Abstract
Understanding the role of epigenetic modifications, e.g. DNA methylation, in the process of aging requires the characterization of methylation patterns in large cohorts. We analysed >480 000 CpG sites using Infinium HumanMethylation450 BeadChip (Illumina) in whole blood DNA of 965 participants of a population-based cohort study aged between 50 and 75 years. In an exploratory analysis in 400 individuals, 200 CpG sites with the highest Spearman correlation coefficients for the association between methylation and age were identified. Of these 200 CpGs, 162 were significantly associated with age, which was verified in an independent cohort of 498 individuals using mixed linear regression models adjusted for gender, smoking behaviour, age-related diseases and random batch effect and corrected for multiple testing by Bonferroni. In another independent cohort of 67 individuals without history of major age-related diseases and with a follow-up of 8 years, we observed a gain in methylation at 96% (52%, significant) of the positively age-associated CpGs and a loss at all (89%, significant) of the negatively age-associated CpGs in each individual while getting 8 years older. A regression model for age prediction based on 17 CpGs as predicting variables explained 71% of the variance in age with an average accuracy of 2.6 years. In comparison with cord blood samples obtained from the Ulm Birth Cohort Study, we observed a more than 2-fold change in mean methylation levels from birth to older age at 86 CpGs. We were able to identify 65 novel CpG sites with significant association of methylation with age.
INTRODUCTION
Alterations in DNA methylation are the most commonly studied epigenetic modification in humans and have been associated with age-related diseases such as diabetes mellitus (type 2) (1), cardiovascular disease (2,3) and cancer (4,5). The relationship of methylation with the aging process itself as a ‘key for understanding the molecular mechanisms of normal and premature ageing, and its applications such as predicting the age of an individual from a forensic sample’ has been highlighted, recently (6). Even though DNA methylation is known to vary between different tissues and cells, aging as a general process affecting all cells might be associated with similar methylation pattern across multiple human tissues. Horvath et al. found robust age-related co-methylation in blood and brain tissue and even concluded that blood, the most widely available and assessable DNA source in large-scale epidemiological studies, is a promising surrogate for brain tissue when studying the effects of age on DNA methylation profiles (7).
Epigenome-wide scans (Infinium HumanMethylation27 BeadChip) have identified age-associated changes in DNA methylation at more than one hundred CpG sites for different human tissues including whole blood/leukocytes (8–10), saliva (11) and skin (12). In healthy adults, Christensen et al. (13) found CpG island-dependent correlations between age and methylation across multiple tissues. Positive correlation with age was found at loci in CpG islands, which were mostly hypomethylated, whereas negative correlation was observed at non-island loci, which were mostly hypermethylated. Most previous studies investigating the association between age and CpG DNA methylation were based on the Infinium HumanMethylation27 BeadChip (Illumina Inc., San Diego, CA, USA), which represents ∼27 000 CpG sites. Recent studies using the Infinum HumanMethylation450 BeadChip (Illumina Inc.) identified 3 regions mapped to the genes ELOVL2, FHL2 and PENK (14) and 70 CpG sites mapped to 58 genes (15), whose methylation status was associated with age. In previous DNA methylation studies based on the Infinum HumanMethylation450 BeadChip, the number of newly identified age-associated CpG sites was limited because of the limited sample size either during screening [64 individuals (14)] or during validation [174 individuals (15)]. Furthermore, these previous analyses were restricted to cross-sectional designs. To enable reliable distinction of aging and birth cohort effects, additional longitudinal analyses are needed.
We aimed to confirm previously found age-associated CpG sites and to identify and validate further age-associated CpG sites in blood DNA samples in a large study cohort using the Infinum HumanMethylation450 BeadChip in both cross-sectional and longitudinal analyses.
RESULTS
Exploratory analysis—epigenome-wide discovery
In this first phase (see sections Materials and methods, and Design of the study), the Spearman correlation coefficient between methylation levels quantified by beta-value and age was calculated for 480 675 CpG sites in 400 individuals. A histogram of all correlation coefficients is presented in Figure 1. The coefficients were normally distributed with a mean of zero. The left window shows an enlargement of the distribution of correlation coefficients below −0.25. In the right window, an enlargement of the distribution of correlation coefficients of >0.25 is presented. Two-hundred CpG sites with the highest absolute value of the correlation coefficient were selected for further analysis. Of these 200 CpG sites, there were one CpG site with an absolute value of the correlation coefficient of >0.6, three CpG sites with values between 0.5 and 0.6, 15 CpG sites with values between 0.4 and 0.5, 146 CpG sites with values between 0.3 and 0.4 and 35 CpG sites with values between 0.291 and 0.3.
Figure 1.
Distribution of Spearman correlation coefficients between age and DNA methylation levels at 480 675 CpG sites calculated in phase 1, ‘Exploratory analysis’. The distribution of correlation coefficients is shown on a larger scale in the left window for correlation coefficients below −0.25 and in the right window for correlation coefficients of >0.25.
Confirmatory analysis I (cross-sectional)
In this second phase, the association of methylation with age at 200 CpG sites, which were selected within the exploratory analysis, was tested for significance using mixed linear regression models adjusting for gender, smoking behaviour, diabetes mellitus, cardiovascular disease, stroke, cancer and random batch effect in 498 individuals.
We first verified the normal distribution of methylation levels at each of the 200 top-ranked CpG sites. Examples of distribution are shown in Supplementary Material, Figure S1. The methylation levels were normally distributed at 75 CpG sites and 42 CpGs for the samples with all participants and with participants free of major age-related diseases, respectively. At 90 and 32 CpG sites, outliers had to be excluded (in most cases <4). Lambda values of the most appropriate transformation are shown for the respective CpG sites in Appendix.
In individuals with single-nucleotide polymorphism (SNP), the methylation levels at the respective CpG site will be influenced (16). In Supplementary Material, Figure S2, the correlation between methylation and age is shown for a CpG site, which is known to be SNP associated. A few individuals show quite different methylation levels, but owing to the applied distribution diagnostics confirming normality before calculating linear regression, these individuals were excluded from further analyses. This is also a good example demonstrating the influence of outliers on linear regression and Pearson correlation coefficient. The Pearson and Spearman correlation coefficients were 0.18 and 0.43 before and 0.43 and 0.45 after exclusion of outliers, respectively (Supplementary Material, Fig. S2A and B). Of all 188 significantly age-associated CpG sites, 26 were SNP associated. None of these 26 CpGs was within the 20 most significantly age-associated CpG sites. A list of theses SNP-associated CpG sites is presented in Supplementary Material, Table S1. As the appropriateness of using of SNP-associated CpG sites is still under discussion, we excluded these 26 CpG sites from further analyses.
For all 200 top-ranked CpG sites, we calculated linear regression models using original methylation levels (columns 2–5 in the table in Appendix). If the methylation level was not normally distributed, we calculated additionally linear regression models using transformed methylation levels (columns 6–10 in the table in Appendix) and then the P-values respectively for transformed values were further considered. After excluding the 26 SNP-associated CpG sites, methylation levels at 162 CpG sites were significantly associated with age with a P-value for the regression coefficient of <2.5 × 10−4 corresponding to the Bonferroni-corrected α-level for 200 tests (Appendix). At all these significant CpG sites, the regression coefficient in the linear model was in the same direction as the Spearman correlation coefficient calculated in the ‘Exploratory analysis’. For 148 CpG sites, the P-value for the regression coefficient between methylation and age was also <10−7, the Bonferroni-corrected α-level testing 480 675 CpG sites.
In Appendix, the ten most significant age-associated CpG sites are marked in grey and CpG sites at the same gene locus are marked in light grey. Several age-associated CpG sites were located on the same gene, e.g. five CpG sites were in the coding region of the Kruppel-like family of transcription factors (KLF14). Three of the four most significant CpG sites were within the coding region of fatty acid elongase 2 (ELOVL2). Among the 10 most significant CpG sites were loci within the coding regions of the proteins four and a half LIM domains 2 (FHL2), OTU domain containing 7 (OTUD7A), solute carrier family 12 (potassium/chloride transporter) member 5 (SLC12A5), zyg-11 homolog A (ZYG11A) and coiled-coil domain containing 102B (CCDC102B). Among the 10 most significant CpG sites were also two sites, which are so far not associated with gene regions (cg08234504 and cg17110586). Of the 162 CpG sites significantly associated with age, 43 sites (26%) showed a negative correlation with age.
After restricting the study sample to 326 participants without history of major age-related diseases like diabetes mellitus, cardiovascular disease, stroke and cancer at baseline, 155 of the 162 CpG sites still showed a significant association with age. Of those CpG sites no longer statistically significant in the restricted sample, 3 had been among the 10 least significant sites in the unrestricted sample. Therefore, the non-significance of these CpG sites might be simply explained by the smaller sample size (326 versus 498). Four CpG sites with P-values of <10−8 were not significant in the population of participants without history of major age-related diseases. These CpG sites were located within coding regions of the protein zinc finger, FYVE domain containing 21 (ZFYVE21), cyclin G-associated kinase (GAK) and chemokine binding protein 2 (CCBP2). These genes play a role in cell adhesion and inflammation (17), and the decreased methylation with age found at these CpG sites might reflect disease-related changes in methylation. To exclude confounding by such disease-related changes in methylation, we restricted the further analysis to the population of 326 participants without major age-related diseases and to the 155 CpG sites identified with the restricted sample.
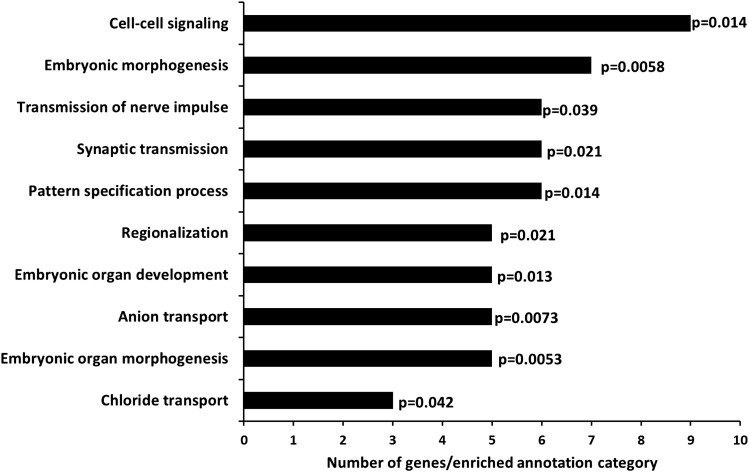
Of the 155 CpG sites, 29 were not located in a known gene region. The remaining 126 mapped to 100 gene regions. For these genes, functional annotation enrichment analysis was performed and the significant annotation categories are presented in Figure 2. Enriched annotations were related to cell communication and developmental processes. Seventy-two genes could not be classified into functional groups among them the most significant CpG sites, ELOVL2, FHL2, ZYG11A and NEFM (neurofilament, medium polypeptide) A special feature of these gene regions is that not only one age-associated CpG site was located in these regions but two or three. Although the correlation between methylation at CpG sites within the same gene locus was mostly strong with correlation coefficients on an average of 0.64 and varying between 0.26 and 0.86, the location of several CpG sites within the same gene region might enhance the relevance of the identified region (18).
Figure 2.
Gene ontology enrichment for genes with significantly age-related differentially methylation.
Confirmatory analysis II (longitudinal)
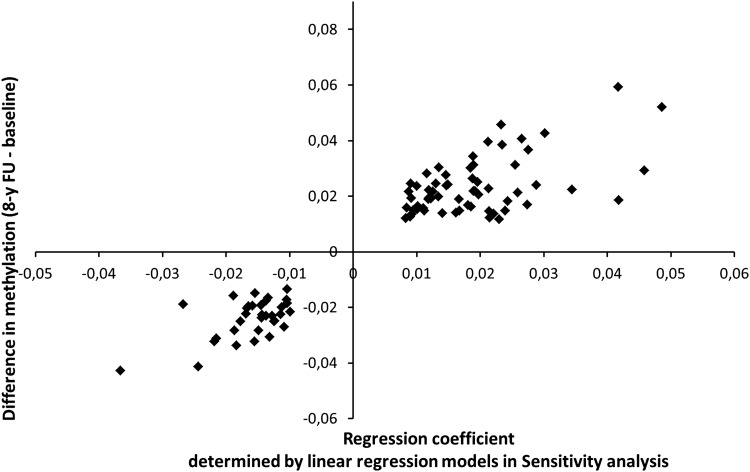
In the third phase, in an independent population of 67 individuals without major age-related diseases, the difference between the methylation level at 8-year follow-up and baseline was calculated for the 155 CpG sites that were significantly associated with age in the restricted sample in confirmatory analysis I. The absolute values of the mean differences ranged from 0.0005 to 0.059. Mean differences, which were below the estimated mean batch effect of 0.0115, were excluded. Among 78 of the remaining 94 CpG sites, the methylation level changed significantly within 8 years of life even after Bonferroni correction (P-value < 0.0002). Methylation increased at 61 CpG sites. At all these CpG sites, a positive regression coefficient for age had also been observed in the linear regression models of the ‘confirmatory analysis I, sensitivity analysis’. At 33 CpG sites, the methylation level decreased within 8 years, and at all these CpG sites, negative regression coefficients had also been observed in the ‘confirmatory analysis I, sensitivity analysis’. This relationship is presented in Figure 3, where the mean difference over all individuals are presented in relation to the respective regression coefficient for age determined in the mixed models calculated in the restricted sample in ‘confirmatory analysis I, sensitivity analysis’. In this Figure, we compared the results obtained in two independent samples, the restricted sample of 326 individuals free of age-related diseases, in which the regression coefficients were determined (‘confirmatory analysis I, sensitivity analysis’), and the cohort of 67 individuals, in which the mean differences between methylation levels at the 8-year follow-up and at baseline were calculated. Furthermore, this Figure shows a clear dose–response relationship between these regression coefficients and the mean difference in methylation over 8 years (Fig. 3).
Figure 3.
Mean difference in DNA methylation between 8-year follow-up (8-y FU) and baseline determined in the longitudinal analysis (‘confirmatory analysis II’) in relation to the regression coefficient for age determined by mixed models in the cross-sectional (‘confirmatory analysis I, sensitivity analysis’) for 94 significant age-associated CpG sites.
Location of significant CpG sites in relation to CpG islands
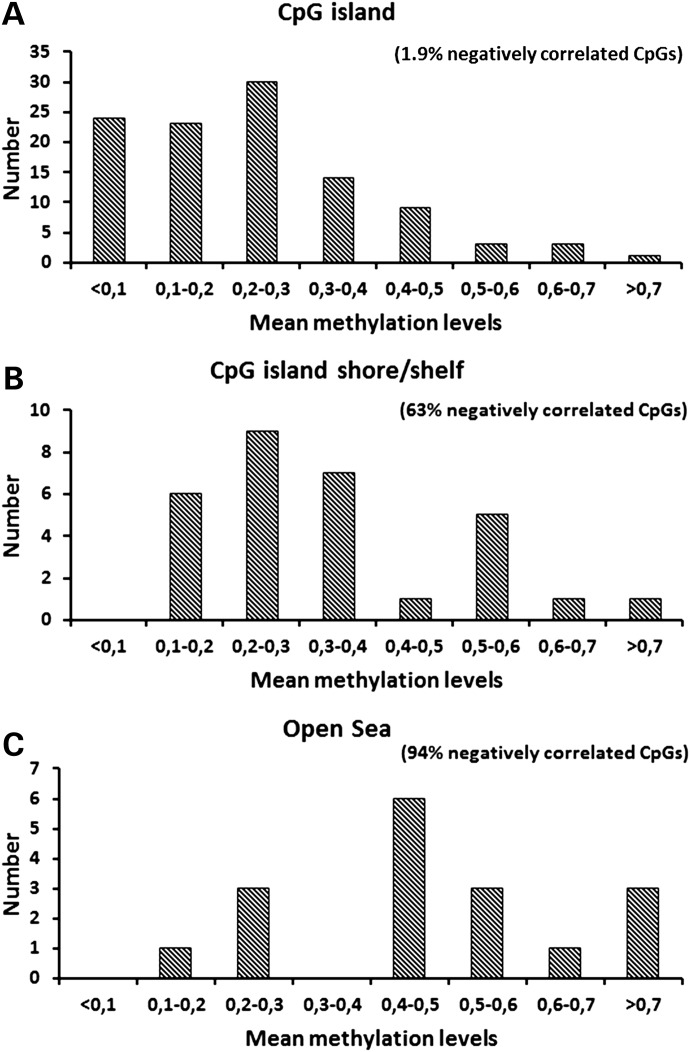
Of the 155 significantly age-associated CpG sites, 70% were located in CpG islands, 19% in CpG island shores or shelves and 11% outside CpG islands (‘open sea’), whereas of all CpG sites on the Infinum HumanMethylation450 BeadChip, 31% were located in CpG islands, 33% in island shores or shelves and 36% in ‘open sea’ (19). In Figure 4A–C, distributions of mean methylation levels at CpG sites located within CpG islands and CpG island shores/shelves and ‘open sea’ are shown, respectively. Within CpG islands, most CpG sites were hypomethylated and methylation was positively associated with age. Only 1.9% of the correlation coefficients were negative. Within CpG island shores/shelves, we found more hypermethylated CpGs than within CpG islands, and at most of these sites, methylation was negatively associated with age (63%). At CpG sites outside CpG islands, we observed more frequently higher methylation levels and mostly a negative association of methylation with age (94%).
Figure 4.
Distribution of mean methylation level for age-associated CpG sites located in CpG islands (A), in CpG shores or shelves (B) or ‘open sea’ (C). In parentheses, the proportion of CpG sites with methylation negatively associated with age is presented.
Figure 5 shows the relation between mean methylation level and direction of association with age. In general, hypomethylated CpG sites were positively associated with age. But there are also several hypermethylated CpG sites, which were positively associated with age. An example is presented in Supplementary Material, Figure S1A. Negative association with age was found more commonly at hypermethylated CpGs but at hypomethylated CpG sites as well, e.g. Supplementary Material, Figure S1D.
Figure 5.
Spearman correlation coefficient in relation to mean methylation level for the significant age-associated CpG sites.
Prediction of age
The determination of predictors of age included a training and a validation step. During training step, 17 CpG sites were included in a linear regression model explaining 78% of the variance of age. Applying this model to the validation set, the explained variance was 71%. Ten of these 17 CpG sites were significantly associated with age regarding a P-value of <0.05. The accuracy of prediction estimated by the average absolute difference between the predicted and the observed age was 2.6 years. The predicted values were highly correlated with the observed age (r = 0.84). Including into the model only cg16867657, the CpG site most significantly associated with age, mapping to the gene region ELOVL2, explained already 47% of the variance of age with an accuracy of 3.7 years. The other 9 CpG sites, which were significant predictors of age in the validation data set, mapped to the gene regions zyg-11 homolog A (ZYG11A), leucine-rich repeat containing 23 (LRRC23), chromobox homolog 4 (CBX4), Scm-like with four mbt domains 1 (SFMBT1), NACHT and WD repeat domain containing 1 (NWD1) and G protein-coupled receptor 62 (GPR62). Three of these age-predicting CpG sites were not located in a known gene region (cg05308819, cg08097417 and cg10804656).
We validated the model in a second cohort, the longitudinal cohort. For age at baseline, the explained variance, accuracy of prediction and correlation between predicted and observed values were 76%, 2.8years and 0.88, respectively. For 8-year follow-up results, explained variance, accuracy of prediction and correlation between predicted and observed value were 62%, 3.5 years and 0.81, respectively. The difference between the predicted age at the 8-year follow-up and at baseline was 8.28 ± 2.52, which is very close to the real age difference of 8.39 ± 0.46 years.
Comparing methylation levels in older adults and in newborn
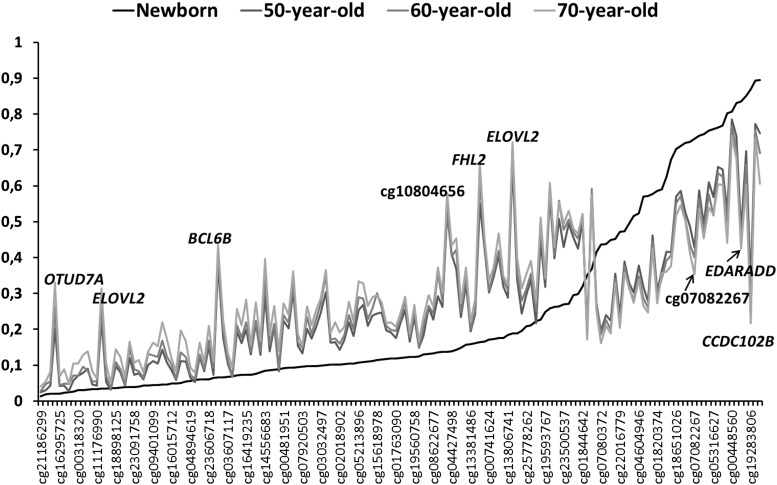
We calculated the mean methylation levels in 50-, 60- and 70-year-old adults of the ESTHER study and in newborn of the Ulm Birth Cohort (UBC) Study for the 155 significantly age-associated CpG sites (Fig. 6). The CpG sites presented at the x-axis were sorted by increasing amount of mean methylation levels in newborn ranging from hypo- (on the left) to hyper-methylation (on the right). The difference in methylation between newborn and adults was multiple times larger than the difference between 50- and 70-year-old adults. An increase in methylation from birth to adulthood was observed at all CpG sites showing positive regression coefficients in confirmatory analysis I (Appendix), whereas a decrease was observed at all CpG sites showing negative regression coefficients. Interestingly, there seems to be a cut point at ∼0.35. At all CpG sites with methylation levels below that cut point in newborn, the methylation increased with aging, whereas at CpG sites with methylation levels above that point in newborn, the methylation decreased with aging.
Figure 6.
Methylation levels measured in newborn and in 50-, 60- and 70-year-old adults sorted by increasing methylation levels at birth. For CpG sites with large differences between the methylation levels in newborn and older adults, the gene location was provided.
CpG sites presenting with peaks in Figure 6 stand for large differences in methylation levels between adults and newborn. For several of them, the respective gene location was presented. Among them were the most significant CpGs found in older adults ELOVL2, FHL2 and OTUD7A, and cg10804656. ELOVL2 and cg10804656 were also significant predictors of age in our above-introduced age-predicting model. For the two CpG sites mapped to the gene ELOV2, the mean methylation level was 0.03 and 0.19 in newborn and 0.31 and 0.72 in 70-year-old participants. Although the absolute difference for one CpG site seems to be small with 0.28, at this CpG sites the methylation level in 70-year-old participants was on average 9-fold higher than that in newborn, whereas for other CpG site with an absolute difference of 0.53 the methylation level was nearly 4-fold higher in adults than in newborn. Overall, at 86 and 132 of the 155 age-associated CpGs, there was 2-fold or 1.5-fold variation between methylation levels in 70-year-old individuals and levels in newborn, respectively.
DISCUSSION
Our large epigenome-wide study with 965 individuals identified 155 significantly age-associated CpG sites mapping to 100 genes. At most of these CpG sites (77%), methylation increased with age and these CpG sites were mainly hypomethylated and within CpG islands. To our knowledge, this is the first epigenome-wide study validating the cross-sectionally identified age-associated CpG sites in a longitudinal analysis. In an independent cohort of 67 individuals, we confirmed that at most of the positively age-associated CpG sites, methylation increased significantly within 8 years. In contrast, at most of the negative age-associated CpG sites, methylation decreased significantly in each individual while getting 8 years older.
Longitudinal changes in methylation in blood DNA have been previously observed in elderly men (20) and in children within the first two years of life (21). In the study of men with a mean age of 72 years, methylation at 9 gene regions was explored (20). Using the Infinum HumanMethylation27 BeadChip, Wang et al. identified 159 CpG sites in 59 male and 149 CpG sites in 46 female Afro-American children, of those 98 CpGs were common in both genders (21).
The function of methylation of CpG sites is determined by its location (22). It is known that the methylation at CpG-island (CGI) transcription start sites (TSS) is associated with long-term silencing of genes (23). Of the presented significantly age-associated CpG sites, 29% could be linked to CGI-TSS and at all of them the methylation was positively associated with age. In contrast to the association in promoter regions, methylation in the gene body is positively correlated with gene expression (23). Of the 155 age-associated CpG sites, 22% were located in gene body, at 38% of them methylation was negatively associated with age and 56% of them were located in CpG islands. At non-CGI-TSS, the association between methylation and gene expression is less clear (23).
The ten CpG sites most significantly associated with age in our study were mapped to genes ELOVL2, FHL2, ZYG11A, SLC12A5, OTUD7A and CCDC102B. Within the gene regions of ELOVL2, FHL2 and ZYG11A, even further one or two significant CpG sites were located, which might enhance the significance of the identified gene region (18). The gene regions ELOVL2 and FHL2 were also identified by the two previous 450K studies in blood DNA (14,15). At all of these CpG sites, the direction of the association between methylation and age was the same in these previous and in our studies. In the study of Garagnani et al. (14), the screening analysis was applied only to 64 individuals therefore limiting the number of CpG sites that could be detected. We identified 37 CpG sites, which were found in the study of Hannum et al. (15) as well, 25 CpGs were found only by Hannum et al. and 118 CpG sites were detected in our study but not by Hannum et al. This apparent discrepancy might be explained by the different statistical analysis applied in each study. In the study of Hannum et al., CpGs were selected, which jointly best predicted age. As a consequence, CpG sites strongly associated with age might not have been included in the model because they did not contribute to the prediction of age independently from other CpG sites already included. Another recent study compared differentially methylated regions (DMR) between a newborn and a centenarian using whole-genome bisulphite sequencing with DMRs between 19 newborn and 19 nonagenarian using the Infinium HumanMethylation450 BeadChip (24). Only 4 of these 3205 CpG sites identified with both methods in this study were found to be significantly associated with age in our study. Although the CpG sites were different, eight of our age-associated CpGs mapped to the same gene regions reported to be differentially methylated in newborn and adults by Heyn et al. (24). In comparison with previous studies using the Infinum HumanMethylation27 BeadChip, 3 of the 131 significantly age-associated CpG sites identified in T-cells and monocytes (9) and 8 of 490 CpGs identified in whole blood (10) were also found in our study showing the same direction of association between methylation and age. On the other hand, none of the CpG sites identified in dermal tissue and blood by Koch et al. proposed for age prediction (12) was significantly age-associated in our study. Considering the provided CpG sites of these previous studies in blood, we were able to identify 107 new age-associated CpG sites. Of them, 16 are not located in a known gene region. The remaining 91 CpG sites mapped to 72 gene regions. The methylation at 23 of them has been previously associated with age although the CpG sites were different. In summary, we identified 65 novel age-associated CpG sites, which were not previously identified by these preceding studies.
In comparison with other tissues, 6 of the 88 CpG sites identified in saliva (11) were also found in our study showing the same direction of association between methylation and age. Among them was a CpG site mapping to NEFM (NEF3) a biomarker for neuronal damage in brain tissue (17,25). In comparison with brain tissue [data thankfully derived from Singleton A and colleagues (26)], we found six CpG sites and four genes showing differentially methylation with age of the same direction in brain and in blood. This is all the more amazing as only 14 of our age-associated CpG sites are present on 27K Infinium array and supports the results of Horvath et al. (7), who reported robust age-related co-methylation in blood and brain tissue. The limited agreement with previous results based on the 27K Infinium array can be explained by the fact that most age-associated CpGs identified using the 450K array in our study (92%) are loci only present on the 450K but not on the 27K platform although the 450K array includes >90% of the content contained on the 27K array. With the 450K array, we were able to identify many new age-associated CpG sites not known before.
All of the previous studies analysing the association of methylation with age were based on study cohorts with a larger range of age, mostly ranging from birth to 80 or 100 years in comparison with our study where the age range was from 50 to 75 years. The wider age range explains why correlation coefficients and differences in methylation were higher in previous studies. This is clearly shown in Figure 6 as well. However, the estimated differences in methylation per increase in age are similar. While finalizing this manuscript, a study in 421 persons ranging in age from 14 to 94 years was published, in which 137 993 CpG sites were associated with cross-sectional age (27).
The 155 age-associated CpG sites mapped to 100 gene regions. Using a list of these genes for functional enrichment analysis, we found enrichment for genes related to cell communication, embryonic morphogenesis and organ development (28). An epigenetic regulation at these loci during the process of aging is quite conceivable. However, most of the genes were not contained in the gene ontology analysis indicating that many genes could not grouped together according to their functional role. Among them are several of the most significant age-associated CpG sites mapping to the genes ELOVL2, FHL2, ZYG11A and NEFM. ELOVL2 (fatty acid elongase 2), belonging to a gene family encoding enzymes (elongases), which are believed to play a role in elongation of very long chain fatty acids and in lipid metabolism (29), FHL2 (four and a half LIM domains 2), encodes a protein that participates in various cellular processes. LIM domains are protein-interaction domains and interact with ∼50 different proteins. Many of these proteins regulate cell proliferation, apoptosis and gene expression (30). ZYG11A [Protein zyg-11 homolog A (Caenorhabditis elegans)] is thought to act as target recruitment subunit in an E3 ubiquitin ligase complex (17). E3 ubiquitin ligase targets specific protein substrates for degradation by the proteasome. NEFM (neurofilament, medium polypeptide) encodes medium neurofilament protein, which is synthesized in neuronal soma, involved in establishing the three-dimensional array of the axoplasm (25) and considered a biomarker for neuronal damage (17). Five of the 155 CpG sites that were significantly associated with age were within the gene encoding a member of the Kruppel-like family of transcription factors (KLF14), which has been linked to diabetes mellitus (31) and which has shown to be a master regulator controlling genes, which are linked to obesity, cholesterol, insulin and glucose levels (32). Further research on the functional role of DNA methylation in this context would be of utmost interest.
The predictability of age by epigenetic markers like DNA methylation at age-associated CpG sites has been previously demonstrated for several tissues (11,12,15). Using 17 CpG sites for prediction, we achieved an explained variance of 71%, which was comparable with previous studies investigating CpG sites in saliva [76%, (11)] and in a mix of different tissues [74%, (12)] with the 27K array. The correlation coefficient between predicted and observed age reported in a recent study analysing CpG sites in whole blood using the 450K array was 0.9 and thus higher than that in our study with 0.84. This high correlation had been achieved by including a much higher number of CpG sites (N = 70) into the model (15). Concerning the predictive accuracy for age, our model (2.6 years) was compared favourably with all previous studies, where the accuracy varied between 4.9 (15) and 11 years (12). Furthermore, applying the prognostic model based on the 17 CpG sites to the longitudinal cohort, the difference in age between baseline and the 8-year follow-up could be predicted very precisely with 8.28 ± 2.52 years in comparison to the real age difference of 8.39 ± 0.46 years. This high predictability might be partly explained by the design of the model containing CpG sites not present on the 27K platform, especially the locus cg16867657 within the gene ELOVL2 explaining alone already 47% of the variance of age.
Our study has specific strengths and limitations. Strengths include the large study cohort facilitating the detection of many so far unknown age-associated CpG sites, cross-sectional and longitudinal validation in independent cohorts and the comparative analyses in subgroups of study participants with and without history of major age-related diseases such as diabetes mellitus, stroke, cardiovascular disease and cancer. Limitations include that a substantial proportion of the >450 000 DNA methylation probes on the 450K array are not aligned to unique, unambiguous loci in the human genome (33).
We used whole blood for DNA extraction resulting in DNA methylation analysis across multiple cell types of leukocytes. Because the analysis is based on stored blood samples from the years 2000 and 2008 and back then the leukocyte cell subtype counts were not documented, adjustment for leukocyte subtype distribution was not possible. However, while the number of neutrophil granulocytes, monocytes and B lymphocytes was shown to be well preserved in healthy elderly persons, only the number of natural killer cells seems to increase and the number of T cells seems to decrease with aging (34). The number of all lymphocytes was reported to decrease from 2320 cells/µl in younger adults (mean age 32 years) to 1760 cells/µl in elderly (mean age 81 years), whereas the number of all leukocytes decreased from 7980 to 6348 cells/µl, respectively (35). As a consequence, the proportion of lymphocytes decreased from 29.1% (=2320/7980) to 27.7% (=1760/6348), which means a decrease by 1.4% units in ∼50 years (and corresponds to a 2% unit decrease in 70 years). It appears extremely unlikely that such a small decrease in the proportion of lymphocytes might explain a 1.5- or 2-fold change in DNA methylation from birth to the age of 70 years. Changes in leukocyte distributions with aging would therefore be expected to account for a very minor share of the change in methylation at age-associated CpG sites. Furthermore, gene ontology enrichment analysis showed completely different categories for genes with age-related differentially methylation identified in our study and for genes showing differentially methylated probes for different leukocytes subtypes in comparison with whole blood (36). Finally, 6 of the 14 significantly age-associated CpG sites in blood present on the 27K Infinium platform were found to be associated with age in brain tissue as well (26).
Although the Infinum HumanMethylation450 BeadChip (Illumina Inc.) offers a reliable cost-effective way of analysing >485 000 sites per sample at single-nucleotide resolution, the microarray is limited to the available probes defined by the company and a Consortium of experts leading to potential selection bias (37,38). Therefore, Harper et al. concluded that the reported statistically significant links to various diseases and biological processes by applying pathway analysis software created for gene expression array to gene methylation arrays might be spurious (39). CpG site selection might also have caused an overrepresentation of sites that are positively associated with age in studies using the Illumina Infinium arrays [100% (9), 100% (14), 76% (11), 77% (present study)] in comparison with whole-genome studies. In a study based on whole-genome bisulphite sequencing, only 13% of 17 930 DMRs were hypermethylated in centenarians in comparison with newborn (24). Even though our sample size was larger than that of previous studies, the power to detect significant associations after correcting for multiple testing was still limited leaving a large number of likely age-associated CpG sites undetected.
Despite these limitations, we identified 155 CpG sites with significant association of DNA methylation with age. We verified the association at these CpG sites in a longitudinal analysis demonstrating a gain or loss of methylation in each individual while getting 8 years older, respectively, for CpG sites positively or negatively correlated with cross-sectional age. To our knowledge, the association of DNA methylation with age at 65 of these CpG sites had not been reported previously. We confirmed the highly CpG island-dependent correlation between DNA methylation and age (13); loci within CpG islands gained methylation with age and loci outside CpG islands lost methylation with age. Furthermore, we were able to predict age from blood DNA with an excellent accuracy, and even the increase in age within the same individual could be predicted very precisely. Thus, DNA methylation at age-associated CpG sites might be used in forensic science to determine a person's age when only a blood sample is available. Interestingly, we found age-associated CpG sites in blood DNA, which were reported to be associated with age also in other human tissues, e.g. CpG sites in the gene NEFM or also known as NEF3 a biomarker for neuronal damage (11). Such patterns of co-methylation might enhance our understanding of the role of epigenetic variations in the process of aging (7) and in the development of age-related diseases and requires further research.
MATERIALS AND METHODS
Study population
The study population consists of 965 participants of the ESTHER study, who were recruited between July and October 2000 and presented the longest follow-up time. The ESTHER study is a general population-based epidemiological study of older adults (age 50–75 years at baseline) conducted in Saarland, a state in southwest Germany. The design of the ESTHER study has been described in detail previously (40). Briefly, participants were recruited by their general practitioners during a routine health check-up between 2000 and 2002, and follow-ups were conducted after 2, 5 and 8 years. Baseline socio-demographic, lifestyle, health and diet information was obtained by a comprehensive questionnaire. Height, weight and history of several diseases, such as hypertension, stroke, diabetes mellitus and cardiovascular disease, were either obtained from a comprehensive participant's questionnaire or from the general practitioner's health check-up report at baseline. History of cancer diseases was obtained from the state-wide Saarland Cancer Registry. Blood samples were taken at baseline and 8-year follow-up. The study was approved by the ethics committees of the University of Heidelberg and of the physician's board of Saarland. Informed consent was obtained from all participants.
Methylation data of the significantly with age-associated CpG sites were compared with methylation data obtained from cord blood samples of 18 newborn of the Ulm Birth Cohort (UBC) Study, which has been described in detail previously (41).
Laboratory methods
DNA was extracted from whole blood samples using a salting-out procedure (42). DNA of the required ESTHER and UBC study samples were randomly plated in a 96-well format. The available DNA concentration was measured using PicoGreen (Invitrogen, Darmstadt, Germany), to assure the required amount of DNA (1.5 µg), for all samples. As quality control, three random DNA samples were placed on each plate as duplicates for replication.
Genome-wide DNA methylation status was measured using the Infinium HumanMethylation450 BeadChip (Illumina Inc.), allowing the simultaneous quantitative measurement of the methylation status at 485 577 CpG sites (37). The laboratory work was done in the Genomics and Proteomics Core Facility at the German Cancer Research Center, Heidelberg, Germany (DKFZ) and has been published previously (43). Methylation levels at each CpG site are indicated by average beta-values where 1 corresponds to complete methylation and 0 to no methylation and were calculated with Illumina's Genomestudio 2011.1 (Modul M Version 1.9.0). According to the manufacturer's protocol, no background correction was done and data were normalized to internal controls provided by the manufacturer. All controls were checked for inconsistencies in each measured plate. Signals of probes with a detection P-value >0.05 were excluded from analysis.
Design of the study
The design of our study is based on the recommendations of Houseman (44). The study consists of three phases: (1) the exploratory study/analysis, which was hypothesis generating (2) a first confirmatory study/analysis (cross-sectional) and (3) a second confirmatory study/analysis (longitudinal). In the first phase among 400 randomly selected participants (from 898 participants with baseline blood samples), we selected 200 CpG sites with the strongest association between methylation and age. In the confirmatory study I (cross-sectional), we tested the association between age and methylation at each of the selected 200 CpG sites in an independent study population of the remaining 498 participants with baseline blood samples. This study therefore included 200 statistical tests.
In order to exclude the possibility that observed associations with age merely reflect disease-related changes in methylation resulting from higher prevalence of major diseases at higher ages, we repeated confirmatory analysis I after restricting the study sample to 326 participants without history of cancer, diabetes mellitus, cardiovascular disease and stroke at baseline.
In confirmatory analysis II, the increase or decrease of methylation with age at age-associated CpG sites, which showed significant associations with age in ‘confirmatory analysis I, sensitivity analysis’, was verified in a longitudinal analysis in an independent sample of 67 individuals, from whom blood samples at baseline and 8 years later were taken. This analysis was also restricted to participants without history of cancer, diabetes mellitus, cardiovascular disease and stroke at baseline. The characteristics of the study populations for the exploratory, confirmatory I and II and sensitivity analyses are presented in Table 1.
Table 1.
Descriptive characteristics of randomly selected non-overlapping samples of ESTHER participants for the exploratory and confirmatory analyses
| Analysis | Cross-sectional |
Longitudinal | ||
|---|---|---|---|---|
| Exploratory | Confirmatory I |
Confirmatory II | ||
| All participants | Participants free of major age-related diseases (sensitivity analysis) | |||
| Number | 400 | 498 | 326/498 | 67 |
| Age | 62 (57, 67) | 62 (58, 67) | 61 (57, 65) | 61 (57, 69)a |
| Men (%) | 47 | 52 | 48 | 50 |
| Smoker | ||||
| Never (%) | 50 | 46 | 47 | 58 |
| Former (%) | 31 | 37 | 34 | 26 |
| Current (%) | 19 | 17 | 19 | 17 |
| Cancer (%) | 6 | 6 | 0 | 0 |
| CVD (%) | 22 | 21 | 0 | 0 |
| Diabetes mellitus (%) | 15 | 15 | 0 | 0 |
| Stroke (%) | 3 | 3 | 0 | 0 |
Values are presented as median [1st, 3rd Quartile] or number in %.
CVD, cardiovascular disease.
aAge at baseline.
Statistical analyses
A sample size calculation indicated that a correlation between age and methylation with strength of 0.2 at 200 CpG sites can be confirmed in ∼500 individuals with a Bonferroni-corrected P-value of 2.5 × 10−4.
In the exploratory analysis (phase I), a non-parametric method was used. The association between age and methylation as quantified by ‘beta value’ at each CpG site was assessed by Spearman correlation coefficients. The 200 CpG sites showing highest Spearman correlation coefficients were selected.
In the confirmatory analysis I, methylation levels were analysed using linear regression, a parametric method with strong normality assumptions, requiring appropriate distribution diagnostics (44). The normal distribution of the methylation levels as quantified by beta-value was verified using the Kolmogorov–Smirnov Test. In case of non-normality, Box–Cox transformation and/or exclusion of outliers was applied. For original and transformed CpG sites, methylation levels at each CpG site were analysed by mixed linear regression with random batch effect (plate/BeadChip). Further independent variables in the models were age, gender, smoking behaviour, history of cancer, diabetes mellitus, cardiovascular disease and stroke. CpG sites with a P-value of <2.5 × 10−4 for the regression coefficient of age in the mixed linear model were considered significant. In the sensitivity analysis restricted to participants free of major age-related diseases, the association with age was estimated by mixed linear regression with random batch effect adjusted for gender and smoking behaviour.
The confirmatory analysis II (longitudinal analysis) was conducted based on blood samples at baseline and at the 8-year follow-up of 67 participants of the ESTHER study, in whom diabetes mellitus, cardiovascular disease, stroke and cancer at baseline was not prevalent. The change in methylation between baseline and 8-year follow-up was estimated by the mean difference between methylation levels at 8-year follow-up and at baseline. To estimate the batch effect, we first calculated the median of the methylation level over all CpG sites for each sample (=individual) and second a median for all samples on the same plate and BeadChip. Then, we calculated the differences between all plate and BeadChip specific medians. The mean of these differences was 0.0115 (SD 0.0086). In further analyses, we considered only mean differences in methylation, which were larger than this estimated batch effect. We corrected for multiple testing according the Bonferroni method.
To predict the age of an individual by epigenetic markers such as DNA methylation, we developed a linear regression model based on the validated CpG sites that were significantly associated with age. For internal validation, we split the data set for the confirmatory analysis I with all participants into a training and a validation set including 249 individuals in each set. Model selection during training step was performed using forward selection by minimization of Akaike's Information Criterion, a measure of relative goodness of model fit starting with all significant age-associated CpG sites (N = 155). After that the model built during training was validated in the independent validation set to obtain unbiased estimates of predictive accuracy for age. Furthermore, we validated the model in the independent longitudinal cohort. We calculated predicted values for age at baseline and for age 8 years later, using the baseline methylation levels and the methylation levels at the 8-year follow-up for prediction, respectively. Then, the differences between predicted values at 8-year follow-up and baseline for each individual and the mean of these differences were calculated.
Median and interquartile range for continuous data or number and percentage for categorical data were calculated for description of population characteristics. Data were analysed with the SAS software package (Version 9.2 and Enterprise Guide 4.2, SAS Institute, Cary, NC, USA), using the procedures PROC POWER, PROC CORR, PROC UNIVARIATE, PROC TRANSREG, PROC MIXED, PROC GLMSELECT and PROC REG.
Gene ontology analysis
Gene ontology enrichment was performed using the Database for Annotation, Visualization and Integrated Discovery Version 6.7 (DAVID, http://david.abcc.ncifcrf.gov) (28). Analysis was done on the list of genes containing significantly age-associated CpG sites using the whole human genome as background. Analyses were based on hypergeometric test and enriched annotations with a P-value of < 0.05 were considered significant and shown in Figure 2.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by grants from the Baden-Württemberg State Ministry of Science, Research and Arts and by the German Federal Ministry of Education and Research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility, especially Matthias Schick and Roger Fischer, for providing the Illumina Human Methylation arrays and related services and Dr Utz Benscheid for helpful comments on programming. We particularly thank Professor Andrew B. Singleton and Dr Michael A. Nalls from the Molecular Genetics Section and Laboratory of Neurogenetics, NIA, NIH, Bethesda, USA for sending us their data on age-associated CpG sites in human brain tissue (24).
Conflict of Interest statement. None declared.
Table 2.
Differentially methylated CpG sites for age with Bonferroni-corrected statistical significance (P < 2.5 × 10−4)
| Models for original methylation levels |
Models for transformed methylation levels |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpGsite | Estimate | Lower | Upper | P-value | Lambda | Estimate | Lower | Upper | P-value | Chr | UCSC_REFGENE |
| cg00484358 | 0.01597 | 0.012475 | 0.019464 | 6.40875E − 18 | 1 | ALX3 | |||||
| cg20822990 | −0.014047 | −0.017906 | −0.010188 | 3.44078E − 12 | 1 | ATP13A2 | |||||
| cg13806070 | 0.007804 | 0.005239 | 0.01037 | 4.47212E − 09 | 1 | BMP8A | |||||
| cg03664992 | 0.005891 | 0.003584 | 0.008197 | 7.49258E − 07 | 0 | 0.037333 | 0.022892 | 0.051774 | 5.44079E − 07 | 1 | BMP8A |
| cg09809672 | −0.027717 | −0.034761 | −0.020672 | 7.00011E − 14 | 0.5 | −0.019063 | −0.024075 | −0.014052 | 3.75201E − 13 | 1 | EDARADD |
| cg19560758 | 0.018868 | 0.013899 | 0.023838 | 4.44369E − 13 | 0 | 0.068926 | 0.053199 | 0.084652 | 1.08582E − 16 | 1 | ERRFI1 |
| cg01281911 | 0.01888 | 0.013303 | 0.024458 | 8.37661E − 11 | −0.25 | 0.309922 | 0.23226 | 0.387585 | 2.95768E − 14 | 1 | FOXE3 |
| cg18815943 | 0.010845 | 0.007622 | 0.014068 | 1.07602E − 10 | 0 | 0.168624 | 0.121592 | 0.215656 | 6.5944E − 12 | 1 | FOXE3 |
| cg23078123 | −0.018333 | −0.024108 | −0.012559 | 1.0147E − 09 | 1 | GPR177 | |||||
| cg01974375 | −0.016748 | −0.02029 | −0.013207 | 6.48498E − 19 | 1 | PI4KB | |||||
| cg25410668 | 0.022775 | 0.016119 | 0.029431 | 5.38061E − 11 | −1.5 | 0.075886 | 0.055045 | 0.096727 | 3.22434E − 12 | 1 | RPA2 |
| cg08426733 | −0.015708 | −0.01916 | −0.012257 | 9.87574E − 18 | 1 | VWA1 | |||||
| cg06784991 | 0.026332 | 0.021059 | 0.031604 | 9.97202E − 21 | −0.25 | 0.11995 | 0.098106 | 0.141794 | 2.06456E − 24 | 1 | ZYG11A |
| cg06335143 | 0.018098 | 0.013472 | 0.022723 | 8.70401E − 14 | 1 | ZYG11A | |||||
| cg16015712 | 0.014645 | 0.009836 | 0.019454 | 4.427E − 09 | 0.25 | 0.074808 | 0.055679 | 0.093936 | 8.96501E − 14 | 1 | ZYG11A |
| cg05308819 | −0.013741 | −0.017976 | −0.009505 | 4.51777E − 10 | 1 | ||||||
| cg18343474 | 0.010438 | 0.006477 | 0.014399 | 3.37437E − 07 | −0.5 | 0.112038 | 0.07346 | 0.150616 | 2.0407E − 08 | 1 | |
| cg24436906 | 0.020205 | 0.015499 | 0.02491 | 4.01706E − 16 | 2 | BOK | |||||
| cg00318320 | 0.015015 | 0.010124 | 0.019907 | 3.37647E − 09 | 0 | 0.156971 | 0.1116 | 0.202341 | 3.19285E − 11 | 2 | BOK |
| cg21117668 | 0.014212 | 0.00966 | 0.018764 | 1.86573E − 09 | −0.25 | 0.292536 | 0.206298 | 0.378774 | 7.35703E − 11 | 2 | BOK |
| cg13388731 | −0.01697 | −0.021401 | −0.012539 | 2.668E − 13 | 2 | DNAJB2 | |||||
| cg22016779 | −0.019078 | −0.023908 | −0.014248 | 5.68083E − 14 | 2 | DNER | |||||
| cg23606718 | 0.024058 | 0.01837 | 0.029745 | 1.01331E − 15 | 2 | FAM123C | |||||
| cg13381486 | 0.009199 | 0.005773 | 0.012625 | 2.01623E − 07 | 2 | FAM123C | |||||
| cg06639320 | 0.030167 | 0.024791 | 0.035544 | 3.47064E − 25 | 0 | 0.062474 | 0.052187 | 0.07276 | 6.76678E − 29 | 2 | FHL2 |
| cg22454769 | 0.043984 | 0.035914 | 0.052054 | 5.3776E − 24 | 2 | FHL2 | |||||
| cg24079702 | 0.034411 | 0.027884 | 0.040938 | 8.34638E − 23 | 2 | FHL2 | |||||
| cg22197050 | 0.017828 | 0.011869 | 0.023787 | 8.08278E − 09 | 0 | 0.055026 | 0.034297 | 0.075756 | 2.7526E − 07 | 2 | LOC100132215 |
| cg10751726 | 0.013869 | 0.008719 | 0.019019 | 1.8941E − 07 | 0 | 0.137967 | 0.089035 | 0.1869 | 5.02695E − 08 | 2 | LOC150786 |
| cg22158769 | 0.010901 | 0.00823 | 0.013571 | 9.31592E − 15 | 0 | 0.215918 | 0.168734 | 0.263102 | 5.93394E − 18 | 2 | LOC375196;LOC100271715 |
| cg12206199 | 0.00891 | 0.006664 | 0.011156 | 4.45932E − 14 | 0 | 0.21947 | 0.170993 | 0.267947 | 1.22522E − 17 | 2 | LOC375196;LOC100271715 |
| cg11176990 | 0.008982 | 0.006642 | 0.011322 | 2.5482E − 13 | 0 | 0.155687 | 0.120167 | 0.191208 | 1.06578E − 16 | 2 | LOC375196;LOC100271715 |
| cg19711579 | 0.011219 | 0.008059 | 0.014378 | 1.07815E − 11 | −0.5 | 0.191564 | 0.140828 | 0.2423 | 5.48858E − 13 | 2 | NEUROD1 |
| cg14674720 | 0.020675 | 0.015082 | 0.026269 | 1.67115E − 12 | −0.5 | 0.12992 | 0.098518 | 0.161323 | 3.78841E − 15 | 2 | |
| cg26158023 | −0.009037 | −0.012228 | −0.005845 | 4.50986E − 08 | 3 | CCBP2 | |||||
| cg01844642 | 0.023465 | 0.018907 | 0.028023 | 8.23392E − 22 | −1.75 | 0.134554 | 0.109322 | 0.159785 | 2.97106E − 23 | 3 | GPR62 |
| cg02336827 | −0.014201 | −0.017663 | −0.010738 | 6.95095E − 15 | 3 | LIMD1 | |||||
| cg13473356 | 0.012142 | 0.008274 | 0.01601 | 1.54644E − 09 | 0 | 0.15189 | 0.113576 | 0.190205 | 4.31388E − 14 | 3 | PEX5L |
| cg04894619 | 0.013831 | 0.009545 | 0.018117 | 5.53825E − 10 | 0 | 0.126066 | 0.089945 | 0.162186 | 2.21296E − 11 | 3 | PEX5L |
| cg12943155 | 0.009342 | 0.006748 | 0.011936 | 5.68221E − 12 | −0.5 | 0.732356 | 0.54263 | 0.922082 | 1.77733E − 13 | 3 | PODXL2 |
| cg03607117 | 0.01073 | 0.007596 | 0.013865 | 5.2631E − 11 | 0 | 0.079814 | 0.057124 | 0.102505 | 1.5501E − 11 | 3 | SFMBT1 |
| cg00481951 | 0.02166 | 0.016903 | 0.026417 | 8.32115E − 18 | 3 | SST | |||||
| cg07806886 | 0.016015 | 0.011161 | 0.020869 | 2.35773E − 10 | 0 | 0.106417 | 0.076401 | 0.136432 | 1.09987E − 11 | 3 | STXBP5L |
| cg07553761 | 0.028278 | 0.022068 | 0.034488 | 9.50133E − 18 | 0 | 0.082935 | 0.067236 | 0.098634 | 6.93228E − 23 | 3 | TRIM59 |
| cg15618978 | 0.029734 | 0.022336 | 0.037132 | 2.18976E − 14 | 0 | 0.096897 | 0.073503 | 0.120291 | 3.52957E − 15 | 3 | TRIM59 |
| cg09401099 | 0.019762 | 0.014387 | 0.025137 | 2.16094E − 12 | 0 | 0.146144 | 0.109358 | 0.182929 | 3.78285E − 14 | 3 | |
| cg15957394 | 0.012489 | 0.007973 | 0.017006 | 9.03936E − 08 | 0.25 | 0.157617 | 0.094686 | 0.220548 | 1.18851E − 06 | 4 | AFAP1 |
| cg03915012 | −0.014954 | −0.019674 | −0.010234 | 1.09543E − 09 | 3 | −0.01042 | −0.013595 | −0.007246 | 2.75189E − 10 | 4 | GAK |
| cg22747507 | −0.020263 | −0.025005 | −0.015521 | 6.01603E − 16 | 3 | −0.003811 | −0.004678 | −0.002945 | 8.42248E − 17 | 4 | GLRA3 |
| cg08262002 | −0.028999 | −0.034416 | −0.023582 | 2.75517E − 23 | 4 | LDB2 | |||||
| cg16295725 | 0.010207 | 0.006697 | 0.013717 | 2.00364E − 08 | −0.25 | 0.340136 | 0.214575 | 0.465698 | 1.57731E − 07 | 4 | ZNF518B |
| cg23995914 | 0.019383 | 0.011324 | 0.027443 | 3.05802E − 06 | 0.25 | 0.060384 | 0.037724 | 0.083044 | 2.4708E − 07 | 4 | ZNF518B |
| cg05991454 | 0.027967 | 0.021684 | 0.034251 | 4.54736E − 17 | 0.25 | 0.097766 | 0.078615 | 0.116916 | 1.38273E − 21 | 4 | |
| cg12765028 | 0.020623 | 0.015304 | 0.025942 | 1.52059E − 13 | −0.5 | 0.070516 | 0.051999 | 0.089033 | 3.5622E − 13 | 4 | |
| cg05369942 | −0.012875 | −0.016977 | −0.008773 | 1.53804E − 09 | 0 | −0.072619 | −0.095301 | −0.049937 | 7.1628E − 10 | 4 | |
| cg25316339 | 0.018533 | 0.012789 | 0.024276 | 5.56906E − 10 | 0 | 0.136729 | 0.100416 | 0.173041 | 6.31616E − 13 | 5 | ANKRD34B |
| cg21800232 | 0.016937 | 0.011498 | 0.022376 | 2.04929E − 09 | 0 | 0.123331 | 0.086681 | 0.159982 | 1.02284E − 10 | 5 | ANKRD34B |
| cg13039251 | −0.026869 | −0.035942 | −0.017797 | 1.12001E − 08 | 3 | −0.013391 | −0.01781 | −0.008973 | 5.06284E − 09 | 5 | PDZD2 |
| cg08234504 | −0.016877 | −0.019934 | −0.01382 | 1.20946E − 24 | 5 | ||||||
| cg06782035 | 0.034653 | 0.018453 | 0.050853 | 3.16753E − 05 | 0 | 0.198402 | 0.16083 | 0.235974 | 8.10229E − 23 | 5 | |
| cg23500537 | 0.027301 | 0.021895 | 0.032707 | 4.04835E − 21 | −0.25 | 0.062103 | 0.050269 | 0.073936 | 1.34346E − 22 | 5 | |
| cg14314729 | −0.019418 | −0.023968 | −0.014869 | 6.43262E − 16 | 5 | ||||||
| cg18651026 | −0.01727 | −0.022044 | −0.012497 | 4.5844E − 12 | 6 | COL11A2 | |||||
| cg16867657 | 0.048095 | 0.043057 | 0.053132 | 2.05436E − 58 | 6 | ELOVL2 | |||||
| cg24724428 | 0.047849 | 0.040871 | 0.054827 | 5.34471E − 35 | 0 | 0.172501 | 0.148263 | 0.196739 | 1.83057E − 37 | 6 | ELOVL2 |
| cg21572722 | 0.022746 | 0.019309 | 0.026183 | 2.80199E − 33 | 6 | ELOVL2 | |||||
| cg06493994 | 0.01693 | 0.012707 | 0.021153 | 2.5137E − 14 | −1.75 | 0.628901 | 0.494918 | 0.762883 | 9.49284E − 19 | 6 | SCGN |
| cg03350900 | 0.020867 | 0.015142 | 0.026592 | 3.25277E − 12 | −1.25 | 0.544736 | 0.41601 | 0.673462 | 9.7699E − 16 | 6 | SOBP |
| cg03236802 | 0.021214 | 0.015319 | 0.027109 | 5.90973E − 12 | 6 | TNXB | |||||
| cg17885226 | 0.025665 | 0.020296 | 0.031035 | 2.50428E − 19 | 6 | ||||||
| cg22273555 | −0.013577 | −0.017608 | −0.009546 | 1.03157E − 10 | 6 | ||||||
| cg05316627 | −0.024637 | −0.031962 | −0.017313 | 1.08838E − 10 | 6 | ||||||
| cg20052760 | −0.011051 | −0.016435 | −0.005667 | 6.44895E − 05 | 6 | ||||||
| cg00292135 | 0.021651 | 0.015081 | 0.02822 | 2.47297E − 10 | 0 | 0.13941 | 0.099822 | 0.178997 | 1.47693E − 11 | 7 | C7orf13;RNF32 |
| cg08097417 | 0.017566 | 0.013688 | 0.021444 | 1.37407E − 17 | 7 | KLF14 | |||||
| cg07955995 | 0.014926 | 0.011041 | 0.01881 | 2.43187E − 13 | 0 | 0.193785 | 0.14299 | 0.24458 | 3.25775E − 13 | 7 | KLF14 |
| cg14361627 | 0.022336 | 0.016206 | 0.028465 | 3.28317E − 12 | 0.5 | 0.045323 | 0.033168 | 0.057477 | 1.01656E − 12 | 7 | KLF14 |
| cg22285878 | 0.008703 | 0.006194 | 0.011212 | 3.00649E − 11 | 0.5 | 0.02946 | 0.019947 | 0.038974 | 2.4163E − 09 | 7 | KLF14 |
| cg20426994 | 0.012153 | 0.007917 | 0.016388 | 3.03605E − 08 | 0 | 0.081902 | 0.052468 | 0.111337 | 7.38964E − 08 | 7 | KLF14 |
| cg12534424 | 0.020149 | 0.015705 | 0.024593 | 1.28324E − 17 | −0.5 | 0.08069 | 0.061736 | 0.099645 | 6.83878E − 16 | 7 | PRRT4 |
| cg18064714 | 0.060344 | 0.046964 | 0.073725 | 1.62468E − 17 | 7 | SP8 | |||||
| cg04084157 | 0.008334 | 0.006318 | 0.010351 | 4.04157E − 15 | 7 | VGF | |||||
| cg21186299 | 0.005357 | 0.003604 | 0.007109 | 3.93611E − 09 | 0 | 0.14231 | 0.100781 | 0.18384 | 4.85258E − 11 | 7 | VGF |
| cg04427498 | 0.021748 | 0.016549 | 0.026947 | 1.97792E − 15 | 7 | ||||||
| cg08540945 | 0.01848 | 0.0131 | 0.02386 | 4.56532E − 11 | 0 | 0.097893 | 0.069357 | 0.12643 | 4.58129E − 11 | 7 | |
| cg23973429 | 0.017205 | 0.011435 | 0.022974 | 8.93971E − 09 | 0.25 | 0.051815 | 0.036293 | 0.067337 | 1.42152E − 10 | 8 | GRHL2 |
| cg07502389 | 0.019666 | 0.01583 | 0.023503 | 1.18108E − 21 | 0 | 0.093603 | 0.076253 | 0.110952 | 1.03319E − 23 | 8 | NEFM |
| cg18898125 | 0.015236 | 0.011653 | 0.01882 | 8.15056E − 16 | 0 | 0.12194 | 0.093198 | 0.150683 | 8.32571E − 16 | 8 | NEFM |
| cg21709871 | 0.02033 | 0.012306 | 0.028354 | 9.11247E − 07 | 8 | NRBP2 | |||||
| cg16419235 | 0.023383 | 0.018043 | 0.028723 | 1.29636E − 16 | 0 | 0.100077 | 0.077846 | 0.122308 | 1.80508E − 17 | 8 | PENK |
| cg13649056 | 0.017773 | 0.014381 | 0.021165 | 1.45792E − 22 | 9 | ||||||
| cg23341182 | −0.023818 | −0.029309 | −0.018327 | 2.35284E − 16 | 1.5 | −0.012658 | −0.015484 | −0.009833 | 2.48786E − 17 | 10 | BLOC1S2 |
| cg06291867 | 0.012629 | 0.008258 | 0.016999 | 2.44285E − 08 | −0.25 | 0.078488 | 0.056459 | 0.100518 | 8.83404E − 12 | 10 | HTR7 |
| cg16541931 | 0.017014 | 0.012064 | 0.021963 | 4.44356E − 11 | −0.5 | 0.226555 | 0.166704 | 0.286407 | 4.85716E − 13 | 10 | LOC100128811;GPR158 |
| cg10804656 | 0.028143 | 0.021715 | 0.034571 | 1.29817E − 16 | 10 | ||||||
| cg25427880 | 0.016584 | 0.01229 | 0.020878 | 1.72886E − 13 | 10 | ||||||
| cg06943835 | −0.015214 | −0.019301 | −0.011126 | 1.19794E − 12 | 11 | ATG2A | |||||
| cg13806741 | 0.012866 | 0.008443 | 0.017289 | 1.93959E − 08 | 11 | FAM181B | |||||
| cg08160331 | 0.021067 | 0.016485 | 0.02565 | 4.90236E − 18 | 0 | 0.040317 | 0.031953 | 0.048681 | 1.30732E − 19 | 11 | KLHL35 |
| cg23091758 | 0.012771 | 0.008008 | 0.017535 | 2.13603E − 07 | 0.25 | 0.072048 | 0.049228 | 0.094868 | 1.20215E − 09 | 11 | NRIP3 |
| cg16061228 | −0.008759 | −0.012435 | −0.005083 | 3.75215E − 06 | −2.75 | −0.018208 | −0.025203 | −0.011214 | 4.55301E − 07 | 11 | SCN4B |
| cg07080372 | −0.01892 | −0.023142 | −0.014699 | 2.77649E − 17 | 1.75 | −0.005162 | −0.006253 | −0.004071 | 5.27736E − 19 | 11 | SLC25A22 |
| cg12189835 | 0.014528 | 0.010251 | 0.018804 | 7.25019E − 11 | −0.75 | 0.216821 | 0.159602 | 0.274041 | 4.58264E − 13 | 11 | SYT7 |
| cg04940570 | 0.026939 | 0.020648 | 0.033229 | 5.25906E − 16 | 0 | 0.166211 | 0.126996 | 0.205426 | 8.94069E − 16 | 11 | TEAD1 |
| cg23744638 | −0.032249 | −0.042052 | −0.022446 | 2.64715E − 10 | 11 | ||||||
| cg11751101 | −0.018751 | −0.024039 | −0.013464 | 1.14213E − 11 | 3 | −0.009019 | −0.011534 | −0.006504 | 6.47386E − 12 | 12 | C1R |
| cg25778262 | 0.019019 | 0.014331 | 0.023707 | 1.18388E − 14 | 12 | CPM | |||||
| cg18473521 | 0.031768 | 0.02511 | 0.038426 | 2.98829E − 19 | 12 | HOXC4 | |||||
| cg01820374 | −0.022865 | −0.027342 | −0.018388 | 1.62092E − 21 | 12 | LAG3 | |||||
| cg04604946 | −0.015974 | −0.020682 | −0.011265 | 7.68402E − 11 | 0 | −0.046764 | −0.059086 | −0.034443 | 4.22786E − 13 | 12 | LRRC23;ENO2 |
| cg08622677 | 0.014418 | 0.011001 | 0.017834 | 1.18272E − 15 | 12 | PRMT8 | |||||
| cg01528542 | −0.03156 | −0.03832 | −0.024799 | 1.65952E − 18 | 3 | −0.009373 | −0.011364 | −0.007382 | 7.61814E − 19 | 12 | |
| cg07920503 | 0.015991 | 0.011244 | 0.020737 | 1.02295E − 10 | −1 | 0.609585 | 0.445423 | 0.773746 | 1.25104E − 12 | 13 | FAM123A |
| cg15480367 | 0.020565 | 0.016365 | 0.024764 | 4.65514E − 20 | 14 | CHGA | |||||
| cg25078444 | 0.020905 | 0.014127 | 0.027683 | 2.86202E − 09 | −0.25 | 0.321249 | 0.243096 | 0.399403 | 5.55999E − 15 | 14 | FOXG1 |
| cg00741624 | 0.019597 | 0.014319 | 0.024874 | 1.34403E − 12 | −2 | 0.534267 | 0.398599 | 0.669935 | 6.12946E − 14 | 14 | KIAA1409 |
| cg10281977 | 0.013688 | 0.007226 | 0.02015 | 3.77239E − 05 | 0 | 0.074611 | 0.040124 | 0.109097 | 2.56108E − 05 | 14 | TMEM179 |
| cg14977938 | −0.017727 | −0.023256 | −0.012198 | 7.06632E − 10 | 14 | ZFYVE21 | |||||
| cg03032497 | 0.025658 | 0.01706 | 0.034256 | 8.74178E − 09 | −1.25 | 0.35327 | 0.277259 | 0.429281 | 2.14674E − 18 | 14 | |
| cg07394446 | 0.011307 | 0.006686 | 0.015927 | 2.07216E − 06 | −0.25 | 0.210841 | 0.131948 | 0.289734 | 2.29015E − 07 | 15 | ADAMTS17 |
| cg02018902 | 0.0133 | 0.009576 | 0.017024 | 8.33772E − 12 | −0.75 | 0.30582 | 0.228375 | 0.383265 | 5.28184E − 14 | 15 | ANKRD34C |
| cg03399905 | 0.012037 | 0.008476 | 0.015598 | 8.87636E − 11 | 0 | 0.081616 | 0.059398 | 0.103834 | 2.1069E − 12 | 15 | ANKRD34C |
| cg21801378 | 0.014239 | 0.010231 | 0.018246 | 1.05304E − 11 | 0.25 | 0.076035 | 0.056293 | 0.095776 | 1.99629E − 13 | 15 | BRUNOL6 |
| cg20809087 | 0.013425 | 0.009583 | 0.017267 | 2.18496E − 11 | 0 | 0.124129 | 0.0866 | 0.161659 | 2.04822E − 10 | 15 | BRUNOL6 |
| cg21296230 | 0.022973 | 0.017016 | 0.02893 | 2.05137E − 13 | 0.75 | 0.028028 | 0.021798 | 0.034257 | 2.01966E − 17 | 15 | GREM1 |
| cg21567504 | 0.014219 | 0.009241 | 0.019196 | 3.4726E − 08 | 0.25 | 0.043991 | 0.027012 | 0.060971 | 5.15632E − 07 | 15 | OCA2 |
| cg04875128 | 0.045425 | 0.037206 | 0.053643 | 1.11293E − 24 | 15 | OTUD7A | |||||
| cg01763090 | 0.013531 | 0.010167 | 0.016895 | 2.08518E − 14 | −1 | 0.351017 | 0.269837 | 0.432196 | 2.53722E − 16 | 15 | OTUD7A |
| cg16717122 | 0.017372 | 0.013352 | 0.021391 | 2.96856E − 16 | −2 | 0.874491 | 0.6864 | 1.062581 | 1.89405E − 18 | 15 | SCG3 |
| cg27569300 | 0.00973 | 0.006567 | 0.012893 | 3.04362E − 09 | 15 | SYNM | |||||
| cg01348004 | 0.002495 | 0.001051 | 0.003938 | 0.000744137 | 0.5 | 0.013955 | 0.006599 | 0.021311 | 0.000216489 | 15 | SYNM |
| cg11220950 | 0.016841 | 0.013343 | 0.020339 | 1.7156E − 19 | 16 | SYNGR3 | |||||
| cg00448560 | −0.015806 | −0.020088 | −0.011525 | 1.77793E − 12 | 3 | −0.009536 | −0.011895 | −0.007178 | 1.42772E − 14 | 16 | ZNF423 |
| cg04208403 | −0.013577 | −0.017271 | −0.009884 | 2.17864E − 12 | 3 | −0.004561 | −0.005727 | −0.003395 | 8.75465E − 14 | 16 | ZNF423 |
| cg07082267 | −0.016468 | −0.020722 | −0.012214 | 1.64915E − 13 | 16 | ||||||
| cg10137837 | 0.022064 | 0.016005 | 0.028122 | 3.38911E − 12 | 17 | BCL6B | |||||
| cg11071401 | 0.023811 | 0.018644 | 0.028977 | 3.64614E − 18 | 17 | CACNA1G | |||||
| cg22353329 | 0.009332 | 0.006027 | 0.012637 | 4.79994E − 08 | 17 | CBX4 | |||||
| cg14973055 | −0.016871 | −0.021829 | −0.011913 | 6.75529E − 11 | 17 | DNAI2 | |||||
| cg22156456 | −0.015896 | −0.019271 | −0.01252 | 8.81012E − 19 | 17 | EIF1 | |||||
| cg00439658 | 0.017956 | 0.01315 | 0.022762 | 9.9394E − 13 | 17 | GRIN2C | |||||
| cg08090640 | −0.027631 | −0.03283 | −0.022432 | 4.20618E − 23 | 17 | IFI35 | |||||
| cg19401340 | 0.007929 | 0.005311 | 0.010547 | 5.34613E − 09 | −0.5 | 0.474585 | 0.332267 | 0.616903 | 1.47739E − 10 | 17 | PPM1E |
| cg26161329 | 0.014363 | 0.009387 | 0.01934 | 2.52689E − 08 | 0.75 | 0.020731 | 0.014361 | 0.0271 | 3.86085E − 10 | 17 | PPM1E |
| cg16477091 | 0.011706 | 0.007341 | 0.01607 | 2.29627E − 07 | 0 | 0.057933 | 0.038484 | 0.077381 | 1.02119E − 08 | 17 | PPM1E |
| cg14692377 | 0.01898 | 0.014022 | 0.023937 | 2.96237E − 13 | 0 | 0.018203 | 0.013954 | 0.022452 | 4.76723E − 16 | 17 | SLC6A4 |
| cg20273670 | 0.032196 | 0.02528 | 0.039112 | 2.02044E − 18 | 17 | ||||||
| cg03259243 | 0.02064 | 0.015415 | 0.025866 | 5.67488E − 14 | 17 | ||||||
| cg19283806 | −0.039403 | −0.046586 | −0.032219 | 2.24176E − 24 | 18 | CCDC102B | |||||
| cg14556683 | 0.030341 | 0.023835 | 0.036847 | 1.78265E − 18 | 19 | EPHX3 | |||||
| cg05213896 | 0.028961 | 0.020297 | 0.037626 | 1.4067E − 10 | 19 | IL4I1 | |||||
| cg19784428 | −0.025963 | −0.034534 | −0.017392 | 5.30294E − 09 | 3 | −0.010931 | −0.014486 | −0.007376 | 3.08211E − 09 | 19 | NWD1 |
| cg16644497 | 0.009985 | 0.0063 | 0.01367 | 1.59858E − 07 | −0.25 | 0.354662 | 0.23657 | 0.472754 | 6.87099E − 09 | 19 | ZNF599 |
| cg17110586 | 0.026352 | 0.021808 | 0.030897 | 1.34319E − 26 | −0.75 | 0.150544 | 0.126514 | 0.174575 | 2.10878E − 30 | 19 | |
| cg05619598 | −0.013875 | −0.018122 | −0.009628 | 3.31198E − 10 | 19 | ||||||
| cg24436715 | 0.00564 | 0.003335 | 0.007945 | 2.07302E − 06 | 0.5 | 0.018749 | 0.01094 | 0.026558 | 3.14475E − 06 | 19 | |
| cg02151301 | −0.016205 | −0.020716 | −0.011695 | 6.33729E − 12 | 1.5 | −0.009358 | −0.011863 | −0.006853 | 9.27052E − 13 | 20 | HM13 |
| cg07547549 | 0.032192 | 0.026015 | 0.03837 | 2.90588E − 22 | −0.5 | 0.10031 | 0.083094 | 0.117526 | 5.94932E − 27 | 20 | SLC12A5 |
| cg19593767 | 0.011985 | 0.006285 | 0.017686 | 4.29871E − 05 | −0.75 | 0.083519 | 0.048304 | 0.118735 | 4.11406E − 06 | 20 | TOX2 |
| cg22682811 | 0.022406 | 0.016562 | 0.028251 | 2.735E − 13 | 0 | 0.0952 | 0.072428 | 0.117972 | 2.03179E − 15 | 20 | |
| cg08415592 | −0.018227 | −0.022597 | −0.013856 | 2.61902E − 15 | 22 | APOL1 | |||||
| cg18448426 | −0.022175 | −0.028365 | −0.015984 | 7.25764E − 12 | 3 | −0.00966 | −0.012375 | −0.006945 | 9.44499E − 12 | 22 | APOL1 |
Rows marked in grey, the ten most significant CpG sites; cells marked in light grey, CpG sites within the same gene region; Chr, chromosome number; estimate, regression coefficient for age in the mixed model adjusted for gender, smoking behavior, diabetes mellitus, cardiovascular disease, stroke, cancer and batch effect; lower, lower limit of the 95% confidence interval of the regression coefficient; upper, upper limit of the 95% confidence interval of the regression coefficient; rho, Spearmann correlation coefficient.
REFERENCES
- 1.Volkmar M., Dedeurwaerder S., Cunha D.A., Ndlovu M.N., Defrance M., Deplus R., Calonne E., Volkmar U., Igoillo-Esteve M., Naamane N., et al. DNA Methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talens R.P., Jukema J.W., Trompet S., Kremer D., Westendorp R.G., Lumey L.H., Sattar N., Putter H., Slagboom P.E., Heijmans B.T. Hypermethylation at loci sensitive to the prenatal environment is associated with increased incidence of myocardial infarction. Int. J. Epidemiol. 2012;41:106–115. doi: 10.1093/ije/dyr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Movassagh M., Choy M.K., Knowles D.A., Cordeddu L., Haider S., Down T., Siggens L., Vujic A., Simeoni I., Penkett C., et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinoue T., Weisenberger D.J., Lange C.P., Shen H., Byun H.M., Van Den Berg D., Malik S., Pan F., Noushmehr H., van Dijk C.M., et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2010;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langevin S.M., Koestler D.C., Christensen B.C., Butler R.A., Wiencke J.K., Nelson H.H., Houseman E.A., Marsit C.J., Kelsey K.T. Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: an epigenome-wide association study. Epigenetics. 2012;7:291–299. doi: 10.4161/epi.7.3.19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess D.J. Human epigenetics: showing your age. Nat. Rev. Genet. 2013;14:6. doi: 10.1038/nrg3391. [DOI] [PubMed] [Google Scholar]
- 7.Horvath S., Zhang Y., Langfelder P., Kahn R.S., Boks M.P.M., van Eijk K., van den Berg L.H., Ophoff R.A. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teschendorff A.E., Menon U., Gentry-Maharaj A., Ramus S.J., Weisenberger D.J., Shen H., Campan M., Noushmehr H., Bell C.G., Maxwell A.P., et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakyan V.K., Down T.A., Maslau S., Andrew T., Yang T.-P., Beyan H., Whittaker P., McCann O.T., Finer S., Valdes A.M., et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell J.T., Tsai P.-C., Yang T.-P., Pidsley R., Nisbet J., Glass D., Mangino M., Zhai G., Zhang F., Valdes A., et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genetics. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bocklandt S., Lin W., Sehl M.E., Sanchez F.J., Sinsheimer J.S., Horvath S., Vilain E. Epigenetic predictors of age. PLoS One. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch C.M., Wagner W. Epigentic-aging-signature to determine age in different tissues. Aging. 2011;3:1018–1027. doi: 10.18632/aging.100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen B.C., Houseman E.A., Marsit C.J., Zheng S., Wrensch M.R., Wiemels J.L., Nelson H.H., Karagas M.R., Padbury J.F., Bueno R., et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genetics. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garagnani P., Bacalli M.G., Pirazzini C., Gori D., Giuliani C., Mari D., Di Blasio A.M., Gentilini D., Vitale G., Collino S., et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- 15.Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.B., Gao Y., et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:1–9. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M. Epigenome-wide association study in European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 17.Stelzer G., Dalah I., Stein T.I., Satanower Y., Rosen N., Nativ N., Oz-Levi D., Olender T., Belinky F., Bahir I., et al. In-silico human genomics with GeneCards. Hum. Genom. 2011;5:709–717. doi: 10.1186/1479-7364-5-6-709. www.genecards.org . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bock C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 19.Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 20.Madrigano J., Baccarelli A., Mittleman M.A., Sparrow D., Vokonas P.S., Tarantini L., Schwartz J. Aging and epigenetics: longitudinal changes in gene-specific DNA methylation. Epigenetics. 2012;7:63–70. doi: 10.4161/epi.7.1.18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D., Liu Y., Xie H., Hong X., Tsai H.-J., Wang G., Liu R., Wang X. Individual variation and longitudinal pattern of genome-wide DNA methylation from birth to the first two years of life. Epigenetics. 2012;7:594–605. doi: 10.4161/epi.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issa J.-P. Age-related variation in DNA methylation. In: Michels K.B., editor. Epigenetic Epidemiology. Springer; 2012. pp. 186–194. [Google Scholar]
- 23.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 24.Heyn H., Li N., Ferreira H.J., Moran S., Pisano D.G., Gomez A., Diez J., Sanchez-Mut J.V., Setien F., Carmona F.J. Distinct DNA methylomes of newborns and centenarians. PNAS. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia M.L., Lobsiger C.S., Shah S.B., Deerinck T.J., Crum J., Young D., Ward C.M., Crawford T.O., Gotow T., Uchiyama Y., et al. NF-M is an essential target for the myelin-directed ‘outside-in’ signaling cascade that mediates radial axonal growth. J. Cell Biol. 2003;163:1011–1020. doi: 10.1083/jcb.200308159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez D.G., Nalls M.A., Gibbs J.R., Arepalli S., von der Brug M., Chong S., Moore M., Longo D.L., Cookson M.R., Traynor B.J., et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson A., Enroth S., Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS one. 2013;8:e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Jakobsson A., Westerberg R., Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid. Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Johannessen M., Moller S., Hansen T., Moens U., Van Ghelue M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Moll. Life Sci. 2006;63:268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small K.S., Hedman A.K., Grundberg E., Nica A.C., Thorleifsson G., Kong A., Thorsteindottir U., Shin S.Y., Richards H.B., et al., editors. GIANT Consortium. Identification of an imprinted master trans-regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Wu W., Thang W. On the analysis of the Illumina 450k array data: probes ambiguously mapped to the Human genome. Front. Genet. 2012;3:73. doi: 10.3389/fgene.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiskopf D., Weinberger B., Grubeck-Loebenstein B. The aging of the immune sytem. Transpl. Int. 2009;22:1041–1050. doi: 10.1111/j.1432-2277.2009.00927.x. [DOI] [PubMed] [Google Scholar]
- 35.Borrego F., Alonso M.C., Galiani M.D., Carracedo J., Ramirez R., Ostos B., Peña J., Solana R. NK Phenotypic markers and IL2 response in NK cells from elderly people. Exp. Gerontol. 1999;34:253–265. doi: 10.1016/s0531-5565(98)00076-x. [DOI] [PubMed] [Google Scholar]
- 36.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlén S.E., Greco D., Söderhäll C., Scheynius A., Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Barault L., Rancourt R.C. Laboratory methods in epigenetic epidemiology. In: Michels K.B., editor. Epigentic Epidemiology. Springer; 2012. p. 45. [Google Scholar]
- 39.Harper K.N., Peters B.A., Gamble M.V. Batch effects and pathway analysis: two potential perils in cancer studies involving DNA methylation array analysis. Cancer Epidemiol. Biomarkers Prev. 2013;22:1052–1060. doi: 10.1158/1055-9965.EPI-13-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raum E., Rothenbacher D., Löw M., Stegmaier C., Ziegler H., Brenner H. Changes of cardiovascular risk factors and their implications in subsequent birth cohorts of older adults in Germany: a life course approach. Eur. J. Cardiovasc. Prev. Rehabil. 2007;14:809–814. doi: 10.1097/HJR.0b013e3282eeb308. [DOI] [PubMed] [Google Scholar]
- 41.Florath I., Kohler M., Weck M.N., Brandt S., Rothenbacher D., Schöttker B., Moß A., Gottmann P., Wabitsch M., Brenner H. Association of pre- and post-natal parental smoking with offspring body mass index: an 8-year follow-up of a birth cohort. Pediatr. Obes. 2013 doi: 10.1111/j.2047-6310.2012.00146.x. doi:10.1111/j.2047-6310.2012.00146.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 42.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch C.M., Reck K., Shao K., Lin Q., Joussen S., Ziegler P., Walenda G., Drescher W., Opalka B., May T., et al. Pluripotent stem cells escape from senescence-associated DNA methylation changes. Genome Res. 2013;23:248–259. doi: 10.1101/gr.141945.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houseman E.A. Biostatistical methods in epigenetic epidemiology. In: Michels K.B., editor. Epigenetic Epidemiology. Springer; 2012. pp. 58–76. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.