Abstract
It is known that the NFκB route is constitutively upregulated in celiac disease (CD), an immune-mediated disorder of the gut caused by intolerance to ingested gluten. Our aim was to scrutinize the expression patterns of several of the most biologically relevant components of the NFκB route in intestinal biopsies from active and treated patients and after in vitro gliadin challenge, and to assess normalization of the expression using an inhibitor of the MALT1 paracaspase. The expression of 93 NFκB genes was measured by RT-PCR in a set of uncultured active and treated CD and control biopsies, and in cultured biopsy series challenged with gliadin, the NFκB modulator, both compounds and none. Methylation of eight genes involved in NFκB signaling was analyzed by conventional pyrosequencing. Groups were compared and Pearson's correlation matrixes were constructed to check for coexpression and co-methylation. Our results confirm the upregulation of the NFκB pathway and show that constitutively altered genes usually belong to the core of the pathway and have central roles, whereas genes overexpressed only in active CD are more peripheral. Additionally, this is the first work to detect methylation level changes in celiac intestinal mucosa. Coexpression is very common in controls, whereas gliadin challenge and especially chronic inflammation present in untreated CD result in the disruption of the regulatory equilibrium. In contrast, co-methylation occurs more often in active CD. Importantly, NFκB modulation partially restores coregulation, opening the door to future therapeutic possibilities and targets.
INTRODUCTION
Celiac disease (CD, MIM: 212750) is a chronic, immune-mediated disorder caused by intolerance to ingested gluten that develops in genetically susceptible individuals and affects ∼1% of Caucasians (1). Histological analysis of intestinal biopsies at diagnosis (when disease is active) shows that CD is characterized by the loss of villi, crypt hyperplasia and lymphocytic infiltration in the gut, and there are circulating IgA and IgG class autoantibodies specific for the enzyme transglutaminase 2 and antibodies against gliadin (2). Clinical presentation varies from typical gastrointestinal symptoms to extraintestinal manifestations, all of which develop in the context of a sustained, chronic inflammation of the gut, but usually revert if gluten is totally excluded from the diet. From a genetic point of view, HLA-DQ2 or HLA-DQ8 are necessary, but yet not sufficient for the development of CD, accounting for ∼35–40% of heritability of the disease (3). More recently, the two genome-wide association studies (GWAS) and the Immunochip fine mapping project have revealed 39 non-HLA regions associated with the genetic risk to CD development (4–6). Several genes within those regions that are related to the immune response have been proposed as etiological candidates, but a systematic functional confirmation is still pending for the identification of the causal variants and causal genes. Altogether those new loci are responsible for an additional 10–15% of the genetic risk to CD, leaving a large proportion of the genetic component of the disease still to be explained. In the post-GWAS era, when increasing resources and efforts are being directed towards huge exploratory projects, it remains important to take into account what is already known about the functional players involved in the disease and adopt a pathophysiology-based point of view in the design of more specific and less costly studies.
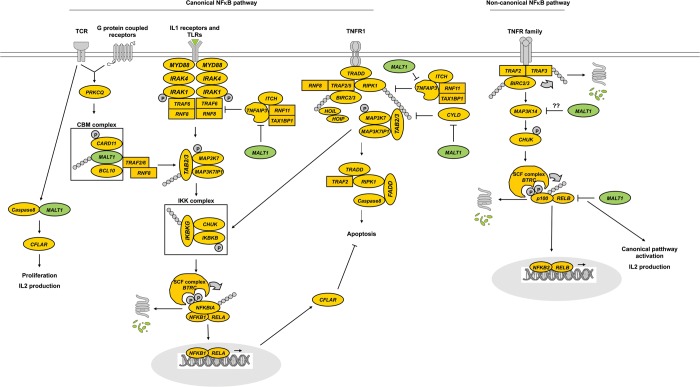
In this sense, based on our previous knowledge on the role of the NFκB pathway in CD, we selected a set of 93 NFκB-related genes because the pathway has been shown to be constitutively upregulated in CD mucosa (7) and also because key mediators such as REL and TNFAIP3 present CD-associated variants (8). The transcription factor NFκB is a crucial regulator of the adaptive immune response and controls lymphocyte activation, proliferation and survival. Additionally, in a recent study, gliadin effects on enterocytes have been shown to be mediated through oxidative stress, NFκB activation and IL15 upregulation (9). Much progress has been made concerning our understanding of the signaling mediators and regulatory mechanisms that govern NFκB activation (10). TCR/CD28 coligation induces canonical NFκB signaling, leading to the transcriptional activity of NFKB1/RELA heterodimers (Fig. 1). This activation involves phosphorylation and degradation of small cytosolic IκB inhibitors, catalyzed by the IκB kinase complex (11). Activation of the atypical protein kinase C (encoded by PRKCQ) has been shown to provide a critical link between TCR/CD28 proximal signaling events and the NFκB pathway (12) through the phosphorylation of CARD11 and subsequent triggering of the recruitment and assembly of the CARD–BCL10–MALT1 (CBM) complex (13). The canonical pathway can also be activated through the signaling of several G protein coupled receptors, IL1- and toll-like receptors and tumor necrosis factor receptor type 1 (TNFR1), whereas the rest of the TNFR family usually activates the non-canonical NFκB pathway, leading to the formation of transcriptionally active NFKB2/RELB heterodimers (14).
Figure 1.
Simplified outline of the core of the NFκB route, focusing on the paracaspase MALT1 (in green) and its targets.
In this work, we studied the expression of a considerable fraction of the most central functional components of the NFκB route, as well as of several of its targets, most of them involved in processes that are relevant to CD, and attempted to normalize the altered expression of the NFκB pathway using Z-VRPR-FMK (Z in this article), a MALT1 inhibitor able to modulate the pathway without completely repressing it (15). Of note, MALT1 is an important paracaspase that can cleave BCL10, TNFAIP3 and CYLD, which respectively trigger the TCR-induced adhesion to fibronectin, the loss of NFκB inhibition downstream of IL1- and toll-like receptor signaling, and the activation of the JNK pathway (Fig. 1). It has also been suggested that MALT1 could take part in the cleavage of MAP3K14, given the fact that the API2-MALT1 fusion protein can do so in the lymphomas of mucosa-associated lymphoid tissue (MALT lymphoma) (14). Additionally, MALT1 controls caspase-8 activation during lymphocyte proliferation (16) and participates in the canonical NFκB activation observed in lymphocytes and lymphoma cell lines after RELB cleavage (17).
We hypothesized that CD could be a good target disease to investigate DNA methylation owing to its inflammatory disease condition. This study is the first one to investigate whether DNA methylation changes are occurring in the celiac gut and if so, whether they are reversible upon treatment with a gluten-free diet (GFD). The fact that several genes involved in the NFκB system have been consistently found to be hypomethylated in some cancers (18) provided us good candidates to study among our set of NFκB-related genes.
RESULTS
Overexpression of NFκB-related genes and in vitro modulation of the pathway
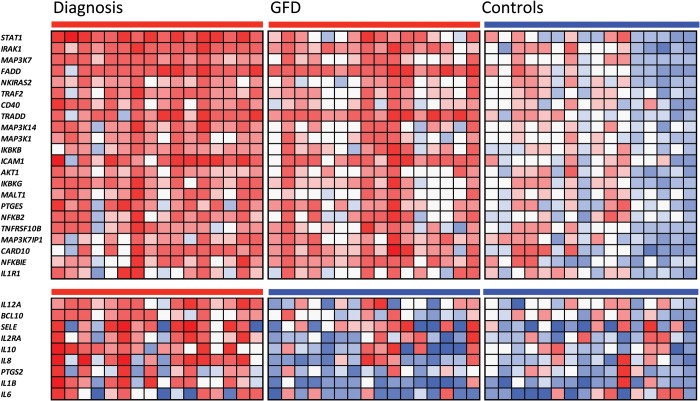
Using a custom-designed Taqman low density array (TLDA), the relative expression of 93 NFκB-related genes (Supplementary Material, Table S1) was assessed in basal (uncultured) biopsies from 16 active and 16 GFD-treated CD patients, as well as in 16 basal biopsies from non-celiac controls without intestinal inflammation. Twenty-two of the 93 genes studied were constitutively upregulated among patients, both at diagnosis (active CD) and after at least two years on GFD (Fig. 2). The only gene showing a significant constitutive downregulation in disease was ENPP2 (data not shown). In turn, 9 of the 93 genes were overexpressed only in the active disease group.
Figure 2.
Heat map showing the 22 constitutively upregulated NFκB-related genes as well as the 9 genes that are overexpressed only in active CD (t–test, P < 0.05). Rows represent samples taken from different patients. The red–blue scale represents up-/down-regulation gradient of each sample for each of the altered genes.
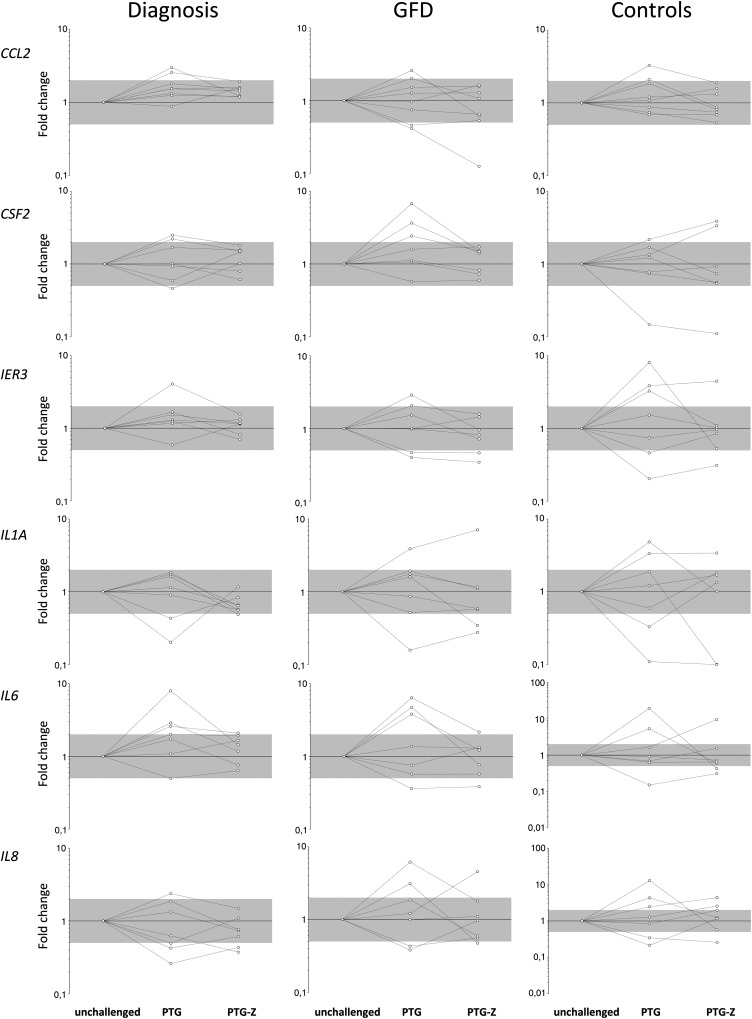
Eight biopsy portions from each group were incubated with either pepsin–trypsin-digested gliadin (PTG), both PTG and the NFκB modulator Z or without any of the compounds (unchallenged), during four hours. Changes in mRNA levels were observed in all groups and culture conditions. In the case of several genes, expression was altered upon stimulation with gliadin but reverted to levels similar to those found in unchallenged cultured biopsy portions when incubated with both PTG and Z (Fig. 3). Intriguingly, both the direction and the intensity of PTG-induced alterations varied notably among samples, whereas Z-mediated normalization was more uniform. Reversion by Z did not affect the group of genes that were constitutively overexpressed in CD patients, so we decided to focus on the differences between active and GFD disease statuses. We performed a class prediction analysis to extract the most meaningful genes indicative of disease activity and inflammation. Active and GFD-treated basal biopsies were randomly divided into training (12 samples from each group) and test subgroups, and the correlation-based feature selection (CFS) method was applied. Four genes (CCL20, IL1B, STAT1 and NOS2) were sufficient to correctly discriminate between disease statuses in seven of the eight test samples; one of the four treated patients was erroneously classified along with the active CD group. When the 16 biopsies from the treated group, cultured either in the presence of PTG or with both PTG and Z, were used as a second test group, PTG-challenged portions were classified into the active disease group, whereas the addition of Z was able to revert this in 5/8 cultured biopsy portions, which were included in the GFD-treated group.
Figure 3.
Relative expression fold change values in biopsies incubated under different conditions (PTG and PTG-Z) with respect to the unchallenged incubated biopsy of each individual. The gray area corresponds to FC values between 0.5 and 2. Lines indicate FC trends of each patient or control.
Coexpression of NFkB-related genes
Pearson's correlation matrixes constructed to check for coexpression between gene pairs showed stronger and more generalized correlations in controls, especially compared with the active CD group (Fig. 4), suggesting a very tight regulatory control of the pathway in non-celiac individuals. The GFD-treated group showed intermediate strength and number of correlations, representing a midway level of regulation in the route.
Figure 4.
Gene pair coexpression matrixes for the different disease statuses. Each small square represents the P value for the correlation of the expression level in a specific gene pair. White, light gray, dark gray and black indicate Pearson's correlation P values of >0.05,<0.05, <0.01 and <0.001, respectively.
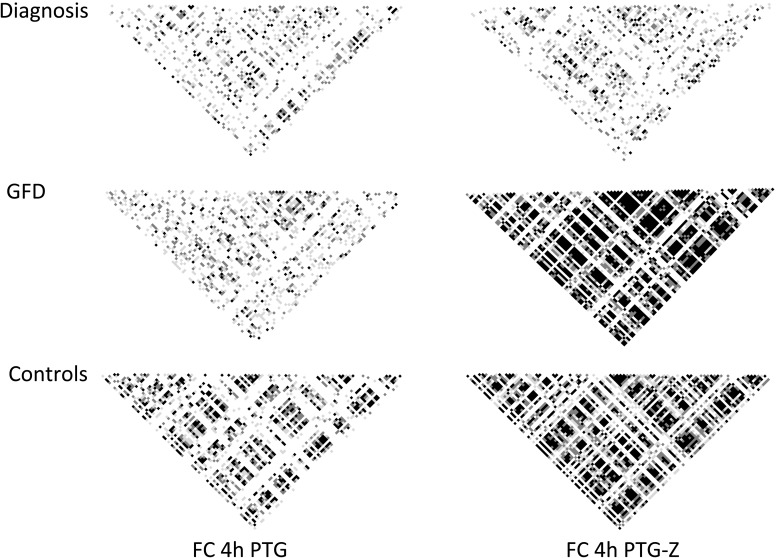
We also tested whether PTG and the combination of PTG and Z could affect the coexpression trends observed in the basal biopsies, so Pearson's correlation matrixes for the expression fold changes provoked by each stimulus (relative to the expression levels of unchallenged, cultured biopsy portions) were constructed. Neither PTG alone nor PTG and Z had remarkable effects on the coordination of expression fold changes in biopsies from the active patient group (Fig. 5). However, in the case of the GFD-treated and control groups, PTG stimulation caused a pronounced loss of the coordination of gene expression trends, and interestingly, coregulation was somewhat restored when the challenge included both PTG and the NFκB modulator Z. Of note, upon PTG challenge, a much lower coordination was observed in the GFD group compared with the control biopsies.
Figure 5.
Correlation matrixes showing P values for the coordinated response (correlation of fold changes with respect to the unchallenged cultured biopsy from each patient) of specific gene pairs under different stimuli (addition to the incubated biopsies of PTG or both PTG and Z modulator). Each small square represents the P value for the correlation of the expression level in a specific gene pair. White, light gray, dark gray and black indicate Pearson's correlation P values of >0.05, <0.05, <0.01 and < 0.001, respectively.
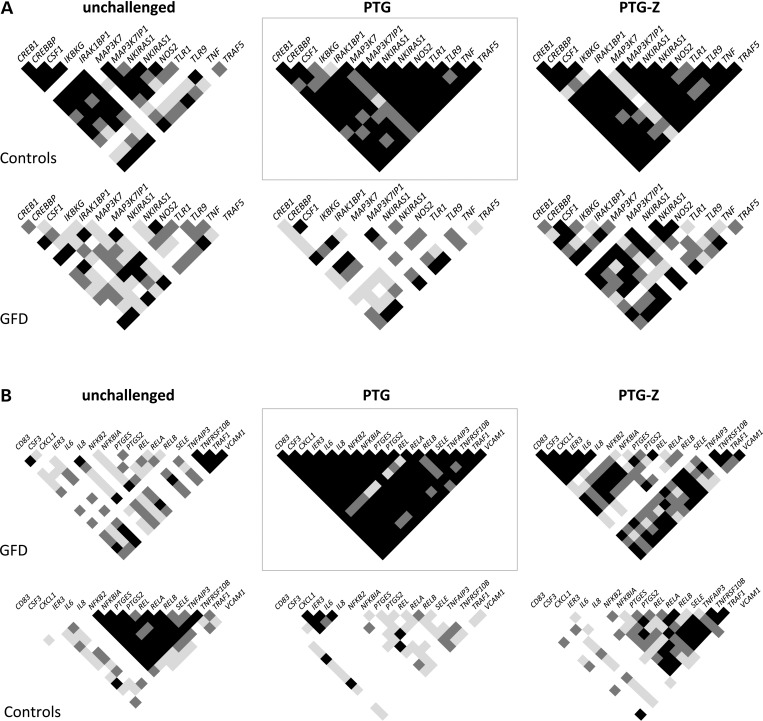
Finally, we identified the more prominent coexpression signatures provoked by PTG challenge in both the control and GFD groups. Gene pairs showing highly significant (P < 0.001) correlations between their expression levels in PTG-stimulated biopsies from controls (but not in the GFD group) were selected as the control-specific coexpression signature, whereas gene pairs showing highly significant correlations after PTG challenge of GFD celiac biopsies (but not in the case of controls) formed the GFD-specific coexpression signature. The correlation matrixes of the genes represented in each of the PTG-response signatures were also constructed in the unchallenged and PTG-Z situations to obtain a global picture of the coexpression profiles (Fig. 6).
Figure 6.
Correlation matrixes showing P values for the coexpression under different stimuli (unchallenged, PTG and PTG-Z) in a subset of gene pairs that constitute coexpression signatures specific to the (a) control and (b) GFD groups after PTG challenge (gray boxes). Each small square represents the P value for the correlation of the expression level in a specific gene pair. White, light gray, dark gray and black indicate Pearson's correlation P values >0.05, <0.05, <0.01 and <0.001, respectively.
The control-specific signature induced by PTG was more stable in every situation, and a strong correlation was still observed in controls in both the unchallenged and in the PTG-Z conditions, suggesting a lower dependence of the coregulation of those genes on the modulation of the NFκB route. The level of correlation of this particular gene set was lower in GFD patients, even though Z seemed capable of reestablishing certain coexpression relationships between several gene pairs. Meanwhile, the set of highly coexpressed genes that make up the GFD-specific response to PTG showed no correlation at all among controls, suggesting an aberrant and coordinated alteration of several genes of the NFκB route in the acute response to gliadin that is mounted in the celiac mucosa. An exception to the lack of coexpression observed in controls was found in CXCL1, IER3, IL6 and IL8 (which were coregulated among them), supporting the idea of a significant, albeit controlled activation in response to gliadin among non-celiac individuals. Again, Z seemed to partially revert this strongly significant coexpression induced by PTG towards the levels observed in unchallenged samples, and the control group adopted a correlation pattern similar to the one shown by unchallenged biopsies.
Partially reversible alteration of promoter methylation levels in active CD
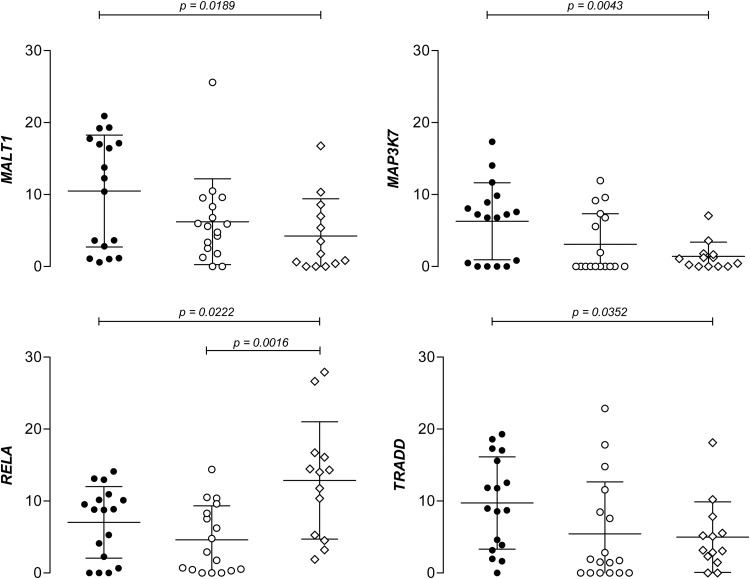
Eight NFκB-related genes (MALT1, MAP3K7, MAP3K14, NFKBIA, RELA, TAB1, TNFAIP3 and TRADD) were selected for the DNA methylation analysis according to the criteria described in section Material and methods, to assess the differences in promoter methylation levels among groups (Supplementary Material, Table S2). We calculated the average methylation percentage (percentage of DNA molecules presenting a methyl group) for each sample in the 4–6 CpG positions that were studied per gene. Four genes presented differences between active disease and control samples, whereas the GFD-treated group showed intermediate methylation levels, suggesting a partial reversion of the trend observed in the active disease status, except in the case of RELA, where methylation differences remained significant after GFD treatment (Fig. 7). MALT1, MAP3K7 and TRADD showed subtle but significantly higher methylation levels than controls and RELA presented the opposite trend. In contrast, LINE-1 elements did not present any methylation differences among groups (data not shown).
Figure 7.
NFκB-related genes showing statistically significant differences among groups in average methylation levels. Black and white circles represent active and treated CD patients, respectively, and diamonds represent non-celiac controls; the y-axis represents methylation values expressed in %; mean and standard deviation values are shown for each of the groups.
Co-methylation and coexpression networks
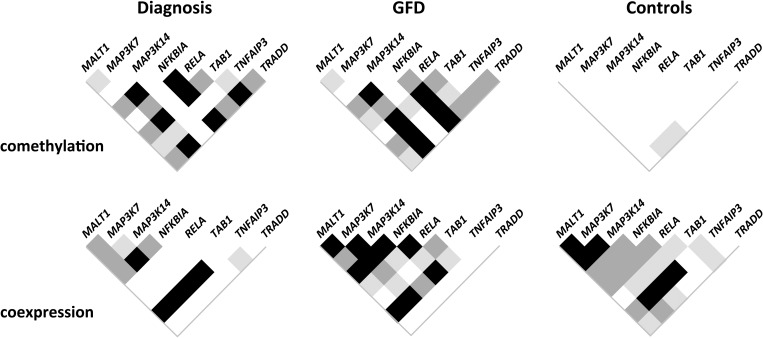
Pearson's correlation test was used to identify possible correlations among genes in both methylation and expression levels. Significant correlations were observed in the groups of celiac patients when methylation levels of different genes were tested, but no correlated patterns were observed at all in the control group or in LINE-1 elements (Fig. 8). In contrast, genes tended to be more strongly coexpressed in the control group, especially compared with patients at diagnosis, where correlations among gene expression levels were shown to be weaker and less frequent, although still significant in several cases. In general, coexpression between two genes was found to be stronger in those cases where co-methylation between them was weaker, particularly in the case of active patients.
Figure 8.
Correlation of methylation and gene expression levels in gene pairs. White squares indicate absence of correlation, whereas shades of gray correspond to P values of <0.05 (light), 0.01 (dark) and 0.001 (black).
MALT1 and RELA, as well as MAP3K7 and TRADD, were coexpressed only in the control group. In contrast, MAP3K7 and TRADD maintained a strong correlation of methylation percentages in both disease statuses, whereas they did not show such behavior in controls. In the case of MALT1 and RELA, they presented only a slight correlation of methylation levels in the treated patient group, even if both genes presented opposite trends when compared with controls.
When the most methylated sites of the above-mentioned genes were analyzed with the online tool ConSite (http:/www.phylofoot.org/consite) (19), both MALT1-RELA and MAP3K7-TRADD gene pairs were shown to share transcription factor binding sites with affinity scores of >80% (default score at the web page) (Supplementary Material, Fig. S1), suggesting a possible functional role for the differential methylation of these CpG islands in CD. In particular, the former gene pair shared a binding site for p65, whereas the latter presented a site in which Snail was predicted to bind.
Finally, a 12-h incubation of celiac biopsies under different conditions (PTG, PTG-Z and without any of the compounds) was performed in the four genes showing the best assay performance (NFKBIA, MAP3K7, TAB1 and TRADD). The biopsy culture caused the coordinated alteration of promoter methylation in active but not in treated disease (Supplementary Material, Fig. S2). Neither PTG nor the NFκB modulator Z had any predictable effects on methylation patterns, but methylation levels were altered, especially in the case of NFKBIA, with fold changes of >1.2 or <0.8 in 63% (15/24) of the samples from patients at diagnosis and 88% (21/24) of those from treated patients. After incubation, the correlation of methylation levels between NFKBIA and TRADD was only seen in biopsies from active patients, suggesting on the one hand the need for longer exposure periods for this phenomenon to take place and, on the other hand, that co-methylations are established in a cell subpopulation migration/selection independent manner, given the fact that biopsy cultures are isolated in vitro and foreign cells cannot be recruited into the site.
DISCUSSION
In this work, we have studied the expression of 93 NFκB-related genes and confirmed the previously reported (7) constitutive activation of a considerable fraction of the most central functional players of this biological route in CD. It is widely accepted that NFκB is a key regulator of inducible gene expression in the immune system. Both innate and adaptive immune responses, as well as the development and maintenance of the cells and organs that comprise the immune system are, at multiple stages, under the control of the NFκB family of transcription factors. Moreover, NFκB is responsible for the transcription of genes encoding a number of proinflammatory cytokines and chemokines (20). It has also been shown that NFκB is a major mediator of IL15, which is able to decrease claudin-2 levels in epithelial tight junction structures and leads to augmented paracellular permeability (21), a phenomenon that is recurrent, relevant and persistent in CD (22). The former characteristics and functions of NFκB, together with the fact that genetic polymorphisms in key NFκB-mediators such as REL and TNFAIP3 have been associated with susceptibility to CD (8), make this pathway an extremely interesting candidate to have a prominent causative role in the development of the disease. The present work supports the idea of a systemic genetic alteration in celiac patients that leads to the constitutive and anomalous activation of the NFκB route, a hallmark of the disease.
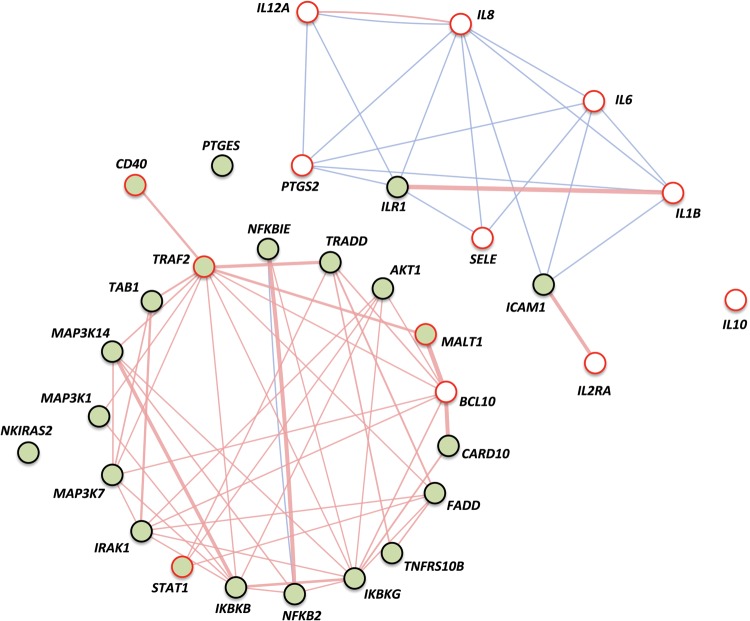
Accumulated information from the literature, functional enrichment web resources and also protein–protein interaction and pathway databases have been recently integrated into a single, comprehensive picture of the proteins that interact with, and participate to the NFκB activation system (23). Among other remarkable results, the work provides a list of the 25 most central proteins in the pathway. Interestingly, our results show that of the 22 genes that are constitutively overexpressed in CD mucosa, 7 (IKBKB, IKBKG, IRAK1, MAP3K14, NFKB2, NFKBIE and TRAF2) are among those core proteins. Another three genes are members of the MAPK family, which has been demonstrated central to the NFκB system by direct interaction, Uniprot annotation and manual curation approaches (23). Two other constitutively activated genes, MALT1 and CARD10, are part of the very central CBM complex (together with BCL10) and, as is the case of FADD and TRADD (also upregulated), interact directly with TRAF2 (Fig. 1).
In contrast, the genes that are overexpressed only in the active stage of the disease were found to be more peripheral to the core of the route, with the exception of BCL10. All these genes were found to be located downstream of at least one NFκB transcription factor family member, including interleukin genes IL1B (24), IL6 (25), IL8 (26), IL10 (27) and IL12A (28); the IL2 receptor alpha chain (IL2RA) (29); PTGS2 (30), a gene encoding the inducible cyclooxygenase; the adhesion mediator and leucocyte recruiting protein SELE (31), and finally, BCL10 (32), a very central regulator and enhancer of NFκB-TFs.
Overall, our results show that genes that are persistently upregulated usually belong to the essential core of the pathway and have crucial, regulatory and central roles in the NFκB signaling system, whereas genes that are overexpressed only in active CD appear to be more peripheral in the route and include mostly NFκB-inducible interleukins, adhesion molecules and receptors, as shown in Figure 9, depicting physical interactions (generally among core genes) and colocalizations (mostly linking peripheral genes, whose products are membrane bound or secreted), according to GeneMANIA (genemania.org) (33).
Figure 9.
Relationships among differentially expressed genes according to GeneMANIA. Constitutively upregulated genes are filled with green whereas genes overexpressed only in active disease are circled in red. Genes showing differential expression among all groups (active and GFD disease statuses as well as controls) are represented with both green background color and red line. Physical interactions occur generally between core genes and are shown by pink lines whereas colocalizations mostly link peripheral genes, whose products are membrane-bound or secreted, and are represented by blue lines. Line thickness depends on the strength of the interaction between genes.
On the other hand, we also found that Z, a modulator of the NFκB signaling route, was able to revert the effects of gliadin stimulation to the point that, after performing a CFS to assign each of the celiac samples to a class, PTG-challenged GFD biopsies were classified together with active patients whereas, more interestingly, the majority of the GFD samples challenged with both PTG and Z were recognized as coming from treated, inactive patients, indicating a certain degree of normalization of expression levels (reversion to the GFD-CD profile) despite an acute exposition to gliadin. Of note, gliadin stimulation provoked very different reactions in each of the different individuals, and several genes appeared both upregulated and downregulated in response to the challenge depending on the sample, illustrating the wide variability of human samples, particularly notable in terms of the effects of external stimuli on genetic expression in the context of a complex disease. However, the influence of Z seemed to be much more homogeneous and was able to revert the PTG effect in several genes.
It is important to stress that we observed strong correlation of expression levels among genes, especially in the case of non-celiac individuals, with GFD-treated patients showing levels of coregulation halfway between controls and active patients. Such degree of coexpression is expected, given the fact that these genes take part in the same biological route and are thus likely to share common regulatory mechanisms. Besides, as has been shown by systems biology approaches, coexpression networks form functional modules that underlie different traits (34). The fact that patients at diagnosis, on a gluten-containing diet, show a remarkably weaker coregulation of the NFκB system, points to the idea of chronic inflammation as the driving force for the altered and less coordinated gene expression profile in symptomatic patients.
To date, there have been few attempts to conduct comprehensive and comparative analyses of gene coexpression at the network level; a recent study aimed to reveal similarities and differences between hepatitis B virus-derived hepatocellular carcinoma (HBV-HCC) and hepatitis C virus (HCV)-derived HCC, focusing on the inflammatory processes driven by the viral infections. (35) This work demonstrated the superiority of a network-based systems biology approach in identifying different oncogenic and dysfunctional modules, over previously employed differential expression analysis and viral protein target-based methods. More recently, a gene coexpression network centered on the response mechanisms to bacterial infection was constructed in the sweet orange (Citrus sinensis) (36). The authors concluded that such gene coexpression network could be very useful to visualize specific subnetworks involved in particular biological processes and aid in the search for gene–gene interactions relevant to the resistance to infection.
Taking into account the potential effectiveness of coexpression-based approaches over classical differential expression-based analyses, we investigated whether the loss of the tight coregulation observed for non-CD controls in the selected NFκB-related genes depended not only on the presence/absence of disease, but also on disease status, gliadin stimulus and/or NFκB pathway modulation by MALT1 inhibition. We therefore studied the effects of both PTG and PTG-Z challenges by constructing correlation matrixes of the expression fold changes induced by the different stimuli. We did not observe coordinated changes in the case of active disease, where coregulation appeared to be completely disrupted and hampered the coordinated response of the different genes. In contrast, in the case of GFD patients and non-CD controls, PTG alone induced fewer coordinated gene expression changes than PTG combined with Z, particularly in the treated CD group. These results suggest that exposition to gliadin may cause the overall deregulation of NFκB-related gene expression, also in non-celiac individuals, but that this lack of coordination in the response is augmented in CD.
Interestingly, PTG-driven loss of coexpression reveals the enormous plasticity of correlated gene expression in the NFκB system as a consequence of the gliadin insult that could have a phenotypic effect, and in the context of other important events, finally translate into the aberrant and augmented response of celiac patients to gluten exposure. The nature of the putative regulators of these coexpressions remains elusive, but it seems plausible that the regulation of the system as a whole, and not just the constitutive and differential expression levels of isolated genes, is the one affected by unidentified genetic variation. Deregulation would therefore occur after an immunotoxic challenge and would only be evident after exposure to gliadin, as has been recently shown in the case of PTPRK and THEMIS gene coexpression in celiac active gut and after in vitro incubation with PTG (37).
The gene coexpression signature specific to the control group upon PTG challenge is somewhat less dependent on NFκB modulation and is tightly regulated. In contrast, this gene set was only faintly coexpressed in PTG-challenged biopsies from GFD celiac patients. For its part, the signature corresponding specifically to the PTG-challenged GFD-treated group was not coregulated at all in non-CD individuals (with several exceptions suggestive of immune activation), indicating an aberrant but coordinated response to the immunogenic insult.
In general, Z again seemed to partially restore the coexpression patterns present in unchallenged biopsies, putting forward that it might be an agent capable of reverting the impact of the gliadin insult in treated celiac patients that are not suffering from chronic inflammation. Once more, the regulatory elements taking part in the coexpressed signatures remain unknown. It will be interesting to study whether, in the future, Z could be used to avoid acute symptomatology in celiac patients that generally comply with the GFD but that might occasionally transgress the only effective treatment for this condition.
This is the first study to investigate changes in the methylation levels of several NFκB related promoters in the celiac gut. The changes observed are somehow maintained in patients even after more than two years on GFD, although most of the time, differences with controls are less pronounced than in active disease, suggesting that CD-related methylation changes may be partially reversible or that more time may be needed in order that methylation levels are normalized. In the recent years, the idea of epigenetic elements as causal contributors to phenotypic variation and common disease has gained credit. For example, several inflammatory diseases of the digestive tract such as inflammatory bowel disease (38) or distal colonic neoplasia (39) as well as other complex diseases such as Type 1 Diabetes (T1D) (40) or psychosis (41) and a long list of cancers (42,43) have been found to have specific DNA methylation signatures.
One of the most interesting findings of the present work relies on the fact that patients in every stage of the disease present strong correlations in the methylation levels of several NFκB-related genes, whereas controls do not at all. Although further research will be needed to confirm this idea, it seems that the observed co-methylations are independent of cell migration and/or selection events, because they can be reproduced in isolated, in vitro-cultured biopsy samples. It has been previously shown in cancer that genes with promoter hypermethylation and hypomethylation are highly consistent in function across different cancer types (44) so that it is intuitively probable that methylation patterns of genes sharing function or participating in the same pathway could be regulated in a coordinated manner in a particular disease status or in response to a specific environmental challenge. Additionally, a recent work on breast cancer has shown that unexpectedly highly correlated DNA methylation levels are found in gene pairs from cancer samples (45). It is important to stress that correlated gene pairs showed strong, combined functional similarities, which led the authors to suggest that these findings may be helpful to annotate unknown genes and to suggest candidate genes that should be closely investigated with respect to particular diseases. Interestingly, in our study, correlation of methylation levels in patients seemed to be somehow associated with the disruption of the coexpression of those same gene pairs, suggesting a coordinated alteration of gene promoter methylation of NFκB-related genes that could affect gene expression, disrupting the regulatory equilibrium of the pathway.
In conclusion, we postulate that genetic and epigenetic variation, together with environmental factors that shape expression and methylation patterns, can affect different levels of the NFκB pathway and provoke the disruption of the tight regulatory equilibrium that ensures its optimal control of the immune response. Maintained disruption of this coordination could constitute a major driving force towards the development of chronic inflammatory disease, as is the case of CD. Although this appealing idea is still very speculative, we believe these results could humbly help configuring a novel point of view, from which the importance of networks, interactions and common regulatory elements or events outweighs the subtle and multiple changes that we observe when interrogating isolated variables, through which we have tried to explain the common complex diseases in the last years.
MATERIAL AND METHODS
Patients and biopsies
Celiac disease was diagnosed according to the ESPGHAN (European Society of Pediatric Gastroenterology Hematology and Nutrition) criteria in force at the time participants were recruited and included anti-gliadin and anti-transglutaminase antibody determinations as well as a confirmatory small bowel biopsy. Individuals without neither CD nor intestinal inflammation that nevertheless underwent endoscopy were recruited as non-celiac controls. Subjects were included in the study, and samples were collected and analyzed after informed consent was obtained from patients or their parents. The study was approved by the local Ethic Committee of the BioCruces Research Institute. Biopsy specimens from the distal duodenum were obtained during routine endoscopy and were immediately frozen (basal tissues) or incubated as described later and were subsequently stored in liquid nitrogen until DNA or RNA was extracted.
Gliadin preparation and biopsy cultures
Gliadin was digested with pepsin and trypsin to obtain PTG as previously described (37). Briefly, 500 mg of gliadin from wheat (Sigma–Aldrich, Cat. No. G3375) was dissolved in 5 ml 0.2 m HCl and incubated with 5 mg pepsin (Sigma–Aldrich, Cat. No. P6887) with continuous stirring at 37°C for 2 h. At the end of this incubation, pH was adjusted to 7.4 with NaOH and the mixture was incubated with 5 mg trypsin (Sigma–Aldrich, Cat. No. T9201) at 37°C for 4 h. To inactivate the enzymes, the solution was heated in a boiling water bath for 30 min. The digest was centrifuged at 2000 g for 10 min, and the supernatant was sterilized by filtering through a 20-µm pore membrane.
We designed an in vitro assay to assess whether PTG or the NFκB modulator Z-VRPR-FMK (Z in this article) (Enzo Life Sciences, Cat. No. ALX-260-166-M001) affects the expression levels or the coexpression patterns in the studied genes. We incubated three biopsy portions, taken from each of the eight active and eight GFD-treated celiac patients as well as from the eight non-CD controls selected for the modulation study, in 150 µl RPMI-1640 10X medium at 37°C and 5% CO2 during 4 h. Apart from the basal biopsy, immediately frozen in liquid nitrogen, a second biopsy portion was cultured during 4 h without any compound (unchallenged), a third was incubated 4 h in the presence of 250 μg/ml PTG (PTG) and a last portion was challenged with the same amount of PTG after a 20-min incubation with 60 µg/ml Z (PTG-Z).
RNA samples for expression analysis
Total RNA was extracted using the NucleoSpin miRNA kit (Macherey-Nagel, Cat. No. 740971.50) from 16 active celiac biopsies, 16 unrelated biopsies taken from GFD-treated celiac patients and from 16 non-celiac controls, as well as from 3 incubated biopsy portion series for each of the 8 patients selected from each group for the in vitro challenge study. After column purification, RNA was eluted in a final volume of 20 μl, quantified using a Nanodrop spectrophotometer (ND-1000, Thermo Scientific) and diluted to a final concentration of 50 ng/μl.
Reverse transcription and quantitative PCR
Reverse transcription (RT) reactions were performed in a C1000 Thermal Cycler (BioRad) using 800 ng RNA in a final reaction volume of 20 μl using SuperScript VILO cDNA Master Mix (Life Technologies, Cat. No. 11754-050), following the manufacturer's protocol. Real-time quantitative PCR experiments of 93 commercially available Taqman Gene Expression Assays targeting NFκB-related genes were performed in a 7900HT RT-PCR system in a TLDA format (assay list in Supplementary Material, Table S1), which functions as an array of reaction vessels for the PCR step. Relative levels of gene expression were determined from the fluorescence data generated during PCR using the ABI PRISM 7900HT Sequence Detection System (all from Life Technologies) and normalized to the expression of the housekeeping gene RPLPO (Life Technologies, Cat. No. 4326314E), which was run simultaneously in each TLDA.
DNA samples for methylation studies
Genomic DNA was isolated from 13 basal biopsies from individuals without intestinal inflammation as well as from basal biopsy pairs from 17 celiac patients, one taken at diagnosis and the other one taken after more than 2 years on GFD. Isolation of DNA was performed using QIAamp DNA Mini kit (QIAGEN, Cat. No. 51304) following the manufacturer's instructions. Samples were quantified using Quant-it PicoGreen dsDNA reagent (Invitrogen, Cat. No. P7581), and DNA concentrations were adjusted to 20 ng/µl in a final volume of 20 µl.
Nine-point methylation curves were used to calibrate the samples run in different plates and to correct for PCR bias using the cubic regression method described previously (46). Curves were constructed combining different amounts of commercially available HeLa Genomic DNA—used as unmethylated DNA (47)—and CpG Methylated HeLa Genomic DNA—representing totally methylated human genomic DNA—both from New England Biolabs (Cat. Nos. N4006S and N4007S, respectively). The resulting methylation levels of the curve were the following: 0, 12.5, 25, 37.5, 50, 62.5, 75, 87.5 and 100%.
Sodium bisulfite conversion of unmethylated cytosines was performed in 400 ng of genomic DNA from each sample using the commercially available Epitect Bisulfite Kit (QIAGEN, Cat. No. 59104) as indicated by the manufacturer, and column-purified into 40 μl. Briefly, the protocol comprises the following steps: bisulfite-mediated conversion of unmethylated cytosines, binding of the converted single-stranded DNA to the membrane of an EpiTect spin column, desulfonation of DNA and washing and elution of the pure, converted DNA from the membrane.
Selection and pyrosequencing of the CpG islands
Eight different genes involved in the NFκB signaling route were selected for methylation analysis (Supplementary Material, Table S2) according to the following criteria: on the one hand, they presented CpG-enriched promoters or first exons, which have been shown to be even more relevant to expression than the former (48). On the other hand, predesigned methylation assays located in the regions of interest were commercially available, and finally, the genes were known to develop regulatory and central functions in the NFκB biological route. Each of the assays selected covered 4–6 CpG islands. Additionally, PyroMark Q24 CpG LINE-1 kit (QIAGEN, Cat. No. 970012) was used to estimate the global methylation of each of the 17 paired celiac and the 13 control basal biopsies.
Amplifications of the loci of interest were performed in a C1000 Thermal Cycler (BioRad) using 1.5 μl of DNA in 25-μl PCR reactions with the PyroMark PCR Kit (QIAGEN Cat. No. 978703) and the forward and the biotinylated reverse specific primers provided with each PyroMark CpG Assay, following the manufacturer's protocol. Thermocycling conditions were as follows: 15 min at 95°C for enzyme activation followed by 45 cycles of denaturation for 30 s at 95°C, annealing for 30 sec at 56°C and extension for 30 s at 72°C, with a final extension of 10 min at 72°C. PCR reactions were tested by agarose gel electrophoresis.
After PCR amplification, 20 μl of each biotinylated PCR product were bound to streptavidin-coated sepharose beads (GE Healthcare Cat. No. 17-5113-01) in 24-well plates and run in a PyroMark Q24 system. PCR products were denatured, and non-biotinylated strands were removed using the PyroMark Q24 vacuum workstations. The beads were then resuspended in 25 μl of annealing buffer containing 1X of the specific sequencing primer from each CpG Assay. Pyrosequencing was performed using PyroGold Q24 reagents (QIAGEN, Cat. No. 970802). Nucleotide, enzyme and substrate dispensation order and volumes, as well as the duration of each step, were planned using PyroMark software based on the target sequence. Following the runs, raw data files were imported into the software for standard quality controls and preliminary analyses.
Data analysis and statistics
The relative expression of each gene was calculated using the accurate ΔΔCt method as previously described (49). t-test was applied in order to assess the differences among groups, and Data Analysis ToolPack from Microsoft Excel 2010 v.10.0 was used to build Pearson's correlation matrixes for each basal or in vitro-cultured biopsy group. Both plots showing fold changes of the incubation series and the heat map containing overexpressed genes were constructed using different Macros for Microsoft Excel 2010 v.10.0.
Class prediction and subsequent assessment of expression normalization by Z were performed using tools available online at the Babelomics website (http://babelomics.bioinfo.cipf.es/) (50). Briefly, gene subset selection was performed by CFS method, using K-fold error estimation (10 repetitions, 5-fold default parameters) and Random Forest algorithm. Samples were randomly divided into a training (12 active and 12 treated CD patient samples) and a test (4 active and 4 treated CD) subgroup. A second test group containing the eight GFD-treated biopsies incubated with PTG and the same biopsies challenged with both PTG and Z was also tested.
In the methylation study, results of the artificially constructed methylation curve included in each plate were fitted into a polynomial cubic equation (y = ax3+ cx2 + dx + e), with the real methylation levels plotted on the x-axis and the observed, experimental values on the y-axis, using Microsoft Excel v.10.0. With this equation, more accurate methylation levels were estimated for each experimental result by solving the x with Cardano's method, as previously described. (51)
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the Instituto de Salud Carlos III of the Spanish Ministry of Economy and Competitiveness (grant no. PI10/0310) and from the Basque Departments of Health and Industry (grant nos. SAN201111034 and SAIO-PE08BF03, respectively) to J.R.B. N.F.-J. and L.P.-I. are predoctoral fellows funded by FPI grants from the Basque Department of Education, Universities and Research (grant nos. BIF-2009-099 and BIF-2010-189, respectively). Technical and human support provided by SGIker (UPV/EHU, MICINN, GV/EJ, ERDF and ESF) is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by the Instituto de Salud Carlos III of the Spanish Ministry of Economy and Competitiveness.
Supplementary Material
REFERENCES
- 1.Green P.H., Cellier C. Celiac disease. N. Engl. J. Med. 2007;357:1731–1743. doi: 10.1056/NEJMra071600. [DOI] [PubMed] [Google Scholar]
- 2.Jabri B., Sollid L.M. Tissue-mediated control of immunopathology in coeliac disease. Nat. Rev. Immunol. 2009;9:858–870. doi: 10.1038/nri2670. [DOI] [PubMed] [Google Scholar]
- 3.Greco L., Romino R., Coto I., Di Cosmo N., Percopo S., Maglio M., Paparo F., Gasperi V., Limongelli M.G., Cotichini R., et al. The first large population based twin study of coeliac disease. Gut. 2002;50:624–628. doi: 10.1136/gut.50.5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M.C., Barnardo M.C., Bethel G., Holmes G.K., et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adány R., Aromaa A., et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trynka G., Hunt K.A., Bockett N.A., Romanos J., Mistry V., Szperl A., Bakker S.F., Bardella M.T., Bhaw-Rosun L., Castillejo G., et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castellanos-Rubio A., Santin I., Martin-Pagola A., Irastorza I., Castaño L., Vitoria J.C., Bilbao J.R. Long-term and acute effects of gliadin on small intestine of patients on potentially pathogenic networks in celiac disease. Autoimmunity. 2010;43:131–139. doi: 10.3109/08916930903225229. [DOI] [PubMed] [Google Scholar]
- 8.Trynka G., Zhernakova A., Romanos J., Franke L., Hunt K.A., Turner G., Bruinenberg M., Heap G.A., Platteel M., Ryan A.W., et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-κB signalling. Gut. 2009;58:1078–1083. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- 9.Parmar A., Greco D., Venäläinen J., Gentile M., Dukes E., Saavalainen P. Gene expression profiling of Gliadin effects on intestinal epithelial cells suggests novel non-enzymatic functions of pepsin and trypsin. PLoS One. 2013;8:e66307. doi: 10.1371/journal.pone.0066307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Düwel M., Welteke V., Oeckinghaus A., Baens M., Kloo B., Ferch U., Darnay B.G., Ruland J., Marynen P., Krappmann D. A20 negatively regulates T cell receptor signaling to NF-κB by cleaving Malt1 ubiquitin chains. J. Immunol. 2009;15:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 11.Hayden M.S., Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Sun Z., Arendt C.W., Ellmeier W., Schaeffer E.M., Sunshine M.J., Gandhi L., Annes J., Petrzilka D., Kupfer A., Schwartzberg P.L., Littman D.R. PKC-φ is required for TCR-induced NF-κB activation in mature but not immature T lymphocytes. Nature. 2000;404:402–407. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- 13.Rawlings D.J., Sommer K., Moreno-Garcia M.E. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat. Rev. Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 14.Rosebeck S., Rehman A.O., Lucas P.C., McAllister-Lucas L.M. From MALT lymphoma to the CBM signalosome: three decades of discovery. Cell Cycle. 2011;10:2485–2496. doi: 10.4161/cc.10.15.16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebeaud F., Hailfinger S., Posevitz-Fejfar A., Tapernoux M., Moser R., Rueda D., Gaide O., Guzzardi M., Iancu E.M., Rufer N., et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 16.Kawadler H., Gantz M.A., Riley J.L., Yang X. The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol. Cell. 2008;31:415–421. doi: 10.1016/j.molcel.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hailfinger S., Nogai H., Pelzer C., Jaworski M., Cabalzar K., Charton J.E., Guzzardi M., Décaillet C., Grau M., Dörken B., et al. Malt1-dependent RelB cleavage promotes canonical NF-kappaB activation in lymphocytes and lymphoma cell lines. Proc. Natl. Acad. Sci. USA. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao C., Li H., Shen X., He Z., He L., Guo Z. Reproducibility and concordance of differential DNA methylation and gene expression in cancer. PLoS One. 2012;7:e29686. doi: 10.1371/journal.pone.0029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandelin A., Wasserman W.W., Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayden M.S., Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone K.P., Kastin A.J., Pan W. NFĸB is an unexpected major mediator of interleukin-15 signaling in cerebral endothelia. Cell. Physiol. Biochem. 2011;28:115–124. doi: 10.1159/000331720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjarnason I., Peters T.J. In vitro determination of small intestinal permeability: demonstration of a persistent defect in patients with coeliac disease. Gut. 1984;25:145–150. doi: 10.1136/gut.25.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tieri P., Termanini A., Bellavista E., Salvioli S., Capri M., Franceschi C. Charting the NF-κB pathway interactome map. PLoS One. 2012;7:e32678. doi: 10.1371/journal.pone.0032678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiscott J., Marois J., Garoufalis J., D'Addario M., Roulston A., Kwan I., Pepin N., Lacoste J., Nguyen H., Bensi G., et al. Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: evidence for a positive autoregulatory loop. Mol. Cell. Biol. 1993;13:6231–6240. doi: 10.1128/mcb.13.10.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Libermann T.A., Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell. Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunsch C., Rosen C.A. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol. Cell. Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L.G., Shu H.B. TNFR-associated factor-3 is associated with BAFF-R and negatively regulates BAFF-R-mediated NF-kappa B activation and IL-10 production. J. Immunol. 2002;169:6883–6889. doi: 10.4049/jimmunol.169.12.6883. [DOI] [PubMed] [Google Scholar]
- 28.Homma Y., Cao S., Shi X., Ma X. The Th2 transcription factor c-Maf inhibits IL-12p35 gene expression in activated macrophages by targeting NF-kappaB nuclear translocation. J. Interferon Cytokine Res. 2007;27:799–808. doi: 10.1089/jir.2007.0006. [DOI] [PubMed] [Google Scholar]
- 29.Ballard D.W., Böhnlein E., Lowenthal J.W., Wano Y., Franza B.R., Greene W.C. HTLV-I tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science. 1988;241:1652–1655. doi: 10.1126/science.241.4873.1652. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto K., Arakawa T., Ueda N., Yamamoto S. Transcriptional roles of nuclear factor kappa B and nuclear factor-interleukin-6 in the tumor necrosis factor alpha-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J. Biol. Chem. 1995;270:31315–31320. doi: 10.1074/jbc.270.52.31315. [DOI] [PubMed] [Google Scholar]
- 31.Schindler U., Baichwal V.R. Three NF-kappa B binding sites in the human E-selectin gene required for maximal tumor necrosis factor alpha-induced expression. Mol. Cell. Biol. 1994;14:5820–5831. doi: 10.1128/mcb.14.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borthakur A., Bhattacharyya S., Anbazhagan A.N., Kumar A., Dudeja P.K., Tobacman J.K. Prolongation of carrageenan-induced inflammation in human colonic epithelial cells by activation of an NFκB-BCL10 loop. Biochim. Biophys. Acta. 2012;1822:1300–1307. doi: 10.1016/j.bbadis.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Fuente A. From ‘differential expression’ to ‘differential networking’ - identification of dysfunctional regulatory networks in diseases. Trends Genet. 2010;26:326–333. doi: 10.1016/j.tig.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 35.He D., Liu Z.P., Honda M., Kaneko S., Chen L. Coexpression network analysis in chronic hepatitis B and C hepatic lesions reveals distinct patterns of disease progression to hepatocellular carcinoma. J. Mol. Cell. Biol. 2012;4:140–152. doi: 10.1093/jmcb/mjs011. [DOI] [PubMed] [Google Scholar]
- 36.Zheng Z.L., Zhao Y. Transcriptome comparison and gene coexpression network analysis provide a systems view of citrus response to ‘Candidatus Liberibacter asiaticus’ infection. BMC Genomics. 2013;14:27. doi: 10.1186/1471-2164-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondar C., Plaza-Izurieta L., Fernandez-Jimenez N., Irastorza I., Withoff S., Wijmenga C., Chirdo F., Bilbao J.R. CEGEC. THEMIS and PTPRK in celiac intestinal mucosa: coexpression in disease and after in vitro gliadin challenge. Eur. J. Hum. Genet. 2013 doi: 10.1038/ejhg.2013.136. doi:10.1038/ejhg.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartnett L., Egan L.J. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis. 2012;33:723–731. doi: 10.1093/carcin/bgs006. [DOI] [PubMed] [Google Scholar]
- 39.Hiraoka S., Kato J., Horii J., Saito S., Harada K., Fujita H., Kuriyama M., Takemoto K., Uraoka T., Yamamoto K. Methylation status of normal background mucosa is correlated with occurrence and development of neoplasia in the distal colon. Hum. Pathol. 2010;41:38–47. doi: 10.1016/j.humpath.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Bell C.G., Teschendorff A.E., Rakyan V.K., Maxwell A.P., Beck S., Savage D.A. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med. Genomics. 2010;5:33. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mill J., Tang T., Kaminsky Z., Khare T., Yazdanpanah S., Bouchard L., Jia P., Assadzadeh A., Flanagan J., Schumacher A., et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell A., Bell D., Weber R.S., El-Naggar A.K. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer. 2011;117:2898–2909. doi: 10.1002/cncr.25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S., Dorsey T.H., Terunuma A., Kittles R.A., Ambs S., Kwabi-Addo B. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012;7:e37928. doi: 10.1371/journal.pone.0037928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen X., He Z., Li H., Yao C., Zhang Y., He L., Li S., Huang J., Guo Z. Distinct functional patterns of gene promoter hypomethylation and hypermethylation in cancer genomes. PLoS One. 2012;7:e44822. doi: 10.1371/journal.pone.0044822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akulenko R., Helms V. DNA co-methylation analysis suggests novel functional associations between gene pairs in breast cancer samples. Hum. Mol. Genet. 2013;22:3016–3022. doi: 10.1093/hmg/ddt158. [DOI] [PubMed] [Google Scholar]
- 46.Fernandez-Jimenez N., Plaza-Izurieta L., Lopez-Euba T., Jauregi-Miguel A., Bilbao J.R. Cubic regression-based degree of correction predicts the performance of whole bisulfitome amplified DNA methylation analysis. Epigenetics. 2012;7:1349–1354. doi: 10.4161/epi.22846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian Z.R., Sano T., Yoshimoto K., Yamada S., Ishizuka A., Mizusawa N., Horiguchi H., Hirokawa M., Asa S.L. Inactivation of RASSF1A tumor suppressor gene by aberrant promoter hypermethylation in human pituitary adenomas. Lab. Invest. 2005;85:464–473. doi: 10.1038/labinvest.3700248. [DOI] [PubMed] [Google Scholar]
- 48.Brenet F., Moh M., Funk P., Feierstein E., Viale A.J., Socci N.D., Scandura J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martín-Pagola A., Ortiz L., Pérez de Nanclares G., Vitoria J.C., Castaño L., Bilbao J.R. Analysis of the expression of MICA in small intestinal mucosa of patients with celiac disease. J. Clin. Immunol. 2003;23:498–503. doi: 10.1023/b:joci.0000010426.16956.cc. [DOI] [PubMed] [Google Scholar]
- 50.Medina I., Carbonell J., Pulido L., Madeira S.C., Goetz S., Conesa A., Tárraga J., Pascual-Montano A., Nogales-Cadenas R., Santoyo J., et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38:W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moskalev E.A., Zavgorodnij M.G., Majorova S.P., Vorobjev I.A., Jandaghi P., Bure I.V., Hoheisel J.D. Correction of PCR-bias in quantitative DNA methylation studies by means of cubic polynomial regression. Nucleic Acids Res. 2011;39:e77. doi: 10.1093/nar/gkr213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.