Summary
Triple-negative breast cancer (TNBC) is defined by a lack of hormone receptor expression as well as lack of overexpression/amplification of HER2/neu. Patients with TNBC show a significantly worse prognosis compared to patients with other breast cancer subtypes. TNBC, however, is a heterogeneous entity both with regard to clinical/pathological characteristics and molecular biology. This review summarizes the current data on TNBC with a particular focus on mutational and gene expression profiling and the association between TNBC and breast cancer stem cells.
KeyWords: Triple negative breast cancer, subtypes; Whole genome sequencing; RNA profiling; Heterogeneity
Zusammenfassung
Das tripelnegative Mammakarzinom (triple negative breast cancer, TNBC) ist definiert durch eine fehlende Hormonrezeptorexpression sowie eine fehlende Amplifikation/Überexpression des HER2/neu-Onkogens. Patientinnen mit einem TNBC zeigen eine signifikant schlechtere Prognose im Vergleich zu Patientinnen mit anderen Mammakarzinomsubtypen. Jedoch handelt es sich beim TNBC durchaus um eine heterogene Erkrankung sowohl im Hinblick auf klinische/pathologische Parameter als auch auf molekularbiologische Faktoren. In dieser Übersichtsarbeit fassen wir die aktuellen Daten zum TNBC zusammen mit einem speziellen Fokus einerseits auf die Ergebnisse von Mutations- und Genexpressionsanalysen, andererseits auf den Zusammenhang zwischen TNBC und Mammakarzinom-Stammzellen.
Definition of Triple-Negative Breast Cancer
Triple-negative breast cancer (TNBC) is defined as a subgroup of breast cancers lacking expression of the estrogen (ER) and progesterone (PR) receptors as well as amplification/overexpression of the HER2/neu oncogene [1]. About 10–15% of patients diagnosed with breast cancer are faced with this diagnosis. Risk factors for TNBC include young age/premenopausal status, African American or Hispanic ethnicity and, most of all, genetic mutation of the BRCA-1 gene [2]. Unfortunately, patients with TNBC carry a more unfavorable prognosis compared to patients with other subtypes of breast cancer, which seems to be due to: (i) a lack of systemic therapies, given that endocrine therapy and HER2-targeted agents are not an option; and (ii) a more aggressive biological behavior that is mirrored by a predominance of grade 3 tumors, high proliferation rate, and visceral and particularly cerebral metastases [3]. This unfavorable prognosis is even more pronounced among patients of young age at diagnosis [4].
Since the increasing use of high-throughput gene expression profiling tools such as gene expression arrays, it is now well established that breast cancer comprises several clinically and biologically distinct subtypes [5, 6]. Basal-like breast cancer (BLBC) has been shown to express particularly basal biomarkers such as cytokeratin 5/6 and vimentin. This breast cancer subtype is often diagnosed among patients with hereditary breast cancer (particularly BRCA-1) and carries a particularly unfavorable prognosis [7, 8]. Although this breast cancer subtype shows a strong clinical and biological correlation with clinically defined TNBC, both definitions are far from being synonymous.
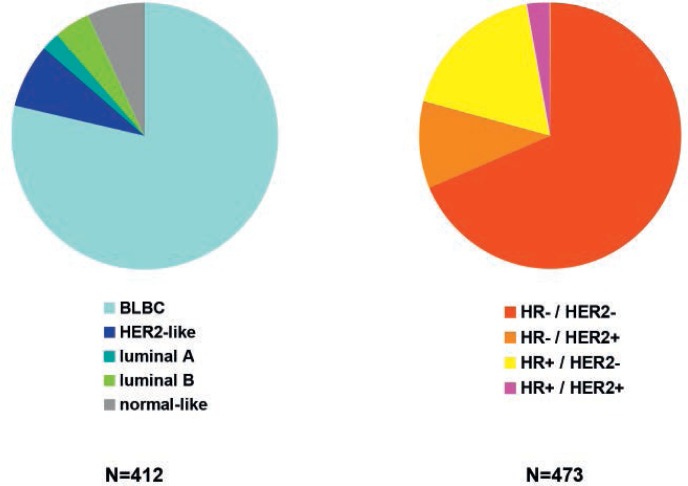
Prat et al. [9] conducted a systematic analysis of both breast cancer subtype definitions. They analyzed 412 breast cancers stratified as triple negative through analysis of ER, PR and HER2 expression, and 473 breast cancers identified as basal-like through PAM-50 analysis. As expected given the results of earlier analyses, 21.4% of TNBC were not profiled as BLBC and 31.5% of BLBC were not stratified as being TNBC (fig. 1). The authors concluded, based on this analysis, that complex gene expression profiles may be so preserved through the disease course of a breast cancer that even loss of ER and/or HER2 expression may not reverse these profiles.
Fig. 1.
Distribution of intrinsic and clinically/immunohistochemically defined breast cancer subtypes within TNBC and BLBC (based on [9]).
In the definition of TNBC it is important to recognize that the cut-off levels for ER and PR have changed within the last decade. While breast cancers were previously stratified as hormone receptor positive if at least 10% of cancer cells expressed ER and/or PR, the cut-off level of hormone receptor positivity is currently set at 1% positive tumor cells. This is particularly important given that earlier reports of the clinical/biological behavior of TNBC were based on the previous definition through which more cases were classified as triple negative than would be the case nowadays; now many would be regarded as hormone receptor positive.
Although there is a clear consequence of more cases being regarded as hormone receptor positive, i.e. extending the indication of endocrine therapy to patients whose tumors express ER and/or PR in 1–9% of tumor cells, there is still ongoing debate as to the biology of these breast cancers with borderline hormone receptor expression.
To shed further light on this area of research, Cheang et al. [10] analyzed breast cancer specimens from more than 1,500 patients included in 3 randomized clinical trials (GEICAM 9906 (n = 820), NCIC CTG MA.5 (n = 476), and NCIC CTG MA.12 (n = 398)). The authors correlated the distribution of intrinsic breast cancer subgroups defined by complex gene expression analysis with the immunohistochemical expression of ER, PR and HER2 [10]. Interestingly, the authors noted a significant amount of ER expression only in a very few cases of BLBC. Among HER2-negative breast cancers with no more that 10% of ER expression (i.e. cases with ‘borderline’ hormone receptor expression based on the previous definition of TNBC), 73% of tumors were classified as BLBC, while 17% of tumors were stratified as HER2-like, 5% as luminal B and 2% as luminal A.
By contrast, Iwamoto et al. [11] determined the ER status of 465 breast cancer cases using both immunohistochemistry and Affymetrix-genechip-based gene expression profiling. The ER expression as determined by genechip analysis (measuring the ESR1 gene) was significantly higher among tumors shown to have ≥ 10% ER-positive tumor cells by immunohistochemistry compared to tumors with 1–9% positive cells or ER-negative tumors. The expression of an ER-like gene signature containing 106 ER-associated genes was similar among tumors with no or 1–9% positive cells and was significantly higher among tumors with ≥ 10% positive cells.
Heterogeneity of TNBC
Patients with TNBC are faced with a particularly unfavorable prognosis given a lack of systemic therapies other than chemotherapy. Although the monoclonal antibody bevacizumab may is licensed in some countries for patients in the first-line metastatic setting in combination with paclitaxel or capecitabine monochemotherapy, targeted treatment options specifically targeting the biology of TNBC are yet to be identified. However, TNBC in itself represents a heterogeneous entity. For instance, patients who respond well to chemotherapy (demonstrated by a pathological complete response (pCR) in the neoadjuvant setting) present with a favorable outcome that is comparable to other breast cancer subtypes [12, 13].
Novel high-throughput analytical methods such as next generation sequencing (NGS) technologies allow a rapid analysis of the mutational spectrum of cancers, thereby providing a means to identify complex mutational profiles. NGS has been used to demonstrate the very heterogeneous nature of TNBC and the large spectrum of gene mutations that occur. The most important mutation was found in the p53 tumor suppressor gene, the second most common mutation was identified in phosphoinositol-3-kinase (PI3K). In addition, there are a large number of mutations that occur only at low frequency but contribute substantially to the heterogeneous nature of TNBC. Using parallel high-throughput mRNA sequencing it was shown that only about 36% of mutations are in fact transcribed to the mRNA level [14].
In another analysis, primary breast cancers were analyzed in parallel using several high-throughput analysis tools (i.e. genomic DNA copy number arrays, DNA methylation, exome sequencing, messenger RNA arrays, microRNA sequencing and reverse-phase protein arrays). The authors found that the expression and mutational profile of BLBC was very similar to that observed in serous ovarian cancer. This suggests a similar etiology, and also provides a basis for the establishment of similar therapeutic concepts [15]. However, clinical studies to demonstrate the true clinical value of these observations are yet to follow.
Molecular Subtypes of TNBC
Since the extent of TNBC heterogeneity has become known, efforts have been made either to simply describe these subgroups or to associate them with disease prognosis or probability of response to certain (systemic) therapies.
Lehmann et al. [16] analyzed and homogenized 587 gene expression datasets from 21 distinct analyses to identify TNBC subtypes using a top-down approach based on hierarchical clustering. The authors were able to identify 6 distinct breast cancer subtypes. These subtypes could be reproduced using a set of breast cancer cell lines, with each TNBC subtype being present in a number of cell lines. Although these subtypes do not yet have implications for standard clinical care, there is increasing evidence that molecular TNBC subtypes may show a clinically distinct behavior. Masuda et al. [17] analyzed gene expression profiles of 130 cases of TNBC. All patients had undergone neoadjuvant chemotherapy. The authors demonstrated that response rates differed between the molecular TNBC subtypes: The BL1 subtype had the highest pCR rate (52%); BL2 and LAR the lowest (0% and 10%, respectively). Similarly, Telli et al. [18] showed that response to a neoadjuvant gemicitabine-carboplatin chemotherapy in combination with the PARP inhibitor iniparib varied among the molecular TNBC subtypes.
Prognostic Gene Expression Signatures
Attempts have been made to define individual biomarkers and complex gene expression signatures of prognostic relevance for patients with TNBC in order to stratify patients for systemic therapy, both in the context of clinical trials and in daily clinical routine.
Rody et al. [19] assembled and homogenized 579 publically available datasets obtained from patients with TNBC. In a subset of 394 cases several metagenes were analyzed for a prognostic function among these patients. The authors observed that an increased expression of immune cell metagenes was correlated with an improved outcome, while expression of metagenes for inflammation and/or angiogenesis was associated with a more unfavorable outcome. A ratio of high B cell and low IL8 metagenes was associated with an improved prognosis among patients with TNBC, and proved to be an independent prognostic factor in multivariate analysis including clinical and pathological variables [19]. While components of the B cell metagene were shown to be expressed by tumor-infiltrating lymphocytes (as could be expected), expression of IL8 was limited to the cancer cells themselves, raising the question whether inhibition of IL8 through small molecules or humanized antibodies might be a therapeutic option for patients with TNBC.
Predictive Gene Expression Signatures
Whereas prognostic signatures may help in determining whether systemic therapies should be applied in light of an unfavorable or more favorable prognosis, predictive signatures are needed not only to define a group of patients at increasing risk of relapse, but also to identify a subgroup of those patients who in addition have a high probability of responding to a given systemic intervention. However, such predictive signatures are lacking for all breast cancer subgroups.
Denkert and colleagues from the German Breast Group [20] analyzed the prognostic and predictive value of KI-67 expression in parallel among breast cancer subgroups based on specimens obtained in the context of a randomized neoadjuvant chemotherapy trial. The role of KI-67 expression is currently an issue of debate, particularly in the context of distinguishing luminal A and B breast cancer subtypes, which are believed to differ particularly with regard to tumor cell proliferation. In this context, increased expression of KI-67 correlates with an adverse prognosis, but also with an increased chance of response to chemotherapy. The routine clinical use of such data is limited at present as the optimal cut-off between tumors of high vs. low proliferation is not yet established and intra-observer reproducibility is limited. The significance of KI-67 expression in TNBC is also not clearly established even though patients and therapists are often faced with very high KI-67 expression values. Denkert et al. [20] analyzed KI-67 expression in 1,166 breast cancer specimens that had been obtained prior to neoadjuvant chemotherapy in the context of the GeparTrio trial. Among TNBC, a significant correlation between KI-67 expression and the probability of a pCR was described; however, no significant correlation between KI-67 expression and prognosis (i.e. disease-free and overall survival) could be demonstrated. The pCR rate for a KI-67 expression of ≤ 15%, 15–35% and ≥ 35% was observed to be 15%, 22% und 38% (p = 0.003). Therefore, although the expression of KI-67 measured using immunohistochemistry is a significant predictive marker for response to neoadjuvant chemotherapy, it does not represent a significant prognostic factor for patients with TNBC.
TNBC and Cancer Stem Cells
The question of the cell of origin of cancer is still controversial. One model describes tumorigenesis as being a product of dysregulation of the process of self-renewal and differentiation of breast stem cells and progenitor cells [21, 22]. As a consequence of this disturbed hierarchical process, extensive proliferation and reduced apoptotic events occur concomitantly with mutational selection of cancer (stem) cells.
Cancer stem cells display a higher resistance against cytotoxic agents compared to other cells. This is probably due to a quiescent state in which they reside, making it difficult to treat these cells with drugs that act on mitotic cells, e.g. highly proliferating cancer cells. On the other hand, cancer stem cells seem to express a set of transporter systems that allow these cells to exclude cytotoxic agents [23]. Thus, when a tumor is targeted using agents against proliferative cells, cancer stem cells might remain inside the tumor and reseed the tumor after chemotherapy has been completed [24, 25]. This phenomenon makes cancer stem cells an interesting and necessary target for future clinical therapies.
Breast cancer stem cells have been shown to be characterized by a specific expression pattern of cell surface markers, i.e. CD44+/CD24-/low [21]. A following paper described a new tool for distinguishing breast cancer stem cells from tissues and cell lines using increased aldehyde dehydrogenase (ALDH) activity [26]. Since then, using both CD44+/CD24-/low expression pattern as well as ALDH activity – besides functional approaches like mammosphere formation or xenograft tumor seeding – has become ‘gold standard’ of breast cancer stem cell characterization in vitro.
Subsequently, these characterization assays have been used to stratify breast cancer subtypes with regard to their cancer stem cell content. In a screening approach using immunohistochemical analysis followed by molecular profiling, Honeth et al. [27] showed a correlation between expression of CD44+/CD24-/low and the BLBC or TNBC subtype. They found that
ALDH activity was linked to poor prognosis as well as younger age of breast cancer patients, both of which are associated with the TNBC subtype [26, 28, 29]. Similar results were shown when breast cancer cell lines representing distinct molecular breast cancer subtypes were analyzed, showing ALDH activity in all basal and HER2-positive cell lines, whereas luminal cell lines showed only about 60% ALDH activity [30, 31]. These ALDH-positive cells also showed characteristics of cancer stem cells as they could form mammospheres in vitro and tumors in vitro [31]. These studies, in which all instruments of molecular profiling of breast cancer tissue specimens and established breast cancer cell lines were used, suggest that a link exists between the TNBC phenotype and the concept of cancer stem cells. How this link accounts for the aggressiveness of TNBC has yet to be analyzed.
Unfortunately, the expression of surface markers or analysis of certain enzyme activities is not transferable to all solid tumors, making it difficult to apply these characterization strategies to other solid tumors. CD44, for example, has been described as a marker for tumor-initiating cells in head and neck squamous cell carcinomas (HNSCC) [32], whereas its expression in breast cancer should be either very low or absent. Another prominent marker for cancer stem cells is expression of CD133, which also occurs in colorectal, pancreatic, brain and lung cancers [33, 34]. In breast cancer, however, CD133 expression has been linked to cancer-initiating cells, but its expression is correlated to HER2-positive breast cancer [35]. Furthermore, whereas CD44+/CD24-/low expression has been reported for both basal-like or triple-negative subtypes of breast cancer, ALDH1 expression was observed for both basal-like and HER2-positive breast [36]. This shows that the concept of cancer stem cell identification using surface markers cannot be generalized to all different types of cancer, not even to different subtypes of breast cancer.
Recently, efforts have been made to try to clarify the nature of cancer stem cells, by analyzing how these cells regulate themselves as well as other cancer cells. Interestingly, many groups have shown similarities between cancer stem cell regulation and metastasis on one hand, and an embryonic process necessary for formation of a proper body plan (epithelial to mesenchymal transition, EMT) on the other. Interestingly, it was shown that by overexpression of EMT-related genes (e.g. Slug, Snail, Vimentin, etc.) the CD44+/CD24-/low phenotype was conferred to formerly CD44+/CD24-/low-negative cells [37, 38, 39, 40, 41, 42]. This suggests that at least the CD44+/CD24-/low phenotype might be associated with the TNBC subtype. This hypothesis would also explain the intrinsic poor prognosis of TNBC compared to other breast cancer subtypes, which might be easier to treat, while breast cancer stem cells still remain responsible for relapses. There is also growing evidence that expression of markers of cancer stem cells is strongly associated with a prognostic and predictive impact [43].
There are additional hints that a paradigm shift might occur in the nature of cellular plasticity of cancer cells. It has been assumed that cancer stem cells either self-renew or differentiate into daughter cells. This has been called the unidirectional hierarchical model of mammary cells. A recent report from the Weinberg group [44], however, showed that cancer cells can spontaneously convert to a cancer stem celllike state. Similarly, Gupta and colleagues [45] claimed that cancer stem-like cells can arise de novo from non-stem-like cells at a low but significant rate.
These findings have a strong impact on clinical research analyzing approaches to specifically target breast cancer stem cells. It has already been shown that certain drugs might specifically target cancer stem cells [24]. If non-stem-like cells can de-differentiate into cancer stem cells, this cancer stem cell population might be regenerated from non-stem-like cells after eradication of existing cancer stem cells [44]. Thus, a combined therapeutic strategy targeting both cancer cells and cancer stem cells at the same time might be a future clinical therapy.
It seems that key players of the tightly regulated process of EMT are highly enriched in the subgroup of BLBC or TNBC [46, 47, 48, 49, 50]. This phenomenon might explain the aggressive nature of – at least some – cases of TNBC in which relapses occur at higher frequencies after chemotherapy compared to other breast cancer subtypes.
The final goal incorporating these findings now is to combine basic and translational research in a way that future therapies benefit from both (1) targeting the cancer cells, which are highly mitotically active, and (2) eradiating cancer stem cells, which can still remain inside a tumor after therapy, and which might be involved in the process of both local relapse and metastasis.
Conclusions
Clinically, TNBC is characterized by an unfavorable prognosis. There is a significant, though limited, correlation between TNBC and BLBC. The clinical significance of this remains to be defined. Similarly, the clinical value of cases with borderline hormone receptor expression (i.e. 1–9% of tumor cells) is not yet clear. There is an increasing body of evidence that TNBC is in itself a both molecularly and clinically heterogeneous entity, which may be one explanation why patients diagnosed with this entity do not yet benefit from the advances made in oncology that are helping patients with other breast cancer subtypes. Recently, molecular subtypes of TNBC have been identified and shown to respond differently to systemic agents, including chemotherapy and targeted agents. Gene expression analysis suggests that certain signatures or gene expression profiles might carry prognostic or predictive information; however, they are yet far from being used in clinical routine.
There might also be an association between the TNBC subtype and the presence of cancer stem cells. Recent studies describe a correlation between TNBC, poor prognosis, expression of EMT-related genes and (cancer) stem cells. However, as TNBC is a heterogeneous entity, the subtype that contains the majority of cancer stem cells has yet to be elucidated. Additionally, future targeted therapies should aim at targeting both cancer stem cells (as the ultimate source for relapses) as well as highly mitotic cancer cells in an efficient way to inhibit future reprogramming events that reseed the population of cancer stem cells.
Disclosure Statement
The authors of this manuscript have no conflicts of interest relevant to the content of this manuscript to disclose.
References
- 1.Gluz O, Liedtke C, Gottschalk N, et al. Triple-negative breast cancer – current status and future directions. Ann Oncol. 2009;20:1913–1927. doi: 10.1093/annonc/mdp492. [DOI] [PubMed] [Google Scholar]
- 2.Carey L, Winer E, Viale G, et al. Triple-negative breast cancer: Disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 3.Heitz F, Harter P, Lueck HJ, et al. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer. 2009;45:2792–2798. doi: 10.1016/j.ejca.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Liedtke C, Hess KR, Karn T, et al. The prognostic impact of age in patients with triple-negative breast cancer. Breast Cancer Res Treat. 2013;138:591–599. doi: 10.1007/s10549-013-2461-x. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carey L.A, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 8.Nofech-Mozes S, Trudeau M, Kahn HK, et al. Patterns of recurrence in the basal and non-basal subtypes of triple-negative breast cancers. Breast Cancer Res Treat. 2009;118:131–137. doi: 10.1007/s10549-008-0295-8. [DOI] [PubMed] [Google Scholar]
- 9.Prat A, Adamo B, Cheang MC, et al. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheang MC, et al. Quantitative hormone receptors, triple-negative breast cancer (TNBC), and molecular subtypes: A collaborative effort of the BIG-NCI NABCG. J Clin Oncol. 2012;30(Suppl) abstr 1008. [Google Scholar]
- 11.Iwamoto T, Booser D, Valero V, et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J Clin Oncol. 2012;30:729–734. doi: 10.1200/JCO.2011.36.2574. [DOI] [PubMed] [Google Scholar]
- 12.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 13.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 14.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda H, Baggerly KA, Wang Y, et al. Differential pathologic complete response rates after neo-adjuvant chemotherapy among molecular subtypes of triple-negative breast cancer. J Clin Oncol. 2013;31(Suppl) abstr 1005. [Google Scholar]
- 18.Telli ML, et al. PrECOG 0105: Final efficacy results from a phase II study of gemcitabine (G) and carboplatin (C) plus iniparib (BSI-201) as neoadjuvant therapy for triple-negative (TN) and BRCA1/2 mutation-associated breast cancer. J Clin Oncol. 2013;31 doi: 10.1200/JCO.2014.57.0085. abstr 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rody A, Karn T, Liedtke C, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denkert C, Loibl S, Müller BM, et al. Ki67 levels in pretherapeutic core biopsies as predictive and prognostic parameters in the neoadjuvant GeparTrio trial. Cancer Res. 2012;72(24 Suppl):102s. doi: 10.1093/annonc/mdt350. [DOI] [PubMed] [Google Scholar]
- 21.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korkaya H, Paulson A, Iovino F, et al. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witkiewicz AK, Dasgupta A, Sotgia F, et al. An absence of stromal caveolin-1 expression predicts early tumor recurrence and poor clinical outcome in human breast cancers. Am J Pathol. 2009;174:2023–2034. doi: 10.2353/ajpath.2009.080873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honeth G, Bendahl PO, Ringnér M, et al. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2008;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 29.Tan EY, Thike AA, Breast Surgical Team at Outram et al. ALDH1 expression is enriched in breast cancers arising in young women but does not predict outcome. Br J Cancer. 2013;109:109–113. doi: 10.1038/bjc.2013.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricardo S, Vieira AF, Gerhard R, et al. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 31.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 34.Hermann PC, Huber SL, Herrler T, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Wang XY, Penalva LO, Yuan H, et al. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakshatri H, Srour EF, Badve S. Breast cancer stem cells and intrinsic subtypes: controversies rage on. Curr Stem Cell Res Ther. 2009;4:50–60. doi: 10.2174/157488809787169110. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 40.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maestro R, Dei Tos AP, Hamamori Y, et al. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morel AP, Lièvre M, Thomas C, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aomatsu N, Yashiro M, Kashiwagi S, et al. CD133 is a useful surrogate marker for predicting chemosensitivity to neoadjuvant chemotherapy in breast cancer. PLoS One. 2012;7:e45865. doi: 10.1371/journal.pone.0045865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta PB, Fillmore CM, Jiang G, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 46.Karihtala P, Auvinen P, Kauppila S, et al. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: Association with an aggressive tumour phenotype. Breast Cancer Res Treat. 2013;138:81–90. doi: 10.1007/s10549-013-2442-0. [DOI] [PubMed] [Google Scholar]
- 47.Liu T, Zhang X, Shang M, et al. Dysregulated expression of Slug, vimentin, and E-cadherin correlates with poor clinical outcome in patients with basal-like breast cancer. J Surg Oncol. 2013;107:188–194. doi: 10.1002/jso.23240. [DOI] [PubMed] [Google Scholar]
- 48.Proia TA, Keller PJ, Gupta PB, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarrió D, Rodriguez-Pinilla SM, Hardisson D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita N, Tokunaga E, Kitao H, et al. Vimentin as a poor prognostic factor for triple-negative breast cancer. J Cancer Res Clin Oncol. 2013;139:739–746. doi: 10.1007/s00432-013-1376-6. [DOI] [PubMed] [Google Scholar]