependent on CpG locations in gene regions (27). While methylation of CpGs in the immediate vicinity of the TSS tends to block transcription initiation, methylation of CpGs in other gene regions (e.g. gene bodies) may not affect gene transcription or even enhance transcription. For example, methylation of CpGs in repeat regions such as centromeres is important for chromosomal stability (28), thus it is favorable for gene transcription. However, further studies are warranted to assess the function of promoter CpG methylation that is positively correlated with gene expression.
Fourthly, genes with sex differences in DNA methylation and gene expression may have specific biological functions that are important for the formation of phenotypic, behavioral or cognitive differences between males and females. Gene functional annotation analysis showed that almost all the top GO terms enriched by some of the 188 genes (involved in the above 585 CpG methylation–gene expression pairs) are related to ribosomal structure and function as well as protein translation (Table 5). Moreover, functional annotation analysis was performed using 54 genes (of the 188 genes) located on sex chromosomes. A majority of the obtained GO terms in the top cluster are relevant to protein translation (see Supplementary Material, Table S4). These findings suggest that sex-biased DNA methylation may result in sex-biased expression of genes that participate in protein synthesis.
Taken together, this study examined the sex influence on methylomic and transcriptomic patterns in postmortem PFC. It provided strong evidence of sex-biased methylation and expression of genes located on either autosomes or sex chromosomes. Given that DNA methylation and gene expression are potentially tissue-specific (29,30), future studies should also examine sex-associated DNA methylation and gene expression in other brain regions and peripheral organs or tissues.
MATERIALS AND METHODS
Human postmortem PFC tissues
Autopsy brain tissue samples were obtained from the New South Wales Tissue Resource Centre (NSW TRC) at the University of Sydney. It has ethics approval from the Sydney Local Health Network and the University of Sydney. Fresh-frozen sections of Brodmann area 9 (BA9, mainly the dorsolateral PFC of the brain) were obtained from 46 European Australians (32 males and 14 females). Subjects were assessed according to criteria in the Diagnostic and Statistical Manual of Mental Disorders 4th Edition (DSM-IV) (31). Among the 46 subjects, 23 (16 males and 7 females) had alcohol use disorders (AUDs). All subjects were not affected with illicit drug abuse and dependence or major psychotic disorders such as schizophrenia and bipolar disorders. The clinical information (including race, age, postmortem intervals, brain weight, brain pH and alcohol daily use) of the 32 male and 14 female samples is presented in Table 1. The male and female samples were not significantly different in subject age, postmortem intervals, brain pH and alcohol daily use (Pt-test > 0.05). However, the mean brain weight (1457.9 g) of males was significantly heavier than that (1254.3 g) of females (Pt-test = 3.3 × 10−6).
Genome-wide DNA methylation assay
Genomic DNA was extracted from postmortem PFC tissues from 32 males and 14 females using the QIAamp DNA Micro Kit (QIAGEN, Valencia, CA, USA). Genomic DNA was checked for quality by 0.8% agarose gel and quantified using a NanoDrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Five hundred nanograms of genomic DNA were treated with bisulfite reagents included in the EZ-96 DNA methylation kit (Zymo Research, Orange, CA, USA) according to the manufacturer's protocol. Genome-wide DNA methylation levels were examined using the Illumina Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA, USA), following the Illumina Infinium HD Methylation protocol (32). This BeadChip interrogates 485 577 CpG sites per sample at single-nucleotide resolution. It covers 99% of RefSeq genes, with 41% of CpG sites in promoter regions [from 1500 bp upstream to 200 bp downstream of the TSS, including all or part of the 5′ UTR and/or the first exon], 31% in gene bodies, 3% in 3′ UTRs and 25% in intergenic regions (32). GenomeStudio software V2011.1 (Methylation Module V1.9.0) from Illumina was used to generate raw β-values for each CpG site, with β-values ranging from 0 (0% methylation or completely unmethylated) to 1 (100% methylation or completely methylated). Methylation data were normalized using data from internal control probes that were designed specifically for Illumina's HumanMethylation450 BeadChip. Ch probes (non-CpG probes), CpG-SNP probes and probes with signal intensities indistinguishable from the background noise (detection P-value > 0.05) were excluded. Additionally, CpGs demonstrating significant sex differences in methylation levels as the result of co-hybridization of cross-reactive probes to both autosomal and sex chromosomes, as described in a previous study (22), were also excluded. The R package sva (33) was applied to compensate for batch effect (due to different chips). Principal variance component analysis (PVCA) (34) was then used to calculate the proportion of total variability explained by batch effect. It was confirmed that batch effect was removed effectively by sva from normalized methyaltion data (Supplementary Material, Fig. S1A). After the above data quality control process, a total of 430 011 CpGs with high-quality methylation data remained for further analysis. Our methyaltion data have been deposited in the NCBI GEO database (Accession Number: GSE49393).
Genome-wide gene expression assay
Total RNA was extracted from postmortem PFC tissue from 32 males and 14 females using the miRNeasy Mini Kit (QIAGEN). Total RNA was quantified by NanoDrop 8000 spectrophotometer (Thermo Scientific). The RNA integrity number (RIN) was determined using the RNA 6000 Nano Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The RIN ranged from 5 to 7 for the 46 RNA samples. Genome-wide expression levels of genes were quantified using the Illumina HumanHT-12 v4 Expression BeadChip (Illumina Inc., San Diego, CA, USA). Each array targets more than 31 000 annotated genes with more than 47 000 probes derived from the National Center for Biotechnology Information Reference Sequence (NCBI) RefSeq Release 38 (November 7, 2009) and other sources. Probe intensity and gene expression data were analyzed using Illumina's GenomeStudio software V2011.1 (Gene Expression Module V1.9.0). Low-level analysis of microarray data was performed in R 2.15.1 using the Bioconductor package lumi (35). The variance stabilizing transformation method (36) and the robust spline normalization (26) method were applied to all arrays. After normalization, annotated genes with intensities indistinguishable from background noise (detection P-value > 0.05) in more than half of the RNA samples were removed. The R package sva (33) was applied to compensate for batch effect. Same as above, PVCA (34) was used to calculate the proportion of total variability explained by batch effect. It was confirmed that batch effect was removed effectively by sva from normalized gene expression data (Supplementary Material, Fig. S1B). After the above data quality control process, a total of 14 851 genes with high-quality expression data remained for the following statistical analysis. Our expression data have been deposited in the NCBI GEO database (Accession Number: GSE49376).
Statistical analysis
Statistical analysis was implemented using the R 2.15.1. To analyze sex-biased methylome in the PFC, the β-values of 430 011 CpGs were compared between males and females using multiple linear regression analysis, with adjustment for covariates age, brain weight, brain pH, PMI and AUDs. Genome-wide gene expression differences between males and females were also examined using multiple linear regression analysis, with adjustment for the above covariates. The availability of genome-wide DNA methylation and gene expression data enabled us to perform integrative analysis of the correlation of CpG methylation and gene expression. To examine the correlation patterns of CpG methylation and gene expression levels, residuals in multiple linear regression models with adjustment for covariates were applied in the Pearson correlation analysis, as described previously (37). To address the multiple testing issue, the q-value was computed for each nominal P-value by controlling the false discovery rate (FDR) at 0.05 using the R package qvalue (38). The q-value method was applied to DNA methylation, gene expression and CpG methylation–gene expression correlation analyses.
Gene set functional enrichment analysis
The above CpG methylation–gene expression correlation analysis generated a list of genes, of which the CpG methylation levels (significantly different between males and females) and the gene expression levels (significantly different between males and females) were significantly correlated. To explore the potential biological function of these genes, they were uploaded to DAVID 6.7 (39) for functional annotation clustering analysis based on GO terms.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by the National Institutes of Health (NIH) grants [K99/R00 DA022891 (H.Z.) and P50 AA12870 (J.G. and H.Z.)].
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the Australian Brain Donor Programs New South Wales Tissue Resource Centre for providing postmortem brain tissue samples for this study. The centre is supported by the University of Sydney, the National Health and Medical Research Council of Australia, and the National Institute on Alcohol Abuse and Alcoholism. We also thank the deceased subjects' next of kin for providing informed written consent for the studies.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gur R.C., Turetsky B.I., Matsui M., Yan M., Bilker W., Hughett P., Gur R.E. Sex differences in brain gray and white matter in healthy young adults: correlations with cognitive performance. J. Neurosci. 1999;19:4065–4072. doi: 10.1523/JNEUROSCI.19-10-04065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein J.M., Jerram M., Poldrack R., Anagnoson R., Breiter H.C., Makris N., Goodman J.M., Tsuang M.T., Seidman L.J. Sex differences in prefrontal cortical brain activity during fMRI of auditory verbal working memory. Neuropsychology. 2005;19:509–519. doi: 10.1037/0894-4105.19.4.509. [DOI] [PubMed] [Google Scholar]
- 3.Tietjen G.E., Peterlin B.L. Childhood abuse and migraine: epidemiology, sex differences, and potential mechanisms. Headache. 2011;51:869–879. doi: 10.1111/j.1526-4610.2011.01906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qureshi I.A., Mehler M.F. Genetic and epigenetic underpinnings of sex differences in the brain and in neurological and psychiatric disease susceptibility. Prog. Brain Res. 2010;186:77–95. doi: 10.1016/B978-0-444-53630-3.00006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman R.E., MacLusky N.J., Sarmiento Y., Frankfurt M., Gordon M., Luine V.N. Sexually dimorphic effects of prenatal stress on cognition, hormonal responses, and central neurotransmitters. Endocrinology. 2004;145:3778–3787. doi: 10.1210/en.2003-1759. [DOI] [PubMed] [Google Scholar]
- 6.Teicher M.H., Dumont N.L., Ito Y., Vaituzis C., Giedd J.N., Andersen S.L. Childhood neglect is associated with reduced corpus callosum area. Biol. Psychiatry. 2004;56:80–85. doi: 10.1016/j.biopsych.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Cooke B.M., Tabibnia G., Breedlove S.M. A brain sexual dimorphism controlled by adult circulating androgens. Proc. Natl Acad. Sci. USA. 1999;96:7538–7540. doi: 10.1073/pnas.96.13.7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenz K.M., McCarthy M.M. Organized for sex-steroid hormones and the developing hypothalamus. Eur. J. Neurosci. 2010;32: 2096–2104. doi: 10.1111/j.1460-9568.2010.07511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranz J.M., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 10.Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vawter M.P., Evans S., Choudary P., Tomita H., Meador-Woodruff J., Molnar M., Li J., Lopez J.F., Myers R., Cox D., et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weickert C.S., Elashoff M., Richards A.B., Sinclair D., Bahn S., Paabo S., Khaitovich P., Webster M.J. Transcriptome analysis of male–female differences in prefrontal cortical development. Mol. Psychiatry. 2009;14:558–561. doi: 10.1038/mp.2009.5. [DOI] [PubMed] [Google Scholar]
- 14.Avner P., Heard E. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 15.Bartolomei M.S. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson K.C., Prescott C.A., Kendler K.S. Sex differences in the genetic and environmental influences on the development of antisocial behavior. Dev. Psychopathol. 2002;14:395–416. doi: 10.1017/s0954579402002110. [DOI] [PubMed] [Google Scholar]
- 17.Ruixing Y., Jinzhen W., Shangling P., Weixiong L., Dezhai Y., Yuming C. Sex differences in environmental and genetic factors for hypertension. Am. J. Med. 2008;121:811–819. doi: 10.1016/j.amjmed.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 18.Rutten B.P., Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr. Bull. 2009;35:1045–1056. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Morgan M., Hutchison K., Calhoun V.D. A study of the influence of sex on genome wide methylation. PloS One. 2010;5:e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Maarri O., Becker T., Junen J., Manzoor S.S., Diaz-Lacava A., Schwaab R., Wienker T., Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum. Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 21.Fuke C., Shimabukuro M., Petronis A., Sugimoto J., Oda T., Miura K., Miyazaki T., Ogura C., Okazaki Y., Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 24.Wijchers P.J., Festenstein R.J. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27: 132–140. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J., et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell J.T., Pai A.A., Pickrell J.K., Gaffney D.J., Pique-Regi R., Degner J.F., Gilad Y., Pritchard J.K. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 28.Moarefi A.H., Chedin F. ICF syndrome mutations cause a broad spectrum of biochemical defects in DNMT3B-mediated de novo DNA methylation. J. Mol. Biol. 2011;409:758–772. doi: 10.1016/j.jmb.2011.04.050. [DOI] [PubMed] [Google Scholar]
- 29.Feng J., Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int. Rev. Neurobiol. 2009;89: 67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 30.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y., et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th edn. Washington, DC: 1994. American Psychiatric Association. [Google Scholar]
- 32.Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 33.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 34.Bremner J.D., Narayan M., Staib L.H., Southwick S.M., McGlashan T., Charney D.S. Neural correlates of memories of childhood sexual abuse in women with and without posttraumatic stress disorder. Am. J. Psychiatry. 1999;156:1787–1795. doi: 10.1176/ajp.156.11.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 36.Lin S.M., Du P., Huber W., Kibbe W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Numata S., Ye T., Hyde T.M., Guitart-Navarro X., Tao R., Wininger M., Colantuoni C., Weinberger D.R., Kleinman J.E., Lipska B.K. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.