Abstract
The six members of the Receptor Expression Enhancing Protein (REEP) family were originally identified based on their ability to enhance heterologous expression of olfactory receptors and other difficult to express G protein-coupled receptors. Interestingly, REEP1 mutations have been linked to neurodegenerative disorders of upper and lower motor neurons, hereditary spastic paraplegia (HSP) and distal hereditary motor neuropathy type V (dHMN-V). The closely related REEP2 isoform has not demonstrated any such disease linkage. Previous research has suggested that REEP1 mRNA is ubiquitously expressed in brain, muscle, endocrine, and multiple other organs, inconsistent with the neurodegenerative phenotype observed in HSP and dHMN-V. To more fully examine REEP1 expression, we developed and characterized a new REEP1 monoclonal antibody for both immunoblotting and immunofluorescent microscopic analysis. Unlike previous RT-PCR studies, immunoblotting demonstrated that REEP1 protein was not ubiquitous; its expression was restricted to neuronal tissues (brain, spinal cord) and testes. Gene expression microarray analysis demonstrated REEP1 and REEP2 mRNA expression in superior cervical and stellate sympathetic ganglia tissue. Furthermore, expression of endogenous REEP1 was confirmed in cultured murine sympathetic ganglion neurons by RTPCR and immunofluorescent staining, with expression occurring between Day 4 and Day 8 of culture. Lastly, we demonstrated that REEP2 protein expression was also restricted to neuronal tissues (brain and spinal cord) and tissues that exhibit neuronal-like exocytosis (testes, pituitary, and adrenal gland). In addition to sensory tissues, expression of the REEP1/REEP2 subfamily appears to be restricted to neuronal and neuronal-like exocytotic tissues, consistent with neuronally restricted symptoms of REEP1 genetic disorders.
Keywords: receptor expression enhancing protein, neurodegeneration, sympathetic ganglion neuron, hereditary spastic paraplegia, distal hereditary motor neuropathy type V
1. Introduction
Many neuronally relevant G protein-coupled receptors (GPCRs) have proven difficult to express in heterologous cell systems (e.g. HEK293), including olfactory (ORs), taste (TRs), and vomeronasal (VRs) receptors (Behrens et al., 2006; Dulac and Axel, 1995; Mombaerts, 2004). Similar to ORs and TRs, α2C adrenergic receptors (ARs) are also not easily expressed in nonneuronal cells, however, α2A ARs do not show such difficulty. However, heterologous expression of α2C ARs in neuronal cell lines (e.g. PC12 cells) led to significant plasma membrane expression, suggesting a role for a neuronal-specific accessory protein(s) (Angelotti et al., 2010; Hurt et al., 2000). Furthermore, α2A and α2C ARs show differential localization within cultured sympathetic ganglion neurons (SGN) (Brum et al., 2006). It was hypothesized that neuronal or sensory cells may express cell-specific accessory proteins necessary for proper membrane targeting of these GPCRs.
While attempting to identify possible accessory proteins, Matsunami and colleagues identified two new families of proteins that appeared to enhance surface expression of ORs, the Receptor Expression Enhancing Protein (REEP) and Receptor Transporting Protein (RTP) families (Saito et al., 2004). In total, six REEPs and four RTPs were identified. The REEP family can be subdivided into two subfamilies REEP1–4 and REEP5–6, with the latter showing the most homology with the yeast homolog Yop1 (Park et al., 2010). Evolutionary analysis has suggested that REEP1–4 evolved from two rounds of whole genome duplication, starting with creation of REEP1/2 and REEP3/4 subfamilies, and eventually REEPs 1–4 (Tinti et al., 2012). Thus, it is plausible that REEP1 and REEP2 evolved to serve tissue-specific functions.
RTP homologs have not been identified outside of vertebrates; however, REEPs belong to a much larger Yip (Ypt interacting protein) family, which has been conserved in invertebrates, yeast, plants, and mammals (Pfeffer and Aivazian, 2004; Tinti et al., 2012). Various Yip family members have been shown to interact directly with Rab GTPases and ER/Golgi vesicle proteins to regulate intracellular trafficking and targeting of cargo proteins within yeast and neurons (Al Awabdh et al., 2012; Brands and Ho, 2002; Calero et al., 2001; Carrel et al., 2008; Heidtman et al., 2003; Martincic et al., 1997). Recent work has demonstrated that REEP isoforms are important determinants of ER tubular structure (Park et al., 2010; Voeltz et al., 2006) and can be further described as ER membrane shaping adapter proteins, based upon their interactions with 14-3-3 proteins and GPCRs, such as α2C ARs (Björk et al., 2013; Johnson et al., 2011). The recent discovery that multiple REEP1 mutations are linked to the neurodegenerative disorders hereditary spastic paraplegia (HSP) and distal hereditary motor neuropathy type V (dHMN-V) has further increased interest in this family of proteins (Beetz et al., 2008; Beetz et al., 2012; Zuchner et al., 2006).
Northern blot, in situ hybridization, RT-PCR, and immunofluorescent analysis has determined REEP expression patterns in various tissues, often with conflicting results. Consistent with enhancement of OR and TR expression, various isoforms were found to be expressed in olfactory and vomeronasal epithelium, circumvallate papillae (tongue), brain, and cultured cortical neurons (Behrens et al., 2006; Ilegems et al., 2010; Park et al., 2010; Saito et al., 2004). Other RT-PCR studies suggested that REEP1 was ubiquitously expressed in brain, muscle, endocrine, and multiple other organs (Zuchner et al., 2006). These latter results ran counter to the original hypothesis that REEPs were tissue-specific accessory proteins necessary for expression of specific GPCRs and appeared counterintuitive to the neurodegenerative phenotypes of HSP and dHMN-V. To date, the only phenotype observed with REEP1 mutations is neurodegenerative motor neuron disease; no other organ system involvement has been observed (Beetz et al., 2008; Beetz et al., 2012). In order to understand cell-type specific roles of REEP1 and REEP2 in neuronal GPCR trafficking and neurological disease, we examined their endogenous expression in neuronal and non-neuronal cell lines, neurons, and tissues. A newly created REEP1 monoclonal antibody (mAb) was first characterized by immunoblotting and immunofluorescent staining, in order to ensure its specificity, as outlined by others (Rhodes and Trimmer, 2006). It was then utilized to examine REEP1 expression in various cell lines and native mouse tissues. Similar studies were done using a commercially available polyclonal REEP2 antisera. DNA microarray analysis revealed that REEP1 and REEP2 mRNA were expressed in murine sympathetic neurons, specifically superior cervical (SCG) and stellate (SG) ganglia, which are major sites of α2 AR expression. Finally, endogenous REEP1 expression in cultured sympathetic ganglion neurons (SGN) was examined by immunofluorescent staining and correlated with RT-PCR data. Together, our results demonstrated that REEP1 and REEP2 were expressed only in neuronal or neuronal-like exocytotic tissues, and that REEP1 expression in cultured SGN is temporally regulated.
2. Results
2.1 REEP1 monoclonal antibody specificity
One limitation of RT-PCR and other mRNA-based methods is that they may demonstrate expression of an mRNA encoding a protein, but not necessarily that the protein is expressed nor correlated with the level of protein expression (Gry et al., 2009; Gygi et al., 1999). Therefore, we developed a monoclonal antibody (mAb) against REEP1 in order to examine REEP1 protein expression in various tissues and cell types by immunoblotting and immunofluorescent analysis. The anti-REEP1 monoclonal antibody was co-developed with the UC Davis/NIH NeuroMab Facility (NIH grant U24NS050606). The antibody was generated against a purified GST-fusion protein encoding amino acids #111-201 of mouse REEP1 carboxyl terminus (GST-REEP1CT). NeuroMab identified multiple clones and clone N345/51 was selected for production based upon its high titer, sensitivity, and selectivity, as characterized by immunoblotting against whole brain protein (data not shown).
To demonstrate specificity of REEP1 mAb clone N345/51 and a commercially available REEP2 antibody, HEK293A cells were transfected with Flag-REEP1, -REEP2, and –REEP6 and analyzed by immunoblot analysis (Figure 1A/B). The REEP1 mAb only detected Flag-REEP1 (calculated Mr = 23.4 kDa); no endogenous REEP1 (calculated Mr = 22.3 kDa) expression was noted. However, the antisera against REEP2 did identify both Flag-REEP2 (calculated Mr = 29.4 kDa) and endogenous REEP2 (calculated Mr = 28.3 kDa) in HEK293A cells, consistent with previous RT-PCR data (Behrens et al., 2006). Despite similar levels of REEP expression (Figure 1C), neither antibody cross-reacted with any other REEPs tested.
Figure 1.
Determination of REEP1 and REEP2 antisera specificity
Whole cell lysates from various Flag-REEP1, -REEP2, -REEP6 transfected HEK293A cells (seventy-five micrograms protein/lane) were analyzed by immunoblot analysis with a new REEP1 mAb (NeuroMab Clone N345/51) or commercially available REEP2 polyclonal antisera to determine their REEP specificity. A. The REEP1 mAb detected only transfected Flag-REEP1 (calculated Mr = 23.4 kDa) (Lane 2); no endogenous REEP1 (calculated Mr = 22.3 kDa) expression was noted in untransfected cells (Lane 1). REEP1 mAb did not cross-react with Flag-REEP2 (Lane 3) or Flag-REEP6 (Lane 4). B. REEP2 polyclonal antisera identified both transfected Flag-REEP2 (calculated Mr = 29.4 kDa) (Lane 3) and endogenous REEP2 (calculated Mr = 28.3 kDa) in HEK293A cells (Lanes 1–4), but not Flag-REEP1 or Flag-REEP6. Note that both REEP1 and REEP2 demonstrate anomalous migration relative to their calculated molecular weights. C. Similar amounts of transfected Flag-REEP1, -REEP2, and –REEP6 were expressed, as detected by anti-Flag epitope mAb (M2). Molecular weight markers (kDa) are shown to right. Representative of three immunoblots and transfections.
2.2 Immunofluorescent characterization of REEP1 monoclonal antibody
HEK293A cells were transfected with cDNAs encoding Flag-REEP1 (Figure 2A) or untagged-REEP1 (Figure 2B) and examined by immunofluorescent imaging (anti-Flag M2 and REEP1 mAb, respectively). Untransfected cells did not show any staining when examined by M2 or REEP1 mAb (data not shown), consistent with immunoblot analysis above and prior RT-PCR data (Behrens et al., 2006; Park et al., 2010). Previously, it was suggested that REEP1 co-localized with mitochondria based upon MitoTracker Red staining (Zuchner et al., 2006), however similar co-localization studies with MitoTracker Red CMXROS (Figure 2A/B) did not suggest co-localization of REEP1 with mitochondria.
Figure 2.
Immunofluorescent characterization of REEP1 mAb
HEK293A cells were transfected with either Flag-REEP1 or native, untagged-REEP1 cDNAs and analyzed by immunofluorescent wide-field microscopy with anti-Flag mAb (M2) and a new REEP1 mAb respectively. Cells were counterstained with either MitoTracker Red CMXROS or anti-calreticulin antisera, as described in Experimental Procedures. A. Immunofluorescent staining for Flag-REEP1 (M2 mAb) expressed in HEK293A cells revealed a fine reticular network that demonstrated a different localization pattern than mitochondria counterstained with MitoTracker Red CMXROS. B. REEP1 mAb detected a similar reticular staining pattern for untagged-REEP1, as seen above for Flag-REEP1. Mitochondria were counterstained with MitoTracker Red CMXROS. C. The fine reticular immunofluorescent pattern seen with M2 staining of Flag-REEP1 showed a similar localization as the ER marker protein calreticulin. D. REEP1 mAb delineated a similar reticular pattern for untagged-REEP1, when compared with anti-calreticulin staining. Bar = 10 μm. Representative of three transfections.
By using a series of biochemical and immunofluorescent methods, we recently demonstrated that REEP1, REEP2, and REEP6 are ER resident proteins (Björk et al., 2013). Therefore, we examined REEP1 localization relative to an ER resident protein (calreticulin) in order to further characterize our new REEP1 mAb. In HEK293A cells transfected with Flag-REEP1, anti-Flag mAb (M2) identified a fine intracellular network that was similar in localization to that identified with anti-calreticulin antisera (Figure 2C), consistent with our previously published ER localization studies (Björk et al., 2013; Park et al., 2010). Likewise, untagged-REEP1 transfected HEK293A cells stained with REEP1 mAb identified a similar intracellular network as calreticulin immunofluorescent staining (Figure 2D). Therefore, the new REEP1 mAb demonstrated specificity for both immunoblot and immunofluorescent applications. Attempts to detect endogenous REEP2 expression in HEK293A cells by immunofluorescence with commercial polyclonal REEP2 antisera were unsuccessful due to non-specific nuclear staining (data not shown).
2.3 Selection of cell lines for study
α2A and α2C ARs are highly homologous GPCRs that traffic to the plasma membrane with different efficiencies, following heterologous expression in different cell lines. For example, transfection of α2A AR cDNA into HEK293 and NRK cells led to robust surface expression (> 90% of total α2A AR expression), whereas similar experiments with α2C AR cDNA led to predominant endoplasmic reticulum (ER) retention (≈ 75% of total α2C AR expression) (Angelotti et al., 2010; Daunt et al., 1997; Hurt et al., 2000). However, α2C ARs exhibited strong plasma membrane localization when heterologously expressed in neuronal cell lines such as PC12 and AtT20 cells (Hurt et al., 2000). Furthermore, α2A and α2C ARs show differential localization within cultured sympathetic ganglion neurons (SGN). α2C ARs are localized exclusively to pre-synaptic sites, whereas α2A ARs are found at extra-synaptic sites (Brum et al., 2006). Therefore, tissue-specific factors may be important determinants of subcellular localization of α2C ARs within SGN as well as plasma membrane expression in neuronal cell lines.
Given the similar difficulties with heterologous expression of ORs/TRs and α2C ARs in non-neuronal cell lines, we determined the expression patterns of REEP1 and REEP2 in cell lines commonly utilized for our previous α2C AR studies (Angelotti et al., 2010; Brum et al., 2006; Daunt et al., 1997; Hurt et al., 2000), including HEK293 (adenovirus transformed human embryonic kidney), NRK (rat renal epithelium), PC12 (rat adrenal pheochromocytoma) and Rat1 (rat fibroblast). In addition, HEK293A cells, a derivative of HEK293 cells, were studied. We have utilized them previously for α2 AR trafficking studies, since they are flatter and larger than wild-type HEK293 cells, enhancing immunofluorescent analysis (Angelotti et al., 2010).
2.4 REEP1 and REEP2 protein expression in various cell lines
An immunoblot of whole cell lysates from various wild-type cell lines was probed with the REEP1 mAb (Figure 3A). None of the cell lines tested (HEK293, HEK293A, NRK, PC12, or Rat1 cells) expressed REEP1. REEP1 was only evident in HEK293A cells transfected with Flag-REEP1 (positive control). REEP1 mAb demonstrated high specificity, detecting a single protein band (Supplemental Figure 1). REEP2 expression in various cell lines was also examined using the commercially available polyclonal antisera (Figure 3B, Supplemental Figure 1). REEP2 was expressed in wild type HEK293 and HEK293A cells and PC12 cells, but was not expressed in NRK or Rat1 cells. Transfected Flag-REEP2 was also detected (positive control).
Figure 3.

Expression of REEP1 and REEP2 proteins in various cell lines
Left. Immunoblot analysis of REEP expression in commonly used cell lines. A. Representative immunoblot analysis of REEP1 expression using a new monoclonal antibody (NeuroMab Clone N345/51). No cell lines examined expressed endogenous REEP1. Immunoreactivity in Lane 6 represents transfected Flag-REEP1 (positive control). B. Representative immunoblot analysis of REEP2 expression in various cell lines using polyclonal REEP2 antisera. Lower molecular weight band correlated with endogenous REEP2, which was found in HEK293, HEK293A, and PC12 cells. Higher molecular weight band in Lane 7 represents transfected Flag-REEP2 (positive control). Larger immunoblots with loading controls are shown elsewhere (Supplemental Figure 1). Molecular weight markers (kDa) are shown to left. One hundred (Lanes 1–5) or forty (Lanes 6–7) micrograms of protein were loaded per lane. Representative of three immunoblots and transfections. Right: Analysis of REEP1 and REEP2 expression in NGF-treated PC12 cells. PC12 cells were treated with NGF for either 0 or 72 hours and immunoblot analysis was performed on whole cell lysates (75 microgram protein/lane). Flag-REEP1 transfected HEK29A or untransfected HEK293A membranes were also loaded as positive controls (50 microgram protein/lane). C. NGF-treatment did not induce REEP1 protein expression. D. REEP2 protein expression levels did not change following NGF treatment. Representative of three immunoblots and separate NGF treatments.
To determine if nerve growth factor (NGF)-induction of PC12 neurite extension enhanced REEP1 or REEP2 expression, cultured PC12 cells were treated for 72 hrs with NGF. Immunoblot analysis with REEP1 mAb did not demonstrate any induction of REEP1 (Figure 3C), whereas the level of REEP2 expression was unchanged (Figure 3D).
2.5 REEP1 and REEP2 murine tissue expression
Whole lysates of multiple mouse tissues were examined by immunoblotting using REEP1 mAb and REEP2 polyclonal antisera, in order to determine their endogenous tissue expression. REEP1 protein was detected only in brain, spinal cord, and testes (Figure 4A), whereas REEP2 protein was detected in brain, spinal cord, adrenal gland, pituitary, and testes (Figure 4B). Neither REEP1 nor REEP2 were detected in skeletal muscle, heart, colon, spleen, pancreas, kidney, liver, or lung. Interestingly, neither REEP1 nor REEP2 were expressed in cultured astrocytes. As described by others (Dittmer and Dittmer, 2006; Ferguson et al., 2005), no protein loading control has been identified to examine loading of different tissues, since no “housekeeping” protein is expressed at equivalent levels between different tissues. Therefore to assess protein loading of our immunoblots, we examined several well-established protein controls including GAPDH (Figure 4C), as well as β-tubulin, β-actin, and calnexin (Supplemental Figure 2). Though GAPDH was expressed at different levels between tissues, it did demonstrate adequate protein loading amongst various tissues, most notably within neuronal tissues and skeletal muscle and solid organs (e.g. lung, liver, kidney). Taken together, the overall loading control immunoblots show somewhat similar protein loading, despite the variable expression levels amongst tissues, as described above.
Figure 4.
REEP1 and REEP2 protein expression in multiple tissues
Representative immunoblot analysis of REEP1 and REEP2 expression in various tissues using REEP1 monoclonal antibody and REEP2 polyclonal antisera. A. REEP1 protein was detected only in brain, spinal cord, and testes. B. REEP2 protein was found in brain, spinal cord, adrenal gland, testes, and pituitary. Note absence of both REEP1 and REEP2 in astrocytes, skeletal muscle, heart, and other tissues. C. GAPDH was utilized as a positive loading control. Note differential expression seen amongst various tissues (Lanes 1–14), as described previously (Dittmer and Dittmer, 2006; Ferguson et al., 2005). Other loading controls were also examined (Supplemental Figure 3). Molecular weight markers (kDa) are shown to right. Representative of two immunoblots each.
2.6 REEP family gene expression analysis in murine sympathetic ganglia tissues
Due to the small size of sympathetic ganglia tissue, a major site of endogenous α2 AR expression, immunoblot analysis of REEP isoform expression was not possible. Therefore, we utilized DNA microarray to examine REEP gene expression profiles in sympathetic cervical (SCG) and stellate (SG) ganglia tissue from adult mice (Figure 5). mRNA was isolated from dissected SCG and SG tissue and examined by DNA microarray following reverse transcription. Consistent with our prior results suggesting a neuronal expression pattern, both REEP1 and REEP2 mRNA were detected in these two sympathetic neuronal tissues, however REEP1 was expressed at higher levels than REEP2. Interestingly, REEP5 mRNA was also detected at similar levels as REEP1, whereas REEP3 mRNA showed moderate expression, similar to REEP2. Neither REEP4 or REEP6 mRNA were appreciably expressed.
Figure 5.
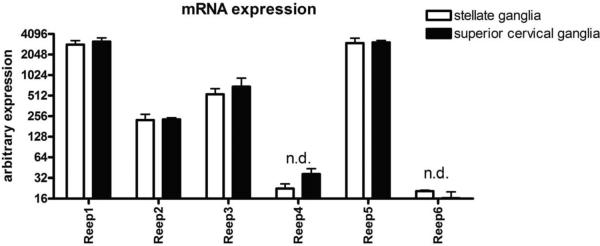
REEP gene expression analysis in superior cervical and stellate sympathetic ganglia
DNA microarray analysis was performed on total RNA isolated from superior cervical (SCG) and stellate (SG) sympathetic ganglia (n = 2, n = 3 respectively), to determine relative mRNA expression of various REEP family members. mRNA expression is represented on the y-axis in arbitrary fluorescence units (± SEM). In both SCG and SG, REEP1 and REEP5 demonstrated high expression, while REEP2 and REEP3 showed moderate expression levels. REEP4 and REEP6 were not detected (n.d.).
2.7 REEP expression in cultured murine SGN
The differential localization of α2A and α2C ARs within cultured SGN is temporally regulated (Brum et al., 2006). α2A ARs are readily found at extra-synaptic sites on the cell membrane at Day 1 of culture and continue to be found at this localization throughout Day 16. However, functional α2C ARs are not found at the plasma membrane until Day 8, and they do not reach their pre-synaptic site of action until Day 16, suggesting differential expression of accessory trafficking proteins. Using heterologous expression in HEK293A cells, we have recently demonstrated that REEP1 and REEP2 interact preferentially with α2C, but not α2A ARs, enhancing the production of a minimally glycosylated form of α2C AR in the ER (Björk et al., 2013).
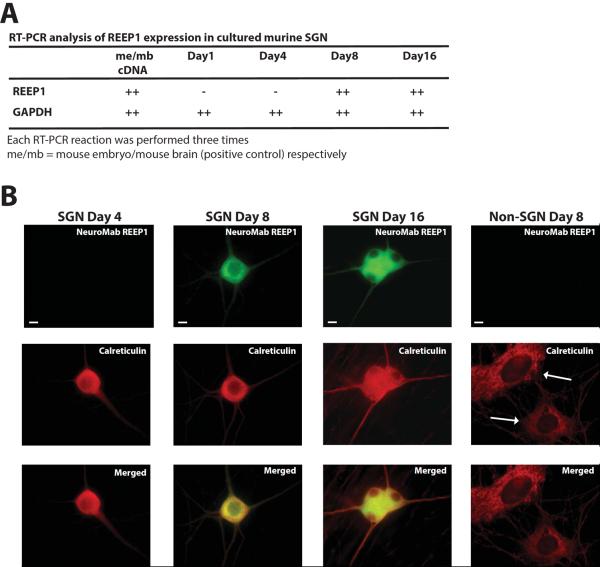
If REEP1 is involved with α2C AR trafficking in cultured SGN, then it should be expressed in these neurons. However, immunoblot analysis was not possible due to the small number of SGNs that can be obtained by dissection (similar to SCG and SG tissues). Therefore, RT-PCR analysis of REEP1 expression was carried out in SGN cultures at Days 1, 4, 8, and 16 (Figure 6A). No REEP1 mRNA expression was observed at Day 1 or Day 4, however REEP1 mRNA was detected at Day 8 and Day 16 of culture. To verify REEP1 protein expression in SGN, immunofluorescence staining was performed with the REEP1 mAb (Figure 6B). At Day 4, no REEP1 immunoreactivity was apparent, however, all SGN exhibited intense REEP1 immunofluorescence at Day 8 and Day 16, consistent with RT-PCR expression analysis. SGN cultures contain a mixture of SGN and supporting cells, predominantly fibroblasts, satellite glial cells, and Schwann cells (Hanani, 2010; Lein et al., 2002). Calreticulin was used to co-stain SGN and other cell types for comparison to REEP1. No REEP1 staining was evident within non-SGN supporting cells at Day 8 and Day 16 (data not shown), though calreticulin staining was noted, thus confirming that the RT-PCR analysis detected REEP1 mRNA expression in SGN only.
Figure 6.
REEP1 expression in cultured SGN
A. RT-PCR analysis of REEP1 expression in cultured SGN. RT-PCR was performed from total RNA isolated at various days of culture (n=3). Note that expression of REEP1 mRNA occurred between Day 4 and Day 8 of culture. GAPDH was utilized as a positive control for all reactions. B. Representative (n=3) immunofluorescent analysis of REEP1 expression in cultured SGN using REEP1 monoclonal antibody. Far Left. At Day 4 of culture, no REEP1 expression was seen in any cultured SGN examined. Calreticulin expression, an ER marker, was seen. Middle Left. At Day 8, extensive expression of REEP1 was noted in all cultured SGN examined. REEP1 was found in cell bodies, axons, and dendrites, revealing a similar localization pattern as the ER marker calreticulin. Middle Right. Similar extensive expression of REEP1 was seen in all cultured SGN examined at Day 16. Far Right. Note absence of REEP1, but expression of calreticulin, in non-SGN supporting cells Day 8. Non-SGN cells present include fibroblasts (arrow), satellite cells, and Schwann cells. Over one hundred cultured SGN were examined at each time point. Bar = 10 μm.
3. Discussion
3.1 Cell line analysis of REEP1 and REEP2 expression
Prior RT-PCR analysis determined that all REEPs (except REEP1) were found in HEK293T cells (Behrens et al., 2006; Park et al., 2010); REEP1 expression in COS7 revealed conflicting results (Park et al., 2010; Zuchner et al., 2006). Our immunoblot analysis suggested that REEP1 protein expression was restricted, since several different cell lines demonstrated no expression of REEP1 (HEK293/HEK293A, Rat1, PC12, NRK). The lack of REEP1 expression in non-neuronal cell lines was not surprising, given that NRK and HEK293/A cells are derived from rat renal epithelium and human embryonic kidney respectively, and that REEP1 was not expressed in kidney. REEP1 expression was not seen in PC12 cells, an adrenal-like neuroendocrine cell line derived from a pheochromocytoma (Greene and Tischler, 1976), however it was not expressed in adrenal gland either.
REEP2 immunoblotting revealed expression in HEK293/A cells, consistent with prior RT-PCR studies (Behrens et al., 2006). This result was interesting, since renal-derived NRK cells did not demonstrate REEP2 expression. Unlike NRK cells, HEK293/A cells are adenovirus transformed cell lines and may not exhibit a pure renal gene/protein expression profile (Graham et al., 1977). Recently, it has been suggested that HEK293 cells may be derived from a neuronal lineage, based upon DNA microarray analysis of neuronal-specific genes (Shaw et al., 2002). This finding may account for the expression of REEP2 in HEK293 cells. Also, REEP2 was not found in Rat1 cells, a cell line derived from rat embryonic fibroblasts (Freeman et al., 1973; Lania et al., 1980). However, REEP2 was expressed in uninduced PC12 cells, which do not possess neurite extensions. Nerve growth factor (NGF) induction of PC12 neurite extension did not alter REEP2 expression levels nor induce REEP1 expression. PC12 cells are derived from an adrenal-derived pheochromocytoma and thus may exhibit a similar gene expression profile as adrenal gland. Overall, it can be concluded that REEP1 and REEP2 expression in transformed cell lines was not ubiquitous and did not always correlate with the tissue of origin.
3.2 Prior tissue expression analysis of REEP1 and REEP2
Originally, REEP1 mRNA was characterized by Northern blot analysis, revealing expression in murine olfactory and vomeronasal epithelium and brain, but not kidney, testis, liver, or heart (Saito et al., 2004). Subsequent, in situ hybridization studies demonstrated expression of REEP1 mRNA in murine olfactory neurons, olfactory bulb, cerebellar, and hippocampal neurons. Similar RT-PCR analysis of human circumvallate papillae (CV) revealed expression of REEP1–6 and human testes expressed REEP1 and REEP4–6 (Behrens et al., 2006). Expression of REEP2 mRNA in murine CV tissue was confirmed by others using RT-PCR (Ilegems et al., 2010). We found REEP1 and REEP2 protein expression in mouse testes by immunoblot (Figure 4), which was not observed with prior Northern blot (murine REEP1) (Saito et al., 2004) nor RT-PCR analysis (human REEP2) (Behrens et al., 2006). However, REEP1 was found in human testis by RT-PCR (Behrens et al., 2006), which may relate to the differences in sensitivity between Northern blot and RT-PCR techniques (Bartlett, 2002; Mocharla et al., 1990). Thus, mRNA expression data (Northern blot, RT-PCR, and in situ hybridization) obtained has not been consistent between studies, which may be due to species variability or the methods utilized.
A multiple tissue RT-PCR analysis revealed human REEP1 mRNA expression in twenty three different tissues, including brain, muscle, testes, liver, kidney, lung, and multiple endocrine organs (Zuchner et al., 2006). However, our immunoblot analysis of similar tissues demonstrated that REEP1 and REEP2 protein expression was restricted to neuronal cells (brain, spinal cord) and other cells that exhibit neuronal-like exocytosis (pituitary, adrenal gland, testes), though REEP1 and REEP2 did not show complete overlap in protein expression. An analysis of the Gene Atlas expression profiles (Wu et al., 2009) for mouse and human REEP1 and REEP2 mRNA demonstrated that they exhibited low level expression in multiple tissues, but much higher levels of mRNA expression were found in mouse tissues that expressed the respective proteins, as seen by immunoblot (Supplemental Figures 3 and 4). For example, cerebral cortex, spinal cord, and cerebellum exhibit high levels of REEP1 mRNA expression that correlated with our REEP1 protein expression data, whereas skeletal muscle, heart, adrenal gland and pituitary expressed low levels of REEP1 mRNA and no REEP1 protein by immunoblotting. Together, the Gene Atlas profile, prior RT-PCR analysis, and various tissue immunoblots demonstrate that protein expression within a tissue cannot be accurately predicted based upon mRNA expression alone.
Recently, a REEP1 gene knockout mouse model of HSP was created (Beetz et al., 2013). In situ hybridization analysis of an E18.5 murine sagittal embryo section demonstrated that REEP1 mRNA was only found in neuronal tissues, including cortical brain structures, brainstem, cerebellum, spinal cord, several major ganglia (e.g. trigeminal, dorsal root, and superior cervical) and olfactory bulb. Their subsequent immunoblot analysis revealed that REEP1 protein was only found in brain, but not liver, lung, eye, kidney, spleen, or glia, consistent with our immunoblot data. Interestingly, REEP1 mRNA was not found in testes. However, its expression may be developmentally regulated and thus not present in an E18.5 embryo.
These recent results are consistent with our currently presented research demonstrating a restricted expression of REEP1 protein to major neuronal structures including brain, spinal cord, and sympathetic ganglia. Additionally, we have demonstrated a lack of REEP1 protein expression in other tissues (e.g. skeletal muscle and adrenal gland) that had previously shown REEP1 mRNA expression by RT-PCR (Zuchner et al., 2006) and Gene Atlas Expression profiles. It is possible that the extreme sensitivity of RT-PCR may allow low copy number mRNA to be identified in multiple tissues, while the translated protein is not detectable. Alternatively, HSP inducing mutations of REEP1 have been identified in the 3' UTR, at a putative miRNA (miR-140) binding site (Zuchner et al., 2006). miR-140 binding to REEP1 mRNA may uncouple mRNA transcription from protein translation, leading to differences between RT-PCR/Northern blot and immunoblot results.
3.3 REEP expression in sympathetic neuron tissues and cultured neurons
Because sympathetic tissues are not abundant, immunoblot analysis could not be performed. However, gene expression analysis of SCG and SG sympathetic tissues revealed that REEP1 and REEP2 mRNA are expressed (along with REEP3 and REEP5), further supporting the neuronal restrictive pattern seen by immunoblotting.
We previously demonstrated that α2A and α2C ARs have differential localizations in cultured SGN and that the time course for their final localization differs (Brum et al., 2006). Additionally, we had demonstrated that several REEP isoforms interact with α2C, but not α2A ARs (Björk et al., 2013), and thus we sought to determine the time course of REEP1 expression in cultured SGN. Interestingly, REEP1 expression appeared in the same time frame as α2C AR localization to the neuron plasma membrane, with both occurring at Day 8 of culture. Unfortunately, proper biochemical analysis of any possible interaction between REEP1 and α2C ARs in cultured SGN is not possible, due to the small amount of protein available from culture SGN.
Analysis of REEP expression in cultured neurons has been limited to REEP1 (Park et al., 2010), where its expression was demonstrated in cultured cerebral cortical neurons by immunofluorescence. However, the time course of REEP1 expression during the duration of the culture was not determined. We have now demonstrated by both RT-PCR and immunofluorescent staining, that REEP1 is temporally expressed in cultured SGN. REEP1 protein was extensively expressed at Day 8 and Day 16, but not at Day 4. It may be that REEP1 expression is dependent upon maturation of cultured SGN, possibly by development of synaptic contacts between neurons.
3.4 Conclusions
Most studies to date have relied upon REEP mRNA expression profiling either by Northern blot or RT-PCR, however, some inconsistencies between various tissues and cell lines have been seen. When REEP1 was identified as a possible causative agent of HSP, an RT-PCR analysis of a large number of tissues suggested that REEP1 was ubiquitously expressed in brain, bone marrow, heart, kidney, liver, prostate, and various endocrine organs, but not peripheral blood or skin fibroblasts (Zuchner et al., 2006). The expression of human REEP1 in non-neuronal tissues as seen by RT-PCR was not consistent with the neuronal-specific symptoms of REEP1 genetic disorders, such as HSP and dHMN-V. Therefore, we developed a new monoclonal antibody (NeuroMab clone N345/51) to assess REEP1 protein expression in various tissues and cells.
Overall, tissue analysis of REEP1 and REEP2 expression by immunoblotting demonstrated that REEP1 is restricted to neurons and testes and that REEP2 is expressed in neurons and other neuronal-like exocytotic organs, including pituitary, adrenal gland, and testes. However, neither was expressed by cultured astrocytes. The observed expression of REEP1 and REEP2 in testes is not unexpected, since it has been shown that brain and testes have the highest similarity of gene expression profiles amongst human tissues (Guo et al., 2003) and sperm exhibit dense core granule exocytosis (Bustos et al., 2012). By combining Northern blot and protein expression analysis, it can be concluded that REEP1 exhibits neuronal/sensory tissue specificity, being expressed in brain, spinal cord, olfactory/vomeronasal epithelium, tongue, and sympathetic ganglion/cerebral cortical neurons. The neuronal specificity of REEP1 protein is more consistent with HSP and dHMN-V pathogenesis by REEP1 mutations.
Our tissue expression data for REEP1 and REEP2 suggests that this subfamily of REEPs may have evolved to serve a cellular function in neuronal or other exocytotic tissues. REEP1–4 all possess a consensus 14-3-3 protein binding site in the carboxyl terminus (Tinti et al., 2012), as well as a series of positively charged residues in the cytoplasmic linker between the dual hairpins, which is necessary for microtubule binding (Schlaitz et al., 2013). As ER shaping adapter proteins, REEPs may have evolved specialized cellular roles that take advantage of their dual hairpin structure, combined with a specific cytoplasmic carboxyl tail function. For example, recent research has demonstrated that REEP3 and REEP4 are ER membrane proteins that regulate mitotic spindle/ER interactions and nuclear envelope assembly (Schlaitz et al., 2013).
Recently, we demonstrated that REEP1 and REEP2 can enhance the expression of and interact with a minimally glycosylated form of α2C ARs, but not α2A ARs (Björk et al., 2013). Similar specialization has been seen with other Yip family members and neuronal proteins. For example, Yif1B has been shown to specifically target 5-HT1A serotonin receptors to neuronal dendrites, whereas Yip6B has been shown to interact with and regulate ER trafficking of multiple glutamate transporters (Al Awabdh et al., 2012; Ruggiero et al., 2008). REEP1 is an ER membrane shaping adapter protein, which interacts with other ER membrane proteins (e.g. atlastin-1, spastin) to regulate ER tubular structure and cytoskeletal interactions in spinal motor neurons (Blackstone et al., 2011). Therefore, REEP1 and REEP2 may have evolved to allow for interactions with specific cargo, ER membrane, and vesicular transport proteins and this specialization may be relevant for neuronal development, function, and disease.
Knowledge of the specific expression patterns of REEPs should aid in further analysis of their function in GPCR expression and cargo trafficking, as well as their role in intracellular transport. It should allow for a more thorough analysis of REEP effects by choosing appropriate tissues and cell lines for future studies. Furthermore, our results should assist future experimentation into the neurodegenerative disorders HSP and dHMN-V. However, RT-PCR results may need to be interpreted with caution, since they may not always correlate with protein based methods such as immunoblotting or immunofluorescent staining (Gry et al., 2009; Gygi et al., 1999), as demonstrated by a new REEP1 monoclonal antibody. Well-characterized antibodies against all REEP isoforms will be required to overcome such limitations.
4. Experimental Procedures
4.1 Materials and antibodies
All chemicals were purchased from Sigma-Aldrich Corp. and Roche. All sera were purchased from Gemini Bio Products. The anti-REEP1 monoclonal antibody (clone N345/51) was co-developed with the UC Davis/NIH NeuroMab Facility, supported by NIH grant U24NS050606 and maintained by the Department of Neurobiology, Physiology and Behavior, College of Biological Sciences, University of California, Davis, CA 95616. Rabbit polyclonal anti-REEP2 antisera was obtained from Proteintech (Chicago, IL). Various mouse tissue total cell lysates were obtained from Zyagen (San Diego, CA). Murine NGF was purchased from Life Technologies (Grand Island, NY).
4.2 Cell culture and transfection methods
HEK293, HEK293A, NRK, Rat1, and PC12 cells were cultured as described previously (Daunt et al., 1997; Hurt et al., 2000). For some experiments, PC12 cells were treated with 50 ng/ml NGF for 72 hours. Mammalian expression vectors encoding untagged REEP1, Flag-REEP1, Flag-REEP2, and Flag-REEP6 have been described previously (Björk et al., 2013) and were transiently expressed in HEK293A cells using Effectene (Qiagen) per manufacturer's instructions. Superior cervical sympathetic ganglia neurons (SGN) were cultured onto poly-D-lysine and laminin coated glass coverslips, as described previously (Brum et al., 2006).
4.3 DNA microarray gene expression analysis
Total RNA was prepared from adult mouse superior cervical (SCG) and stellate (SG) ganglia with the RNeasy Mini Kit (Qiagen, Hilden, Germany) (n = 2, n = 3 respectively). RNA quality was assessed on RNA LabChips (Agilent, Böblingen, Germany) according to manufacturers' protocols. Microarray experiments were carried out using GeneChip Mouse Genome 430A 2.0 arrays (Affymetrix, Santa Clara, CA, USA), as described previously (Gilsbach et al., 2010). Results were analyzed with ArrayAssist 5.0 software (Stratagene, Amsterdam, The Netherlands) using the MAS5 algorithm. In case of multiple probes per transcript, results of the probe with the highest signal intensities are shown. Microarray data for stellate ganglia have been deposited in NCBI's Gene Expression Omnibus (accession number GSE18004). The following murine gene sequences were used: REEP1 (NM_178608), REEP2 (NM_144865), REEP3 (NM_178606), REEP4 (NM_180588), REEP5 (NM_007874), REEP6 (NM_139292).
4.4 RNA isolation and RT-PCR
For RT-PCR, SGN were cultured and a fixed number of detached cells (50,000–250,000) were utilized. Cultured SGN were plated onto glass coverslips as described previously (Brum et al., 2006). At Day 1, 4, 8 and 16 of culture, 25,000 plated SGN were rinsed three times with 1xPBS (phosphate buffered saline) and scrapped into 1.5 ml microcentrifuge tubes. Total RNA was stabilized for storage with RNAlater (Ambion) and cDNA was synthesized by reverse transcription using the Cells-to-cDNA™ II kit (Ambion), according to manufacturer's instructions. The standard protocol for the kit was followed, except that the DNase I reaction was prolonged to 60 min at 37°C. Reverse transcription was done in a two-step protocol using random decamers, according to the manufacturer's instructions. Samples were preheated at 70°C for 3 min before adding enzymes, then incubated at 42°C for 45 min and inactivated at 94°C for 10 min.
Primers specific for the mouse REEP1 and GAPDH sequences were synthesized based on GenBank sequences NM_178608 and NM_008084.2, respectively, with DNAStar Primer Select software (Madison, WI). The specific primer pairs were designed in part to have similar Tm values (REEP1 Forward Primer: TATTTGGCACCCTTTATCCTG; REEP1 Reverse Primer: CATCTTGGGCTGGCTGTGTTT; GAPDH Forward Primer: GTCATCATCTCCGCCCCTTCT; GAPDH Reverse Primer: TGCCTGCTTCACCACCTTCTT). PCR amplifying conditions were optimized using either commercial mouse embryo (Ambion), or mouse brain (Maximbio, CA) cDNA. REEP1 Protocol: An annealing temperature of 60.1 C was chosen and MgSO4 (1 mM) was added. GAPDH Protocol: An annealing temperature of 60.0 C was chosen and MgSO4 (2 mM) was added. For both reactions, the final PCR protocol consisted of forty 30 sec cycles each of denaturation (94 C), annealing, and extension (72 C), preceded by a 3 min denaturation at 94 C, and followed by a 3 min final extension at 72 C. PCR products were analyzed with standard DNA agarose gel electrophoresis methods and confirmed by either restriction enzyme digestion or DNA sequencing. Cultured SGN RT-PCR reactions were performed three times and to be considered positive, an appropriate PCR product had to be present at least twice.
4.5 Immunofluorescent staining
HEK293A cells were cultured on poly-D-lysine-coated glass coverslips for 48 h prior to transfection and maintained in culture for 48 h after transfection, prior to immunofluorescent staining as described previously (Angelotti et al., 2010). Cultured SGN were fixed and prepared for immunofluorescent staining, as per previous methods (Brum et al., 2006). Endogenous REEP1 was detected by either anti-REEP1 monoclonal hybridoma supernatant (1:2 dilution) or purified antibody (mAb) (0.1 mcg/ml). Heterologously expressed Flag-REEP1 was detected with the M2 anti-Flag mAb (1:500, Sigma). Cultured SGN and HEK293A cells were counterstained with rabbit polyclonal anti-calreticulin antisera (1:500, Abcam). Goat anti-mouse Alexa Fluor 488 and donkey anti-rabbit Alexa Fluor 596 secondary antibodies were utilized (1:1000, Invitrogen). Mitochondria were counterstained with 200 nM MitoTracker Red CMXROS (Invitrogen), per manufacturer's instructions. Coverslips were mounted with DAPI-containing Vectashield™ (Vector Labs). Stained cells were observed using a fluorescence microscope (Zeiss Axioplan 2 imaging; Carl Zeiss, Inc.) with a 63 × 1.4 NA oil immersion apochromatic lens (Carl Zeiss, Inc.), as described previously (Angelotti et al., 2010). The cell images were acquired by digital camera (Roper Scientific RTE/CCD) using IPlabs software (Macintosh version). The image brightness and contrast were adjusted using Photoshop 6 (PC version; Adobe).
4.6 Immunoblotting
Whole cell lysates (RIPA buffer) were made from untransfected PC12, NRK, Rat1, wild-type HEK293, HEK293A cells and transfected HEK293A cells (forty eight hrs post-transfection) (Angelotti et al., 2010; Hurt et al., 2000). Cultured mouse astrocyte membranes (Day 21) were the gift of Dr. R. White (Stanford, CA) (Ouyang et al., 2011). One hundred micrograms of mouse tissue protein and seventy-five micrograms of whole cell lysates were separated by 10% SDS-PAGE, transferred to PVDF (Biorad) and handled as described previously (Angelotti et al., 2010). REEPs were detected using undiluted monoclonal anti-REEP1 hybridoma supernatant or rabbit polyclonal anti-REEP2 (1:500 dilution, Proteintech). Protein loading controls were detected with either mouse anti-GAPDH (1:2000 dilution, Abcam), mouse anti-β-actin (1:5000 dilution, Sigma), mouse anti-β-tubulin (1:5000 dilution, Abcam), or rabbit anti-calnexin (1:2000 dilution, Abcam). Either horseradish peroxidase conjugated goat anti-mouse or anti-rabbit IgG were used for detection (1:100,000, GE Healthcare) or IRDye 800CW Goat anti-Mouse IgG and IRDye 680 Goat anti-Rabbit IgG (1:10,000; LI-COR Biosciences) were used as secondary antibodies. Immunoblot images were obtained using ECL Plus (GE Healthcare) and autoradiography per manufacturer's instructions or Odyssey scanner (LiCOR Biosciences).
Supplementary Material
Highlights
A new specific REEP1 monoclonal antibody was developed and characterized
REEP1 tissue expression is restricted to brain, spinal cord, and testes
REEP2 expression is restricted to neuronal and neuronal-like exocytotic tissues
REEP1 expression is temporally regulated in cultured sympathetic ganglion neurons
Neuronally restricted REEP1 expression is consistent with HSP disease phenotypes
Acknowledgments
We thank Dr. H. Matsunami for the generous gift of REEP cDNAs. In addition, we thank Drs. C. Blackstone and B. Kobilka for helpful discussions and reagents. This work was supported in part by grants from NINDS K08NS050654-01A1 and K08 Supplement (NOT-NS-08-009) (TA). The anti-REEP1 monoclonal antibody was co-developed with the UC Davis/NIH NeuroMab Facility, supported by NIH grant U24NS050606 and maintained by the Department of Neurobiology, Physiology and Behavior, College of Biological Sciences, University of California, Davis, CA 95616.
Abbreviations
- AR
adrenergic receptor
- dHMN-V
distal hereditary motor neuropathy type V
- GPCR
G protein-coupled receptor
- HSP
hereditary spastic paraplegia
- mAb
monoclonal antibody
- NGF
nerve growth factor
- OR
olfactory receptor
- REEP
receptor expression enhancing protein
- RTP
receptor transporting protein
- RT-PCR
reverse transcription polymerase chain reaction
- SCG
superior cervical ganglia
- SG
stellate ganglion
- SGN
sympathetic ganglion neurons
- Yip
Ypt interacting protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al Awabdh S, et al. A New Vesicular Scaffolding Complex Mediates the G-Protein-Coupled 5-HT1A Receptor Targeting to Neuronal Dendrites. J Neurosci. 2012;32:14227–14241. doi: 10.1523/JNEUROSCI.6329-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelotti T, et al. Regulation of G-protein coupled receptor traffic by an evolutionary conserved hydrophobic signal. Traffic. 2010;11:560–78. doi: 10.1111/j.1600-0854.2010.01033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JM. Approaches to the analysis of gene expression using mRNA: a technical overview. Mol Biotechnol. 2002;21:149–60. doi: 10.1385/MB:21:2:149. [DOI] [PubMed] [Google Scholar]
- Beetz C, et al. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain. 2008;131:1078–86. doi: 10.1093/brain/awn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetz C, et al. Exome sequencing identifies a REEP1 mutation involved in distal hereditary motor neuropathy type V. Am J Hum Genet. 2012;91:139–45. doi: 10.1016/j.ajhg.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetz C, et al. A spastic paraplegia mouse model reveals REEP1-dependent ER shaping. J Clin Invest. 2013;123:4273–4282. doi: 10.1172/JCI65665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, et al. Members of RTP and REEP Gene Families Influence Functional Bitter Taste Receptor Expression. J Biol Chem. 2006;281:20650–9. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- Björk S, et al. REEPs are membrane shaping adapter proteins that modulate specific G protein-coupled receptor trafficking by affecting ER cargo capacity. PLoS One. 2013;8:e76366. doi: 10.1371/journal.pone.0076366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C, O'Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12:31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands A, Ho TH. Function of a plant stress-induced gene, HVA22. Synthetic enhancement screen with its yeast homolog reveals its role in vesicular traffic. Plant Physiol. 2002;130:1121–31. doi: 10.1104/pp.007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brum PC, et al. Differential targeting and function of alpha(2A) and alpha(2C) adrenergic receptor subtypes in cultured sympathetic neurons. Neuropharmacology. 2006;51:397–413. doi: 10.1016/j.neuropharm.2006.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos MA, et al. Rab27 and Rab3 sequentially regulate human sperm dense-core granule exocytosis. Proc Natl Acad Sci U S A. 2012;109:E2057–66. doi: 10.1073/pnas.1121173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero M, Whittaker GR, Collins RN. Yop1p, the yeast homolog of the polyposis locus protein 1, interacts with Yip1p and negatively regulates cell growth. J Biol Chem. 2001;276:12100–12. doi: 10.1074/jbc.M008439200. [DOI] [PubMed] [Google Scholar]
- Carrel D, et al. Targeting of the 5-HT1A serotonin receptor to neuronal dendrites is mediated by Yif1B. J Neurosci. 2008;28:8063–73. doi: 10.1523/JNEUROSCI.4487-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daunt DA, et al. Subtype-specific intracellular trafficking of alpha2-adrenergic receptors. Mol Pharmacol. 1997;51:711–20. doi: 10.1124/mol.51.5.711. [DOI] [PubMed] [Google Scholar]
- Dittmer A, Dittmer J. Beta-actin is not a reliable loading control in Western blot analysis. Electrophoresis. 2006;27:2844–5. doi: 10.1002/elps.200500785. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Ferguson RE, et al. Housekeeping proteins: a preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005;5:566–71. doi: 10.1002/pmic.200400941. [DOI] [PubMed] [Google Scholar]
- Freeman AE, et al. 5-Bromo-2'-deoxyuridine potentiation of transformation of rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci U S A. 1973;70:2415–9. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilsbach R, et al. Sympathetic alpha(2)-adrenoceptors prevent cardiac hypertrophy and fibrosis in mice at baseline but not after chronic pressure overload. Cardiovasc Res. 2010;86:432–42. doi: 10.1093/cvr/cvq014. [DOI] [PubMed] [Google Scholar]
- Graham FL, et al. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976;73:2424–8. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gry M, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, et al. In silico analysis indicates a similar gene expression pattern between human brain and testis. Cytogenet Genome Res. 2003;103:58–62. doi: 10.1159/000076290. [DOI] [PubMed] [Google Scholar]
- Gygi SP, et al. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–30. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanani M. Satellite glial cells in sympathetic and parasympathetic ganglia: in search of function. Brain Res Rev. 2010;64:304–27. doi: 10.1016/j.brainresrev.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Heidtman M, et al. A role for Yip1p in COPII vesicle biogenesis. J Cell Biol. 2003;163:57–69. doi: 10.1083/jcb.200306118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt CM, Feng FY, Kobilka B. Cell-type specific targeting of the alpha 2ca-drenoceptor. Evidence for the organization of receptor microdomains during neuronal differentiation of PC12 cells. J Biol Chem. 2000;275:35424–31. doi: 10.1074/jbc.M006241200. [DOI] [PubMed] [Google Scholar]
- Ilegems E, et al. REEP2 enhances sweet receptor function by recruitment to lipid rafts. J Neurosci. 2010;30:13774–83. doi: 10.1523/JNEUROSCI.0091-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, et al. Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome. Mol Cell Proteomics. 2011;10:1–15. doi: 10.1074/mcp.M110.005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lania L, et al. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980;101:217–32. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Lein PJ, et al. Glia induce dendritic growth in cultured sympathetic neurons by modulating the balance between bone morphogenetic proteins (BMPs) and BMP antagonists. J Neurosci. 2002;22:10377–87. doi: 10.1523/JNEUROSCI.22-23-10377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincic I, Peralta ME, Ngsee JK. Isolation and characterization of a dual prenylated Rab and VAMP2 receptor. J Biol Chem. 1997;272:26991–8. doi: 10.1074/jbc.272.43.26991. [DOI] [PubMed] [Google Scholar]
- Mocharla H, Mocharla R, Hodes ME. Coupled reverse transcription-polymerase chain reaction (RT-PCR) as a sensitive and rapid method for isozyme genotyping. Gene. 1990;93:271–5. doi: 10.1016/0378-1119(90)90235-j. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–78. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Ouyang YB, et al. Overexpressing GRP78 influences Ca2+ handling and function of mitochondria in astrocytes after ischemia-like stress. Mitochondrion. 2011;11:279–86. doi: 10.1016/j.mito.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, et al. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120:1097–110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–96. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Trimmer JS. Antibodies as valuable neuroscience research tools versus reagents of mass distraction. J Neurosci. 2006;26:8017–20. doi: 10.1523/JNEUROSCI.2728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero AM, et al. The endoplasmic reticulum exit of glutamate transporter is regulated by the inducible mammalian Yip6b/GTRAP3–18 protein. J Biol Chem. 2008;283:6175–83. doi: 10.1074/jbc.M701008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, et al. RTP Family Members Induce Functional Expression of Mammalian Odorant Receptors. Cell. 2004;119:679–91. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Schlaitz AL, et al. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell. 2013;26:315–323. doi: 10.1016/j.devcel.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, et al. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–71. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Tinti M, et al. Evolution of signal multiplexing by 14-3-3-binding 2R-ohnologue protein families in the vertebrates. Open Biol. 2012;2:120103. doi: 10.1098/rsob.120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, et al. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–86. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79:365–9. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.