Abstract
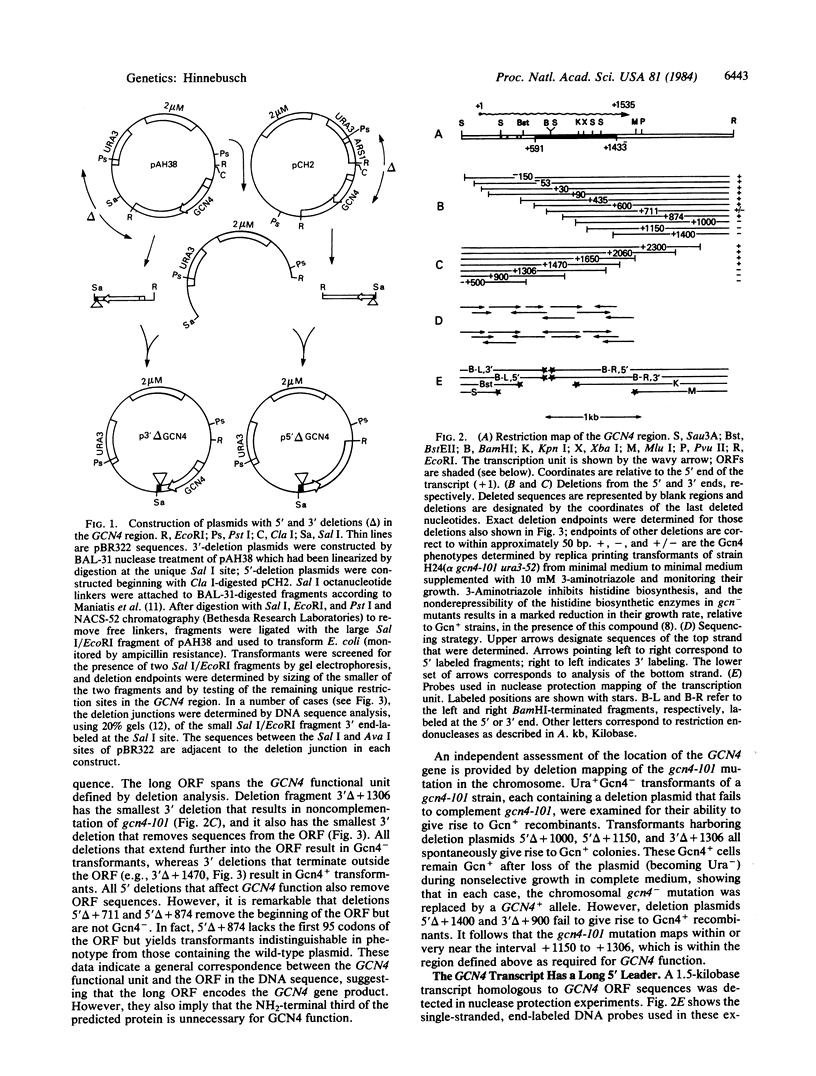
The GCN4 gene encodes a positive regulator of unlinked amino acid biosynthetic genes in yeast. I present evidence that the GCN4 gene is itself regulated by amino acid availability and that the regulation occurs at the translational level. A GCN4-lacZ fusion was used as a measure of the expression of GCN4 gene product. Starvation for histidine leads to derepression of the fusion enzyme in the wild type but not in a gcn2- strain. The gcn2- mutation does not reduce fusion transcript levels relative to wild type, suggesting that the product of GCN2 functions as an activator of GCN4 translation. The GCN4 transcript has a 5' leader that is approximately equal to 600 nucleotides long and contains four small open reading frames. A deletion of the small open reading frames results in constitutive derepression of fusion enzyme levels as the result of an approximately equal to 10-fold increase in the efficiency of translation of the fusion transcript. The deletion suppresses the requirement for GCN2 function. These results suggest that the GCN4 5' leader acts in cis to repress GCN4 translation and that GCN4 translation increases in response to amino acid starvation as the result of GCN2 antagonism of the repressing sequences in the GCN4 5' leader.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Penn M. D., Galgoci B., Greer H. Identification of AAS genes and their regulatory role in general control of amino acid biosynthesis in yeast. Proc Natl Acad Sci U S A. 1983 May;80(9):2704–2708. doi: 10.1073/pnas.80.9.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Regulatory sites for his3 gene expression in yeast. Nature. 1982 Nov 18;300(5889):285–286. doi: 10.1038/300284a0. [DOI] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]