Abstract
Background
The high molecular weight isoform of the actin-binding protein Caldesmon (h-CaD) regulates smooth muscle contractile function by modulating cross-bridge cycling of myosin heads. The normal inhibitory activity of h-CaD is regulated by the enteric nervous system; however, the role of h-CaD during intestinal peristalsis has never been studied.
Methods
We identified a zebrafish paralog of the human CALD1 gene that encodes an h-CaD isoform expressed in intestinal smooth muscle. We examined the role of h-CaD during intestinal peristalsis in zebrafish larvae by knocking down the h-CaD protein using an antisense morpholino oligonucleotide. We also developed transgenic zebrafish that express inhibitory peptides derived from the h-CaD myosin and actin-binding domains, and examined their effect on peristalsis in wild-type zebrafish larvae and sox10colourless mutant larvae that lack enteric nerves.
Key Results
Genomic analyses identified two zebrafish Caldesmon paralogs. The cald1a ortholog encoded a high molecular weight isoform generated by alternative splicing whose intestinal expression was restricted to smooth muscle. Propulsive intestinal peristalsis was increased in wild-type zebrafish larvae by h-CaD knockdown and by expression of transgenes encoding inhibitory myosin and actin-binding domain peptides. Peristalsis in the non-innervated intestine of sox10colourless larvae was partially restored by h-CaD knockdown and expression of the myosin-binding peptide.
Conclusions & Inferences
Disruption of the normal inhibitory function of h-CaD enhances intestinal peristalsis in both wild-type zebrafish larvae and mutant larvae that lack enteric nerves, thus confirming a physiologic role for regulation of smooth muscle contraction at the actin filament.
Keywords: enteric nervous system, gastrointestinal motility, hypomotility, myosin light chain, zebrafish
INTRODUCTION
Peristalsis in the mammalian intestine is regulated by the enteric nervous system (ENS). ENS stimulation initiates smooth muscle contraction through a calcium-mediated signaling cascade that culminates in phosphorylation of the regulatory Myosin light chain (Mlc) which is physically associated with the myosin filament.1,2 Phosphorylation of Mlc by Myosin light chain kinase (Mlck) induces a conformational change in myosin that activates its ATPase. This induces cross-bridge cycling within the actomyosin complex, and ultimately, the generation of contractile force. Biochemical analyses have shown that endogenous actomyosin interactions can generate and sustain smooth muscle contractile force when levels of phospho-Mlc are greatly reduced.3–6 To account for this, it has been proposed that contraction is also regulated by actin-binding proteins that alter actomyosin interactions independent of phospho-Mlc.7
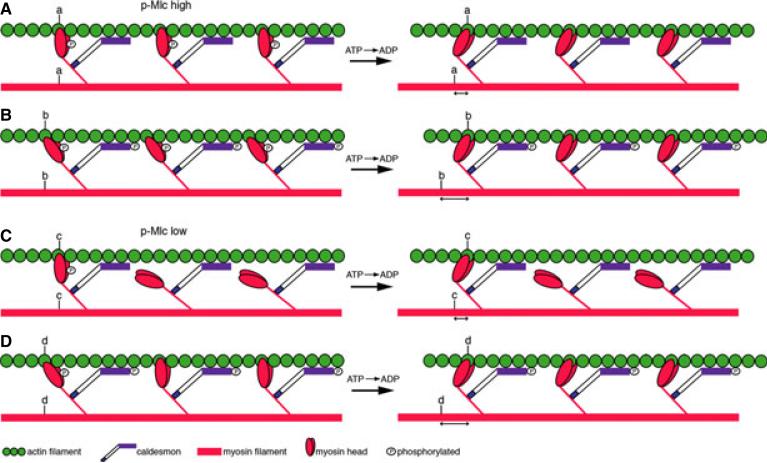
One smooth muscle actin-binding protein that has been extensively studied is Caldesmon (CaD).8 CaD exists as two predominant isoforms that are generated by alternative splicing of a single mRNA transcript.9 The low molecular weight isoform (l-CaD) is expressed in most cell types, including at low levels in smooth muscle, where it mediates actin and non-muscle myosin interaction in the cortical cytoskeleton.10 The high molecular weight isoform (h-CaD) is expressed specifically in smooth muscle. It is distinguished from l-CaD by a peptide spacer domain in the middle of the protein that is thought to enable h-CaD's simultaneous binding to smooth muscle actin and myosin filaments.11 Binding of h-CaD to the phosphorylated actomyosin complex reduces contractile force by inhibiting myosin ATPase activity,12–14 possibly by restricting myosin binding to actin, or by stabilizing a less active configuration of the actin filament15 (Fig. 1A based on prior model16). h-CaD's inhibition of the smooth muscle actomyosin complex can be reversed by phosphorylation or through its interaction with Calmodulin in the presence of calcium.17–19 This results in increased contractile force by enhancing myosin binding to actin20 (Fig. 1B), which in turn may stabilize an active configuration of the actin filament.15 In smooth muscle with low levels of phospho-Mlc, phospho-h-CaD is thought to enhance contraction by promoting interaction of non-phosphorylated myosin heads with actin21 (Fig. 1C,D).
Figure 1.
Simplified model of h-CaD function. Smooth muscle contraction depicted by sliding of actin (green) on myosin (red) filaments, following ATP hydrolysis (derived from previously described model16). (A) In smooth muscle with high levels of phospho-Mlc (p-Mlc), non-phosphorylated h-CaD restricts binding of myosin heads to the actin filament such that force generation is inhibited. (B) When h-CaD is phosphorylated, the myosin heads are able to bind the actin filament in a manner that enhances actomyosin interaction (b). This leads to increased force generation (distance b > distance a). (C) In smooth muscle with low levels of p-Mlc, non-phosphorylated h-CaD prevents binding of non-phosphorylated myosin heads to actin. (D) phospho-h-CaD promotes binding of the non-phosphorylated myosin heads to actin, thereby enhancing contraction; (distance d > distance c). Distances a, b and c, d are not drawn to scale.
Both the inhibitory role of h-CaD and the molecular signaling pathways that regulate its activity have been characterized in smooth muscle cells from the digestive tract and other mammalian tissues.13,22–26 Despite these advances in understanding h-CaD function and regulation, its role in modulating complex physiologic responses, such as intestinal peristalsis, has not been directly examined in vivo. Isoform specific knockout of h-CaD in mice has been reported; however, its effect on smooth muscle function was not examined.27 Here, we address the role of h-CaD in intestinal smooth muscle in vivo using the zebrafish system. We show that the zebrafish genome contains two Caldesmon gene paralogs, cald1a and cald1b, that are orthologs of the human CALD1 gene. The cald1a paralog encodes an h-CaD isoform expressed in intestinal smooth muscle. Disruption of h-CaD function enhances propulsive peristalsis in wild-type zebrafish larvae and in sox10colourless mutants that lack enteric nerves thus confirming an in vivo regulatory role for h-CaD in smooth muscle with normal and low levels of phospho-Mlc.
MATERIALS AND METHODS
Fish stocks
Maintenance and breeding were performed as previously described.28 Colourless mutants were purchased from the Zebra-fish International Resource Center (Eugene, OR, USA).
Morpholino knockdown
Fertilized embryos were injected at one-cell stage with 20 pg of the Cald1a-i4e5 morpholino. (Data S1).
RT-PCR
RNA was isolated from dissected 5 days post fertilization (dpf) larval intestines, and RT-PCR was carried out using standard procedures (Data S1). Primer sequences are listed in Table S1.
Generation of CALD1 myosin- and actin-binding peptide transgenic fish
Primers corresponding to the human CALD1 myosin-binding peptide29 were identified in the zebrafish cald1cDNA. Transgenic constructs were generated with the peptide fragment by Tol2kit site-specific recombination-based cloning (Data S1).
Intestinal microsphere assay
Larvae were fed paramecia in media containing fluorescent latex beads, and bead motility was monitored (Data S1).
Cell dissociation and FACS
Dissociation and cell sorting were performed using standard procedures (Data S1).
Western blot/ immunoprecipitation
Dissected intestines from 30 larvae at 4 dpf and 6 dpf were homogenized in 1× sample buffer, and protein was recovered and used for Western analysis using standard methods30 (Data S1).
RESULTS
The zebrafish genome contains two gene paralogs with high sequence homology to human CALD1
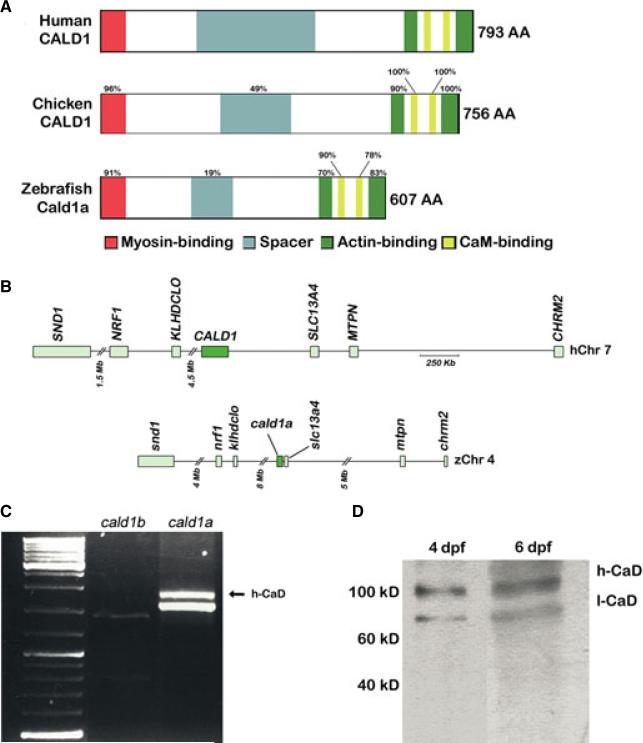
To identify a zebrafish h-CaD ortholog, we first performed BLAST analyses of the most recent zebrafish genome assembly using human CALD1 coding sequence. Two loci encoding genes with relatively high sequence homology to CALD1 were identified on chromosome 4 and chromosome 25 (hereafter cald1a and cald1b, respectively). The predicted protein encoded by the cald1a locus had the greatest degree of homology to human CALD1, particularly in the actin and myosin-binding domains (Fig. 2A, and Figure S1).
Figure 2.
Zebrafish smooth muscle Caldesmon (h-CaD). (A) Schematic representation of the human, chicken and zebrafish h-CaD orthologs. Conserved protein domains and percent amino acid homology are indicated. (B) Conserved syntenic relationships surrounding the human CALD1 locus on chromosome 7 and the zebrafish cald1a locus on chromosome 4. (C) RT-PCR showing amplification of the full length cDNA corresponding to the high and low molecular weight zebrafish cald1a isoforms from intestinal cDNA. A correctly sized transcript for the predicted low molecular weight cald1b isoform is also detected. (D) Western blot showing intestinal levels of CaD isoforms. Molecular weight standards indicated. Mb, megabase; Kb, kilobase; dpf, days post fertilization.
We also compared syntenic relationships surrounding the cald1a and cald1b loci with human CALD1. We first identified the map positions of the zebrafish orthologs of 65 genes immediately surrounding human CALD1 (Fig. 2B, and Figure S2). Of 34 genes located 5′ of CALD1, 26 were located in a comparable position with respect to the cald1a locus, whereas only six mapped to a comparable position with respect to cald1b. For 31 genes 3′ of CALD1, 14 were located surrounding cald1a, whereas none were located adjacent to cald1b. Next, we performed the reciprocal experiment and mapped the position of the human orthologs of genes surrounding the two zebrafish cald1 loci. Twelve of 40 genes 5′ of cald1a, but only 7 of 38 genes 5′ of cald1b mapped to comparable regions of CALD1. Similarly, nine of 43 genes 3′ of cald1a, but only 3 of 38 genes 3′ of cald1b mapped nearby CALD1. Paralogs for 17 zebrafish genes were identified in the region surrounding the two cald1 loci (Figure S2).
All together, these findings argue that cald1a and cald1b are gene paralogs that arose from a whole genome duplication which occurred in an ancestral species of ray-finned fish.31 We predicted that the Cald1a protein was likely to be a functional ortholog of human CALD1 protein based on amino acid sequence homology with CALD1, and because of more highly conserved gene synteny surrounding the cald1a locus.
cald1a encodes high and low molecular weight isoforms generated by alternative splicing
The cald1b locus encodes an l-CaD-like ortholog that was recently reported to play a role in cardiovascular development.32 Genomic sequence analysis of this locus did not identify a potential h-CaD transcript nor did BLAST searches of the zebrafish EST database. To determine whether an alternatively spliced h-CaD mRNA transcript was encoded by cald1a, we amplified its full length cDNA from whole 5 dpf larvae that have functional intestinal smooth muscle. These primers amplified only a fragment corresponding to the predicted low molecular weight cald1a isoform (data not shown). When the same primers were used to amplify RNA recovered from dissected intestines (that were enriched for smooth muscle) the low molecular weight band and a higher molecular weight band were recovered (Fig. 2C). In contrast, amplification of intestinal cDNA using comparable primers from the cald1b ortholog only amplified low levels of a corresponding low molecular weight band (Fig. 2C). DNA sequence analysis showed that the larger band amplified from cald1a encoded a predicted h-CaD ortholog that included an 86 amino acid spacer domain. Genomic DNA analyses indicated that the spacer was encoded by two exons, 36 basepairs (bp) and 225 bp respectively. We observed only limited amino acid homology with spacer domains from other vertebrate h-CaD proteins (Fig. 2A, and Figure S1). This was expected, as the spacer sequence is not highly conserved in human, mouse or chicken h-CaD. Outside of the spacer domain, the h-CaD cDNA sequence was identical to the full length cald1a cDNA amplified from RNA derived from whole larvae, although a splice variant involving the terminal exon was infrequently detected (Figure S3). These findings show that the cald1a locus encodes two alternatively spliced transcripts that are orthologs of the mammalian l-CaD and h-CaD isoforms.
To determine whether the h-CaD transcript encoded a functional protein, we performed Western blot analysis of 5 dpf intestinal extracts using an antibody directed against mouse CaD protein. This antibody detected both high and low molecular weight bands (Fig. 2D). The high molecular weight band was first detected around the onset of peristaltic contractions (~78 h post fertilization; data not shown) when only circular smooth muscle is present,28 and protein levels increased between 4 dpf and 6 dpf. At these later stages, longitudinal smooth muscle develops, and peristaltic contractions are more pronounced.28 The estimated molecular mass of the high molecular weight band (125 kD) was significantly greater than the predicted h-CaD molecular mass (70 kD). A comparable discrepancy between predicted and observed molecular mass of mammalian h-CaD proteins has been reported and attributed to its relatively large number of acidic amino acids.33 The molecular weight of the presumptive l-CaD protein was similarly affected.
h-CaD expression in the zebrafish intestine is restricted to smooth muscle
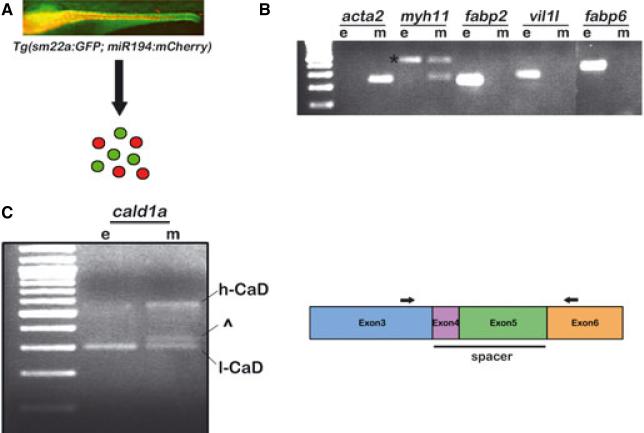
High molecular weight Caldesmon isoform expression in mammals and chicken is restricted to smooth muscle, whereas l-CaD is ubiquitously expressed. RNA in situ hybridization using an isoform specific zebrafish h-CaD probe showed only low level background staining in 5 dpf larvae (data not shown). To assay h-CaD expression in intestinal smooth muscle, we recovered epithelial cell and smooth muscle cell RNAs from bigenic larvae that express cell-type specific fluorescent reporters (Fig. 3A). The bigenic larvae express GFP from the smooth muscle sm22a promoter beginning around 72 h post fertilization34 and mCherry from a miR194 promoter fragment that is activated in the epithelium beginning around the same time. RT-PCR amplification of established epithelial (fabp2, vil11, and fabp6) and smooth muscle (acta2, myh11) markers confirmed purity of the sorted cell populations (Fig. 3B). The RT-PCR experiments also confirmed that zebrafish h-CaD expression was restricted to smooth muscle, whereas l-CaD was expressed in both smooth muscle and epithelial cells (Fig. 3C).
Figure 3.
Expression of the zebrafish h-CaD ortholog in intestinal smooth muscle. (A) Scheme to isolate intestinal smooth muscle (green) and epithelial (red) cells from Tg(sm22a:GFP; miR194:mCherry) larvae. Fluorescent image of the intestine of a 5 dpf bigenic larva is shown. Smooth muscle of the anterior intestine is not in plane of focus. (B) RT-PCR amplification showing expression of intestinal smooth muscle and epithelial markers in sorted cells. The myh11 primers amplify a band (*) from contaminating epithelial and smooth muscle genomic DNA (confirmed by sequencing). (C) Expression of the high molecular weight cald1a transcript is restricted to smooth muscle. In some experiments, an additional band was amplified from smooth muscle that migrated near the low molecular weight cald1 transcript (^); however, this was detected in only a minority of experiments. Schematic indicates the location of PCR primers (arrows) in exon 3 and exon 6 that were used to amplify the cald1a isoforms. e, epithelial; m, smooth muscle.
Knockdown of zebrafish h-CaD enhances propulsive intestinal peristalsis
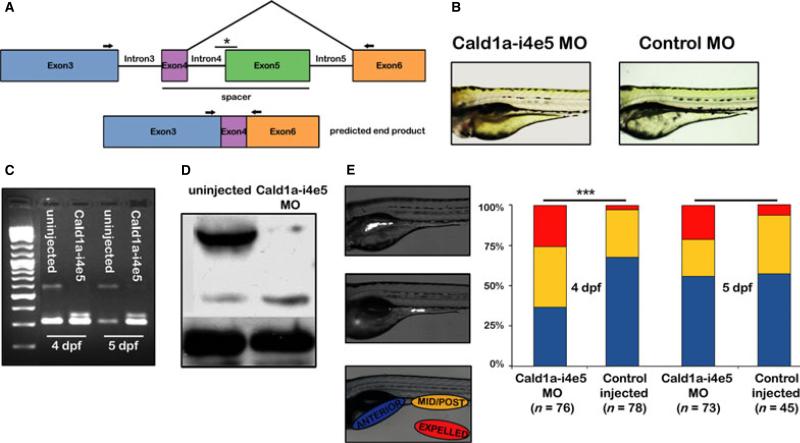
To investigate zebrafish h-CaD function in vivo, we generated a cald1a splice blocking morpholino (Cald1a-i4e5 MO) that enabled us to specifically disrupt h-CaD translation without affecting translation of the l-CaD isoform. The specificity of Cald1a-i4e5 MO also prevented any disruption of the normal function of cald1b. It was crucial not to disrupt l-CaD function as this isoform is predicted to be essential for a wide range of non-muscle phenotypes. For this study, we designed a morpholino complementary to the exon 5 splice acceptor of the h-CaD pre-mRNA (Fig. 4A). This was predicted to generate a novel Caldesmon transcript that encoded only 12 of 87 h-CaD spacer region amino acids.
Figure 4.
h-CaD deficiency enhances intestinal peristalsis in zebrafish larvae. (A) Schematic depicting isoform specific targeting of the high molecular weight cald1a transcript by a splice blocking morpholino (Cald1a-i4e5 MO). The exon 5 splice acceptor is targeted (*). (B) Normal morphology of 4 dpf larvae injected with Cald1a-i4e5 MO and control morpholinos. (C) RT-PCR of intestinal cDNA from Cald1a-i4e5 MO larvae using exon 3 and exon 6 primers (arrows in panel 3A) shows markedly reduced expression of the high molecular weight cald1a transcript. (D) Western blot using intestinal protein from Cald1a-i4e5 MO larvae shows reduced levels of h-CaD with slightly increased l-CaD levels. Increased protein corresponding to l-CaD likely reflects additional protein translated from modified transcript only 12 amino acids larger than l-CaD. (E) Images of live 5 dpf larvae that ingested fluorescent microspheres located in anterior and mid-posterior intestine, respectively. Color scheme in bar graph depicts bead location indicated in lateral image of larva (lower panel). Cald1a-i4e5 MO larvae show increased propulsive peristalsis at 4dpf (chi-squared test, ***P < 001). 5dpf P-value = 0.08, likely due to transient effect of morpholino knockdown.
Zebrafish embryos and larvae injected with Cald1a-i4e5 MO had normal morphology other than mild developmental delay that was also seen in control larvae (Fig. 4B). In RT-PCR experiments using intestinal RNA and primers surrounding exons 4 and 5, a fragment of the truncated h-CaD transcript that lacked exon 5, but retained exon 4, and the fragment corresponding to l-CaD were both amplified (Fig. 4C). The truncated h-CaD transcript, which was 36 bp larger than endogenous l-CaD transcript, encoded 12 amino acids in-frame with exon 6. Successful inhibition of translation of the h-CaD transcript was confirmed using Western blot (Fig. 4D). The apparent increase in l-CaD protein detected by this blot is likely due to the presence of the truncated h-CaD protein generated by the Cald1a-i5e5 MO (as it is only 12 amino acids longer than endogenous l-CaD).
To determine whether h-CaD knockdown affected smooth muscle contraction, we assayed propulsive peristalsis in the intestine of control and Cald1a-i4e5 MO injected larvae that were fed paramecia combined with fluorescent microspheres (Fig. 4E). Following their ingestion, the microspheres form an aggregate in the intestinal bulb as a result of mixing peristalsis. Thereafter, the aggregate is transported into the mid and posterior intestine via propulsive peristalsis, and eventually expelled. We measured peristalsis in 4 dpf and 5 dpf control and Cald1a-i4e5 MO injected larvae 2 h after bead ingestion. As shown in Fig. 4E, a higher percentage of h-CaD deficient larvae had transported beads to the mid-posterior intestine and/or expelled the beads. Video recordings of live wild type and Cald1a-i4e5 MO injected larvae showed that the rate of peristaltic contractions was unaffected by h-CaD knockdown (Figure S4). The effect of Cald1a-i4e5 MO on peristalsis was less pronounced at 5 dpf than at 4 dpf, most likely because of the transient nature of the morpholino knockdown. In addition, because levels of phospho-h-CaD have increased at this stage,30 the overall level of inhibition imparted by h-CaD is likely to be reduced. Together these data indicate that the normal function of h-CaD at this developmental stage is to negatively regulate the force of phasic intestinal smooth muscle contraction, as intestinal transit is accelerated in the larvae injected with Cald1a-i4e5 MO without a concomitant effect on contractile rate.
Interference with h-CaD binding to smooth muscle myosin and actin enhances intestinal propulsive peristalsis
h-CaD tethers myosin and actin filaments through interactions with its N- and C-terminal domains, respectively.35,36 The h-CaD – smooth muscle myosin interaction can be disrupted in vitro by expression of cDNA encoding a peptide fragment from the myosin-binding domain common to l-CaD and h-CaD isoforms.29 By disrupting endogenous h-CaD binding to myosin, actin–myosin interactions are enhanced, thereby increasing basal contractile tone. A comparable effect has been reported with a peptide derived from the CaD actin-binding domain.20,37
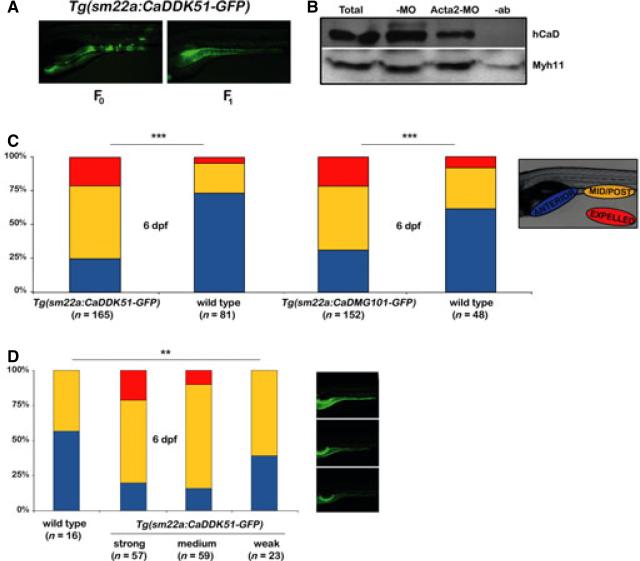
We generated a transgenic line, Tg(sm22a:CADDK51-GFP), with stable smooth muscle expression of a zebrafish ortholog of mammalian myosin-binding peptide (CADDK51) to assay h-CaD function in older zebrafish larvae that have a well-developed enteric neuromuscular system. This was not possible in larvae injected with the Cald1a-i4e5 MO, because its inhibitory effect on translation is transient. In addition to the cDNA encoding the peptide, the expression vector used to generate the transgenic line encoded GFP downstream of a viral 2A recognition motif.38 This allowed us to identify fluorescent smooth muscle cells expressing the CADDK51 peptide following its cleavage from GFP. F0 larvae that had mosaic expression of CADDK51 in smooth muscle under control of the zebrafish sm22a promoter and F1 germline transgenic progeny with ubiquitous intestinal smooth muscle GFP expression were both viable (Fig. 5A). Variable transgene expression was detected in F1 larvae, most likely resulting from position effects arising from independent insertion sites of the trans-gene into the F0 germline. Collectively, F1 transgenic larvae were used to further assay the in vivo function of h-CaD in intestinal smooth muscle.
Figure 5.
Expression of peptides blocking h-CaD interaction at myosin and actin filaments increases intestinal propulsive peristalsis. (A) Lateral images of live 5 dpf F0 and F1 Tg(sm22a: CaDDK51-GFP) larvae showing mosaic and widespread smooth muscle GFP expression, respectively. (B) IP-Western blot confirms that CaDDK51 myosin-binding domain peptide blocks interaction of h-CaD with Myh11. Total intestinal protein was IP'ed with Myh11 antibody and blotted as indicated. (C) Bar graph shows increased propulsive peristalsis in unsorted Tg(sm22a: CaDDK51 -GFP) larvae and Tg (sm22a: CaDMG101 -GFP) larvae, in which peptides block h-CaD interaction with myosin and actin, respectively (chi-squared test, ***P < 001). (D) Enhanced peristalsis in Tg (sm22a: CaDDK51 -GFP) larvae correlates with the level of transgene expression (determined by GFP fluorescence). Images show larvae with strong, medium and weak GFP expression (Fisher's exact test, **P < 05). -MO – control larvae; Acta2-MO – larvae injected with smooth muscle actin morpholino; -ab – control IP without antibody; Myh11 – smooth muscle myosin.
The effect of CADDK51 on h-CaD–myosin filament interaction was examined by measuring levels of h-CaD bound to smooth muscle myosin protein (Myh11) in the transgenic larvae (Fig. 5B). As h-CaD remains bound to Acta2 in the actomyosin complex, transgenic larvae injected with a morpholino targeting smooth muscle actin protein (Acta2)30 were used for this immunoprecipitation experiment. This allowed us to determine the effect of CADDK51 on h-CaD–myosin interaction independent of h-CaD–Acta2 interaction. Indeed, Western blot analysis showed that there was significantly less h-CaD bound to Myh11 in Acta2 deficient transgenic larvae compared with control transgenic larvae. These findings support binding of the peptide fragment to Myh11 with subsequent displacement of endogenous h-CaD.
To determine the effect of CADDK51 peptide on smooth muscle contraction in vivo, we assayed intestinal peristalsis in the transgenic larvae. Using the intestinal microsphere assay, we found a significant increase in the propulsive peristalsis of 6 dpf F1 transgenic larvae compared with non-transgenic siblings, as measured by transport of ingested beads for 2 h to the mid-posterior intestine and their complete expulsion (Fig. 5C). When larvae were sorted prior to bead feeding based upon the intensity of GFP expression, we were able to show that increased propulsion was dose dependant in that GFP expression levels, which are a surrogate for the level of CADDK51, correlated with changes in bead motility (Fig. 5D). Increased intestinal propulsion in transgenic larvae was not related to altered contractile rate (Figure S4).
In addition to the Tg(sm22a:CADDK51-GFP) line, we generated a second line, Tg(sm22a:CaDMG101-GFP) with smooth muscle expression of an inhibitory peptide derived from the h-CaD actin-binding domain.37 The CaDMG101-GFP peptide also had a pronounced enhancing effect on intestinal motility similar to that of the CADDK51 peptide (Fig. 5C).
h-CaD modulates endogenous smooth muscle contraction independent of phospho-Mlc
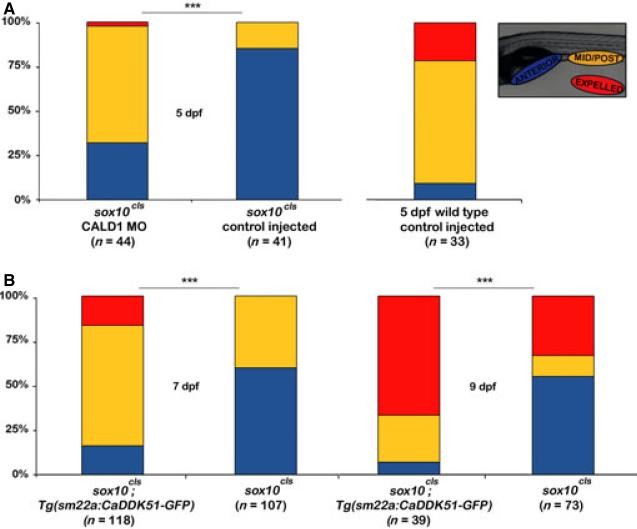
To address the role of thin filament regulation of intestinal peristalsis independent of phospho-Mlc, we took advantage of a zebrafish mutant, sox10colourless (cls) that lacks enteric nerves.39,40 In previous work, we showed that cls mutants have nearly undetectable levels of intestinal phospho-Mlc and phospho-h-CaD, but normal total h-CaD levels.30 Surprisingly, even though cls larvae had little if any intestinal phospho-Mlc, they had a significant amount of residual propulsive peristaltic activity.30 This suggested that peristalsis was driven by smooth muscle contraction arising from intrinsic actomyosin interactions. Low phospho-h-CaD levels in cls suggested that baseline peristalsis was inhibited and might be enhanced by h-CaD knockdown or by disrupting h-CaD interaction with either myosin or actin filaments. To determine if the lack of phospho-h-CaD inhibited peristalsis in the absence of neuronal input, we repeated the h-CaD morpholino experiments in cls mutant larvae (Fig. 6A). In cls larvae injected with Cald1a-i4e5 MO, intestinal motility was assayed at 5 dpf. As a result of reduced uptake of the beads and slower intestinal transit by cls mutants, larvae in this assay were exposed to the beads for approximately 6 h and assayed 20 h later. This experiment showed enhanced propulsive peristalsis in h-CaD deficient cls larvae (Fig. 6A). While fewer of the Cald1a-i4e5 MO cls larvae expelled the beads (2%, compared to 21% of wild type larvae), many more had transported the beads to the mid-posterior intestine compared with control cls larvae (68% vs. 15%, respectively).
Figure 6.
Intestinal peristalsis is increased in h-CaD deficient colourless larvae. (A) Bar graph shows partial rescue of intestinal peristalsis in h-CaD deficient 5 dpf cls larvae. (B) Tg (sm22a: CaDDK51 -GFP) cls larvae show partial rescue of propulsive peristalsis at 7 dpf and 9 dpf (chi-squared test, ***P < 001).
Next, we assayed peristalsis in 7 dpf and 9 dpf cls larvae that express the myosin-binding peptide in intestinal smooth muscle. As these larvae were older than the Cald1a-i4e5 MO injected larvae, a 2-h feeding was sufficient, and peristalsis was assayed 20 h after feeding. This experiment showed that both transport of the fluorescent beads to the mid-posterior intestine and their expulsion was significantly greater in the transgenic cls larvae (Fig. 6B). The increased propulsion in transgenic cls larvae was not related to altered contractile rate (Fig. S4). The peristaltic defect in cls was not fully rescued by h-CaD disruption, as wild-type larvae at both stages completely cleared all ingested beads (data not shown). This was expected, given that the enteric nervous system is the primary stimulus for smooth muscle contraction. However, these findings do show that h-CaD-mediated inhibition of intrinsic actomyosin interactions contributes to reduced intestinal motility in the non-innervated intestine of zebrafish larvae.
DISCUSSION
Although regulation of smooth muscle contraction principally occurs at the myosin (thick) filament through phosphorylation of regulatory Mlc, a potentially important role for regulation of contraction at the actin (thin) filament has long been recognized. However, the physiologic importance of this mode of regulation has never been directly assayed in vivo. Here, we show that the smooth muscle actin-binding protein h-CaD regulates intestinal peristalsis in the wild type and non-innervated intestine of zebrafish larvae.
The h-CaD isoform that we identified arises from alternative splicing of a pre-mRNA transcribed from cald1a, one of two cald1 paralogs we show are present in the zebrafish genome. The cald1a locus also encodes an l-CaD isoform expressed in intestinal smooth muscle. Predicted translation of the h-CaD protein showed high sequence homology of the actin and myosin-binding domains compared with other vertebrate h-CaD proteins. In contrast, there is less sequence homology between the various spacer domains, possibly because of its proposed structural role as a charged single α-helix.41,42 The cald1b locus encodes an l-CaD isoform expressed at low levels in intestinal smooth muscle that was previously reported to play a role in vascular development.32 Expression of a cald1b h-CaD ortholog was not detected. Together with functional analyses described below, these findings show that cald1a encodes the zebrafish ortholog of the h-CaD isoform expressed in mammalian intestinal smooth muscle. These findings also argue that cald1a and cald1b have non-overlapping functions in smooth muscle, similar to other zebrafish gene paralogs that arose from genome duplication events.
We examined the role of h-CaD in intestinal peristalsis and smooth muscle contraction using a previously described simple in vivo assay.30 Propulsive peristalsis was increased in h-CaD deficient wild-type larvae generated via isoform specific antisense targeting. Similarly, peristalsis was increased in larvae that express peptide transgenes designed to interfere with h-CaD binding to either the myosin or actin filament. As the rate of smooth muscle contraction was not increased in transgenic larvae, our findings argue that h-CaD normally functions to inhibit the force of smooth muscle contraction, as previously reported.25 Whether h-CaD affects contraction velocity or the distance of contraction in the intestine could not be determined using our methods. Although these results are consistent with the predicted role of h-CaD in modulating smooth muscle contractile force from phosphorylated cross-bridges, to our knowledge, they are the first evidence for this phenomenon in vivo. h-CaD deficient mice have been generated through gene targeting; however, the effect of h-CaD disruption on intestinal motility has not been reported.27 Given the similarities of intestinal anatomy and physiology in zebrafish and other vertebrates, a comparable effect of h-CaD deficiency on mammalian intestinal motility seems likely.28,43,44
h-CaD function has been studied extensively in vascular (tonic) smooth muscle. Besides functioning as a brake on contraction arising from phosphorylated myosin heads, h-CaD is also thought to sustain contrac-tile force generated by non- or de-phosphorylated myosin heads, thus accounting for force generation at low levels of phospho-Mlc. The mechanism of this aspect of h-CaD function remains unknown, but it has been postulated to occur through either physical stabilization of the actomyosin complex or the cooperative activation of nonphosphorylated and phosphorylated myosin heads.45 This presumably takes place when h-CaD itself has been inhibited, such as occurs with phosphorylation.
Whether h-CaD functions comparably in phasic smooth muscle of the intestine and colon remains unknown. Analyses of cls larvae provided an opportunity to examine this question in vivo, as they have nearly undetectable levels of intestinal phospho-Mlc.30 Surprisingly, cls larvae retain a significant degree of peristaltic function,30 presumably from ENS-independent smooth muscle contraction, which has been observed in the embryonic mouse and zebrafish intestine.46–48 In addition to very low phospho-Mlc levels, cls larvae also have undetectable levels of h-CaD phosphorylated at a serine residue corresponding to a major regulatory motif of human CALD1, serine-789.30 Low phospho-h-CaD to total h-CaD ratio in cls suggested that the basal level of smooth muscle contraction might be inhibited. Our experiments here indicate that this is indeed the case as both Cald1a-i4e5 MO injection and transgenic expression of the CADDK51 inhibitory peptide enhanced propulsive peristalsis in cls. Although it is difficult to make mechanistic inferences regarding h-CaD function from this study, our findings are consistent with the idea that h-CaD restricts binding of non-phosphorylated myosin heads to actin (Fig. 1C), as h-CaD knockdown enhanced peristalsis in cls mutants that have low levels of p-Mlc. The knockdown experiments in wild-type larvae (with high levels of p-Mlc) argue that h-CaD restricts binding of phosphorylated myosin heads to their preferred site on the actin filament (Fig. 1A) and that this restriction is relieved in the absence of h-CaD.
h-CaD is widely expressed in human smooth muscle and has been used extensively as a tumor marker.49,50 Its best characterized physiologic role is during pregnancy when total h-CaD levels increase,51 thus inhibiting premature stretch induced uterine contraction.52 At the time of delivery, phospho-h-CaD levels increase dramatically. This is thought to enhance the force of uterine contractions. A role for premature phosphorylation of h-CaD in a rodent model of preterm labor has also been reported.53
Relatively little is known about the regulation or function of h-CaD in the human intestine. In rabbit colonic smooth muscle, h-CaD serine-789 is phosphor-ylated upon exposure to the enteric neurotransmitter acetylcholine, which also drives phosphorylation of Mlc.54 This argues that h-CaD's normal function is to enhance ENS-mediated smooth muscle contraction. Furthermore, it suggests that in the setting of ENS dysfunction, levels of both phospho-Mlc and phosphoh-CaD are reduced, as occurs in zebrafish cls mutants. Surprisingly, cls larvae retain peristaltic activity, even though they have few, if any enteric nerves. This is likely a function of smooth muscle physiology and intestinal anatomy at larval stages. By contrast, a comparable level of ENS disruption in the adult mammalian intestine, which can be modeled by conditional Mlck disruption in mice,55 is predicted to cause irreversible intestinal motility defects. Hypomotility caused by a more modest reduction in ENS function, could conceivably be enhanced by modulation of h-CaD. Targeting h-CaD could therefore be a therapeutic strategy to treat hypomotility caused by enteric neuropathies and related disorders associated with reduced phospho-Mlc, such as is likely to occur with aging and disorders affecting Cajal pacemaker cells.56,57
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Jie He and Mani Muthumani for technical assistance and Kristin Lorent for generating figures.
FUNDING
This work was supported by R01-DK54942 (M.P.), and core facilities were provided by P30-DK50306.
Abbreviations
- h-CaD
high molecular weight Caldesmon isoform
- l-CaD
low molecular weight Caldesmon isoform
- ENS
enteric nervous system
- Mlc
myosin light chain
- Mlck
myosin light chain kinase
- dpf
days post fertilization
Footnotes
AUTHOR CONTRIBUTIONS
JA, GD, and CS performed the experiments, and all authors contributed to analysis and interpretation of the data; JA and MP conceived the study and designed the experiments; JA and MP drafted the manuscript.
DISCLOSURES
No competing interests declared.
REFERENCES
- 1.Kamm KE, Stull JT. The function of myosin and myosin light chain kinase phosphorylation in smooth muscle. Annu Rev Pharmacol Toxicol. 1985;25:593–620. doi: 10.1146/annurev.pa.25.040185.003113. [DOI] [PubMed] [Google Scholar]
- 2.Murphy RA. Contraction in smooth muscle cells. Annu Rev Physiol. 1989;51:275–83. doi: 10.1146/annurev.ph.51.030189.001423. [DOI] [PubMed] [Google Scholar]
- 3.Siegman MJ, Butler TM, Mooers SU, Michalek A. Ca2+ can affect Vmax without changes in myosin light chain phosphorylation in smooth muscle. Pflugers Arch. 1984;401:385–90. doi: 10.1007/BF00584340. [DOI] [PubMed] [Google Scholar]
- 4.Gerthoffer WT. Dissociation of myosin phosphorylation and active tension during muscarinic stimulation of tracheal smooth muscle. J Pharmacol Exp Ther. 1987;240:8–15. [PubMed] [Google Scholar]
- 5.Haeberle JR, Hott JW, Hathaway DR. Regulation of isometric force and isotonic shortening velocity by phosphorylation of the 20,000 dalton myosin light chain of rat uterine smooth muscle. Pflugers Arch. 1985;403:215–9. doi: 10.1007/BF00584103. [DOI] [PubMed] [Google Scholar]
- 6.Moreland S, Moreland RS. Effects of dihydropyridines on stress, myosin phosphorylation, and V0 in smooth muscle. Am J Physiol. 1987;252:H1049–58. doi: 10.1152/ajpheart.1987.252.6.H1049. [DOI] [PubMed] [Google Scholar]
- 7.Gusev NB. Some properties of caldesmon and calponin and the participation of these proteins in regulation of smooth muscle contraction and cytoskeleton formation. Biochemistry. 2001;66:1112–21. doi: 10.1023/a:1012480829618. [DOI] [PubMed] [Google Scholar]
- 8.Sobue K, Muramoto Y, Fujita M, Kakiuchi S. Purification of a calmodulin-binding protein from chicken gizzard that interacts with F-actin. Proc Natl Acad Sci USA. 1981;78:5652–5. doi: 10.1073/pnas.78.9.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kordowska J, Huang R, Wang CL. Phosphorylation of caldesmon during smooth muscle contraction and cell migration or proliferation. J Biomed Sci. 2006;13:159–72. doi: 10.1007/s11373-005-9060-8. [DOI] [PubMed] [Google Scholar]
- 10.Helfman DM, Levy ET, Berthier C, et al. Caldesmon inhibits nonmuscle cell contractility and interferes with the formation of focal adhesions. Mol Biol Cell. 1999;10:3097–112. doi: 10.1091/mbc.10.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CL, Chalovich JM, Graceffa P, Lu RC, Mabuchi K, Stafford WF. A long helix from the central region of smooth muscle caldesmon. J Biol Chem. 1991;266:13958–63. [PMC free article] [PubMed] [Google Scholar]
- 12.Horiuchi KY, Miyata H, Chacko S. Modulation of smooth muscle actomyosin ATPase by thin filament associated proteins. Biochem Biophys Res Commun. 1986;136:962–8. doi: 10.1016/0006-291x(86)90426-2. [DOI] [PubMed] [Google Scholar]
- 13.Earley JJ, Su X, Moreland RS. Caldesmon inhibits active crossbridges in unstimulated vascular smooth muscle: an antisense oligodeoxynucleotide approach. Circ Res. 1998;83:661–7. doi: 10.1161/01.res.83.6.661. [DOI] [PubMed] [Google Scholar]
- 14.Szpacenko A, Wagner J, Dabrowska R, Ruegg JC. Caldesmon-induced inhibition of ATPase activity of actomyosin and contraction of skinned fibres of chicken gizzard smooth muscle. FEBS Lett. 1985;192:9–12. doi: 10.1016/0014-5793(85)80032-6. [DOI] [PubMed] [Google Scholar]
- 15.Ansari S, Alahyan M, Marston SB, El-Mezgueldi M. Role of caldesmon in the Ca2+ regulation of smooth muscle thin filaments: evidence for a cooperative switching mechanism. J Biol Chem. 2008;283:47–56. doi: 10.1074/jbc.M706771200. [DOI] [PubMed] [Google Scholar]
- 16.Wang CL. Caldesmon and smooth-muscle regulation. Cell Biochem Biophys. 2001;35:275–88. doi: 10.1385/cbb:35:3:275. [DOI] [PubMed] [Google Scholar]
- 17.Smith CW, Marston SB. Disassembly and reconstitution of the Ca2+ -sensitive thin filaments of vascular smooth muscle. FEBS Lett. 1985;184:115–9. doi: 10.1016/0014-5793(85)80665-7. [DOI] [PubMed] [Google Scholar]
- 18.Ikebe M, Reardon S. Phosphorylation of smooth muscle caldesmon by calmodulin-dependent protein kinase II Identification of the phosphorylation sites. J Biol Chem. 1990;265:17607–12. [PubMed] [Google Scholar]
- 19.Childs TJ, Watson MH, Sanghera JS, Campbell DL, Pelech SL, Mak AS. Phosphorylation of smooth muscle caldesmon by mitogen-activated protein (MAP) kinase and expression of MAP kinase in differentiated smooth muscle cells. J Biol Chem. 1992;267:22853–9. [PubMed] [Google Scholar]
- 20.Katsuyama H, Wang CL, Morgan KG. Regulation of vascular smooth muscle tone by caldesmon. J Biol Chem. 1992;267:14555–8. [PubMed] [Google Scholar]
- 21.Horiuchi KY, Chacko S. Caldesmon inhibits the cooperative turning-on of the smooth muscle heavy meromyosin by tropomyosin-actin. Biochemistry. 1989;28:9111–6. doi: 10.1021/bi00449a023. [DOI] [PubMed] [Google Scholar]
- 22.Lu C, Liu Y, Tang X, Ye H, Zhu D. Role of 15-hydroxyeicosatetraenoic acid in phosphorylation of ERK1/2 and caldesmon in pulmonary arterial smooth muscle cells. Can J Physiol Pharmacol. 2006;84:1061–9. doi: 10.1139/y06-057. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Zhang L. Regulation of alpha1-adrenoceptor-mediated contractions of the uterine artery by protein kinase C: role of the thick- and thin-filament regulatory pathways. J Pharmacol Exp Ther. 2007;322:1253–60. doi: 10.1124/jpet.107.124313. [DOI] [PubMed] [Google Scholar]
- 24.Chacko S, Chang S, Hypolite J, Disanto M, Wein A. Alteration of contractile and regulatory proteins following partial bladder outlet obstruction. Scand J Urol Nephrol Suppl. 2004;215:26–36. doi: 10.1080/03008880410015147. [DOI] [PubMed] [Google Scholar]
- 25.Smolock EM, Trappanese DM, Chang S, Wang T, Titchenell P, Moreland RS. siRNA-mediated knockdown of h-caldesmon in vascular smooth muscle. Am J Physiol Heart Circ Physiol. 2009;297:H1930–9. doi: 10.1152/ajpheart.00129.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerthoffer WT, Pohl J. Caldesmon and calponin phosphorylation in regulation of smooth muscle contraction. Can J Physiol Pharmacol. 1994;72:1410–4. doi: 10.1139/y94-203. [DOI] [PubMed] [Google Scholar]
- 27.Guo H, Wang CL. Specific disruption of smooth muscle caldesmon expression in mice. Biochem Biophys Res Commun. 2005;330:1132–7. doi: 10.1016/j.bbrc.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 28.Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–73. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Lee YH, Gallant C, Guo H, Li Y, Wang CA, Morgan KG. Regulation of vascular smooth muscle tone by N-terminal region of caldesmon. Possible role of tethering actin to myosin. J Biol Chem. 2000;275:3213–20. doi: 10.1074/jbc.275.5.3213. [DOI] [PubMed] [Google Scholar]
- 30.Davuluri G, Seiler C, Abrams J, Soriano AJ, Pack M. Differential effects of thin and thick filament disruption on zebrafish smooth muscle regulatory proteins. Neurogastroenterol Motil. 2010;22:1100–e1285. doi: 10.1111/j.1365-2982.2010.01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amores A, Force A, Yan YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 32.Zheng PP, Severijnen LA, van der Weiden M, Willemsen R, Kros JM. A crucial role of caldesmon in vascular development in vivo. Cardiovasc Res. 2009;81:362–9. doi: 10.1093/cvr/cvn294. [DOI] [PubMed] [Google Scholar]
- 33.Bryan J. Caldesmon, acidic amino acids and molecular weight determinations. J Muscle Res Cell Motil. 1989;10:95–6. doi: 10.1007/BF01739964. [DOI] [PubMed] [Google Scholar]
- 34.Seiler C, Abrams J, Pack M. Characterization of zebrafish intestinal smooth muscle development using a novel sm22alpha-b promoter. Dev Dyn. 2010;239:2806–12. doi: 10.1002/dvdy.22420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikebe M, Reardon S. Binding of caldesmon to smooth muscle myosin. J Biol Chem. 1988;263:3055–8. [PubMed] [Google Scholar]
- 36.Wang Z, Jiang H, Yang ZQ, Chacko S. Both N-terminal myosin-binding and C-terminal actin-binding sites on smooth muscle caldesmon are required for caldesmon-mediated inhibition of actin filament velocity. Proc Natl Acad Sci USA. 1997;94:11899–904. doi: 10.1073/pnas.94.22.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan QQ, Wong SS, Wang CL. A calmodulin-binding peptide of caldesmon. J Biol Chem. 1991;266:21810–4. [PubMed] [Google Scholar]
- 38.Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–9. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- 39.Kelsh RN, Eisen JS. The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development. 2000;127:515–25. doi: 10.1242/dev.127.3.515. [DOI] [PubMed] [Google Scholar]
- 40.Dutton KA, Pauliny A, Lopes SS, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–25. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- 41.Mabuchi K, Wang CL. Electron microscopic studies of chicken gizzard caldesmon and its complex with calmodulin. J Muscle Res Cell Motil. 1991;12:145–51. doi: 10.1007/BF01774033. [DOI] [PubMed] [Google Scholar]
- 42.Suveges D, Gaspari Z, Toth G, Nyi-tray L. Charged single alpha-helix: a versatile protein structural motif. Proteins. 2009;74:905–16. doi: 10.1002/prot.22183. [DOI] [PubMed] [Google Scholar]
- 43.Rich A, Leddon SA, Hess SL, et al. Kit-like immunoreactivity in the zebrafish gastrointestinal tract reveals putative ICC. Dev Dyn. 2007;236:903–11. doi: 10.1002/dvdy.21086. [DOI] [PubMed] [Google Scholar]
- 44.Shepherd I, Eisen J. Development of the zebrafish enteric nervous system. Methods Cell Biol. 2011;101:143–60. doi: 10.1016/B978-0-12-387036-0.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albrecht K, Schneider A, Liebetrau C, Ruegg JC, Pfitzer G. Exogenous caldesmon promotes relaxation of guinea-pig skinned taenia coli smooth muscles: inhibition of cooperative reattachment of latch bridges? Pflugers Arch. 1997;434:534–42. doi: 10.1007/s004240050433. [DOI] [PubMed] [Google Scholar]
- 46.Roberts RR, Murphy JF, Young HM, Bornstein JC. Development of colonic motility in the neonatal mouse-studies using spatiotemporal maps. Am J Physiol Gastrointest Liver Physiol. 2007;292:G930–8. doi: 10.1152/ajpgi.00444.2006. [DOI] [PubMed] [Google Scholar]
- 47.Roberts RR, Ellis M, Gwynne RM, et al. The first intestinal motility patterns in fetal mice are not mediated by neurons or interstitial cells of Cajal. J Physiol. 2010;588:1153–69. doi: 10.1113/jphysiol.2009.185421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmberg A, Olsson C, Hennig GW. TTX-sensitive and TTX-insensitive control of spontaneous gut motility in the developing zebrafish (Danio rerio) larvae. J Exp Biol. 2007;210:1084–91. doi: 10.1242/jeb.000935. [DOI] [PubMed] [Google Scholar]
- 49.Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med. 2006;130:1466–78. doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- 50.McCluggage WG. A critical appraisal of the value of immunohistochemistry in diagnosis of uterine neoplasms. Adv Anat Pathol. 2004;11:162–71. doi: 10.1097/00125480-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 51.Word RA, Stull JT, Casey ML, Kamm KE. Contractile elements and myosin light chain phosphorylation in myometrial tissue from nonpregnant and pregnant women. J Clin Invest. 1993;92:29–37. doi: 10.1172/JCI116564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Reznichenko M, Tribe RM, et al. Stretch activates human myometrium via ERK, caldesmon and focal adhesion signaling. PLoS ONE. 2009;4:e7489. doi: 10.1371/journal.pone.0007489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Je HD, Malek S, Morgan KG. Role of ERK1/2 in uterine contractility and preterm labor in rats. Am J Physiol Regul Integr Comp Physiol. 2004;287:R328–35. doi: 10.1152/ajpregu.00042.2004. [DOI] [PubMed] [Google Scholar]
- 54.Somara S, Bitar KN. Phosphorylated HSP27 modulates the association of phosphorylated caldesmon with tropomyosin in colonic smooth muscle. Am J Physiol Gastrointest Liver Physiol. 2006;291:G630–9. doi: 10.1152/ajpgi.00350.2005. [DOI] [PubMed] [Google Scholar]
- 55.He WQ, Peng YJ, Zhang WC, et al. Myosin light chain kinase is central to smooth muscle contraction and required for gastrointestinal motility in mice. Gastroenterology. 2008;135:610–20. doi: 10.1053/j.gastro.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somara S, Gilmont RR, Martens JR, Bitar KN. Ectopic expression of cave-olin-1 restores physiological contrac-tile response of aged colonic smooth muscle. Am J Physiol Gastrointest Liver Physiol. 2007;293:G240–9. doi: 10.1152/ajpgi.00064.2007. [DOI] [PubMed] [Google Scholar]
- 57.Farrugia G. Interstitial cells of Cajal in health and disease. Neurogastroenterol Motil. 2008;20:54–63. doi: 10.1111/j.1365-2982.2008.01109.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.