Abstract
Definition of the molecular pathogenesis of lung cancer allows investigators an enhanced understanding of the natural history of the disease, thus fostering development of new prevention strategies. In addition to regulating epithelial-to-mesenchymal transition (EMT), the transcription factor Snail exerts global effects on gene expression. Our recent studies reveal that Snail is upregulated in non-small cell lung cancer (NSCLC), is associated with poor prognosis, and promotes tumor progression in vivo. Herein, we demonstrate that overexpression of Snail leads to upregulation of Secreted Protein, Acidic and Rich in Cysteine (SPARC) in models of premalignancy and established disease, as well as in lung carcinoma tissues in situ. Snail overexpression leads to increased SPARC-dependent invasion in vitro, indicating that SPARC may play a role in lung cancer progression. Bioinformatic analysis implicates TGF-β, ERK1/2, and miR-29b as potential intermediaries in Snail-mediated upregulation of SPARC. Both the TGF-β1 ligand and TGF-βR2 are upregulated following Snail overexpression. Treatment of human bronchial epithelial cell (HBEC) lines with TGF-β1 and inhibition of TGF-β1 mRNA expression modulated SPARC expression. Inhibition of MEK phosphorylation downregulated SPARC. MiR-29b is downregulated in Snail overexpressing cell lines, while overexpression of miR-29b inhibited SPARC expression. In addition, miR-29b was upregulated following ERK inhibition, suggesting a Snail-dependent pathway by which Snail activation of TGF-β and ERK signaling results in downregulation of miR-29b and subsequent upregulation of SPARC. Our discovery of pathways responsible for Snail-induced SPARC expression contributes to the definition of NSCLC pathogenesis.
Keywords: NSCLC, Snail, SPARC, invasion, parallel progression
Introduction
Lung cancer is the leading cause of cancer death worldwide, with a median survival of eight months following diagnosis and only 16% of patients surviving more than five years (1). Understanding early lung cancer pathogenesis will facilitate targeted approaches for chemoprevention. Inflammatory mediators, such as transforming growth factor-β (TGF-β), eicosanoids, and interleukin 1-β (IL-1β) are overexpressed in the pulmonary microenvironment of smokers and patients with emphysema or pulmonary fibrosis; these patients have a heightened risk of developing lung cancer (2, 3). Recently described as one of the hallmarks of cancer, chronic inflammation is now considered a risk factor for the development of lung cancer (4). Although an active area of investigation, the molecular mechanisms underlying the association between inflammation and lung cancer initiation and progression remain largely undefined.
The zinc-finger transcription factor Snail, encoded by the SNAI1 gene, has been shown to be upregulated following exposure to inflammatory mediators such as prostaglandin E2 (PGE2) (5) and TGF-β (6). Snail exerts global effects on epithelial cell gene expression profiles, including regulation of EMT (7, 8). Snail plays a pivotal role in inducing EMT in several malignancies (9–11) and is expressed in a stem cell-like subpopulation within immortalized human mammary epithelial cells that are capable of transformation (12). Recent studies suggest that Snail may play a broader role in carcinogenesis (13, 14). We have shown that Snail is upregulated in human NSCLC tissues, is associated with poor prognosis, and promotes NSCLC tumor progression in vivo (15). Furthermore, Snail overexpression in NSCLC is associated with differential gene expression related to diverse aspects of lung cancer progression, including angiogenesis (15). Identification of the mechanisms by which the inflammation-induced transcriptional repressor Snail contributes to lung cancer pathogenesis, specifically to the invasive phenotype, would be a step forward in understanding the contribution of inflammation to lung cancer development and progression.
SPARC, also known as osteonectin, is an extracellular matrix glycoprotein first identified as a major non-collagenous component of bovine bone (16). Its expression modulates reversible interactions between cells and the extracellular matrix (17). Upregulation of SPARC is associated with metastatic potential of melanomas and gliomas, as well as an invasive phenotype in breast, prostate, and colorectal carcinomas (18). Expression of SPARC in the NSCLC stroma is associated with poor prognosis (19), though its role in lung tumor progression, especially in relation to epithelial cell Snail expression, has not been evaluated.
To understand the molecular changes that occur during NSCLC initiation and development, we overexpressed Snail in both immortalized HBECs and NSCLC cells; the HBECs were previously established as a robust model of the pulmonary airway epithelium and its associated malignant transformation (20–24). We demonstrate that Snail upregulates SPARC and drives SPARC-dependent invasion in both models of premalignancy and established NSCLC. We also examined the mechanism for Snail-induced SPARC expression and identified key signaling pathways critical to this relationship.
Materials and Methods
Human cell lines and reagents
Lung cancer cell lines A549, H292, H358, H441, and H1437 were obtained from American Type Culture Collection(ATCC) (Rockville, MD). All HBEC lines were provided by Drs. John D. Minna and Jerry W. Shay at the University of Texas, Southwestern Medical Center. The cells were immortalized in the absence of viral oncoproteins via ectopic expression of human telomerase (hTERT) and cyclin-dependent kinase 4 under control of puromycin and geneticin, respectively (22). Four parental cell lines derived from four patients, HBEC2, HBEC3, HBEC4, and HBEC7, were used. We also utilized a mutated HBEC3 cell line designated H3mutP53/KRAS (or H3mut). This cell line was derived by stably transfecting HBEC3 with an shRNA construct targeting the tumor suppressor P53 and a construct overexpressing the oncogenic protein KRAS with an activating mutation under control of zeocin and blasticidin, respectively. All cell lines were routinely tested for the presence of Mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza, Walkerville, CA). Cell lines were authenticated in the UCLA Genotyping and Sequencing Core utilizing Promega’s (Madison, WI) DNA IQ System and Powerplex 1.2 system according to the manufacturer’s instructions. All cells were utilized within 10 passages of genotyping. Lung cancer cell lines were grown in RPMI 1640 (Mediatech Inc, Herndon, VA) supplemented with 10% fetal bovine serum (Gemini Biological Products, Calabasas, CA), 1% penicillin/streptomycin (Life Technologies, Grand Island, NY), and 2mM glutamine (Life Technologies). HBEC lines were grown in Keratinocyte Serum-Free Media (Life Technologies) supplemented with 30ug/mL Bovine Pituitary Extract and 0.2ng/mL recombinant Epidermal Growth Factor 1-53 (Life Technologies). Treatments were carried out in 6 well plates at a density of 1.25×105 cells per well in serum-free medium, unless stated otherwise. The MEK/ERK inhibitor U0126 (Cell Signaling, Danvers, MA) was prepared in sterile DMSO at a concentration of 10mg/mL, and cells were treated at a final concentration of 15ug/mL for 24 hours. Recombinant human TGF-β1 (PeproTech, Minneapolis, MN) was prepared in sterile 0.1% BSA at a concentration of 5ng/μL, and cells were treated at a final concentration of 5ng/mL for 24 hours.
Stable overexpression of Snail
Cells were stably transduced as follows: wild-type Snail cDNA pcDNA3 (a gift from Dr. E. Fearon, University of Michigan, Ann Arbor, MI) was excised from the plasmid with HindIII and EcoRV and subcloned into the retroviral vector pLHCX (Clontech, Mountain View, CA) that includes a drug resistance (hygromycin B) marker. All constructs were verified by restriction endonuclease digestion. For virus production, 70% confluent 293T cells were cotransfected with pLHCX-Snail or pLHCX (vector alone). Tumor cells were then transduced with high-titer supernatants producing either Snail or pLHCX virus. Following transduction, the cells were selected with hygromycin B (Life Technologies).
Stable genetic inhibition of SPARC
SPARC shRNA plasmids on the pLKO.1-Puro vector backbone and relevant controls were obtained from Sigma-Aldrich (St. Louis, MO). Using the viral transduction method described above, NSCLC cell lines were stably transduced with one of five shRNA sequences: -shSPARC1, -shSPARC2, -shSPARC3, a nonsilencing sequence (-shNS), or the pLKO.1 vector backbone sequence (-shV). The cell lines were selected with optimized concentrations of puromycin (EMD Chemicals, Billerica, MA). For the HBEC lines, as the overexpression of hTERT was under puromycin selection, the puromycin selection marker in the shSPARC vectors was replaced with the inosine-5'-monophosphate dehydrogenase (IMPDH) gene, encoding resistance to mycophenolic acid (MPA). After viral transduction, the cells were selected with optimized concentrations of MPA (Alfa Aesar, Ward Hill, MA).
Transient transfection of microRNA mimics and siRNA
Cells were plated in 6 well plates at 1.25×105 cells per well and cultured for 24 hours in growth media. For transient overexpression of miR-29b, complete media was replaced prior to transfection with the miRVana™ miR-29b mimic (Life Technologies) or miRVana™ miRNA mimic Negative Control #1 (Life Technologies). For transient inhibition of TGF-β1, complete media was replaced with serum-free media overnight prior to transfection with target siRNA (Integrated DNA Technologies, Coralville, IA) or Silencer® Negative Control #1 siRNA (Life Technologies). Transfections were carried out using the Lipofectamine RNAiMAX Transfection Reagent (Life Technologies) in serum-free media for 4 hours before replacement with fresh serum-free media and an additional 20-hour incubation.
Western blot analysis
Cells were washed with PBS and whole-cell lysates were collected over ice using lysis buffer prepared according to standard methods (25). Protein concentrations were measured with a BCA proteinassay reagent (Pierce, Rockford, IL). Proteins were resolved by 10% SDS-PAGE and analyzed by western blot using polyvinylidene difluoride membranes (Millipore, Bedford, CA) according to standard methods. Membranes were blocked with 5% nonfat dry milk or 5% BSA in TBS plus 0.05% Tween 20. The membranes were probedovernight at 4°C with anti-Snail, anti-SPARC, anti-TGFBR2, anti-ERK1/2, and anti-phospho-ERK1/2 (all from Cell Signaling). Secondary antibodies, goat anti-mouse (Bio-Rad, Hercules, CA) and goat anti-rabbit (Santa Cruz Biotechnology, Dallas, TX), were incubated at room temperature for 60 minutes. Membraneswere developed using Supersignal Chemiluminescence System (Pierce) or Western Lightning Plus-ECL (Perkin-Elmer, Waltham, MA) and exposed to X-ray film(Life Sciences Products, Inc, Frederick, CO). Equal loading of samples was confirmed by probing the membranes with alpha-tubulin (Cell Signaling). Antibody catalog numbers and blotting conditions are detailed in Supplementary Table S1. Each experiment was performed at least three times, and one representative experiment or image is shown.
TGF-β ELISA
Secreted TGF-β1 levels were quantified using the eBioscience Human/Mouse TGF beta1 ELISA Ready-SET-Go! Kit (Cat #88-8350; San Diego, CA). Cells were plated in 6-well plates at a density of 1.25×105 cells per well; triplicates of each condition were plated. Once adherent, cells were washed and media were replaced with 1mL serum-free media. Media supernatants and cell lysates were collected after 24 hours. Supernatant TGF-β levels were evaluated following the manufacturers’ instructions and normalized to cell lysate protein concentrations. Each experiment was performed at least three times, and one representative experiment or image is shown.
Invasion Assay
Cells were serum-starved and plated at a density of 2×104 (cancer cells) or 1×104 (HBEC) cells per well in Corning HTS Transwell-96 Permeable Support Plate (Sigma-Aldrich); six replicates of each condition were plated. Prior to plating, transwells were coated with Type I Rat Tail Collagen (BD Biosciences) to create a “membrane” for the cells to degrade and invade. Cells were allowed to invade for 48 hours into a lower chamber containing media with 20% FBS (cancer cells) or 2% FBS (HBEC) as a chemoattractant. The upper chamber was aspirated and washed with PBS to remove noninvasive cells. The lower chamber was washed with PBS and invasive cells were released from the underside of the top chamber with Cell Dissociation Solution (Trevigen, Gaithersburg, MD). Calcein AM (Life Technologies) was used to stain viable cells in the lower chamber only, and fluorescence was quantified and compared to measurements from Day 0 control plates that contained the total number of cells plated for each cell line to generate “% input invasion” values. Each experiment was performed at least three times, and one representative experiment or image is shown.
Total RNA preparation, cDNA synthesis and real-time PCR
RNA was isolated using the miRNeasy Mini kit (Qiagen, Valencia, CA), and cDNA was prepared using the High Capacity RNA-to-cDNA Kit (Life Technologies) according to the manufacturers’protocols. Transcript levels of miR-29b and SPARC were measured by quantitative real-time reverse transcription polymerase chain reaction (q-RT-PCR) using the TaqMan Probe-based Gene Expression system (Life Technologies) in a MyiQ Cycler (Bio-Rad) following the manufacturer's protocol. Amplification was carried out for 40 cycles of 15 seconds at 95°C and 60 secondsat 60°C. All samples were run in triplicate, and their relative expression levels were determined by normalizingthe expression of each target to RNU6b (miR-29b) or GUSB (SPARC). These levels were then compared to the normalized expression levels in a reference sample using the 2(−Delta Delta C(T)) method to calculate fold-change values (26). Each experiment was performed at least three times, and one representative experiment or image is shown.
Immunohistochemistry
Serial sections obtained from human NSCLC clinical specimens archived in the University of California at Los Angeles (UCLA) Lung Cancer Specialized Programs of Research Excellence tissue bank were collected from patients under informed consent in accordance with the Declaration of Helsinki (IRB #10-001096). Antigen retrieval was accomplished with sodium citrate 10mmol/L (pH 6.0). Sections were blocked with 10% normal goat serum and then probed with an antibody against Snail (Cat #ab85931, Abcam, Cambridge, MA) or SPARC (Cat #AON-5031, Heamatologic Technologies, Essex Junction, VT) using a working dilution of 1:500 for tissue staining. Primary antibodies were incubated overnight at 4°C. After incubation with secondary antibody (Vector Laboratories, Burlingame, CA), staining was developed using DAB Substrate kit for Peroxidase (Cat #SK-4100, Vector Laboratories). Snail and SPARC expression was evaluated by a pathologist (MCF) specializing in cardiopulmonary disease. Evaluation of the clinical specimens was based on epithelial cell staining intensity and correlation of staining between serial sections; stromal and immune cell staining patterns were also evaluated but were of secondary importance to the current study. Photomicrographs were obtained using an Olympus BX50 microscope, with Plan APO objective lenses. An Olympus DP11 camera and Olympus Camedia software were used to produce the images.
Differential miRNA expression analysis
Single samples of RNA were collected from Snail-overexpressing and Vector control H292, H358, H441, and H1437 cell lines with the miRNeasy Mini kit (Qiagen). One microgram of total RNA was labeled using miRCURY LNA™ microRNA Array Power Labeling kit by the UCLA Clinical Microarray Core. The labeled miRNAs were hybridized to Exiqon miRCURY LNA microRNA Array-6th Generation according to the manufacturer’s instructions. This array includes 927/648/351 human/mouse/rat miRNAs, as well as 438 miRPlus miRNAs. The miRNA array slides were scanned using Axon GenePix 4000B scanner (Axon Instruments, Foster City, CA) and processed by using the GenePix Pro 6.0 software (Axon Instruments). The raw data were normalized by using a combination of housekeeping miRNA, spike-in miRNA, and invariant endogenous miRNAs. Harvard dChip software (27) was used for data analysis. Microarray data discussed in this publication are deposited in National Center for Biotechnology Information's Gene Expression Omnibus (accession #GSE48922) (28).
Statistical analysis
Samples were plated and run in triplicate, unless otherwise indicated, and all experiments were performed at least three times. Results from one representative experiment or image are shown. Probability values were calculated using two-tailed non-paired Student's t-test for most count-based data, and analysis of variance (a.k.a, ANOVA) models were utilized where multiple pairwise comparisons were made. Tests of statistical significance were considered significant as follows: * if p ≤ 0.05, ** if p ≤ 0.001, and *** if p ≤ 0.0001.
Results
SPARC is upregulated in Snail-overexpressing models of premalignancy and established NSCLC
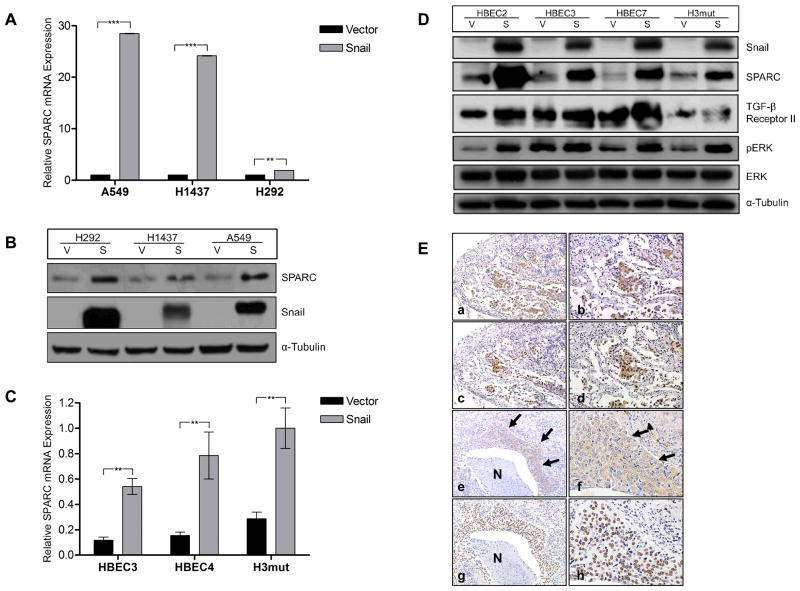
Recently, we have found that forced overexpression of Snail in NSCLC cell lines leads to global expression changes, including increased expression of genes implicated in angiogenesis (15). Gene expression profiling and bioinformatic analysis indicated that a panel of Snail overexpressing NSCLC cell lines have significantly increased expression of SPARC (15). Fold change (FC) values of >10 were observed in three of four cell lines tested (H358 FC=32, H441 FC=193, and H1437 FC=1276). Q-RT-PCR and immunoblot analyses of NSCLC cell lines with and without Snail overexpression confirmed the relationship between Snail and SPARC in established cancer cell lines (Fig. 1A, B, respectively). To model early pathogenesis of lung cancer, the same Snail overexpression plasmid was introduced into HBECs. Q-RT-PCR and immunoblot analyses of these cell lines show that Snail overexpression leads to enhanced SPARC expression in this model as well (Fig. 1C, D). Taken together, these data suggest that SPARC expression is positively associated with Snail in models of both premalignancy and established lung cancer.
Figure 1. Snail overexpression is correlated with upregulation of SPARC.
(A) The NSCLC cell lines A549, H1437, and H292 were stably transfected with either a Vector control plasmid (V) or a Snail expression plasmid (S). Total RNA was isolated from the cell lines and expression levels of SPARC were evaluated by q-RT-PCR using TaqMan primers. mRNA levels were normalized to GUSB. (B) The same NSCLC cell lines were evaluated for protein level expression of Snail and SPARC by western blotting. Protein levels were normalized to α-Tubulin. (C) Total RNA was isolated from HBEC3-V/S, HBEC4-V/S, and H3mut-V/S cell lines. Expression levels of SPARC were evaluated by q-RT-PCR using TaqMan primers. mRNA levels were normalized to GUSB. A similar pattern was also observed for HBEC7 (data not shown). (D) The same HBEC cells were evaluated for protein level expression of Snail and SPARC by western blotting. Protein levels were normalized to α-Tubulin. (E) Serial sections of ADC (a–d). Positive (brown staining) cytoplasmic expression of SPARC (row 1; a and b) and positive nuclear expression of Snail (row 2; c and d) were observed in the epithelial component of the neoplasm; × 100 low magnification (left; a and c) and × 200 high magnification (right; b and d) images of the same specimen and staining condition. Serial sections of SCC (e–h). Positive cytoplasmic expression of SPARC (row 3; e and f, arrows) and positive nuclear expression of Snail (row 4; g and h) were observed in the epithelial component of the neoplasm; × 100 low magnification (left; e and g) and × 400 high magnification (right; f and h) images of the same specimen and staining condition. Necrotic areas of the neoplasm (N) do not stain for either protein. (** = p < 0.001; *** = p < 0.0001)
To confirm the relationship between Snail and SPARC in human NSCLC neoplasms in situ, we stained serial sections of paraffin-embedded human lung adenocarcinoma (ADC; n = 10) and squamous cell carcinoma (SCC; n = 9) for Snail and SPARC protein expression. Immunolocalization revealed a pattern of uniform cytoplasmic SPARC staining of the epithelial component of the neoplasm in association with intense nuclear Snail staining of the epithelia (Fig. 1E and Table 1). The immunostained sections were reviewed by a pathologist (MCF) and scored based on Snail or SPARC staining intensity (low, medium, or high) and the Snail:SPARC correlation (yes or no; ADC = 9/10 yes, SCC = 7/9 yes). In regions of more poorly differentiated and infiltrative tumor, there tended to be increased SPARC expression, while necrotic neoplasm and benign airways were not positive for either protein. The stromal and immune cell components of the tumor microenvironment were characterized by positive cytoplasmic SPARC staining of stromal spindle cells (likely fibroblasts and myofibroblasts) and alveolar macrophages and absence of staining of the infiltrating lymphocytes. In contrast, large numbers of infiltrating lymphocytes were positive for nuclear Snail staining, as were alveolar macrophages and stromal spindle cells. No significant difference was noted when comparing ADC and SCC staining of the three compartments (epithelial, stromal, and immune). These results suggest a relationship between epithelial cell Snail and SPARC in human non-small cell lung cancers.
Table 1.
Staining intensity and correlation between epithelial Snail (nuclear) and SPARC (cytoplasmic) immunostaining of paraffin-embedded lung ADC (n=10) and SCC (n=9) clinical specimens.
| Adenocarcinoma (9/10) | |||
|---|---|---|---|
| Sample ID | Snail Grade | SPARC Grade | Correlation (Y/N) |
| ADC-1 | High | High | Y |
| ADC-2 | Medium | Medium | Y |
| ADC-3 | High | Medium | Y |
| ADC-4 | High | High | Y |
| ADC-5 | High | High | Y |
| ADC-6 | Low | Low | Y |
| ADC-7 | Low | Low | Y |
| ADC-8 | Medium | Low | N |
| ADC-9 | Low | Low | Y |
| ADC-10 | Low | Low | Y |
| Squamous Cell Carcinoma (7/9) | |||
|---|---|---|---|
| Sample ID | Snail Grade | SPARC Grade | Correlation (Y/N) |
| SCC-1 | High | High | Y |
| SCC-2 | High | Low | N |
| SCC-3 | Medium | Low | N |
| SCC-4 | Low | Low | Y |
| SCC-5 | Low | Low | Y |
| SCC-6 | Low | Low | Y |
| SCC-7 | Low | Low | Y |
| SCC-8 | Medium | Medium | Y |
| SCC-9 | Low | Low | Y |
Snail-dependent invasion of HBEC and NSCLC cell lines is SPARC-mediated
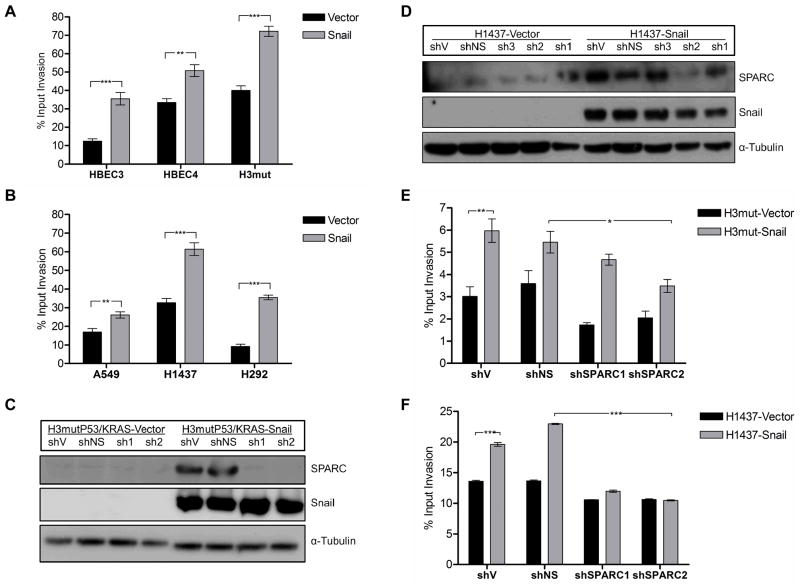
As SPARC is known to be associated with invasive cancers and is correlated with poor patient prognosis in NSCLC (18, 19), we evaluated both the HBEC and NSCLC cell lines utilizing in vitro invasion assays. In both the HBEC and the NSCLC cell lines, Snail overexpression led to increased invasion (Fig. 2A, B). Our previous studies demonstrated that Snail overexpression confers a proliferative disadvantage to NSCLC cells in vitro (15), suggesting that the increased invasion is independent of proliferation. Having established that Snail overexpression leads to both elevated SPARC expression and invasion, we hypothesized that the increased invasion is mediated by SPARC. The HBEC line HBEC3mutP53/KRAS and cancer cell line H1437 with stable Snail overexpression (-S) or vector control (-V) were stably transfected with a plasmid containing an shRNA sequence specific to the 3’UTR of SPARC or non-silencing controls (-NS). Coordinate protein level expression of Snail and SPARC was confirmed in all cell lines by western analysis (Fig. 2C, D). Utilizing the modified Boyden chambers described in Materials and Methods, we plated the cells and allowed them to invade for 48 hours. SPARC knockdown by shRNA in both HBEC and NSCLC cells reversed Snail-mediated invasion (Fig. 2E, F), indicating that SPARC is at least partially responsible for increased invasion downstream of Snail. Similar results were obtained for HBEC3mutP53/KRAS and HBEC3 (data not shown), suggesting that the SPARC effect in HBECs in not mutation-dependent for these mutations. The observation that Snail continues to drive low level invasion of HBEC cells in the absence of SPARC, while SPARC knockdown totally abrogates Snail-driven invasion of NSCLC cells, suggests a differential reliance on Snail and SPARC for the invasion program at different timepoints during lung carcinogenesis.
Figure 2. Snail overexpression leads to SPARC-dependent increased invasion in premalignant and established NSCLC.
(A) The invasive capacity of the HBEC lines HBEC3, HBEC4, and H3mutP53/KRAS (H3mut) with and without Snail overexpression were evaluated in a modified Boyden chamber assay for invasion through a collagen matrix over 48 hrs. Fluorescence values were divided by maximum input fluorescence measured on Day 0 for each cell line to derive a percent input invasion value. (B) The NSCLC cell lines A549, H1437, and H292 with and without Snail overexpression were evaluated as in (A). (C–F) SPARC shRNA sequences (sh1, sh2, sh3) were stably transfected into H3mut and H1437 vector control and Snail-overexpressing cell lines along with a nonsilencing (NS) shRNA control. Protein level expression of SPARC, Snail, and α-Tubulin were evaluated in the HBEC (C) and NSCLC (D) lines by western blot. The HBEC (E) and NSCLC lines (F) were also evaluated in a modified Boyden chamber assay for invasion through a collagen matrix over 48 hrs. The following comparisons were made for both cell types: (1) Vector versus Snail cells transduced with shV and (2) Snail cells transduced with shNS versus shSPARC2. (* = p < 0.05; ** = p < 0.001; *** = p < 0.0001)
TGF-β is upregulated in Snail overexpressing cell lines
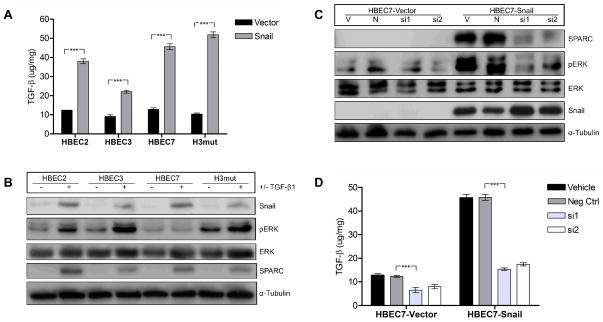
The promoter region of SPARC does not contain a binding site for Snail, indicating that Snail must upregulate SPARC by an indirect mechanism. Total RNA was isolated from a panel of Snail overexpressing NSCLC cell lines and vector controls and subjected to microRNA array. Array results were combined with mRNA array results previously published (15) and analyzed by Ingenuity Pathway Analysis Software. Combinatorial analysis revealed multiple potential intermediary molecules and signaling pathways that could be responsible for Snail-mediated upregulation of SPARC; most prominently that Snail may interact with SPARC via TGF-β, ERK1/2, and miR-29b. We evaluated the secreted protein levels of one potential candidate, the cytokine TGF-β1, utilizing ELISA. TGF-β1 secretion was significantly increased (HBEC2 FC=3.1, HBEC3 FC=2.4, HBEC7 FC=3.6, H3mut FC=5.0; p ≤ 0.0001) in Snail-overexpressing HBEC lines compared to vector controls (Fig. 3A), indicating that TGF-β is upregulated by Snail and may be upstream of SPARC. Treatment of the parental HBEC lines with recombinant TGF-β1 resulted in increased expression of Snail, SPARC, and phosphorylation of ERK1/2 (Fig. 3B), suggesting an autocrine signaling mechanism for Snail and TGF-β1 expression. Inhibition of TGF-β1 expression by siRNA abrogated Snail-mediated ERK1/2 phosphorylation and SPARC expression (Fig. 3C), further suggesting that TGF-β1 is necessary for Snail-mediated SPARC expression. The knockdown efficiency of the TGF-β siRNAs was measured by ELISA following inhibition (Fig. 3D). Immunoblot analysis of a panel of HBEC lines indicated that the TGF-β Receptor 2 (TGFβR2) is also elevated in Snail overexpressing lines (Fig. 1D), suggesting that both the ligand and the receptor are upregulated by Snail and are intermediates in the Snail-to-SPARC pathway.
Figure 3. TGF-β1 is upregulated by Snail upstream of ERK1/2 and SPARC.
(A) The secreted protein levels of TGF-β1 were measured by ELISA from supernatants of Snail overexpressing HBEC lines and compared to appropriate vector controls. (B) Parental HBEC lines were treated with recombinant TGF-β1 (5ng/mL) or vehicle control for 24 hours in serum-free media. Lysates were collected and protein expression of Snail, phosphorylated ERK1/2 (pERK), total ERK1/2, SPARC, and α-tubulin were measured by western blot. Protein levels were normalized to α-tubulin. (C) Snail-overexpressing HBEC lines and vector controls were treated with single siRNA sequences targeting TGF-β1 (si1 and si2), a negative control siRNA (N), or left untreated (V) for 24 hours in serum-free media. Lysates were collected and protein expression was measured as in (B). (D) Efficiency of TGF-β1 knockdown was measured by ELISA 24 hours following transfection. (*** = p < 0.0001)
ERK1/2 signaling is activated in Snail overexpressing cell lines
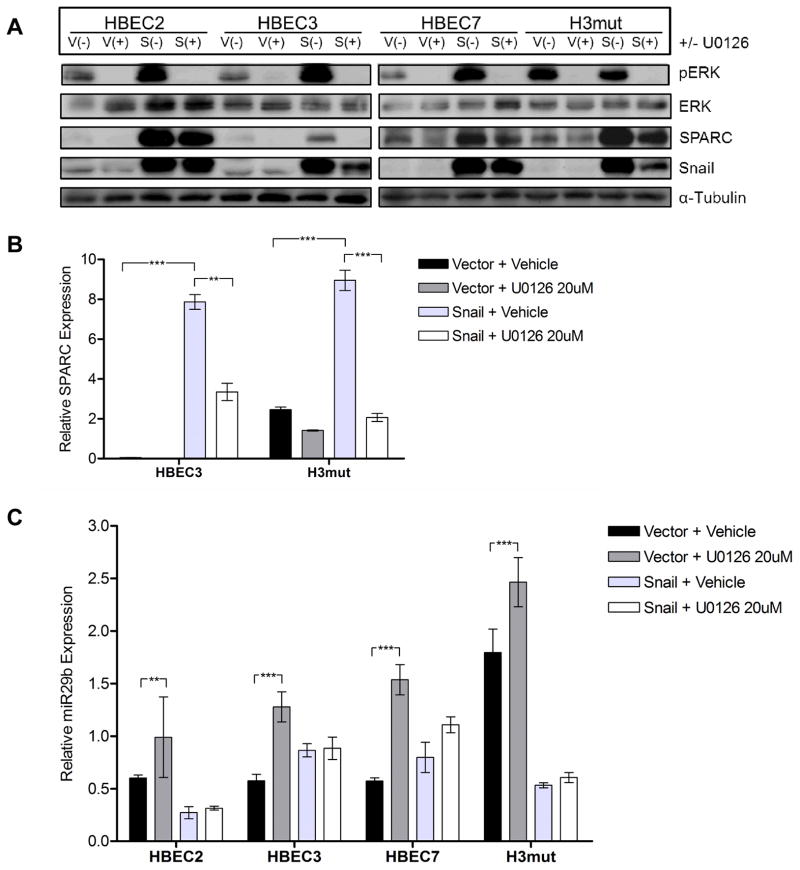
ERK1/2 was also indicated as a potential intermediate between Snail and SPARC (29–31). Immunoblot analysis of a panel of HBEC lines indicated that ERK phosphorylation was increased in Snail overexpressing lines (Fig. 1D) and may be an intermediate in the Snail-to-SPARC pathway. Suppression of ERK activation by the MEK1/2 phosphorylation inhibitor U0126 led to decreased mRNA and protein expression of SPARC in Snail overexpressing HBEC lines (Fig. 4A, B). In addition, U0126 exposure led to increased expression of the microRNA miR-29b (Fig. 4C).
Figure 4. ERK1/2 is phosphorylated downstream of Snail and upstream of SPARC.
(A) The cell lines HBEC2, HBEC3, HBEC7, and H3mutP53/KRAS (H3mut) with and without Snail overexpression (-V/-S) were treated with the MEK1/2 phosphorylation inhibitor U0126 (15uM) and evaluated for SPARC protein expression. Membranes were incubated with antibodies against Snail, pERK, total ERK1/2, SPARC, and α-tubulin. Protein levels were normalized to α-tubulin. (B) SPARC mRNA expression was evaluated following U0126 treatment as in (A). mRNA expression was normalized to GUSB. (C) miR-29b miRNA expression was evaluated following U0126 treatment as in (A). miRNA expression was normalized to RNU6b. (** = p < 0.001; *** = p < 0.0001)
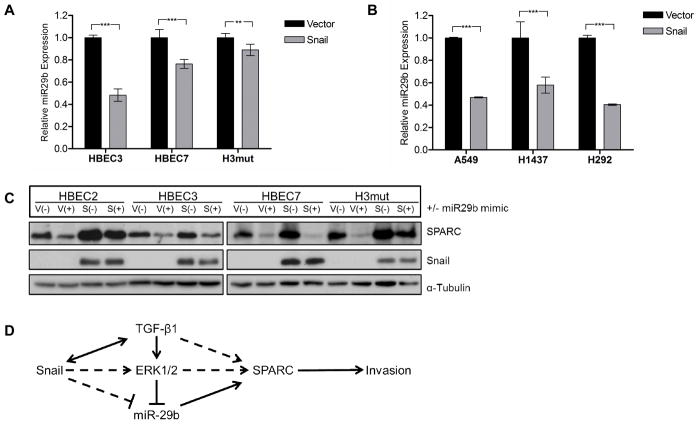
miR-29b is downregulated in Snail overexpressing cell lines
Our miRNA array data analysis indicated that miR-29b levels were significantly reduced in H441 and H1437 Snail overexpressing cell lines compared to vector controls (FC=2.3 and 2.6, respectively). Based on these findings, we hypothesized that miR-29b could be an intermediate regulator in the Snail to SPARC pathway. Analysis of the 3’UTR of SPARC mRNA by TargetScan software revealed three putative binding sequences for miR-29b. We compared the levels of miR-29b in three NSCLC and four HBEC cell lines ectopically expressing Snail to levels in the corresponding vector control lines by q-RT-PCR using a TaqMan® miRNA assay. In both types of cell lines, miR-29b expression levels were lower in Snail-overexpressing lines compared to vector controls (Fig. 5A, B), indicating that miR-29b is downregulated by Snail and may be involved in SPARC regulation. Furthermore, transfection of the cells with a miR-29b precursor mimic led to downregulation of SPARC protein in a panel of Snail-overexpressing HBEC lines (Fig. 5C). In addition, miR-29b expression was significantly upregulated following MEK/ERK inhibition (Fig. 4C), indicating a direct link between Snail, TGF-β, MEK/ERK, and miR-29b upstream of SPARC. We propose a regulatory pathway wherein Snail upregulates TGF-β in an autocrine or paracrine fashion, leading to activation of the MEK/ERK pathway, downregulation of miR-29b, and finally upregulation of SPARC (Fig. 5D).
Figure 5. miR-29b is downregulated in NSCLC cell lines overexpressing Snail.
(A) Total RNA was isolated from HBEC3-V/S, HBEC4-V/S, and H3mut-V/S cell lines. Expression levels of miR-29b were evaluated by q-RT-PCR using TaqMan primers. miRNA levels were normalized to RNU6b. (B) Total RNA was isolated from A549-V/S, H1437-V/S, and H292-V/S cell lines. Expression levels of miR-29b were evaluated by q-RT-PCR using TaqMan primers. miRNA levels were normalized to RNU6b. (C) The HBEC lines in (A) were stably transfected with a miR-29b precursor sequence and evaluated for expression of Snail and SPARC. Protein levels were normalized to α-tubulin. (D) We propose a regulatory pathway wherein Snail upregulates TGF-β in an autocrine or paracrine fashion, leading to activation of the MEK/ERK pathway, downregulation of miR-29b, and finally upregulation of SPARC. Snail may suppress miR-29b in both an ERK-dependent or -independent manner. (** = p < 0.001; *** = p < 0.0001)
Discussion
This is the first report demonstrating that Snail upregulates SPARC in models of both pulmonary premalignancy and established NSCLC. The necessity of SPARC for robust Snail-mediated invasion in both models suggests a role for both Snail and SPARC in the pathogenesis of NSCLC. Furthermore, we have identified a number of critical intermediates in this pathway. The deregulation of both the TGF-β and MEK/ERK pathways in malignancy is well known (32–34), although this is the first description of their deregulation in the context of Snail-mediated lung carcinogenesis.
To progress from in situ to metastatic disease, tumors must acquire characteristics that allow them to degrade and invade their local basement membrane and then migrate before forming a micro-metastatic focus. As part of this process, tumors undergo a series of events comprising the EMT program, wherein cells may transiently lose epithelial characteristics and become more mesenchymal in phenotype and molecular profile. The classical model of “linear” tumor progression proposes that a small population of cells within the invasive edge of an established tumor acquires characteristics necessary for EMT. However, recent findings suggest that micro-metastases may occur very early in the course of the disease, supporting a recently proposed model of “parallel” tumor progression in which metastatic dissemination occurs throughout the course of primary tumor development (35–39). This phenomenon may be especially relevant in the clinical subset of lung cancers that apparently disseminate early, leading to the clinical problem of metastatic disease following surgery for early stage disease. According to this model, metastases arise from a subpopulation of stem cell-like cells present at tumor initiation, which express proteins that induce EMT and confer putative stem cells with migratory and invasive capacity (36). The clinical course of patients presenting with metastatic disease following surgery appears to be consistent with this model. Parallel progression appears to be a frequent and major clinical problem in the treatment of NSCLC; as many as 40% of patients will have recurrence of lung cancer at metastatic sites following lung cancer resection (1), thought to be due to micro-metastatic disease that is below the level of detection by imaging studies at the time of surgery (35, 37, 40). Thus, our finding of the necessity of SPARC for Snail-mediated invasion in premalignant and established NSCLC models suggests that the Snail-SPARC axis may play a role in this process.
While our observation of the highly motile and invasive capacity of epithelial cells relative to tumor cells may appear counterintuitive, these findings are actually consistent with those in the recent literature that document the profoundly motile capacity of epithelial cells (38, 39, 41). This motile capacity is consistent with the movement characteristics of epithelial cells in embryonic development and wound closure. In a teleological view, these profound capacities endowed by the genetic programs of EMT are inherent in embryonic development and wound closure, which are requisite phenotypes whose evolutionary histories apparently far exceed that of malignancy.
The studies described here delineate a Snail-to-SPARC pathway involving a set of intermediates, several of which have been implicated in carcinogenesis. The molecules and pathways described here are likely not the only intermediaries in the Snail-to-SPARC pathway and further studies will be required to identify additional pathways as well as possible epigenetic changes involved. While we propose a largely linear pathway for Snail to SPARC expression, it is likely that the interactions among all of the involved intermediates are more complex. For example, SPARC is known to increase expression of Snail and therefore repress E-cadherin in melanoma progression (42). In hepatic stellate cells, TGF-β and SPARC are known to cooperate in an autocrine-feedback loop revealed by the finding that SPARC knockdown reduces TGF-β1 secretion while TGF-β1 treatment increases SPARC gene expression (43). As we demonstrate here, TGF-β1 and Snail also cooperate in an autocrine or paracrine feedback loop in lung cancer. Furthermore, miR-29b likely has many targets in addition to SPARC, with as many as 7,000 predicted by computational algorithms. These signaling pathways are known to have numerous effects on processes independent of invasion and metastatic progression, including cell cycle regulation, proliferation, and survival. Understanding the contribution of the Snail-SPARC axis to these phenotypes may yield a more thorough understanding of the pathogenesis of NSCLC.
Here we have identified molecular pathways in which altered genetic expression or activation in an in vitro system lay the groundwork for additional investigation of the potentially important Snail-SPARC pathway in human lung cancer.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Ying Lin and Zhe Jing for their technical assistance. We also acknowledge the following UCLA core facilities and their funding sources: the UCLA Genotyping and Sequencing Core and the Clinical Microarray Core.
Grant support: Research reported in this publication was supported in part by funding from the National Cancer Institute (#T32-CA009120-36; S. Dubinett, J. Grant) (#U01CA152751; S. Dubinett, M. Fishbein, T. Walser), Department of Defense (#W81XWH-10-1-1006; S. Dubinett, M. Fishbein, K. Krysan, J. Minna, T. Walser), UCLA Clinical and Translational Science Institute (#UL1TR000124; S. Dubinett), Merit Review Research Funds from the Department of Veteran Affairs (#5I01BX000359; S. Dubinett), Lung Cancer SPORE (#P50CA70907; J. Minna, J. Shay), NASA NSCOR (#NNX11AC54G; J. Minna, J. Shay) and Tobacco-Related Disease Research Program (#18FT-0060; T. Walser) (#20KT-0055; T. Walser) (#21DT-0013; J. Grant).
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest to disclose.
Literature Cited
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013 Jan;63(1):11–30. doi: 10.3322/caac.21166. Epub 2013/01/22.eng. [DOI] [PubMed] [Google Scholar]
- 2.Heinrich EL, Walser TC, Krysan K, Liclican EL, Grant JL, Rodriguez NL, et al. The inflammatory tumor microenvironment, epithelial mesenchymal transition and lung carcinogenesis. Cancer Microenviron. 2012 Apr;5(1):5–18. doi: 10.1007/s12307-011-0089-0. Epub 2011/09/17.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee G, Walser TC, Dubinett SM. Chronic inflammation, chronic obstructive pulmonary disease, and lung cancer. Curr Opin Pulm Med. 2009 Jul;15(4):303–7. doi: 10.1097/MCP.0b013e32832c975a. Epub 2009/05/07.eng. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. Epub 2011/03/08.eng. [DOI] [PubMed] [Google Scholar]
- 5.Dohadwala M, Yang SC, Luo J, Sharma S, Batra RK, Huang M, et al. Cyclooxygenase-2-dependent regulation of E-cadherin: prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res. 2006 May 15;66(10):5338–45. doi: 10.1158/0008-5472.CAN-05-3635. Epub 2006/05/19.eng. [DOI] [PubMed] [Google Scholar]
- 6.Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of Ras signaling in the induction of snail by transforming growth factor-beta. J Biol Chem. 2009 Jan 2;284(1):245–53. doi: 10.1074/jbc.M804777200. eng. [DOI] [PubMed] [Google Scholar]
- 7.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004 Jan;24(1):306–19. doi: 10.1128/MCB.24.1.306-319.2004. Epub 2003/12/16.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003 Jun 6;278(23):21113–23. doi: 10.1074/jbc.M211304200. Epub 2003/04/01.eng. [DOI] [PubMed] [Google Scholar]
- 9.Dhasarathy A, Phadke D, Mav D, Shah RR, Wade PA. The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer. PLoS ONE. 2011;6(10):e26514. doi: 10.1371/journal.pone.0026514. Epub 2011/10/27.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav A, Kumar B, Datta J, Teknos TN, Kumar P. IL-6 Promotes Head and Neck Tumor Metastasis by Inducing Epithelial-Mesenchymal Transition via the JAK-STAT3-SNAIL Signaling Pathway. Mol Cancer Res. 2011 Dec;9(12):1658–67. doi: 10.1158/1541-7786.MCR-11-0271. Epub 2011/10/07.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng P, Meng HM, Gao WZ, Chen L, Liu XH, Xiao ZQ, et al. Snail as a key regulator of PRL-3 gene in colorectal cancer. Cancer Biol Ther. 2011 Oct 15;12(8):742–9. doi: 10.4161/cbt.12.8.15981. Epub 2011/08/04.eng. [DOI] [PubMed] [Google Scholar]
- 12.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008 May 16;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. Epub 2008/05/20.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Li Y, Wang K, Wang Y, Yin W, Li L. P38/NF-kappaB/Snail Pathway Is Involved in Caffeic Acid-Induced Inhibition of Cancer Stem Cells-Like Properties and Migratory Capacity in Malignant Human Keratinocyte. PLoS ONE. 2013;8(3):e58915. doi: 10.1371/journal.pone.0058915. Epub 2013/03/22.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii G, Hashimoto H, Atsumi N, Hoshino A, Ochiai A. Morphophenotype of floating colonies derived from a single cancer cell has a critical impact on tumor-forming activity. Pathol Int. 2013 Jan;63(1):29–36. doi: 10.1111/pin.12018. Epub 2013/01/30.eng. [DOI] [PubMed] [Google Scholar]
- 15.Yanagawa J, Walser TC, Zhu LX, Hong L, Fishbein MC, Mah V, et al. Snail promotes CXCR2 ligand-dependent tumor progression in non-small cell lung carcinoma. Clin Cancer Res. 2009 Nov 15;15(22):6820–9. doi: 10.1158/1078-0432.CCR-09-1558. Epub 2009/11/06.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Termine JD, Kleinman HK, Whitson SW, Conn KM, McGarvey ML, Martin GR. Osteonectin, a bone-specific protein linking mineral to collagen. Cell. 1981 Oct;26(1 Pt 1):99–105. doi: 10.1016/0092-8674(81)90037-4. Epub 1981/10/01.eng. [DOI] [PubMed] [Google Scholar]
- 17.Lane TF, Sage EH. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 1994 Feb;8(2):163–73. Epub 1994/02/01.eng. [PubMed] [Google Scholar]
- 18.Framson PE, Sage EH. SPARC and tumor growth: where the seed meets the soil? J Cell Biochem. 2004 Jul 1;92(4):679–90. doi: 10.1002/jcb.20091. Epub 2004/06/24.eng. [DOI] [PubMed] [Google Scholar]
- 19.Koukourakis MI, Giatromanolaki A, Brekken RA, Sivridis E, Gatter KC, Harris AL, et al. Enhanced expression of SPARC/osteonectin in the tumor-associated stroma of non-small cell lung cancer is correlated with markers of hypoxia/acidity and with poor prognosis of patients. Cancer Res. 2003 Sep 1;63(17):5376–80. Epub 2003/09/23.eng. [PubMed] [Google Scholar]
- 20.Sato M, Larsen JE, Lee W, Sun H, Shames DS, Dalvi MP, et al. Human Lung Epithelial Cells Progressed to Malignancy through Specific Oncogenic Manipulations. Mol Cancer Res. 2013 Jun;11(6):638–50. doi: 10.1158/1541-7786.MCR-12-0634-T. Epub 2013/03/02.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004 Dec 15;64(24):9027–34. doi: 10.1158/0008-5472.CAN-04-3703. Epub 2004/12/18.eng. [DOI] [PubMed] [Google Scholar]
- 22.Vaughan MB, Ramirez RD, Wright WE, Minna JD, Shay JW. A three-dimensional model of differentiation of immortalized human bronchial epithelial cells. Differentiation. 2006 Apr;74(4):141–8. doi: 10.1111/j.1432-0436.2006.00069.x. Epub 2006/05/11.eng. [DOI] [PubMed] [Google Scholar]
- 23.Minna JD, Girard L, Sato M, et al. Molecular pathogenesis of lung cancer with translation to the clinic. J Thorac Oncol. 2007 Aug;2(8, Supplement 4):S178–S86. [Google Scholar]
- 24.Araya J, Cambier S, Markovics JA, Wolters P, Jablons D, Hill A, et al. Squamous metaplasia amplifies pathologic epithelial-mesenchymal interactions in COPD patients. J Clin Invest. 2007 Nov;117(11):3551–62. doi: 10.1172/JCI32526. Epub 2007/10/30.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazra S, Dubinett SM. Ciglitazone mediates COX-2 dependent suppression of PGE2 in human non-small cell lung cancer cells. Prostaglandins Leukot Essent Fatty Acids. 2007 Jul;77(1):51–8. doi: 10.1016/j.plefa.2007.05.006. Epub 2007/08/21.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001 Jan 2;98(1):31–6. doi: 10.1073/pnas.011404098. Epub 2001/01/03.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olmeda D, Moreno-Bueno G, Flores JM, Fabra A, Portillo F, Cano A. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res. 2007 Dec 15;67(24):11721–31. doi: 10.1158/0008-5472.CAN-07-2318. Epub 2007/12/20.eng. [DOI] [PubMed] [Google Scholar]
- 29.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010 Apr 9;38(1):114–27. doi: 10.1016/j.molcel.2010.02.020. Epub 2010/04/14.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenouille N, Puissant A, Dufies M, Robert G, Jacquel A, Ohanna M, et al. Persistent activation of the Fyn/ERK kinase signaling axis mediates imatinib resistance in chronic myelogenous leukemia cells through upregulation of intracellular SPARC. Cancer Res. 2010 Dec 1;70(23):9659–70. doi: 10.1158/0008-5472.CAN-10-2034. Epub 2010/11/26.eng. [DOI] [PubMed] [Google Scholar]
- 31.Shields MA, Dangi-Garimella S, Krantz SB, Bentrem DJ, Munshi HG. Pancreatic cancer cells respond to type I collagen by inducing snail expression to promote membrane type 1 matrix metalloproteinase-dependent collagen invasion. J Biol Chem. 2011 Mar 25;286(12):10495–504. doi: 10.1074/jbc.M110.195628. Epub 2011/02/04.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vicent S, Lopez-Picazo JM, Toledo G, Lozano MD, Torre W, Garcia-Corchon C, et al. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br J Cancer. 2004 Mar 8;90(5):1047–52. doi: 10.1038/sj.bjc.6601644. Epub 2004/03/05.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT) Respir Res. 2005;6:56. doi: 10.1186/1465-9921-6-56. Epub 2005/06/11.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashida T, Decaestecker M, Schnaper HW. Cross-talk between ERK MAP kinase and Smad signaling pathways enhances TGF-beta-dependent responses in human mesangial cells. FASEB J. 2003 Aug;17(11):1576–8. doi: 10.1096/fj.03-0037fje. Epub 2003/06/26.eng. [DOI] [PubMed] [Google Scholar]
- 35.Varlotto JM, Recht A, Flickinger JC, Medford-Davis LN, Dyer AM, Decamp MM. Factors associated with local and distant recurrence and survival in patients with resected nonsmall cell lung cancer. Cancer. 2009 Mar 1;115(5):1059–69. doi: 10.1002/cncr.24133. Epub 2009/01/20.eng. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Garcia I. The crossroads of oncogenesis and metastasis. N Engl J Med. 2009 Jan 15;360(3):297–9. doi: 10.1056/NEJMcibr0808031. Epub 2009/01/16.eng. [DOI] [PubMed] [Google Scholar]
- 37.Varlotto JM, Medford-Davis LN, Recht A, Flickinger J, Yao N, Hess C, et al. Identification of Stage I Non-small Cell Lung Cancer Patients at High Risk for Local Recurrence Following Sublobar Resection. Chest. 2013 May 1;143(5):1365–77. doi: 10.1378/chest.12-0710. Epub 2013/05/30.eng. [DOI] [PubMed] [Google Scholar]
- 38.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008 Jan;13(1):58–68. doi: 10.1016/j.ccr.2007.12.003. Epub 2008/01/03.eng. [DOI] [PubMed] [Google Scholar]
- 39.Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008 Sep 26;321(5897):1841–4. doi: 10.1126/science.1161621. Epub 2008/08/30.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dijkman BG, Schuurbiers OC, Vriens D, Looijen-Salamon M, Bussink J, Timmer-Bonte JN, et al. The role of (18)F-FDG PET in the differentiation between lung metastases and synchronous second primary lung tumours. Eur J Nucl Med Mol Imaging. 2010 Nov;37(11):2037–47. doi: 10.1007/s00259-010-1505-2. Epub 2010/06/10.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung WC, Chen SH, Paul CD, Stroka KM, Lo YC, Yang JT, et al. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J Cell Biol. 2013 Sep 2;202(5):807–24. doi: 10.1083/jcb.201302132. Epub 2013/08/28.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert G, Gaggioli C, Bailet O, Chavey C, Abbe P, Aberdam E, et al. SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res. 2006 Aug 1;66(15):7516–23. doi: 10.1158/0008-5472.CAN-05-3189. Epub 2006/08/04.eng. [DOI] [PubMed] [Google Scholar]
- 43.Atorrasagasti C, Aquino JB, Hofman L, Alaniz L, Malvicini M, Garcia M, et al. SPARC downregulation attenuates the profibrogenic response of hepatic stellate cells induced by TGF-beta1 and PDGF. Am J Physiol Gastrointest Liver Physiol. 2011 May;300(5):G739–48. doi: 10.1152/ajpgi.00316.2010. Epub 2011/02/12.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.