Abstract
Nipah virus has caused recurring outbreaks in central and northwest Bangladesh (the “Nipah Belt”). Little is known about roosting behavior of the fruit bat reservoir, Pteropus giganteus, or factors driving spillover. We compared human population density and ecological characteristics of case villages and control villages (no reported outbreaks) to understand their role in P. giganteus roosting ecology and Nipah virus spillover risk. Nipah Belt villages have a higher human population density (P < 0.0001), and forests that are more fragmented than elsewhere in Bangladesh (0.50 versus 0.32 patches/km2, P < 0.0001). The number of roosts in a village correlates with forest fragmentation (r = 0.22, P = 0.03). Villages with a roost containing Polyalthia longifolia or Bombax ceiba trees were more likely case villages (odds ratio [OR] = 10.8, 95% confidence interval [CI] = 1.3–90.6). This study suggests that, in addition to human population density, composition and structure of the landscape shared by P. giganteus and humans may influence the geographic distribution of Nipah virus spillovers.
Introduction
Nipah virus was recognized in Bangladesh in 2001 and has caused recurring outbreaks clustered seasonally, during December to April, and spatially, within central and northwest Bangladesh, which we here term, the “Nipah Belt.”1 The question of what drives spatial patterning of outbreaks remains unanswered. Principal routes of transmission to humans are consumption of raw date palm sap contaminated by the natural bat reservoir in Bangladesh, Pteropus giganteus, and person-to-person contact.2 Because no other reservoir has been identified in Bangladesh,3 index cases likely result from spillover transmission from the bat reservoir.4 Outbreaks separated by many months in different locations in Bangladesh and genetic characterization of the virus in Bangladeshi patients suggests repeated spillovers from bats to humans.4–6 Little is known about the impact of landscape patterns on the roosting ecology of Pteropus spp. or why there are Nipah virus spillovers in only a subset of villages where P. giganteus roost. We compared human population density and ecological characteristics of case and control villages to understand the role of landscape on P. giganteus roosting ecology and likelihood of Nipah virus spillover to humans.
Material and Methods
This study is a component of a larger village-level investigation of Nipah virus spillover conducted by the International Center for Diarrheal Disease Research, Bangladesh (icddr,b). We compared characteristics of P. giganteus roosts, landscape structure, and human population density of villages within and outside the Nipah Belt and in spillover case villages to control villages within the Nipah Belt. A case village was defined as any village where a Nipah virus case-patient was identified between 2001 and 2011, and where the infection was apparently introduced from a spillover event from the bat reservoir. We defined the Nipah Belt using a 50 km buffer around case villages. This distance is farther than the average Pteropus spp. foraging range7 so that a region outside the Nipah Belt would likely not be visited by bats roosting inside the Nipah Belt. To generate our control sample of villages with no reported Nipah virus cases, we drew 5 km buffers around case villages, and then selected a geographically random sample of points inside and outside the Nipah Belt, avoiding areas within the buffers.
We conducted two case-control comparisons. First, we used random sampling to select approximately half of our case and control villages for field visits (referred to as the “field-validated sample”) to assess detailed information on P. giganteus roosting ecology. We also created a second study sample using the full spillover case list and 20,000 geographically random control points (10,000 inside and 10,000 outside the Nipah Belt).8 We used this group (referred to as the “remotely sensed sample”) to conduct a second case-control analysis that used satellite and human population modeling data. The large sample size of the second study sample gave us the power to detect smaller effects of human population density and village forest structure on risk of Nipah virus spillover than would have been possible using only the field-validated sample.
Locating study villages and conducting roost ecological assessment.
We limited field data collection to December through early February to measure the environment in villages during the season when most human cases have been identified.4 Field teams used Global Positioning System (GPS) devices and GoogleEarth to identify and then enroll a control village near each set of random coordinates. They located P. giganteus roosts that were inhabited at some point in the past 5 years inside and within 5 km of the village boundaries through interviews with community leaders. They collected GPS coordinates at all roosts and counted the number of bats in active roosts (i.e., at least one bat present when the roost was located; inactive roosts were inhabited within the last 5 years but were not inhabited when the roost was located). A subset of roosts was selected for an intensive ecological assessment because of the restricted study duration. This assessment was conducted on the two largest, active roosts nearest the village center. Field teams used transects to delineate a 20 × 20 m plot around the central roost tree. Within the plot, they measured tree species, height (using a Suunto clinometer, Vantaa, Finland), and diameter at breast height (DBH) (using a diameter tape) for all trees with a DBH > 4 cm.
Satellite data derivation.
We used satellite and human population distribution data to measure forest structure and human population density within a 20 × 20 km square-shaped buffer around the center of each study village (distance from village center to center of the buffer's side was 10 km; referred to as a 10 km buffer, henceforth).
The land cover map used to calculate forest metrics was derived from GoogleMap and several satellites including MODIS (April 2012, 250 m), IRS Pan (2000–2005, 5.8 m), ASTER (2003–2008, 15 m), and IRS P6 LISS (2005, 23.5 m). Classification results were resampled at 250 m for the final map. We measured forest fragmentation by calculating forest patch density (number of patches/10 km village buffer), edge density (total length of forest pixels bordering non-forest pixels/10 km village buffer), and the largest patch index (percent of the 10 km village buffer occupied by the largest contiguous forest patch) using FRAGSTATS.9
Human population density was derived using the Landscan dataset, a global population surface created using an algorithm that integrates census data and ancillary datasets including land use, topography, and transportation.10
Data analysis.
Assessing tree species composition of roost sites.
We used non-parametric ordination methods and PC-ORD software11 to derive a measure of tree species composition at each roost site. First, we calculated the average basal area of each tree species within a plot using our DBH measurements. Next, we calculated Bray Curtis dissimilarity12 on the log-transformed matrix and ran a hierarchical cluster analysis to group roosts into five significant clusters based on similarity of the tree species biomass in each roost site. We used Multi-Response Permutation Procedures13 to ensure clusters were mutually exclusive. Finally, we conducted Indicator Species Analysis13 to identify diagnostic tree species for each roost cluster.
Comparing characteristics of roosts sites and villages in the field-validated sample.
Using the field-validated sample, we compared human population density, number of households, village area, bat population density and number of bats, roost characteristics, and forest fragmentation metrics of villages inside and outside the Nipah belt region to identify unique ecological characteristics of the Nipah Belt. We also compared characteristics of case villages to control villages inside the Nipah Belt to identify possible risk factors for Nipah virus spillover. To test for significant differences, we used t tests for continuous data, and χ2 or Fisher exact tests for categorical data. Standard deviations around mean estimates were calculated to assess the magnitude of these differences. We considered P ≤ 0.05 significant, but we used a highly conservative step-down procedure as a multiple comparison adjustment to assess the strength of the relationships of our significant variables.
We constructed a correlation matrix to assess relationships between human population density, number of roosts (active and inactive), and landscape characteristics of villages. Only uncorrelated variables (r < 0.7) were analyzed in the same regression model to prevent multicollinearity.14 We built Firth logistic regression models (to correct for quasi-separation) using significant variables from the univariate analyses, and we calculated odds ratios (ORs) with 95% confidence intervals (CIs) to assess the increased odds of being a case village associated with each environmental risk factor when controlling for human population density. We constructed models that compared cases to each of the control groups (inside and outside the Nipah Belt) and a model comparing the control groups. The comparison of control groups allowed us to verify that significant variables in the comparison of cases versus controls outside the Nipah Belt were in fact related to case status rather than differences inside and outside the Nipah Belt.
Comparing human population and village landscape characteristics in the remotely sensed sample.
We compared human population density and forest landscape characteristics of villages inside and outside the Nipah Belt and of case villages and Nipah Belt controls using the remotely sensed sample. We used logistic regression to compare forest metrics while controlling for human population density. We estimated a Satterthwaite-adjusted χ2 in the regression comparing case villages and Nipah Belt controls to account for the large difference in sample size.
Ethical considerations.
Each interviewee gave verbal informed consent for participation. We asked permission of the landowner at roost sites before taking field measurements. The study protocol was approved by the icddr,b and University of Wisconsin-Madison Institutional Review Boards.
Results
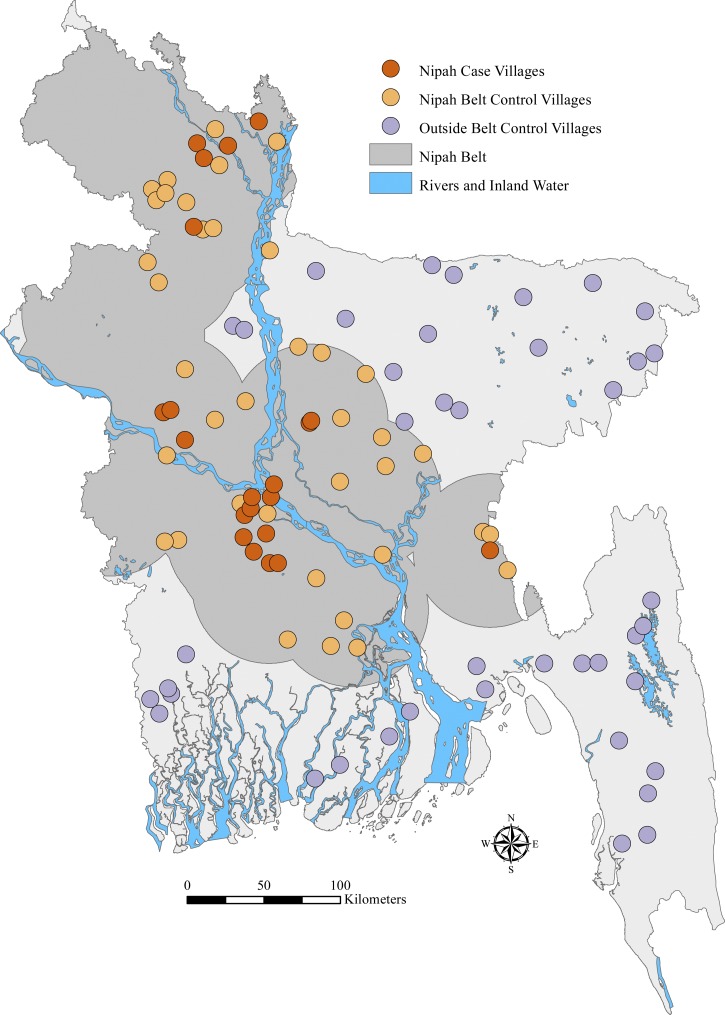
Our field-validated sample contained 100 villages: 21 case villages, 38 control villages inside the Nipah Belt, and 41 control villages outside the belt region (Figure 1). Our field teams located 222 active and inactive roosts, and 87 of the 100 study villages had at least one roost inside the village or within 5 km of the village boundary. The GPS coordinates and bat counts were taken at 221 of the roosts (99.5%), and an ecological assessment in addition to the coordinates and bat counts was conducted at 143 (65%) of these roosts.
Figure 1.
Location of field-validated villages (N = 100).
Comparing characteristics of roost sites and villages in the field-validated sample.
Nipah Belt villages had a higher human population density within a 10 km buffer of the village center (1,311 people/km2 ± 489) than villages outside the Nipah Belt (895 people/km2 ± 440, P < 0.0001) (Table 1 and Figure 2). Within the Nipah Belt, there was no significant difference in human population density between case and control villages. There were no significant differences in the number of households or village area among the study groups.
Table 1.
Characteristics of laboratory-confirmed Nipah spillover case villages and control villages inside and outside the Nipah Belt, Bangladesh (“field-validated sample,” n = 100)
| Characteristics* | Units | Case villages (n = 21) | Nipah belt controls (n = 38) | All Nipah Belt villages (case villages and Nipah Belt controls) (n = 59) | Outside belt controls (n = 41) | Case villages vs. Nipah Belt controls p-value† | Nipah Belt villages vs. outside belt controls p-value† |
|---|---|---|---|---|---|---|---|
| Human population characteristics | |||||||
| Human population density (10 km buffer) | people/km2 | 1333 ± 566 | 1299 ± 448 | 1311 ± 489 | 895 ± 440 | 0.80 | < 0.0001‡ |
| Number of households | 232 ± 157 | 173 ± 150 | 194 ± 154 | 252 ± 308 | 0.16 | 0.27 | |
| Village area | km2 | 4.0 ± 13 | 1.2 ± 2.1 | 2.2 ± 8.1 | 1.1 ± 1.0 | 0.36 | 0.31 |
| Pteropus population characteristics | |||||||
| Total bats roosting within 5 km of village boundary | 618 ± 806 | 659 ± 683 | 646 ± 716 | 603 ± 994 | 0.85 | 0.82 | |
| Bat population density | bats/km2 | 86 ± 270 | 147 ± 503 | 128 ± 441 | 11 ± 28 | 0.56 | 0.06 |
| Roost characteristics | |||||||
| Villages with at least one identified roost | 20/21 (95%) | 35/38 (92%) | 55/59 (93%) | 32/41 (78%) | 1.00§ | 0.03 | |
| Number of roosts (active and inactive) identified within 5 km of village boundary | 3.0 ± 1.9 | 2.4 ± 1.7 | 2.6 ± 1.8 | 1.6 ± 1.5 | 0.18 | 0.004 | |
| Number of active roosts identified within 5 km of village boundary | 1.6 ± 1.3 | 1.8 ± 1.3 | 1.7 ± 1.3 | 1.2 ± 1.0 | 0.53 | 0.05 | |
| Mean tree height | m | 13 ± 3.7 | 14 ± 2.6 | 13 ± 3.0 | 17 ± 4.4 | 0.24 | < 0.0001‡ |
| Mean tree diameter at breast height (DBH) | cm | 32 ± 20 | 40 ± 28 | 38 ± 25 | 38 ± 22 | 0.29 | 0.96 |
| Mean percent canopy cover | 53 ± 12 | 64 ± 13 | 60 ± 14 | 61 ± 22 | 0.002 | 0.91 | |
| Mean tree species richness | 7.4 ± 2.7 | 6.4 ± 3.5 | 6.8 ± 3.2 | 6.0 ± 2.9 | 0.30 | 0.26 | |
| Mean distance to village boundary | km | 1.5 ± 1.0 | 1.6 ± 1.1 | 1.6 ± 1.1 | 1.7 ± 1.5 | 0.91 | 0.62 |
| Mean distance to nearest household | m | 300 ± 744 | 127 ± 332 | 183 ± 504 | 74 ± 208 | 0.37 | 0.17 |
| Tree species composition of roosts§ | |||||||
| Silk cotton and Indian mast tree roosts | 6/18 (33%) | 1/34 (3%) | 7/52 (14%) | 1/32 (3%) | 0.01 | 0.15 | |
| Bamboo roosts | 10/18 (56%) | 26/34 (77%) | 36/52 (69%) | 17/32 (53%) | 0.12 | 0.14 | |
| Banyan roosts | 4/18 (22%) | 4/34 (12%) | 8/52 (15%) | 2/32 (6%) | 0.42 | 0.31 | |
| Raintree and Mahagony roosts | 9/18 (50%) | 15/34 (44%) | 24/52 (46%) | 18/32 (56%) | 0.69 | 0.37 | |
| Teak roosts | 0/18 (0%) | 1/34 (3%) | 1/52 (2%) | 1/32 (3%) | 1.00 | 1.00 | |
| Village land cover characteristics | |||||||
| Percent forest cover (10 km buffer) | 24 ± 4.3 | 27 ± 12 | 26 ± 10 | 33 ± 22 | 0.20 | 0.04 | |
| Forest patch density (10 km buffer) | No. patches/km2 | 0.52 ± 0.14 | 0.48 ± 0.20 | 0.50 ± 0.18 | 0.32 ± 0.21 | 0.40 | < 0.0001‡ |
| Forest edge density (10 km buffer) | edge length (m)/km2 | 22 ± 3.3 | 21 ± 6.2 | 21 ± 5.3 | 18 ± 6.4 | 0.75 | 0.01 |
| Largest forest patch index (10 km buffer) | % of village | 3.6 ± 1.5 | 9.0 ± 13 | 7.1 ± 11 | 19 ± 26 | 0.02 | 0.01 |
Data presented as means ± 1 SD unless otherwise noted.
Based on two-tailed, independent groups t tests unless noted; Satterwaite method was used when P < 0.05 for the F-test for equality of variances; unadjusted P values are shown.
Highly significant based on the adjusted step-down procedure P value.
Shows number of villages where a roost in that species cluster was identified/total villages in the case or control group where a roost was found and an ecological assessment was conducted; P value results are based on χ2 test or two-tailed Fisher exact test when tabular cell counts were < 5.
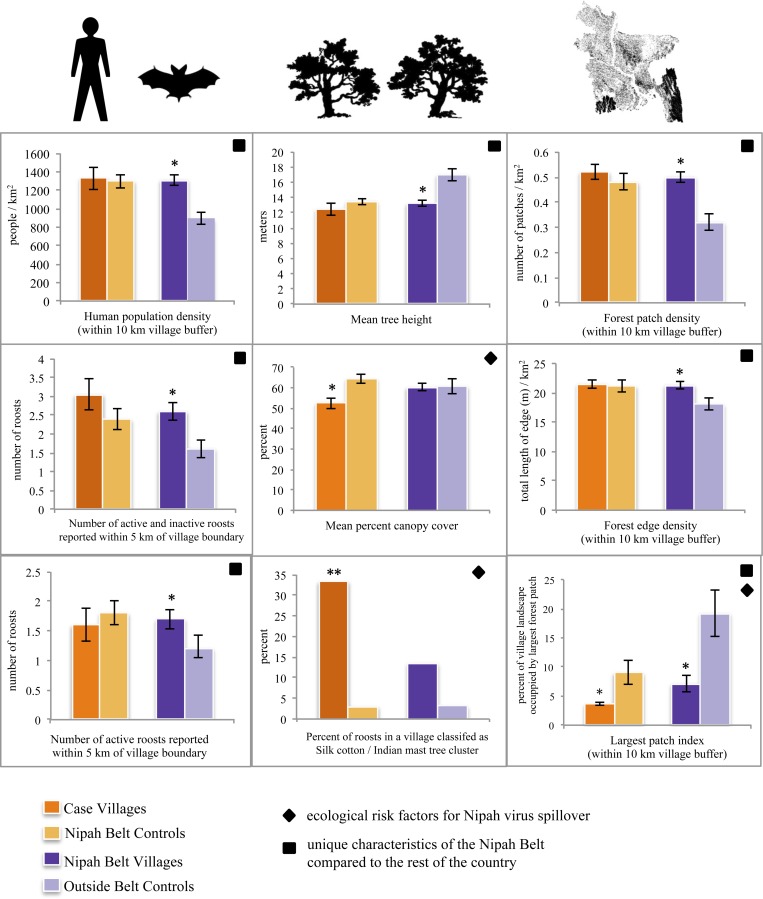
Figure 2.
Comparing human and bat population indicators, Pteropus giganteus roost characteristics, and village forest metrics in Nipah case and control villages, Bangladesh, field-validated sample (N = 100). Data are presented as means with error bars showing ± SE. An (*) or (**) over the Case Villages bar indicates significant t test or Fisher's exact test results, respectively, at the α = 0.05 level compared with Nipah Belt Controls, an indicator of an ecological risk factor for Nipah virus spillover (♦). An (*) over the Nipah Belt Villages bar indicates a significant t test result at the α = 0.05 level compared with the Outside Belt controls, an indicator of a unique ecological characteristic of the Nipah Belt compared with the rest of the country (■). The colors of the columns correspond to the key in Figure 1.
There were no significant differences in the total number of bats, bat population density, or proximity of roosts to human settlements among the study groups. However, inside the Nipah Belt, a greater proportion of villages had an active or inactive P. giganteus roost (93% versus 78%, P = 0.03). Respondents from the Nipah Belt villages reported more roosts (inactive and active) per village (2.6 ± 1.8) than respondents from villages outside the Nipah Belt (1.6 ± 1.5, P = 0.004). Additionally, field teams identified more active roosts within 5 km of the village boundary in Nipah Belt villages (1.7 ± 1.3) compared with villages outside the belt (1.2 ± 1.0, P = 0.05). The percent canopy cover in the roost sites was denser in roosts located in Nipah Belt control compared with case villages (64% ± 13 versus 53% ± 12, P = 0.002). Case villages were more likely to have at least one roost that was categorized by the presence of silk cotton (Bombax spp.) or Indian mast trees (Polyalthia longifolia) compared with controls inside the Nipah Belt (33% versus 3%, P = 0.01) based on the species biomass found in each roost site.
Bat colony size differed by type of roost tree species cluster from the ordination analysis (F = 2.54, degrees of freedom [df] = 4, P = 0.04). Roosts characterized by raintree (Albzia spp.) or mahogany (Swietenia mahagoni) supported the largest bat colonies (623 ± 708), whereas cotton silk or Indian mast tree roosts supported the smallest P. giganteus colonies (170 ± 283).
Nipah Belt villages had less forest cover (26% ± 10), higher forest patch density and edge density (0.50 patches/km2 ± 0.18; 21 m/km2 ± 5.3), and a lower largest patch index (7.1% ± 11) compared with villages outside the Nipah Belt (33% ± 22, P = 0.04; 0.32 patches/km2 ± 0.21, P < 0.0001; 18 m/km2 ± 6.4, P = 0.01; 19% ± 26, P = 0.01). The forest around Nipah virus case villages was the most fragmented of the study groups, but the only significantly different forest metric between case villages and Nipah Belt controls was the largest patch index (3.6% ± 1.5 versus 9.0% ± 13, P = 0.02).
Identifying environmental risk factors for Nipah virus spillover.
In the model comparing cases to Nipah Belt control villages, the odds of being a case was 2.0 times higher (95% CI = 1.1–3.6, P = 0.02) for every 10% decrease in the average canopy cover of roosts sites in the village after controlling for human population density and other significant covariates from the univariate analysis (Table 2). Additionally, villages with at least one silk cotton/Indian mast roost were 10.8 times more likely (95% CI = 1.3–90.6, P = 0.03) to be a case village. In the comparison of cases to controls outside the Nipah Belt, decreased mean tree height in roosts and increased forest patch density in the village were both significant risk factors for Nipah virus spillover. In the model comparing the control groups, a decrease in mean tree height was associated with Nipah Belt control villages.
Table 2.
Multivariate logistic regression models showing the odds of being a Nipah spillover village compared with control villages inside and outside the Nipah Belt, and odds of being a Nipah Belt control village compared with outside belt control villages, Bangladesh (field-validated sample, N = 100)*
| Village characteristics | Unit | Case villages vs. Nipah Belt controls OR (95% CI)† | P value | Case villages vs. outside belt controls OR (95% CI)† | P value | Nipah Belt controls vs. outside belt controls OR (95% CI)† | P value |
|---|---|---|---|---|---|---|---|
| Human population density | Increase of 100 people per km2 | 1.1 (0.9, 1.2) | 0.30 | 1.3 (1.0, 1.6) | 0.06 | 1.1 (1.0, 1.3) | 0.10 |
| Number of active roosts identified within 5 km of village boundary | Increase of 1 roost | 0.7 (0.4, 1.4) | 0.37 | 1.4 (0.4, 4.7) | 0.56 | 1.4 (0.8, 2.6) | 0.24 |
| Mean tree height of roosts located within 5 km of village boundary | Decrease of 1 m | 1.1 (0.8, 1.3) | 0.62 | 1.7 (1.1, 2.5) | 0.01 | 1.3 (1.0, 1.6) | 0.03 |
| Mean percent canopy cover of roosts located within 5 km of village boundary | Decrease of 10% | 2.0 (1.1, 3.6) | 0.02 | 2.1 (0.9, 5.3) | 0.10 | 0.8 (0.6, 1.2) | 0.26 |
| At least one roost in a village classified as Silk cotton and Indian mast tree roost | Presence of cluster | 10.8 (1.3, 90.6) | 0.03 | 5.8 (0.4, 75.4) | 0.18 | 0.3 (0.0, 9.0) | 0.46 |
| Forest patch density (10 km buffer) | Increase of 1 patch per km2 | 1.1 (0.8, 1.7) | 0.53 | 2.8 (1.1, 7.1) | 0.03 | 1.1 (0.8, 1.5) | 0.44 |
Results are from Firth conditional logistic regression, used to correct for quasi-separation in data.
Adjusted for all predictors in the table.
OR = odds ratio; CI = confidence interval.
Assessing relationships between P. giganteus roosting ecology, forest structure, and human population density.
Human population density was positively correlated with forest fragmentation using a variety of indicators (Table 3). Both the number of active and inactive P. giganteus roosts that were reported by community members and the number of active roosts identified by our field teams increased as the human population density and the degree of forest fragmentation increased. As the amount of contiguous forest in a village increased, the mean tree height and percent canopy cover in roosts sites increased, but the tree species diversity decreased.
Table 3.
Correlation matrix of characteristics of villages from field-validated sample (N = 100)
| Village characteristics* | Human population density† | Number of active and inactive roosts reported‡ | Number of active roosts identified‡ In a 10 km buffer from village center‡ | Mean tree height in roosts identified‡ | Mean percent canopy cover in roosts identified‡ | Species richness in roost sites identified‡§ | Percent forest cover† | Forest patch density† | Forest edge density† |
|---|---|---|---|---|---|---|---|---|---|
| Number of active and inactive roosts reported‡ | 0.30 (0.003) | ||||||||
| Number of active roosts identified‡ | 0.30 (0.002) | 0.78 (< 0.0001) | |||||||
| Mean tree height in roosts identified‡ | −0.21 (0.06) | −0.07 (0.50) | 0.02 (0.86) | ||||||
| Mean percent canopy cover in roosts identified‡ | 0.002 (0.98) | −0.03 (0.77) | −0.06 (0.62) | 0.18 (0.11) | |||||
| Species richness in roost sites identified‡§ | 0.09 (0.40) | 0.02 (0.86) | −0.19 (0.09) | −0.35 (0.001) | −0.09 (0.41) | ||||
| Percent forest cover† | −0.20 (0.05) | −0.18 (0.07) | −0.13 (0.19) | 0.28 (0.01) | 0.37 (0.001) | −0.30 (0.01) | |||
| Forest patch density† | 0.32 (0.001) | 0.24 (0.02) | 0.22 (0.03) | −0.22 (0.04) | −0.01 (0.96) | 0.05 (0.63) | −0.54 (< 0.0001) | ||
| Forest edge density† | 0.40 (< 0.0001) | 0.21 (0.04) | 0.17 (0.09) | −0.06 (0.58) | 0.33 (0.002) | −0.09 (0.43) | 0.38 (< 0.0001) | 0.15 (0.13) | |
| Largest forest patch index†¶ | −0.40 (< 0.0001) | −0.27 (0.01) | −0.20 (0.05) | 0.38 (0.000) | 0.26 (0.02) | −0.36 (0.001) | 0.94 (< 0.0001) | −0.65 (< 0.0001) | 0.12 (0.25) |
Data presented as Pearson's correlation coefficient (P value); bolded values are significant.
In a 10 km buffer from village center.
Within 5 km of village boundary.
Mean number of tree species found in roost sites.
Percent of village area occupied by the largest forest patch.
Comparing human population and village landscape characteristics in the remotely sensed sample.
Univariate comparisons within the remotely sensed sample yielded similar results to the field-validated sample, showing that human population density is higher and the forest is more fragmented inside the Nipah Belt compared with the rest of the country (Table 4). However, the larger sample size increased our statistical power to detect differences in the forest structure of case villages compared with Nipah Belt control points. Cases had less forest cover (22% ± 4.1 versus 23% ± 9.8, P = 0.04), higher forest patch density (0.55 patches/km2 ± 0.14 versus 0.49 patches/km2 ± 0.20), higher forest edge density (21 m/km2 ± 3.6 versus 20 m/km2 ± 5.3, P = 0.004), and a smaller contiguous forest area (3.5% ± 1.9 versus 6.3% ± 10, P = 0.000) than remotely sensed Nipah Belt controls after controlling for human population density.
Table 4.
Characteristics of laboratory-confirmed Nipah spillover case villages (identified from 2001 to 2011) and 20,000 remotely sensed control sites inside and outside the Nipah Belt, Bangladesh (“remotely sensed sample,” N = 20,057)
| Village characteristics* | Units | Case villages (N = 57) | Nipah Belt remotely sensed controls (N = 10,000) | All Nipah Belt villages (case villages and remotely sensed Nipah Belt controls) (N = 10,057) | Outside belt remotely sensed controls (N = 10,000) | Case villages vs. Nipah Belt remotely sensed controls P value† | Nipah Belt villages vs. outside belt remotely sensed controls P value† |
|---|---|---|---|---|---|---|---|
| Human population density‡ | People/km2 | 1572 ± 2524 | 1381 ± 1355 | 1382 ± 1364 | 873 ± 760 | 0.57 | < 0.0001 |
| Percent forest cover‡ | % | 22 ± 4.1 | 23 ± 9.8 | 23 ± 9.8 | 38 ± 27 | 0.04§ | < 0.0001 |
| Forest patch density‡ | No. patches/km2 | 0.55 ± 0.14 | 0.49 ± 0.20 | 0.49 ± 0.20 | 0.30 ± 0.21 | 0.001§ | < 0.0001 |
| Forest edge density‡ | Edge length (m)/km2 | 21 ± 3.6 | 20 ± 5.3 | 20 ± 5.3 | 16 ± 8.3 | 0.004§ | < 0.0001 |
| Largest forest patch index‡¶ | % of village | 3.5 ± 1.9 | 6.3 ± 10 | 6.3 ± 10 | 24 ± 31 | 0.000§ | < 0.0001 |
Data presented as means ± 1 SD.
Human population density comparison is based on a two-tailed, independent group t test; forest metric comparisons are based on χ2 results from logistic regression controlling for human population density.
In a 10 km buffer from village center.
The comparison of case villages to Nipah Belt controls is a Satterwaite-adjusted ±2 to account for the large difference in sample size.
Percent of village area occupied by the largest forest patch.
Discussion
Our study shows that villages inside the Nipah Belt of Bangladesh have significantly higher human population density and forests that are more fragmented than in the rest of the country. The finding that human population density correlates with spillover risk is not unexpected; but has not been previously shown. However, our finding that landscape factors also correlate with Nipah virus spillover risk is novel and suggests that these factors could be used to understand the distribution of Nipah cases in other regions.
Our study also shows that the roosting ecology of P. giganteus is associated with forest fragmentation, and that canopy density and tree species composition in roosts and degree of forest fragmentation in a village are associated with Nipah virus spillover. The number of roosts was higher in villages with more fragmented forests, although the number of bats in a village was not. This suggests that roost colony size is limited by tree availability and that P. giganteus can occupy fragmented landscapes. Studies have found that other bats with generalist diets thrive in fragmented forests with numerous small patches of remnant forest due to their diverse diet and ability to travel long foraging distances that allow them to use patchy landscapes that other forest-obligate species cannot15; likely the same applies to P. giganteus.
We also found that biodiversity of tree species in roosts increased as the degree of forest fragmentation increased. Because villages with high human population density had the most fragmented forests in our study, the biodiversity in these Nipah Belt roosts is likely caused by species-rich homegardens.16 A survey of homegardens found that on average, a single Bangladeshi household is growing 34 tree species.16 A benefit for Pteropus living in fragmented forests is that these mixed landscapes may provide a more consistent food source than habitats comprised solely of pristine forest species.15 Some have suggested that agroforestry may support larger Pteropus populations than would be viable without human cultivated crops,17 suggesting that the P. giganteus population in the Nipah Belt may expand alongside the human population.18
Within the Nipah Belt, villages where there have been Nipah virus spillovers had more fragmented forests compared with neighboring communities. In these fragmented forests, P. giganteus tended to settle in several small roosts scattered throughout the villages rather than in one large roosting colony. This roosting behavior, perhaps in response to the forest structure, could have implications for Nipah virus spillover. In these fragmented forests, the combination of more people and sporadic P. giganteus colonies could increase the likelihood that P. giganteus will feed on human food resources, both fruits from homegardens and date palm sap collection containers. Future assessments should assess differences in P. giganteus feeding behavior across gradients of forest fragmentation. Second, forest fragmentation could affect Nipah transmission within the P. giganteus population if roost colony size affects intra-population virus transmission dynamics. Future research should assess virus prevalence in roosting colonies with different sized populations and whether these roosting colonies represent distinct metapopulations or artificial divisions within a single colony to understand these relationships.
Two tree species have potential importance for Nipah virus spillover. The presence of a roost distinguished by silk cotton (Bengali: shimul) or Indian mast trees (Bengali: debdaru) was associated with an increased risk of Nipah virus spillover. Both trees are known Pteropus food resources,19–21 have short flowering durations in the winter, and silk cottons (“morphologically a perfect bat plant”22) have chiropterophilous23 traits that facilitate pollination by Pteropus20,21 (e.g., brightly colored, open and increased nectar production at night when bats are feeding21). A possible link between these tree species and Nipah virus dynamics is that because these trees flower during the season of limited fruit resources in Bangladesh, they are visited by high concentrations of P. giganteus during their short flowering periods.21 As a result of this “watering hole effect” where bats congregate around limited resources, saliva or other bodily fluids containing Nipah virus could be more likely exchanged when drinking from the same flower or during defensive behavior, behaviors that have been observed in Pteropus populations feeding on silk cottons in the Pacific Islands.21 The result could be an increase in the prevalence of acute infections of bats living near these food resources and increased likelihood of spillover to humans when bats drink from date palm sap containers. However, roosts with these indicator species only occurred in one-third of case villages, suggesting that although these species may be indicators of spillover risk, they are not a prerequisite. In addition to the trees that are seeded by foraging bats, these trees could also be indicators of forest fragmentation. Indian mast trees are often planted as ornamental border trees24 so their presence could be associated with human impact on the landscape. Future studies are needed to assess the infection prevalence of bat colonies in relation to their roosting habitat.
Several village and roost characteristics were not associated with Nipah virus spillover in this study including geographic size of a village and distance of roosts to the village boundary or nearest household. This finding supports the idea that although presence of P. giganteus near human activity is a likely precursor of viral spillover, there are other processes that determine the probability that spillover occurs. Our study suggests that the composition and structure of the landscape shared by P. giganteus and humans may be two drivers.
We relied on community members to identify and locate roosts. It is possible that we missed isolated roosts of which respondents were not aware, which would underestimate the number of roosts in less populated areas, like those in regions outside the Nipah Belt, perhaps making the number of roosts inside and outside the belt more comparable. However, this bias is unlikely because most roosts are highly conspicuous as a result of the bats' size and noisy demeanor. Roosts without bats present during our study (inactive roosts) present another possible bias. Our results were the same when we assessed total roosts or restricted our analysis to only active roosts, so this bias is also unlikely to affect our conclusions.
The resolution of our satellite-derived forest cover map was 250 m, which made it difficult to identify small forest patches. If small patches were misclassified as non-forest, particularly in areas with highly fragmented forests, then our fragmentation measures would be underestimated and the association between fragmentation, spillover status, and bat roost abundance would have been conservative.
Although it is important to note that human population density appears to be a key driver of Nipah virus risk in rural Bangladesh, this study supports the hypothesis that the geographic distribution of Nipah virus spillover is also influenced by the configuration of suitable P. giganteus habitat and the roosting ecology of these bats. Although bats are present in similar numbers throughout Bangladesh, the abundance of roosts is higher in the region where all Nipah outbreaks have occurred, suggesting that the distribution of these bats across the village landscape may increase risk. Even though the precise mechanism is unknown, perhaps the fragmented forest landscape increases overlap between human and bat food resources, specifically in a region of high human population density. Although there have been attempts to track Pteropus spp. movements away from the roost,25–27 more research is needed to quantify P. giganteus foraging behavior in fragmented and biologically diverse landscapes as well as seasonal foraging distance to test this hypothesis. Additionally, tree species composition in roosts is associated with risk of Nipah virus spillover and may influence P. giganteus interactions and viral transmission within bat communities.
As the Bangladeshi population continues to grow to over 200 million by 2050,18 more undisturbed forest areas will become sites for homesteads and expanding villages. Ecosystem and land-use changes have played a significant role in infectious disease emergence and re-emergence in humans, including malaria, yellow fever, hantavirus, leishmaniasis, and hemorrhagic fevers.28–32 Understanding how these changes alter the forest structure, and in turn, the habitats and the ecology of disease reservoirs may provide insights to reduce the spillover of zoonotic disease agents.33,34
ACKNOWLEDGMENTS
We thank Hossain M. S. Sazzad, Golam Dostogir Harun, A. K. M. Dawlat Khan, and Sonia T. Hegde for their invaluable administrative and logistical support during the field component of this study. Our sincerest gratitude to the icddr,b field staff for their dedication to collecting accurate and complete data. Thank you to M. A. Yushuf Sharker and Ron Gangnon for their statistical support. Finally, many thanks to the study respondents for their time and for welcoming us into their homes and communities.
Footnotes
Financial support: This research investigation was funded by the NSF/NIH Ecology and Evolution of Infectious Diseases grant no. 2R01-TW005869 (Daszak, PI) from the Fogarty International Center and the NSF IGERT grant no. 0549407 (Patz, PI): the CHANGE-IGERT in the Nelson Institute at the University of Wisconsin-Madison.
Authors' addresses: Micah B. Hahn and Jonathan A. Patz, Nelson Institute, SAGE (Center for Sustainability and the Global Environment), Department of Population Health Sciences, School of Medicine and Public Health, University of Wisconsin, Madison, WI, E-mails: mbhahn@wisc.edu or micah.hahn@gmail.com and patz@wisc.edu. Emily S. Gurley, International Center for Diarrheal Disease Research, Bangladesh (icddr,b), Dhaka, Bangladesh, E-mail: egurley@icddrb.org. Jonathan H. Epstein and Peter Daszak, EcoHealth Alliance, E-mails: epstein@ecohealthalliance.org and daszak@ecohealthalliance.org. Mohammad S. Islam, Center for Environmental and Geographic Information Services, E-mail: sislam@cegisbd.com. Stephen P. Luby, Stanford University, E-mail: sluby@stanford.edu.
References
- 1.Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis. 2009;15:1229–1235. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luby SP, Rahman M, Hossain MJ, Blum LS, Husain MM, Gurley E, Khan R, Ahmed B-N, Rahman S, Nahar N, Kenah E, Comer JA, Ksiazek TG. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu VP, Hossain MJ, Parashar UD, Ali MM, Ksiazek TG, Kuzmin I, Niezgoda M, Rupprecht C, Bresee J, Breiman RF. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis. 2004;10:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luby SP, Gurley ES, Hossain MJ. Transmission of human infection with Nipah virus. Clin Infect Dis. 2009;49:1743–1748. doi: 10.1086/647951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harcourt BH, Lowe L, Tamin A, Liu X, Bankamp B, Bowden N, Rollin PE, Comer JA, Ksiazek TG, Hossain MJ, Gurley ES, Breiman RF, Bellini WJ, Rota PA. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg Infect Dis. 2005;11:1594–1597. doi: 10.3201/eid1110.050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo MK, Lowe L, Hummel KB, Sazzad HM, Gurley ES, Hossain MJ, Luby SP, Miller DM, Comer JA, Rollin PE, Bellini WJ, Rota PA. Characterization of Nipah virus from outbreaks in Bangladesh, 2008–2010. Emerg Infect Dis. 2012;18:248–255. doi: 10.3201/eid1802.111492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tidemann CR, Nelson JE. Long-distance movements of the grey-headed flying fox (Pteropus poliocephalus) J Zool. 2004;263:141–146. [Google Scholar]
- 8.Wisz MS, Guisan A. Do pseudo-absence selection strategies influence species distribution models and their predictions? An information-theoretic approach based on simulated data. BMC Ecology. 2009;9:8. doi: 10.1186/1472-6785-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGarigal K, Cushman SA, Ene E. FRAGSTATS v4: spatial pattern analysis program for categorical and continuous maps. 2012. http://www.umass.edu/landeco/research/fragstats/fragstats.html Computer software program produced by the authors at the University of Massachusetts, Amherst. Available at. Accessed October 15, 2012.
- 10.Oak Ridge National Laboratory, LandScan Global Population Database. 2011. http://www.ornl.gov/sci/landscan/index.shtml Available at. Accessed September 1, 2012.
- 11.McCune B, Mefford M. PC-ORD: Software for Multivariate Analysis of Ecological Data. 2010. Version 6. [Google Scholar]
- 12.Bray JR, Curtis TP. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–349. [Google Scholar]
- 13.McCune B, Grace J, Dean L. Analysis of Ecological Communities. 2002. p. 75. Mjm Software Design. [Google Scholar]
- 14.Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, Marquéz JR, Gruber B, Lafourcade B, Leitão PJ, Münkemüller T, McClean C, Osborne PE, Reineking B, Schröder B, Skidmore AK, Zurell D, Lautenbach S. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography. 2012;35:001–020. [Google Scholar]
- 15.Gorresen PM, Willig MR. Landscape responses of bats to habitat fragmentation in Atlantic forest of Paraguay. J Mammal. 2004;85:688–697. [Google Scholar]
- 16.Kabir E, Webb EL. Can homegardens conserve biodiversity in Bangladesh? Biotropica. 2008;40:95–103. [Google Scholar]
- 17.Stier SC, Mildenstein TL. Dietary habits of the world's largest bats: the Philippine Flying Foxes, Acerodon Jubatus and Pteropus Vampyrus Lanensis. J Mammal. 2005;86:719–728. [Google Scholar]
- 18.Streatfield PK, Karar ZA. Population challenges for Bangladesh in the coming decades. J Health Popul Nutr. 2008;26:261–272. [PMC free article] [PubMed] [Google Scholar]
- 19.Vendan SE, Kaleeswaran B. Plant dispersal by Indian flying fox Pteropus giganteus in Madurai region, India. Elixir Bio. Div. 2011;30:1810–1813. [Google Scholar]
- 20.Singaravelan N, Marimuthu G. Nectar feeding and pollen carrying from Ceiba pentandra by Pteropodid bats. J Mammal. 2004;85:1–7. [Google Scholar]
- 21.Elmqvist T, Cox PA, Rainey WE, Pierson ED. Restricted pollination on Oceanic islands: pollination of Ceiba pentandra by flying foxes in Samoa. Biotropica. 1992;24:15–23. [Google Scholar]
- 22.Pierson ED, Rainey WE. The biology of flying foxes of the Genus Pteropus: a review. Pacific Island Flying Foxes: Proceedings of an International Conservation Conference. U.S. Department of the Interior. 1992;90:1–17. [Google Scholar]
- 23.Marshall AG. Bats, flowers and fruit: evolutionary relationships in the Old World. Biological Journal of the Linnean Society. 1983;20:115–135. [Google Scholar]
- 24.Nagendra H, Gopal D. Tree diversity, distribution, history and change in urban parks: studies in Bangalore, India. Urban Ecosyst. 2013;14:211–223. [Google Scholar]
- 25.Epstein JH, Olival KJ, Pulliam JRC, Smith C, Westrum J, Hughes T, Dobson AP, Zubaid A, Rahman SA, Basir MM, Field HE, Daszak P. Pteropus vampyrus, a hunted migratory species with a multinational home-range and a need for regional management. J Appl Ecol. 2009;46:991–1002. [Google Scholar]
- 26.Walton R, Trowbridge BJ. The use of radio-tracking in studying the foraging behavior of the Indian flying fox (Pteropus giganteus) J Zool. 1983;201:575–579. [Google Scholar]
- 27.Mildenstein TL, Stier SC, Nuevo-Diego CE, Mills LS, Nuevodiego C. Habitat selection of endangered and endemic large flying-foxes in Subic Bay, Philippines. Biol Conserv. 2005;126:93–102. [Google Scholar]
- 28.Lindblade KA, Walker ED, Onapa AW, Katungu J, Wilson ML. Land use change alters malaria transmission parameters by modifying temperature in a highland area of Uganda. Trop Med Int Health. 2000;5:263–274. doi: 10.1046/j.1365-3156.2000.00551.x. [DOI] [PubMed] [Google Scholar]
- 29.Poletto DW, Rodas LA, Koyanagui PH, Botti MV, Mucci LF, Gomes AD. Entomological investigation of a sylvatic yellow fever area in São Paulo State, Brazil. Cad Saude Publica. 2005;21:1278–1286. doi: 10.1590/s0102-311x2005000400031. [DOI] [PubMed] [Google Scholar]
- 30.Suzán G, Armién A, Mills JN, Marcé E, Ceballos G, Ávila M, Salazar-bravo J, Ruedas L, Armién B, Yates TL, Armien B, Marce E. Epidemiological considerations of rodent community composition in fragmented landscapes in Panama. J Mammal. 2008;89:684–690. [Google Scholar]
- 31.Campbell-Lendrum D, Dujardin JP, Martinez E, Feliciangeli MD, Perez JE, Silans LN, Desjeux P. Domestic and peridomestic transmission of American cutaneous leishmaniasis: changing epidemiological patterns present new control opportunities. Mem Inst Oswaldo Cruz. 2001;96:159–162. doi: 10.1590/s0074-02762001000200004. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe ND, Daszak P, Kilpatrick AM, Burke DS. Bushmeat hunting, deforesation, and prediction of zoonotic disease emergence. Emerg Infect Dis. 2004;11:1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halpin K, Hyatt AD, Plowright RK, Epstein JH, Daszak P, Field HE, Wang L, Daniels PW. Emerging viruses: coming in on a wrinkled wing and a prayer. Clin Infect Dis. 2007;44:711–717. doi: 10.1086/511078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood JL, Leach M, Waldman L, Macgregor H, Fooks AR, Jones KE, Restif O, Dechmann D, Hayman DT, Baker KS, Peel AJ, Kamins AO, Fahr J, Ntiamoa-Baidu Y, Suu-Ire R, Breiman RF, Epstein JH, Field HE, Cunningham A. A framework for the study of zoonotic disease emergence and its drivers: spillover of bat pathogens as a case study. Philos Trans R Soc Lond B Biol Sci. 2012;367:2881–2892. doi: 10.1098/rstb.2012.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]