Abstract
Spinal muscular atrophy results from diminished levels of survival motor neuron (SMN) protein in spinal motor neurons. Low levels of SMN also occur in models of amyotrophic lateral sclerosis (ALS) caused by mutant superoxide dismutase 1 (SOD1) and genetic reduction of SMN levels exacerbates the phenotype of transgenic SOD1G93A mice. Here, we demonstrate that SMN protein is significantly reduced in the spinal cords of patients with sporadic ALS. To test the potential of SMN as a modifier of ALS, we overexpressed SMN in 2 different strains of SOD1G93A mice. Neuronal overexpression of SMN significantly preserved locomotor function, rescued motor neurons, and attenuated astrogliosis in spinal cords of SOD1G93A mice. Despite this, survival was not prolonged, most likely resulting from SMN mislocalization and depletion of gems in motor neurons of symptomatic mice. Our results reveal that SMN upregulation slows locomotor deficit onset and motor neuron loss in this mouse model of ALS. However, disruption of SMN nuclear complexes by high levels of mutant SOD1, even in the presence of SMN overexpression, might limit its survival promoting effects in this specific mouse model. Studies in emerging mouse models of ALS are therefore warranted to further explore the potential of SMN as a modifier of ALS.
Keywords: Amyotrophic lateral sclerosis, Spinal muscular atrophy, Superoxide dismutase 1, Survival motor neuron
1. Introduction
Motor neuron diseases are primary disorders of corticospinal, corticobulbar, cranial, and spinal efferents leading to secondary muscle atrophy. In the case of amyotrophic lateral sclerosis (ALS), upper and lower motor neurons degenerate in adults, resulting in muscle wasting and fatal paralysis. Although most ALS is sporadic, mutations in superoxide dismutase 1 (SOD1) are the best understood cause of inherited ALS (Ferraiuolo et al., 2011). In spinal muscular atrophy (SMA), there is lower motor neuron loss in spinal cords of infants, causing muscle atrophy, weakness, and death. Autosomal recessive SMA results from inactivating mutations in the survival motor neuron 1 (SMN1) gene, which leads to reduced dosage of full-length survival motor neuron (SMN) protein (Burghes and Beattie, 2009). The severity of SMA is determined according to retention and copy number of the almost identical SMN2 gene, which produces a transcript lacking exon 7 (SMNΔ7) and only approximately 10% full-length SMN protein.
The vulnerability of lower motor neurons in the spinal cord in both ALS and SMA suggests shared disease mechanisms and susceptibility factors. Genetic association studies provide evidence for SMN1 and SMN2 deletions and duplications as risk factors for sporadic ALS. Abnormal SMN1 copy numbers of 1 (Corcia et al., 2002, 2006; Veldink et al., 2005) or 3 (Blauw et al., 2012; Corcia et al., 2002, 2006) have been associated with ALS. SMN2 absence (Lee et al., 2012; Veldink et al., 2001) or single copy (Veldink et al., 2005) were also associated with increased risk for ALS, and paradoxically, homozygous SMN2 deletion was recently reported as protective (Corcia et al., 2012). In contrast to SMA, no point mutations in SMN1 were found in ALS (Blauw et al., 2012). Despite these apparently conflicting genetic association data, it was speculated that either deletions or duplications of SMN1 or SMN2 would predict abnormal SMN protein levels in ALS that render spinal motor neurons susceptible to degeneration (Blauw et al., 2012; Veldink et al., 2005). However, SMN expression studies have not yet been performed on ALS tissue to verify this hypothesis.
A second line of evidence that SMN loss contributes to ALS pathogenesis comes from genetic models of ALS. In motor neuronal cell lines and spinal cords of transgenic mice expressing mutant superoxide dismutase (SOD1), levels of SMN protein were significantly reduced and SMN was mislocalized from the nucleus, where it usually forms structures known as gems (Turner et al., 2008; Turner et al., 2009). Disruption of nuclear SMN complexes was also recently reported in spinal motor neurons of other mutant SOD1 mouse lines before symptom onset (Gertz et al., 2012; Kariya et al., 2012). Furthermore, partial ablation of mouse SMN accelerates disease progression and paralysis in SOD1G93A mice (Turner et al., 2009), demonstrating that reduced SMN expression can enhance the ALS-like phenotype in mice. Most recently, disruption of SMN gems by ALS-linked mutant TAR DNA binding protein 43 (TDP-43) and fused in sarcoma (FUS) was shown (Groen et al., 2013; Yamazaki et al., 2012), reinforcing evidence that loss of SMN function might be a common pathogenic pathway in ALS and SMA.
Collectively, these studies in patients with ALS and genetic models raise 2 salient questions. First, is SMN protein expression aberrant in human sporadic ALS, as it is in models of mutant SOD1-induced disease? In this study, we demonstrated that SMN protein levels are lower in the spinal cords of patients with sporadic ALS compared with age-matched control subjects. Second, can increased expression of SMN in mutant SOD1 mice ameliorate disease, as measured according to motor neuron pathology, age of disease onset, or rate of progression? Here, we showed that increased SMN expression in neurons of 2 different strains of SOD1G93A mice delayed onset of motor deficits and robustly protected spinal motor neurons, and did not prolong life span. This may result from specific displacement of SMN complexes from the nucleus of motor neurons in spinal cord, mediated by the very high levels of mutant SOD1 expression characteristic of this mouse model. The outcomes of our study have important ramifications for future trials of SMN therapeutic agents in ALS models that aim to replace or upregulate SMN protein. Although SMN is neuroprotective in the spinal cord and can slow symptom onset in mutant SOD1 mice, it may require additional therapeutic agents and strategies to promote an overall increase in survival in the presence of high levels of mutant SOD1 protein expression.
2. Methods
2.1. Human tissue
This study was approved by the Howard Florey Institute Human Ethics Committee. Human lumbar spinal cord segments (L3–L5) from 9 patients who died from respiratory failure caused by ALS were provided by the Motor Neuron Disease Research Tissue Bank of Victoria (Table 1). The clinical diagnosis of ALS was confirmed at postmortem and there was no family history of ALS. Control lumbar spinal cords were obtained from 5 individuals without evidence for neurologic or psychiatric disease. The mean age and postmortem interval for ALS and control cases did not significantly differ. Tissues were fresh-frozen in liquid nitrogen and stored at −80 °C.
Table 1.
Patient information
| Control cases | ||||
|---|---|---|---|---|
| Case | Sex | Age (y) | PMI (h) | Cause of death |
| 1 | M | 52 | 33 | Aortic rupture |
| 2 | M | 75 | 50 | Acute myocardial infarction |
| 3 | M | 64 | 68 | Ischemic heart disease |
| 4 | M | 85 | 68 | Ischemic heart disease |
| 5 | M | 86 | 71 | Multiple myeloma |
| ALS patients | ||||
|---|---|---|---|---|
| Case | Sex | Age (y) | PMI (h) | Disease duration (y) |
| 1 | M | 42 | 8 | 3.5 |
| 2 | M | 79 | 25 | 7.0 |
| 3 | F | 56 | 45 | 1.0 |
| 4 | F | 60 | 90 | 1.6 |
| 5 | M | 72 | 7 | 8.7 |
| 6 | F | 63 | 72 | 1.4 |
| 7 | M | 72 | 67 | 3.8 |
| 8 | M | 38 | 21 | 7.3 |
| 9 | M | 64 | 14 | 2.0 |
Key: Age, age at death; F, female; M, male; PMI, postmortem interval.
2.2. Mouse lines
All experiments conformed to the Australian National Health and Medical Research Council published code of practice and were approved by the Howard Florey Institute Animal Ethics Committee (permit number 10-024) and Home Office and University of Oxford. Transgenic SOD1G93A mice derived from the B6SJL-Tg(SOD1*G93A)1Gur/J line (Jackson Laboratory, USA, strain 002726) were maintained on a mixed B6SJL background (Florey Institute of Neuroscience and Mental Health) or congenic C57BL/6 background (University of Oxford). PrP-SMN mice (line 92) maintained on an FVBn background were provided by Prof Arthur Burghes (Ohio State University). Heterozygous PrP-SMN mice carry 8–9 copies of human SMN1 and express 2–3-fold SMN protein in brain and spinal cord which rescues a severe SMA mouse model (Gavrilina et al., 2008). Male SOD1G93A mice were intercrossed with female PrP-SMN heterozygotes to generate F1 littermates with identical genetic background for analysis with the following 4 genotypes: SOD1G93A; PrP-SMN, SOD1G93A, PrP-SMN, and wild type (WT). Only male progeny mice were analyzed. Primers used for detection of the PrP-SMN transgene were SMN-Comm_F 5′- GAT GAT TCT GAC ATT TGG GAT G-3′ and SMNd7_R 5′-CCA GCA TGA TAG TAA GTG GGG-3′. Standard SOD1 and internal control interleukin-2 primers were used to genotype the SOD1 transgene.
2.3. Behavioral analysis
Mice were weighed 3-times a week and analyzed for locomotor function weekly from 60 days using an accelerating Mouse-Rota MK-2 rotarod (Monash University) or Rota-Rod 47600 (Ugo Basile Italy). Mice were trained in 3 ramping sessions at 3–30 rpm for 5 minutes each with 10 minute-rest intervals. Mice were tested twice at 3–30 rpm for 5 minutes and the average time latency to fall was recorded. Disease onset was determined retrospectively using the age of peak body weight as a measure of denervation muscle atrophy onset as previously described (Boillee et al., 2006). The onset of rotarod deficit was calculated using the age of peak rotarod activity preceding first impairment. For survival analysis, SOD1G93A; PrP-SMN and SOD1G93A mice were killed at the time of clinical end point onset defined by failure to splay hind limbs because of paralysis. Mice were killed by lethal injection (sodium pentobarbitone, 100 mg/kg, intraperitoneal).
2.4. Histology
Mice were transcardially perfused with phosphate buffered saline (PBS) followed by 4% paraformaldehyde in 0.1 M phosphate buffer. Lumbar spinal cords were dissected out, postfixed in 4% paraformaldehyde for 2 hours and cryoprotected in 30% sucrose in PBS overnight at 4 °C. Cords were embedded in optimal cutting temperature medium by freezing in isopentane cooled by liquid nitrogen. Horizontal 20 μm sections were cut and stained with 0.5% cresyl violet using a standard protocol, dehydrated, and coverslipped. Nissl-positive motor neurons in every third section were counted from a total of 30 ventral horns per mouse identified according to neuronal morphology and size exceeding 20-μm diameter with a distinct nucleolar profile.
2.5. Immunohistochemistry
Immunohistochemistry was performed in mice derived from SOD1G93A B6SJL line. Spinal cord sections were permeabilized in 0.4% TX-100 in PBS for 10 minutes, blocked in 5% skim milk in PBS for 30 minutes, and incubated with mouse SMN (1:50; BD Biosciences, Australia, 610647), rabbit neuronal nuclear antigen (NeuN) (1:500, Abcam, UK, ab104225), mouse glial fibrillary acidic protein (GFAP) (1:200, Millipore, Australia MAB360), and rabbit ionised calcium binding adaptor molecule 1 (Iba1) (1:1000, Wako Industries, Japan, #019-19741) antibodies overnight at 4 °C. Sections were incubated with Alexa Fluor conjugated secondary antibodies (1:1000, Life Technologies, Australia) for 2 hours, stained with Hoechst 33342 (1:10,000, Life Technologies) for 15 minutes and mounted on slides using fluorescent mounting medium (Dako, Australia) for microscopy using an Olympus FV 1000 confocal microscope. Images were captured using identical exposure and gain settings. Negative control samples without primary antibodies produced no staining. Nuclear SMN gems were counted from at least 25 motor neurons per genotype.
2.6. Immunoblot analysis
Fresh-frozen lumbar spinal cords were homogenized in RIPA lysis buffer containing 50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.1% SDS, 1% sodium deoxycholate, 1% TX-100, and 1% protease inhibitor cocktail (Sigma, Australia) by sonication at 50% output for 15 seconds, stored on ice for 20 minutes, and centrifuged at 15,800g for 20 minutes to collect supernatants. Proteins (20 μg) were electrophoresed through 12.5% SDS polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad, Australia). Membranes were blocked with 5% skim milk in TBST, pH 8.0, for 30 minutes and incubated with mouse SMN (1:2000), sheep SOD1 (1:4000, Calbiochem, Australia, 574597), rabbit NeuN (1:1000), or mouse β-actin (1:2000, Sigma, A5316) antibodies overnight at 4 °C. Blots were incubated with HRP-conjugated secondary antibodies (1:10,000, Millipore) then enhanced chemiluminescence reagents (GE Healthcare, Australia) for detection. Blots were quantified using Image J software v 1.4 (http://rsb.info.nih.gov/ij/) by taking the mean gray value of SMN bands normalized to β-actin level after subtracting background intensity.
2.7. Statistical analysis
Blot densitometry, weight loss, and rotarod deficit onset were analyzed using an unpaired t test. Motor neuron and SMN gem counts were compared using 1-way analysis of variance with Tukey's post hoc test. Survival data were analyzed using Kaplan-Meier survival analysis with the log-rank test. Analyses were performed using GraphPad Prism 5.0 software (GraphPad, La Jolla, CA, USA).
3. Results
3.1. SMN protein levels are reduced in ALS spinal cord
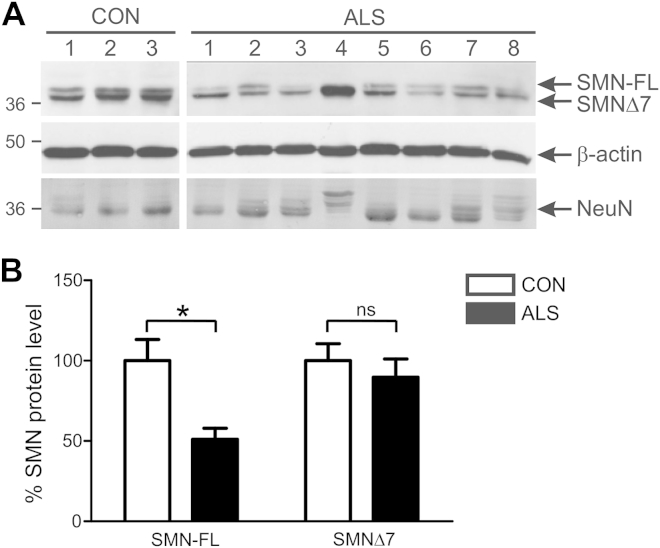
SMN protein expression was measured in postmortem spinal cord tissue obtained from patients with sporadic ALS and age-matched controls without neurologic disease. The details of patients and controls are provided in Table 1. Western blot analysis using high-resolving gels of control tissues showed 2 major SMN bands at approximately 36 kDa, corresponding to full-length SMN (SMN-FL) and spliced SMN (SMNΔ7) (Fig. 1A). SMN-FL levels were approximately 50% lower in ALS tissue than in control tissue (Fig. 1B, p < 0.05). In some cases, SMN-FL was almost completely absent in spinal cords of patients with ALS. SMNΔ7 levels were slightly reduced in ALS than in control tissue, although this difference was not statistically significant. SMN loss did not result from neuron loss because neuronal marker NeuN levels were similar in ALS and control tissues.
Fig. 1.
Survival motor neuron (SMN) protein level is diminished in spinal cords of sporadic amyotrophic lateral sclerosis (ALS) patients. (A) Immunoblot analysis of SMN products in lumbar spinal cord tissue of ALS and control (CON) subjects shows presence of full-length SMN (SMN-FL) and spliced survival motor neuron (SMNΔ7) bands. β-actin and NeuN are used as protein and neuronal loading controls, respectively. (B) Quantification of SMN-FL and SMNΔ7 protein levels normalized to β-actin from immunoblots. Data represent mean ± SEM; n = 5 CON and n =9 ALS subjects; * p < 0.05 compared with CON group using unpaired t test.
3.2. SMN overexpression delays motor deficit onset in 2 strains of SOD1G93A mice
We next investigated whether SMN upregulation ameliorates clinical features and motor neuron pathology in mutant SOD1 mice. PrP-SMN mice, expressing 8–9 human SMN1 transgene copies driven by the mouse prion protein promoter (PrP), were bred with 2 strains of SOD1G93A mice on either mixed B6SJL or congenic C57BL/6 genetic backgrounds. The PrP promoter directs neuronal SMN expression in these mice which is 2–3-fold higher than endogenous mouse SMN (Anderton et al., 2012; Gavrilina et al., 2008). The growth, locomotor activity, and lifespan of PrP-SMN mice were normal and SMN overexpression did not lead to any pathologic consequences (Supplementary Fig. 1). The resulting genotypes from this cross were analyzed: (1) double transgenic SOD1G93A; PrP-SMN; (2) SOD1G93A; (3) PrP-SMN; and (4) WT mice.
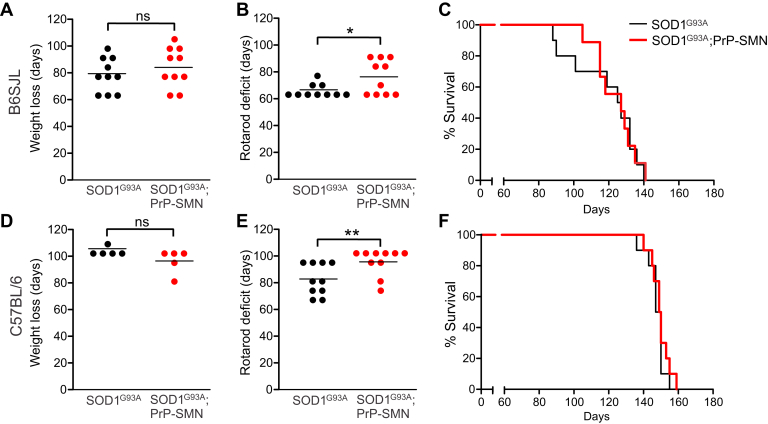
We first examined the effect of increased SMN expression on SOD1G93A B6SJL mice. Disease onset, determined by the age of peak body weight preceding muscle wasting, was not significantly different between SOD1G93A; PrP-SMN mice (84 ± 5 days, mean ± SEM) and SOD1G93A mice (78 ± 4 days) (Fig. 2A, Table 2). The onset of locomotor deficit shown by peak rotarod performance, which is a more sensitive measure of neuromuscular function than muscle wasting, was significantly delayed in SOD1G93A; PrP-SMN mice (76 ± 4 days) compared with SOD1G93A mice (66 ± 2 days; p < 0.01; Fig. 2B), equating to a 15% delay in age at onset of motor deficit. However, survival defined by the onset of hind limb paralysis was similar in SOD1G93A; PrP-SMN mice (121 ± 5 days) and SOD1G93A mice (119 ± 6 days, Fig. 2C). Thus, SMN upregulation delays the onset of motor deficits in SOD1G93A B6SJL mice without prolonging lifespan.
Fig. 2.
Neuronal survival motor neuron (SMN) overexpression delays onset of motor deficits in 2 different strains of transgenic SOD1G93A mice. (A) Disease onset determined by body weight loss, (B) motor deficit onset determined by rotarod deficit and (C) survival in mice derived from the SOD1G93A B6SJL line. (D) Disease onset determined by body weight loss, (E) motor deficit onset determined by rotarod deficit and (F) survival in mice derived from the SOD1G93A C57BL/6 line. Lines represent means, n = 10 mice per genotype, * p < 0.05 and ** p < 0.01 compared with SOD1G93A mice using unpaired t-test.
Table 2.
Effects of survival motor neuron (SMN) overexpression on phenotype of mutant SOD1 mice
| Genotype | Onset of weight loss (d) | Onset of rotarod deficit (d) | Survival (d) |
|---|---|---|---|
| B6SJL background | |||
| SOD1G93A; PrP-SMN | 84 ± 5 | 76 ± 4a | 121 ± 5 |
| SOD1G93A | 78 ± 4 | 66 ± 2 | 119 ± 6 |
| C57BL/6 background | |||
| SOD1G93A; PrP-SMN | 96 ± 4 | 96 ± 3b | 150 ± 2 |
| SOD1G93A | 103 ± 1 | 82 ± 4 | 148 ± 2 |
Mean ± SEM.
p < 0.05 compared with SOD1G93A B6SJL mice.
p < 0.01 compared with SOD1G93A C57BL/6 mice.
We confirmed the effects of SMN overexpression on a second strain of SOD1G93A C57BL/6 mice, which have a longer lifespan with less variation in mortality than the SOD1G93A B6SJL line (Heiman-Patterson et al., 2005). Disease onset shown by peak body weight was similar in SOD1G93A; PrP-SMN and SOD1G93A mice (Fig. 2D, Table 2). However, onset of motor dysfunction measured according to rotarod deficit was significantly delayed by 2 weeks in SOD1G93A; PrP-SMN mice (96 ± 3 days) compared with SOD1G93A mice (82 ± 4 days; p < 0.05, Fig. 2E), representing an approximately 20% delay in motor deficit age. As with SOD1G93A B6SJL mice, survival of SOD1G93A; PrP-SMN mice was not extended (150 ± 2 days) compared with SOD1G93A mice (148 ± 2 days, Fig. 2F) on the C57BL/6 background.
3.3. SMN overexpression protects motor neurons in SOD1G93A mice
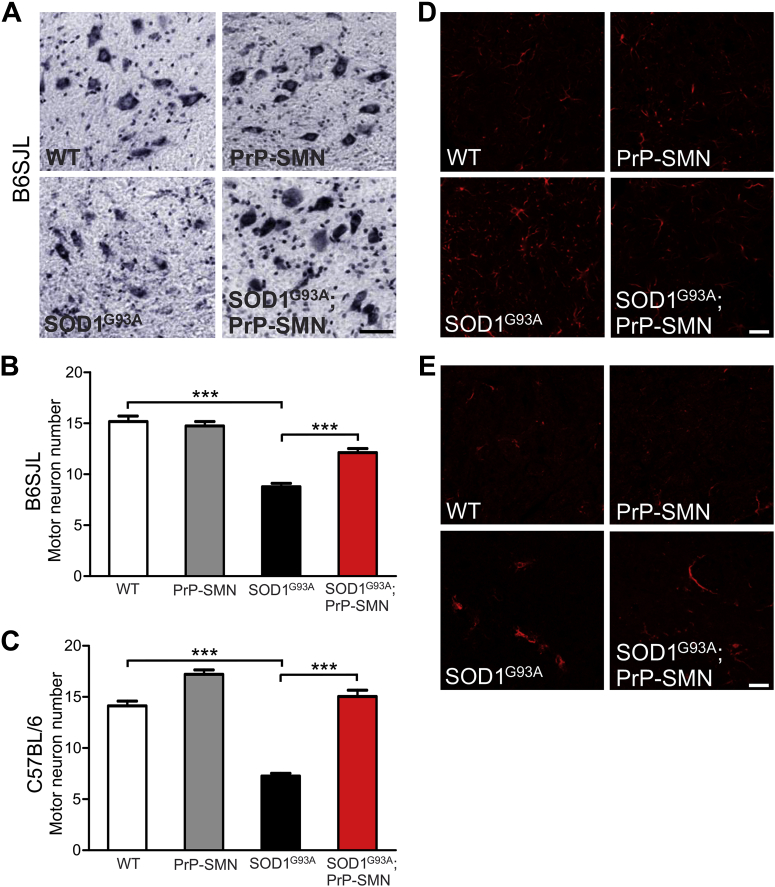
We next examined spinal cord pathology in these mice to account for the delay in motor deficits conferred by SMN overexpression. Spinal motor neuron numbers were quantified in mice at postnatal day 100 (P100). Motor neuron counts were similar in PrP-SMN and WT mice (Fig. 3A and B). In SOD1G93A B6SJL mice, there was a ∼50% loss of motor neurons compared with WT animals and SMN overexpression significantly rescued motor neurons in SOD1G93A; PrP-SMN mice (Fig. 3B, p < 0.001). In SOD1G93A C57BL/6 mice, there was 50% drop out of motor neurons relative to WT mice and increased SMN expression completely rescued motor neurons in SOD1G93A; PrP-SMN mice (Fig. 3C, p < 0.001).
Fig. 3.
Survival motor neuron (SMN) overexpression rescues motor neuron loss and attenuates astrogliosis in spinal cords of transgenic SOD1G93A mice at postnatal day 100. (A) Micrographs of ventral horns stained with cresyl violet in lumbar spinal cords of mice. Scale bar = 50 μm. (B) Spinal motor neuron counts in mice derived from SOD1G93A B6SJL line. (C) Spinal motor neuron counts in mice derived from SOD1G93A C57BL/6 line. Data represent means ± SEM, n = 3–7 mice per genotype, *** p < 0.001 using 1-way analysis of variance (ANOVA) with Tukey's posttest. Immunohistochemical analysis of (D) GFAP and (E) Iba1 in spinal cords of mice. Astrocyte and microglial activation is increased in SOD1G93A mice and astrogliosis is reduced by SMN overexpression in SOD1G93A; PrP-SMN mice. Scale bar = 50 μm.
The effect of SMN overexpression on neuroinflammatory cells in mice at P100 was examined using the markers GFAP and Iba1 for astrocytes and microglia, respectively. Astrocyte morphology was normal in spinal cords of WT and PrP-SMN mice (Fig. 3D). In SOD1G93A B6SJL mice, there was pronounced astrocyte activation and interestingly, SMN overexpression attenuated astrogliosis in SOD1G93A; PrP-SMN mice. Microglia appeared normal in spinal cords of WT and PrP-SMN mice (Fig. 3E). Microglial activation was evident in spinal cords of SOD1G93A B6SJL mice, however similar microgliosis was observed in SOD1G93A; PrP-SMN mice. Hence, SMN overexpression improved rotarod performance by rescuing motor neurons and reducing astrocyte activation in spinal cords of SOD1G93A mice.
3.4. Mutant SOD1 disrupts nuclear localization of SMN in spinal motor neurons
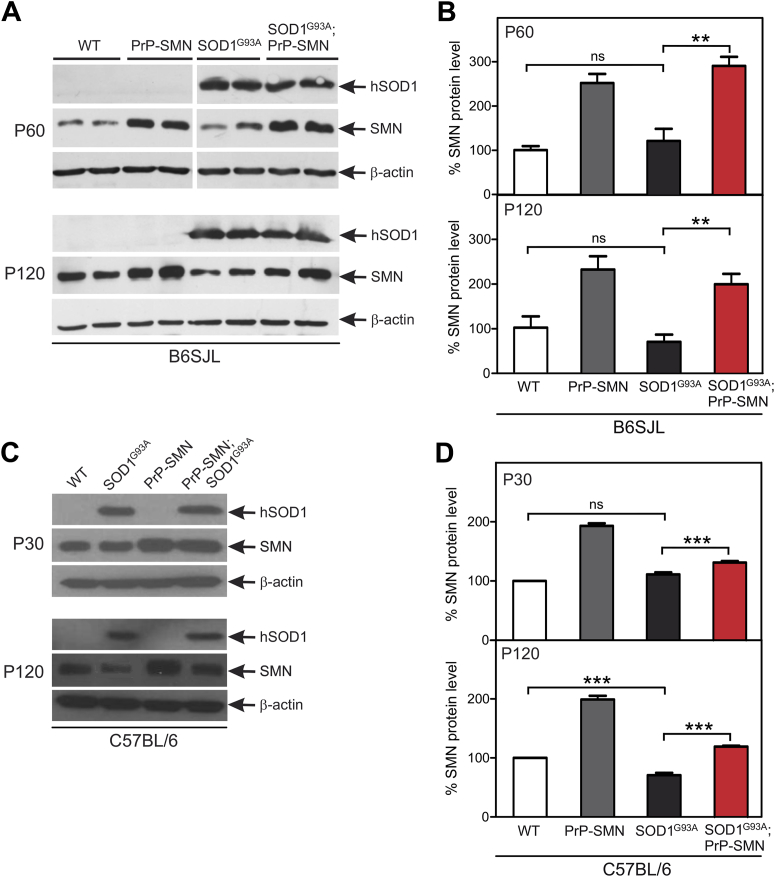
Despite rescuing motor neurons and delaying early symptoms, SMN overexpression did not prevent late symptoms or paralysis. We therefore analyzed SMN expression level and cellular localization in spinal cords of mice at different disease stages. We first studied SOD1G93A B6SJL mice at presymptomatic (P60) and symptomatic (P120) time points. SMN levels were 2.5-fold higher in PrP-SMN mice than in WT animals at P60 (Fig. 4A and B). SMN expression level was similar in SOD1G93A and WT mice at this age. Importantly, SMN expression was 3-fold greater in SOD1G93A; PrP-SMN mice than in WT mice at the presymptomatic disease stage (p < 0.01). At P120, SMN levels were still 2.5-fold higher in PrP-SMN mice than in WT mice. SMN levels were decreased by ∼30% in spinal cords of symptomatic SOD1G93A mice compared with WT, although not statistically significant. In SOD1G93A; PrP-SMN mice, SMN expression was reduced to 2-fold of WT mice (p < 0.01), suggesting that mutant SOD1 disrupts both endogenous and transgenic SMN expression with disease progression.
Fig. 4.
Mutant superoxide dismutase (SOD1) disrupts endogenous and transgenic survival motor neuron (SMN) expression in spinal cords of SOD1G93A mice. (A) Immunoblot analysis of SMN and SOD1 expression in lumbar spinal cords of mice derived from SOD1G93A B6SJL line, n = 2 different mice shown per genotype. (B) Quantification of SMN protein levels from immunoblots. (C) Immunoblot analysis of SMN and SOD1 expression in spinal cords of mice derived from SOD1G93A C57BL/6 line. (D) Quantification of SMN protein levels from immunoblots. Data represent means ± SEM, n = 5 mice per genotype, ** p < 0.01 and *** p < 0.001 using 1-way analysis of variance (ANOVA) with Tukey's posttest, P = postnatal day.
SMN expression was then measured in spinal cords from SOD1G93A C57BL/6 mice. SMN levels in PrP-SMN mice were increased 2-fold at P30 and this level was maintained at P120, compared with WT animals (Fig. 4C and D). SMN level was also elevated in SOD1G93A; PrP-SMN mice compared with SOD1G93A mice at P30 (p < 0.001). However, at P120, SMN expression was reduced in both SOD1G93A; PrP-SMN (p < 0.001) and SOD1G93A mice (p < 0.001). These results confirm that downregulation of endogenous and exogenous SMN protein occurs in spinal cords of symptomatic mutant SOD1 mice.
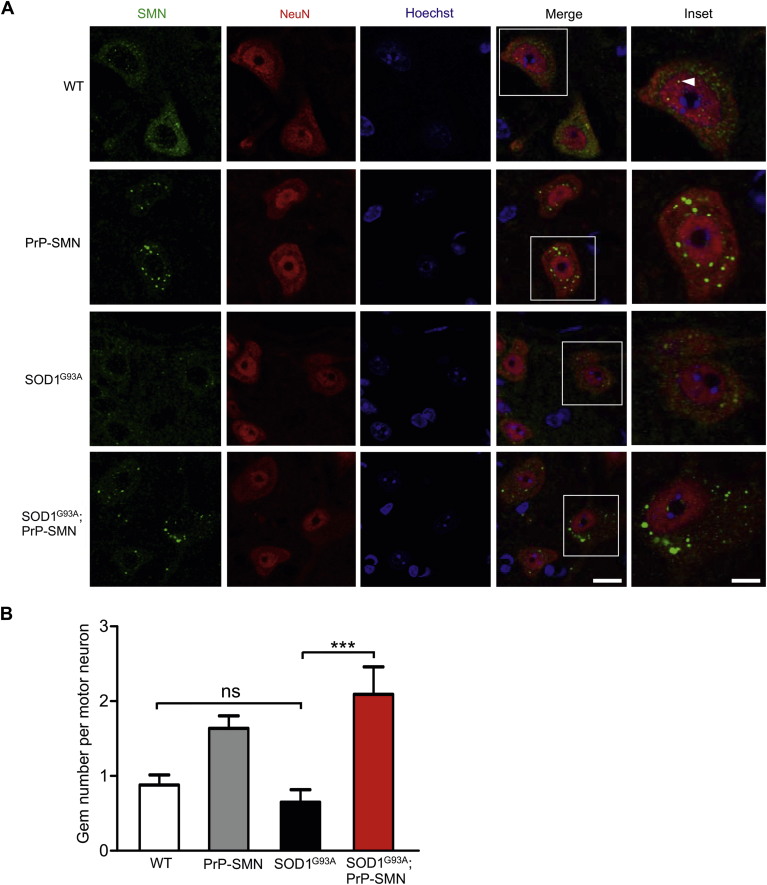
The cellular distribution of SMN was examined using immunohistochemistry in P60 mice. In WT mice, SMN was distributed diffusely throughout the cytoplasm and concentrated in nuclear gems of spinal motor neurons identified by size, location, and NeuN staining (Fig. 5A). In PrP-SMN mice, SMN formed multiple cytoplasmic aggregates and nuclear gems. These cytoplasmic SMN foci did not occur in nonneuronal cells lacking NeuN staining, but identified with Hoechst staining. In motor neurons of presymptomatic SOD1G93A B6SJL mice, there was reduced SMN staining in cells. In SOD1G93A; PrP-SMN mice, there were multiple cytoplasmic and nuclear foci of SMN, although some clustering of cytoplasmic aggregates was evident in motor neurons. We quantified SMN nuclear gem number in motor neurons, showing similar gem levels in WT and SOD1G93A mice (Fig. 5B). In both PrP-SMN and SOD1G93A; PrP-SMN mice, gem counts were increased by approximately 2-fold, confirming high SMN levels.
Fig. 5.
Survival motor neuron (SMN) localization and nuclear complex formation in motor neurons of mice at postnatal day 60. (A) SMN immunohistochemical analysis of lumbar spinal cords from mice derived from SOD1G93A B6SJL mice. In wild-type (WT) mice, SMN shows cytoplasmic and nuclear localization in NeuN-positive motor neurons, the latter forming gems (arrowhead). In PrP-SMN mice, there are multiple cytoplasmic aggregates of SMN. In SOD1G93A mice, SMN staining is reduced. In SOD1G93A; PrP-SMN mice, SMN forms multiple cytoplasmic aggregates with clustering. Scale bar = 50 μm, inset scale bar = 10 μm. (B) Quantification of SMN nuclear gems in motor neurons of mice. Data represent means ± SEM, n = 3 mice per genotype, *** p < 0.001 using 1-way analysis of variance (ANOVA) with Tukey's posttest.
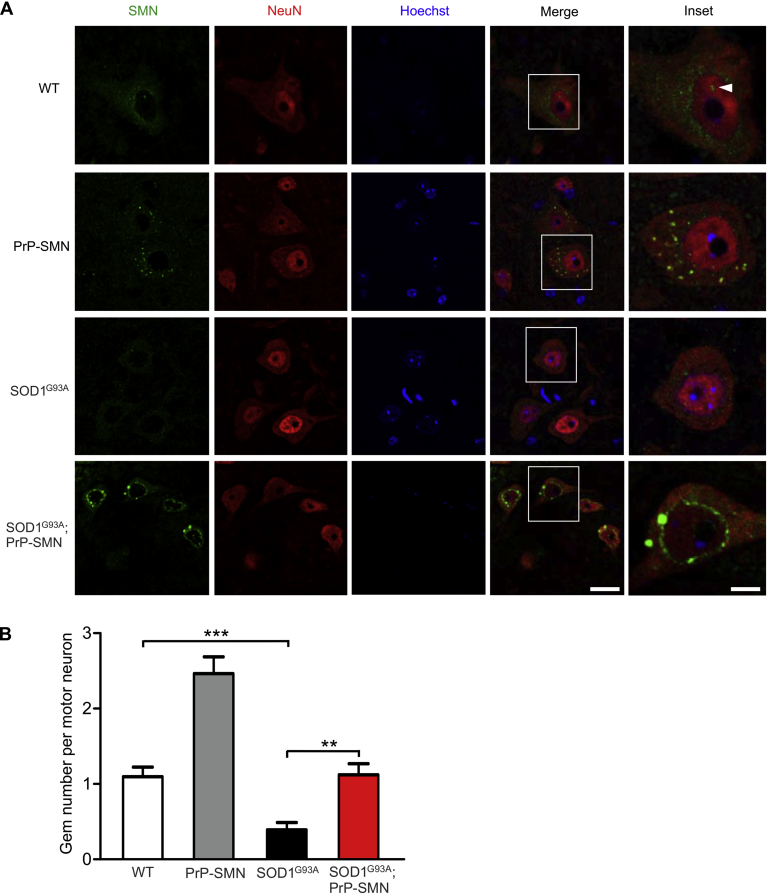
SMN localization was next assessed in spinal motor neurons of P100 mice. In WT mice, SMN was cytoplasmic and nuclear (Fig. 5A). In PrP-SMN mice, SMN occurred in cytoplasmic aggregates and nuclear gems. In motor neurons of symptomatic SOD1G93A B6SJL mice, there was a paucity of SMN throughout the cell, including a loss of nuclear gems, consistent with our Western blot analysis data (Fig. 4). In contrast to PrP-SMN mice, there was a markedly abnormal perinuclear accumulation of SMN in SOD1G93A; PrP-SMN mice, in some instances seemingly encasing the nucleus, suggesting a defect in nuclear import of SMN. SMN gem counts were reduced by ∼60% in SOD1G93A mice compared with WT (Fig. 6B, p < 0.001). Furthermore, SMN gem number was reduced by ∼50% in SOD1G93A; PrP-SMN mice compared with PrP-SMN mice, confirming impaired SMN nuclear transport. Thus, transgenic SMN is aberrantly localized in spinal motor neurons and may be sequestrated in the cytoplasm, leading to depletion of SMN from the nucleus or the axon, where it has also been identified as having a putative neuronal-specific function. This may explain why SMN overexpression cannot extend lifespan in SOD1G93A mice, despite reproducible beneficial effects on early disease signs and motor neuron survival.
Fig. 6.
Survival motor neuron (SMN) localization and nuclear complex formation in motor neurons of mice at postnatal day 100. (A) SMN immunohistochemical analysis of lumbar spinal cords from mice derived from SOD1G93A B6SJL mice. In wild-type (WT) mice, SMN shows cytoplasmic and nuclear distribution in NeuN-positive motor neurons, the latter forming gems (arrowhead). In PrP-SMN mice, there are multiple cytoplasmic aggregates of SMN. In SOD1G93A mice, SMN levels are severely reduced and nuclear gems disrupted. In SOD1G93A; PrP-SMN mice, SMN is abnormally localized around the nucleus and gems are disrupted. Scale bar = 50 μm, inset scale bar = 10 μm. (B) Quantification of SMN nuclear gems in motor neurons of mice. Data represent means ± SEM, n = 3 mice per genotype, ** p < 0.01 and *** p < 0.001 using 1-way analysis of variance (ANOVA) with Tukey's posttest.
4. Discussion
This study is the first to show that SMN protein is diminished in the spinal cords of patients with sporadic ALS. We determined an approximately 50% reduction in SMN-FL level. Previous studies correlating SMN protein level with SMA severity reported SMN reductions of 80%–95% in type I patients and 80% in type II patients (Lefebvre et al., 1997). This supports the threshold hypothesis in SMA that reduction of SMN protein levels to 0% results in lethality, severe reduction to 10%–20% causes SMA types I and II, and moderate reduction to 35%–45% causes SMA types III and IV (Sleigh et al., 2011). Approximately 50% reduction in SMN-FL in patients with ALS and mutant SOD1 mice would therefore be consistent with permitting disease progression and argues that SMN loss occurs early in pathogenesis.
The second major finding from this study is that SMN overexpression delays motor deficit onset and enhances spinal motor neuron survival in 2 different strains of mutant SOD1 mice, suggesting that the role of SMN in promoting motor neuron viability may extend to non-SMA disease models. We addressed this question by exploiting transgenic PrP-SMN mice, which robustly overexpress SMN1 transgenes producing only SMN-FL (Gavrilina et al., 2008), in preference to transgenic mice expressing SMN2 transgenes, which produce little SMN-FL (Monani et al., 2000). We extended characterization of PrP-SMN mice, showing accumulation of cytoplasmic and nuclear foci of SMN in motor neurons, but not non-neuronal cells. These subcellular structures conform to the morphology and diameter of SMN cytoplasmic bodies and nuclear gems described by others (Le et al., 2000; Liu and Dreyfuss, 1996). PrP-SMN mice showed normal development, growth, survival, and spinal motor neuron complement in our study. It was recently proposed that SMN1 duplications in ALS could produce higher levels of SMN protein that are toxic to motor neurons (Blauw et al., 2012); we found no evidence of pathology here.
We previously demonstrated that both dismutase active and inactive SOD1 mutants impair SMN entry into the nucleus causing its accumulation in the cytoplasm (Turner et al., 2009). Because nuclear depletion and cytoplasmic redistribution of mutant, misfolded or pathologic forms of SOD1, TDP-43, and FUS occurs in affected neurons in ALS (Neumann et al., 2006; Sau et al., 2007; Vance et al., 2009), it suggests that defective nuclear transport is a feature common to ALS, regardless of the etiology. In the nucleus, SMN gems colocalize with coilin, which is a marker of Cajal bodies involved in small nuclear RNA and ribonuclear protein assembly (Liu and Dreyfuss, 1996). In keeping with other reports (Gertz et al., 2012; Kariya et al., 2012), we found that nuclear gems were disrupted in spinal motor neurons of SOD1G93A mice. As Cajal bodies are preserved in motor neurons of mutant SOD1 mice (Kariya et al., 2012; Turner et al., 2008), it is likely that targeting of SMN to Cajal bodies is impaired. We predict that SMN abnormalities in these mice affect nuclear gem assembly with consequences that may include small nuclear RNA dysregulation and defective pre-mRNA splicing as proposed for mouse models of SMA deficient for SMN (Baumer et al., 2009; Zhang et al., 2008). The effects of SMN nuclear complex disruption on transcription and alternative splicing could be more formally evaluated by profiling spinal cords of presymptomatic mutant SOD1 mice with splicing-sensitive junction arrays focusing on known SMN target transcripts. Indeed, aberrant exon splicing was reported in spinal cords of mutant SOD1 mice at disease onset, implicating diverse cell pathways in pathogenesis (Chen et al., 2010). Interestingly, TDP-43 depletion in brain causes deficiency of long pre-messenger RNAs and missplicing of transcripts tightly linked to neuronal and synaptic function (Polymenidou et al., 2011; Tollervey et al., 2011). It is also relevant that SMN nuclear gems are altered by TDP-43 overexpression or knockout in spinal motor neurons of mice (Shan et al., 2010). Furthermore, SMN gems are disrupted by mutant TDP-43 and FUS in transfected cells and fibroblasts of patients with ALS (Groen et al., 2013; Yamazaki et al., 2012), pointing to SMN mislocalization as a common disease pathway in ALS. We speculate that similar missplicing of transcripts essential for motor neuron survival result from nuclear SMN complex disruption in mutant SOD1 mice. The disruption to the cellular distribution and expression of SMN also provides a molecular basis for selective neuronal vulnerability in ALS and we argue that SMN protein deficiency confers susceptibility to spinal motor neurons in ALS, and SMA.
The mechanisms that disrupt SMN nuclear gems in mutant SOD1 mice remain unclear. Transport through the nuclear membrane is mediated by the nuclear pore complex, which permits passive diffusion of small molecules and active transport of <40-kDa proteins (McLane and Corbett, 2009). Cytoplasmic proteins bearing an N-terminal nuclear localization sequence bind to the soluble nuclear import receptors, importin-α and importin-β, before transit through the nuclear pore complex. Although SMN does not carry a nuclear localization sequence, it binds to the zinc finger protein ZPR1, which mediates its nuclear entry (Gangwani et al., 2001). The nuclear envelope is abnormal in spinal motor neurons of patients with sporadic ALS and presymptomatic SOD1G93A mice (Kinoshita et al., 2009). Moreover, importins are depleted from the nucleus of motor neurons in ALS and mutant SOD1 mice (Kinoshita et al., 2005; Zhang et al., 2006), suggesting defective nuclear transport. Interestingly, knockdown of importin-β triggers cytoplasmic TDP-43 accumulation in cell culture and recapitulates ALS pathology (Nishimura et al., 2010; Sato et al., 2009). We speculate that loss of importins or some other factor(s) that prevent nuclear entry underlies disruption of SMN nuclear gems in mutant SOD1 mice.
We showed that neuronal SMN overexpression improved neuromuscular function and protected against motor neuron loss in 2 strains of mutant SOD1 mice, but did not prolong survival. This most likely results from disruption of nuclear SMN complexes in motor neurons with disease progression. It was recently shown that SMN overexpression achieved by crossing SOD1G86R and transgenic SMN2 mice also slowed the symptom onset and spared spinal motor neurons, without increasing survival (Kariya et al., 2012). This effect was attributed to progression of muscle denervation despite elevated SMN levels, although weight loss was not slowed by SMN overexpression in this study. Thus, an alternative explanation for the failure to extend mouse survival here is that SMN exerts protection in early disease, but exclusion of SMN from the nucleus and axon due to its sustained mislocalization at cell bodies may deprive motor neurons of SMN complexes required to maintain distal synapses. As evidence of this, we show that SMN overexpression rescues or even normalizes motor neuron counts in spinal cords of SOD1G93A mice despite transiently delaying rotarod deficits, implicating ongoing pathology distal to the spinal cord resulting from SMN sequestration at the cell body. As the PrP-SMN transgene corrects the phenotype of SMN deficient mice (Gavrilina et al., 2008) and even SMN2 and/or SMNΔ7 overexpression rescues the survival of SMN deficient mice (Le et al., 2000; Monani et al., 2000), we predict that nuclear transport defects present in ALS are not present in SMA. Our data might suggest caution in the design and application of SMN therapeutics for ALS, including viral and nonviral mediated gene delivery, oligonucleotide-mediated splicing modification, and small molecules that stimulate SMN expression (Sleigh et al., 2011). Strategies that upregulate or replace SMN might therefore need to be combined with therapeutics that stabilize neuromuscular junctions to be efficacious in patients with ALS and its preclinical animal models. However, it is also possible that the extremely high levels of mutant SOD1 protein present in these SOD1G93A models, which may not reflect human ALS, mask potentially greater effects of SMN in promoting survival of motor neurons, which could be tested by crossing SMN overexpressing mice with more physiologically relevant models based on TDP-43 and FUS mutations when these become available.
Disclosure statement
All authors declare no conflicts of interest.
Acknowledgements
This work was supported by grants from the Australian National Health & Medical Research Council (Project Grant 1008910), MND Research Institute of Australia (Mick Rodger Benalla MND Research Grant), Bethlehem Griffiths Research Foundation Grant and Victorian Government through the Operational Infrastructure Scheme (BJT), Motor Neurone Disease Association (KT), and UK Medical Research Council (KED and JNS). The authors thank Prof Arthur Burghes for generously providing PrP-SMN mice, Dr Nicholas Parkinson, Dr Dirk Bäumer, Fiona Christensen and Rian MacKenzie for mouse breeding and genotyping, Dr Tobias Merson for Iba1 antibody and Dr Julie Atkin for helpful discussions. Human spinal cord samples were received from Ms Fairlie Hinton, from the MND Research Tissue Bank of Victoria, supported by the Victorian Brain Bank Network, Mental Health Research Institute, Alfred Hospital, Victorian Forensic Institute of Medicine and the University of Melbourne. Studies were approved by the Howard Florey Institute Human and Animal Ethics Committees.
Contributor Information
Bradley J. Turner, Email: bradley.turner@florey.edu.au.
Kevin Talbot, Email: kevin.talbot@ndcn.ox.ac.uk.
Appendix A. Supplementary data
Survival motor neuron overexpression is not harmful to mice. (A) Body weight and (B) rotarod activity assessed weekly in transgenic prion protien promoter-survival motor neuron (PrP-SMN) and wild-type (WT) mice. Data represent means means ± standard error of mean, n = 10 mice per genotype.
References
- Anderton R.S., Meloni B.P., Turner B.J., Mitrpant C., Mastaglia F.L., Goh C., Boulos S. Co-regulation of survival motor neuron and Bcl-xL expression: implications in spinal muscular atrophy. Neuroscience. 2012;220:228–236. doi: 10.1016/j.neuroscience.2012.06.042. [DOI] [PubMed] [Google Scholar]
- Baumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E., Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:18. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauw H.M., Barnes C.P., van Vught P.W., van Rheenen W., Verheul M., Cuppen E., Veldink J.H., van den Berg L.H. SMN1 gene duplications are associated with sporadic ALS. Neurology. 2012;78:776–780. doi: 10.1212/WNL.0b013e318249f697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S., Vande Velde C., Cleveland D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Burghes A.H., Beattie C.E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Guo Y., Hu M., Duan W., Chang G., Li C. Differential expression and alternative splicing of genes in lumbar spinal cord of an amyotrophic lateral sclerosis mouse model. Brain Res. 2010;22:52–69. doi: 10.1016/j.brainres.2010.03.075. [DOI] [PubMed] [Google Scholar]
- Corcia P., Camu W., Halimi J.M., Vourc'h P., Antar C., Vedrine S., Giraudeau B., de Toffol B., Andres C.R. SMN1 gene, but not SMN2, is a risk factor for sporadic ALS. Neurology. 2006;67:1147–1150. doi: 10.1212/01.wnl.0000233830.85206.1e. [DOI] [PubMed] [Google Scholar]
- Corcia P., Ingre C., Blasco H., Press R., Praline J., Antar C., Veyrat-Durebex C., Guettard Y.O., Camu W., Andersen P.M., Vourc'h P., Andres C.R. Homozygous SMN2 deletion is a protective factor in the Swedish ALS population. Eur. J. Hum. Genet. 2012;20:588–591. doi: 10.1038/ejhg.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcia P., Mayeux-Portas V., Khoris J., de Toffol B., Autret A., Muh J.P., Camu W., Andres C. Abnormal SMN1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann. Neurol. 2002;51:243–246. doi: 10.1002/ana.10104. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo L., Kirby J., Grierson A.J., Sendtner M., Shaw P.J. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2011;7:616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- Gangwani L., Mikrut M., Theroux S., Sharma M., Davis R.J. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat. Cell Biol. 2001;3:376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz B., Wong M., Martin L.J. Nuclear localization of human SOD1 and mutant SOD1-specific disruption of survival motor neuron protein complex in transgenic amyotrophic lateral sclerosis mice. J. Neuropathol. Exp. Neurol. 2012;71:162–177. doi: 10.1097/NEN.0b013e318244b635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen E.J., Fumoto K., Blokhuis A.M., Engelen-Lee J., Zhou Y., van den Heuvel D.M., Koppers M., van Diggelen F., van Heest J., Demmers J.A., Kirby J., Shaw P.J., Aronica E., Spliet W.G., Veldink J.H., van den Berg L.H., Pasterkamp R.J. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum. Mol. Genet. 2013;22:3690–3704. doi: 10.1093/hmg/ddt222. [DOI] [PubMed] [Google Scholar]
- Heiman-Patterson T.D., Deitch J.S., Blankenhorn E.P., Erwin K.L., Perreault M.J., Alexander B.K., Byers N., Toman I., Alexander G.M. Background and gender effects on survival in the TgN(SOD1-G93A)1Gur mouse model of ALS. J. Neurol. Sci. 2005;236:1–7. doi: 10.1016/j.jns.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Kariya S., Re D.B., Jacquier A., Nelson K., Przedborski S., Monani U.R. Mutant superoxide dismutase 1 (SOD1), a cause of amyotrophic lateral sclerosis, disrupts the recruitment of SMN, the spinal muscular atrophy protein to nuclear Cajal bodies. Hum. Mol. Genet. 2012;21:3421–3434. doi: 10.1093/hmg/dds174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N., Hashimoto K., Yamamura T., Teranuma H., Koizumi T., Satoh K., Katayama T., Sakagami H. Protection by antioxidants of copper-induced decline of proliferation and SOD activity. Anticancer Res. 2005;25:283–289. [PubMed] [Google Scholar]
- Kinoshita Y., Ito H., Hirano A., Fujita K., Wate R., Nakamura M., Kaneko S., Nakano S., Kusaka H. Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2009;68:1184–1192. doi: 10.1097/NEN.0b013e3181bc3bec. [DOI] [PubMed] [Google Scholar]
- Le T.T., Coovert D.D., Monani U.R., Morris G.E., Burghes A.H. The survival motor neuron (SMN) protein: effect of exon loss and mutation on protein localization. Neurogenetics. 2000;3:7–16. doi: 10.1007/s100480000090. [DOI] [PubMed] [Google Scholar]
- Lee J.B., Lee K.A., Hong J.M., Suh G.I., Choi Y.C. Homozygous SMN2 deletion is a major risk factor among twenty-five Korean sporadic amyotrophic lateral sclerosis patients. Yonsei Med. J. 2012;53:53–57. doi: 10.3349/ymj.2012.53.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Liu Q., Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- McLane L.M., Corbett A.H. Nuclear localization signals and human disease. IUBMB Life. 2009;61:697–706. doi: 10.1002/iub.194. [DOI] [PubMed] [Google Scholar]
- Monani U.R., Coovert D.D., Burghes A.H. Animal models of spinal muscular atrophy. Hum. Mol. Genet. 2000;9:2451–2457. doi: 10.1093/hmg/9.16.2451. [DOI] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., McCluskey L.F., Miller B.L., Masliah E., Mackenzie I.R., Feldman H., Feiden W., Kretzschmar H.A., Trojanowski J.Q., Lee V.M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nishimura A.L., Zupunski V., Troakes C., Kathe C., Fratta P., Howell M., Gallo J.M., Hortobagyi T., Shaw C.E., Rogelj B. Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain. 2010;133:1763–1771. doi: 10.1093/brain/awq111. [DOI] [PubMed] [Google Scholar]
- Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.-C., Sun E., Wancewicz E., Mazur C., Kordasiewicz H., Sedaghat Y., Donohue J.P., Shiue L., Bennett C.F., Yeo G.W., Cleveland D.W. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Takeuchi S., Saito A., Ding W., Bamba H., Matsuura H., Hisa Y., Tooyama I., Urushitani M. Axonal ligation induces transient redistribution of TDP-43 in brainstem motor neurons. Neuroscience. 2009;164:1565–1578. doi: 10.1016/j.neuroscience.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Sau D., De Biasi S., Vitellaro-Zuccarello L., Riso P., Guarnieri S., Porrini M., Simeoni S., Crippa V., Onesto E., Palazzolo I., Rusmini P., Bolzoni E., Bendotti C., Poletti A. Mutation of SOD1 in ALS: a gain of a loss of function. Hum. Mol. Genet. 2007;16:1604–1618. doi: 10.1093/hmg/ddm110. [DOI] [PubMed] [Google Scholar]
- Shan X., Chiang P.M., Price D.L., Wong P.C. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh J.N., Gillingwater T.H., Talbot K. The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy. Dis. Model Mech. 2011;4:457–467. doi: 10.1242/dmm.007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V., Patani R., Chandran S., Rot G., Zupan B., Shaw C.E., Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.J., Baumer D., Parkinson N.J., Scaber J., Ansorge O., Talbot K. TDP-43 expression in mouse models of amyotrophic lateral sclerosis and spinal muscular atrophy. BMC Neurosci. 2008;9:1471–2202. doi: 10.1186/1471-2202-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.J., Parkinson N.J., Davies K.E., Talbot K. Survival motor neuron deficiency enhances progression in an amyotrophic lateral sclerosis mouse model. Neurobiol. Dis. 2009;34:511–517. doi: 10.1016/j.nbd.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K.L., Tripathi V., Al-Saraj S., Al-Chalabi A., Leigh P.N., Blair I.P., Nicholson G., de Belleroche J., Gallo J.-M., Miller C.C., Shaw C.E. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldink J.H., Kalmijn S., Van der Hout A.H., Lemmink H.H., Groeneveld G.J., Lummen C., Scheffer H., Wokke J.H., Van den Berg L.H. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology. 2005;65:820–825. doi: 10.1212/01.wnl.0000174472.03292.dd. [DOI] [PubMed] [Google Scholar]
- Veldink J.H., van den Berg L.H., Cobben J.M., Stulp R.P., De Jong J.M., Vogels O.J., Baas F., Wokke J.H., Scheffer H. Homozygous deletion of the survival motor neuron 2 gene is a prognostic factor in sporadic ALS. Neurology. 2001;56:749–752. doi: 10.1212/wnl.56.6.749. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Chen S., Yu Y., Yan B., Haertlein T.C., Carrasco M.A., Tapia J.C., Zhai B., Das R., Lalancette-Hebert M., Sharma A., Chandran S., Sullivan G., Nishimura A.L., Shaw C.E., Gygi S.P., Shneider N.A., Maniatis T., Reed R. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ito H., Wate R., Ohnishi S., Nakano S., Kusaka H. Altered distributions of nucleocytoplasmic transport-related proteins in the spinal cord of a mouse model of amyotrophic lateral sclerosis. Acta Neuropathol. 2006;112:673–680. doi: 10.1007/s00401-006-0130-4. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Survival motor neuron overexpression is not harmful to mice. (A) Body weight and (B) rotarod activity assessed weekly in transgenic prion protien promoter-survival motor neuron (PrP-SMN) and wild-type (WT) mice. Data represent means means ± standard error of mean, n = 10 mice per genotype.