Abstract
The aryl hydrocarbon receptor (AhR) is an important mediator of toxic responses after exposure to xenobiotics including 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and dioxin-like polychlorinated biphenyls (PCBs). Activation of AhR responsive genes requires AhR dimerization with the aryl hydrocarbon receptor nuclear translocator (ARNT), a heterodimeric partner also shared by the hypoxia-inducible factor-1α (HIF-1α) protein. TCDD-stimulated AhR transcriptional activity can be influenced by hypoxia; however, it less well known whether hypoxia interferes with AhR transcriptional transactivation in the context of PCB-mediated AhR activation in human cells. Elucidation of this interaction is important in liver hepatocytes which extensively metabolize ingested PCBs and experience varying degrees of oxygen tension during normal physiologic function. This study was designed to assess the effect of hypoxia on AhR transcriptional responses after exposure to 3,3',4,4',5-pentachlorobiphenyl (PCB 126). Exposure to 1% O2 prior to PCB 126 treatment significantly inhibited CYP1A1 mRNA and protein expression in human HepG2 and HaCaT cells. CYP1A1 transcriptional activation was significantly decreased upon PCB 126 stimulation under conditions of hypoxia. Additionally, hypoxia pre-treatment reduced PCB 126 induced AhR binding to CYP1 target gene promoters. Importantly, ARNT overexpression rescued cells from the inhibitory effect of hypoxia on XRE-luciferase reporter activity. Therefore, the mechanism of interference of the signaling crosstalk between the AhR and hypoxia pathways appears to be at least in part dependent on ARNT availability. Our results show that AhR activation and CYP1A1 expression induced by PCB 126 were significantly inhibited by hypoxia and hypoxia might therefore play an important role in PCB metabolism and toxicity.
Keywords: AhR, HIF-1α, ARNT, PCB 126, CYP1A1, hypoxia, oxygen effects
Introduction
Polychlorinated biphenyls (PCBs) are a group of synthetic organic chemicals consisting of 209 congeners that accumulate and persist in the environment (Safe, 1993). PCBs that are more highly chlorinated have much slower rates of metabolism, and therefore quickly accumulate in the human body. Human blood serum levels of PCBs in a population in Alabama, USA range from 0.003 µM to 6.5 µM (Hansen et al., 2003). Exposure to PCBs is known to cause a broad range of adverse health effects and due to their persistence and ubiquitous distribution, PCBs pose a serious hazard to human and animal health (Silberhorn et al., 1990; Ludewig et al., 2008; Crinnion, 2011).
3,3',4,4',5-pentachlorobiphenyl (PCB 126) is a coplanar and the most potent dioxin-like PCB. As such, PCB 126 can bind to and activate the aryl hydrocarbon receptor (AhR), an immediate early cellular response to xenobiotics and mediator of many of the toxic effects of PCB exposure. Upon ligand binding, the AhR translocates to the nucleus and forms a heterodimer with the aryl hydrocarbon receptor nuclear translocator (ARNT). This complex binds to and activates the promoters of genes containing xenobiotic response elements (XREs). Binding to these regulatory sequences induces transcription and expression of AhR target genes such as the cytochrome P450 (CYP) 1 family, including CYP1A1 and CYP1B1 (Beischlag et al., 2008).
Similar to the AhR, hypoxia-inducible factor-1α (HIF-1α) heterodimerization with ARNT is a prerequisite for transcriptional activation of hypoxia-inducible genes (Kewley et al., 2004). HIF-1α is an important sensor of oxygen environments and regulates responses to reduced oxygen availability (hypoxia). In the context of sufficient oxygen concentrations (normoxia), HIF-1α is continuously hydroxylated by oxygen-dependent prolyl hydroxylase domain-containing enzymes (PHDs) which target HIF-1α for subsequent ubiquitination and degradation. Under limiting oxygen concentrations, HIF-1α is stabilized allowing it to translocate to the nucleus where it binds to ARNT. The HIF-1α:ARNT heterodimer binds to hypoxia response element (HRE) sequences in hypoxia-inducible target genes, and subsequently induces gene transcription (Majmundar et al., 2010).
Since ARNT is critical for both the AhR and hypoxia pathways, it seems likely that a crosstalk exists that can affect the regulation of gene expression. This may be important in the context of organs such as the liver, which contains zones of various oxygen tensions, and is responsible for a large proportion of PCB metabolism (Oinonen and Lindros, 1998). Interestingly, regional induction of CYP1A1 has been observed in rat livers. Rats exposed to low doses of PCB 126 induced CYP1A1 mainly in the perivenous zone where oxygen is scarce (Chubb et al., 2004). Numerous studies have analyzed the effect of hypoxia on AhR target genes in different cell lines after exposure to exogenous ligands (Kim and Sheen, 2000; Nie et al., 2001). However, these studies have focused on using 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) as an AhR ligand and used chemical compounds such as cobalt chloride (CoCl2) to stabilize HIF-1α and induce a hypoxic response. Only one study in topminnow hepatocarcinoma cells showed that hypoxia could inhibit a PCB 126 induced AhR response (Fleming et al., 2009). Overall, the crosstalk between HIF-1α and the AhR using PCBs as AhR ligands and genuine hypoxia has not been extensively studied in human cell lines.
In an effort to understand PCB-induced AhR function and target gene expression in different oxygen environments, we exposed human hepatocellular carcinoma and human keratinocyte cell lines to PCB 126 in normal oxygen (21% O2) or hypoxia (1% O2). Our results show that hypoxia significantly interfered with AhR signaling and could inhibit AhR target gene expression and function.
Materials and Methods
Chemicals and PCB treatment
3,3',4,4',5-pentachlorobiphenyl (PCB 126), 2,2',4,4',5,5'-hexachlorobiphenyl (PCB 153), and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) were provided by Dr. H. J. Lehmler of the Iowa Superfund Research Program at The University of Iowa. 6,2',4'-trimethoxyflavone (TMF) was purchased from Sigma (St. Louis, MO). All stock solutions were made in dimethyl sulfoxide (DMSO) and the appropriate amount of stock solution was added to the cell culture medium. PCBs were used at concentrations ranging from 2 nM to 3 µM. Cells were left untreated or DMSO was used as the vehicle control in control cultures. The final concentration of DMSO was adjusted to less than 0.3%. All exposures were done in serum-free medium.
Cell lines and cell culture conditions
Human hepatocellular carcinoma cells (HepG2) and human keratinocyte cells (HaCaT) were purchased from the American Tissue Culture Collection (ATCC) (Manassas, VA). Cell lines were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin and were maintained in a humidified 37°C incubator with 5% CO2. All cell lines were routinely passaged before reaching confluence by washing and detaching cells with TrypLE Express. All experiments were done using exponentially growing cell cultures at 60% up to 90% confluence. Hypoxic cell culture was carried out in a 37°C, humidified Bactron anaerobic/environmental chamber (SHEL LAB, Cornelius, OR) flooded with a 1% O2, 94% N2 and 5% CO2 certified gas mixture. Cell lines were incubated in hypoxia for 8 or 16 hours prior to PCB 126 treatment and were kept at 1% O2 for the duration of the treatments. The accuracy of hypoxic exposure was verified by measuring HIF-1α protein expression in normoxic and hypoxic cells using an anti-HIF-1α antibody (ab2185, Abcam, Cambridge, MA) at 1:500 dilution (data not shown).
Quantitative real-time RT-PCR
After treatment, total RNA was extracted from individual cell lines at > 60% confluence using TRIzol reagent (Invitrogen, Grand Island, NY) and quantified using a ND1000 NanoDrop spectrophotometer. 500 ng of total RNA was reverse transcribed using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) for 2 hours at 37°C. The cDNA was subsequently used for quantitative real-time RT-PCR (qRT-PCR) using the Universal ProbeLibrary (UPL) system (Roche Applied Science, Indianapolis, IN). Primers and probes specific to CYP1A1, AhR, ARNT, and ribosomal protein, large, P0 (RPLP0), were designed using the online Universal ProbeLibrary Assay Design Center. CYP1A1 forward primer: 5'-GGTCAAGGAGCACTACAAAACC-3', reverse primer: 5'-TGGACATTGGCGTTCTCAT-3', UPL probe: #2; AhR forward primer: 5'-AGCCGGTGCAGAAAACAG-3', reverse primer: 5'-CTATGCCGCTTGGAAGGAT-3', UPL probe: #33; ARNT forward primer: 5'-CTACCCGCTCAGGCTTTTC-3', reverse primer: 5'-CACCAAACTGGGAAGTACGAG-3', UPL probe: #3; RPLP0 forward primer: 5'-TCTACAACCCTGAAGTGCTTGAT-3', reverse primer: 5'-CAATCTGCAGACAGACACTGG-3', UPL probe: #6. The qRT-PCR was set up as follows: 10 ng of cDNA was used as template for each real-time PCR reaction (10 µL reaction volume) with primer pairs at 0.5 µM and UPL probes at 0.2 µM. For all reactions TaqMan Universal PCR Master Mix (Applied Biosystems) was used. The DNA polymerase was heat-activated at 95°C for 10 minutes followed by 40 cycles, denaturing at 95°C for 15 seconds, annealing and elongating at 60°C for 1 minute. Data were collected with the ABI PRISM 7500 sequence detection system (Applied Biosystems). An amplification threshold in the linear range of each sample was selected to calculate the cycle threshold (CT) for each sample. The relative mRNA levels were calculated as follows: ΔCT (sample) = CT (mRNA of interest) - CT (RPLP0); ΔΔCT = ΔCT (treatment) - ΔCT (control); relative expression = 2−ΔΔCT.
Western Blot analysis
Treated cells at 90% confluence were washed with ice cold 1x phosphate-buffered saline (1x PBS), lysed on the plate in 150 µl RIPA cell-lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Na Deoxycholate, 1% TX-100) containing 10 mM NaF, 1 mM NaVO4, 1 mM PMSF, and 1/100 protease inhibitor cocktail (Sigma), and sonicated briefly in a cup sonicator. Protein was quantified using a Bradford assay and stabilized with SDS sample buffer. Prior to electrophoresis, samples were boiled for 2 minutes. SDS-polyacrylamide gels (10%) were used for protein electrophoresis. Proteins were electrotransferred onto nitrocellulose membranes (BioRad, Hercules, CA) and incubated with an anti-CYP1A1 antibody (ab3568, Abcam) at 1:500 dilution overnight at 4°C. Equal protein loading was confirmed on all immunoblots using human beta-tubulin antibody (Developmental Studies Hybridoma Bank, The University of Iowa, IA) at a dilution of 1:1000. Goat anti-rabbit IgG and goat anti-mouse IgG (Millipore, Billerica, MA) were used as secondary antibodies against all primary antibodies. Bands were visualized by chemiluminescence with ECL plus reagent (Pierce, Rockford, IL) on a Typhoon FLA 7000 and quantified using ImageJ image analysis software (Schneider et al., 2012). Intensities of CYP1A1 were normalized to beta-tubulin and are depicted relative to PCB 126 treated cells in normal oxygen.
CYP1A1 activity assay
HepG2 and HaCaT cells were seeded in a 96 well plate (10,000 cells/ well) and CYP1A1 enzyme activity was measured using the luminogenic substrate Luciferin-1A1 (6-(2-chloroethoxy)benzo[d]thiazole-2-carbonitrile) and Luciferin Detection Reagent (Promega, Madison, WI). Exposed cells were washed with Krebs-Henseleit Buffer (Sigma), and 4 µM CYP1A1 substrate diluted in Krebs-Henseleit Buffer containing 3 mM salicylamide (Sigma) was added to each well. After 30 minutes incubation at 37°C, 50 µl supernatant was transferred to a 96 well opaque white luminometer plate and 50 µl Luciferin Detection Reagent with D-cysteine was added to each well. Cells treated in hypoxia were transferred to normal oxygen at this point. The reactions were incubated at room temperature for 20 minutes and luminescence was measured with a Tecan SpectraFluor Plus luminometer.
Promoter reporter assay
A XRE-luciferase reporter vector containing the 5'-flanking region (−1566 to +73) of the human CYP1A1 gene sub-cloned into the pGL3-Basic vector (Promega) upstream of the firefly luciferase reporter gene (Morel and Barouki, 1998) was kindly provided by Dr. Linh Chi Bui (INSERM UMR-S 747, Paris, France). Cell lines ~80% confluent in 60 mm dishes were transfected with the XRE-luciferase reporter vector (4 µg) or HRE-luciferase reporter vector (Salnikow et al., 1999) (4 µg) and Renilla luciferase vector (4 µg) according to the Lipofectamine 2000 transfection reagent protocol (Invitrogen). Transfection media was removed after 6 hours and replaced with fresh medium. Transfected cells were split the next day and treated. Cells transfected with a XRE-luciferase reporter vector were kept in normoxia or were transferred to 1% O2 16 hours prior to PCB 126 treatment. Cells transfected with a HRE-luciferase reporter vector were treated with PCB 126 for 4 hours before transferring cells to hypoxia for 4 hours. Cells were lysed according to the dual-luciferase reporter assay system protocol (Promega) and luminescence was measured from whole cells lysates twice per sample using a Tecan SpectraFluor Plus luminometer.
ARNT overexpression
An ARNT expression vector (EX-C0312-M68) and empty control vector (EX-NEG-M68) were obtained from GeneCopoeia (Rockville, MD). The ARNT expression vector consists of the human ARNT open reading frame cloned into the mammalian expression vector pReceiver-M68. HepG2 cells ~80% confluent in 60 mm dishes were transfected with the XRE-luciferase reporter vector (3 µg), Renilla luciferase vector (3 µg), and ARNT expression vector (2 µg) or empty control vector (2 µg) using Lipofectamine 2000 transfection reagent as described earlier. Transfected cells were kept in normoxia or were transferred to 1% O2 16 hours prior to PCB 126 treatment for 6 hours. Luminescence was measured from whole cells lysates twice per sample. ARNT overexpression was verified using qRT-PCR with primers specific for ARNT. ARNT protein expression in transfected cells was measured using an anti-ARNT1 antibody (H-172, sc-5580X, Santa Cruz Biotechnology, Dallas, TX) at 1:200 dilution.
Isolation of nuclear extracts
Untreated and PCB 126 treated HepG2 cells in normoxia and hypoxia at ~90% confluence were washed with 1x PBS, harvested with 1x PBS containing 1 mM PMSF, and collected by centrifugation. The pellet was resuspended in 5×packed cell volume equivalent of hypotonic lysis buffer (10 mM Tris HCl, pH 7.5, 2 mM MgCl2, 3 mM CaCl2, and 0.32 M sucrose) plus 10 mM NaF, 1 mM NaVO4, 1 mM PMSF, and 1/100 protease inhibitor cocktail (Roche; Complete Mini) and incubated on ice for 15 minutes. Swollen cells were lysed by addition of 0.87% NP-40 and centrifuged. The supernatant was collected (cytoplasmic extract) and the remaining nuclear pellet was washed with hypotonic lysis buffer containing 1 mM PMSF and 0.8% NP-40. Nuclei were resuspended in 2×packed cell volume equivalent of RIPA cell-lysis buffer plus protease inhibitors and centrifuged, and the supernatant was collected (nuclear extract).
Electrophoretic mobility shift assay (EMSA)
A 24 bp double-stranded oligonucleotide probe (5'-TCCCGGCTCGCGTGAGAAGCGCTG-3', GRCh37/hg19 ch15:75018359-75018382) containing a consensus XRE binding site was end-labeled with γ32P ATP (Perkin-Elmer, Waltham, Massachusetts) using T4 Polynucleotide kinase (New England Biolabs, Ipswich, Massachusetts) and purified using G25 Quick Spin Columns for radiolabeled DNA purification (Roche). A 24 bp mutant probe (TCGCGTG in the XRE-probe was changed to CCATAGG) was labeled in the same manner. Gel shift reactions were performed as follows: 1 µg of nuclear proteins were incubated with the radiolabeled XRE-probe in 5×gel shift buffer (Promega) in the presence of 0.25 µg poly(deoxyinosinic-deoxycytidylic) acid (Sigma) for 45 minutes at room temperature. 20 µM of cold competitor (unlabeled XRE-probe) or 10 µM mutant probe were added as needed. For supershift reactions, 1 µl anti-ARNT antibody (ab2, Abcam) was added to the gel shift reaction after 10 minutes incubation. Reactions were run on a 6% native polyacrylamide gel and subsequently dried in a gel drier. The gel was exposed to x-ray film (Kodak) for 3 days at room temperature. The film was scanned and bands were quantified using ImageJ.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the MAGnify Chromatin Immunoprecipitation System (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol with slight modifications. HaCaT cells at 80–90% confluence in a 10 cm dish were washed with cold 1x Hank’s Balanced Salt Solution (HBSS) and DNA-protein complexes were crosslinked with 1% formaldehyde for 12 minutes at room temperature. Samples in hypoxia were handled in the hypoxic chamber until after DNA-protein complexes were crosslinked. Crosslinked samples were transferred to normal oxygen for the subsequent steps. After the crosslinking reaction was stopped with 1.25 M glycine, cells were washed with cold 1x HBSS and harvested in 1x HBSS containing 0.1% EDTA, 2% FBS and protease inhibitors (1 mM PMSF, 1 µg/ml Aprotinin, 1 µg/ml Pepstatin A). Cells were centrifuged, washed with cold 1x HBSS containing protease inhibitors, and lysed in the appropriate amount of lysis buffer on ice for 20 minutes. Cell lysates were transferred to 0.5 ml Bioruptor tubes and sonicated for 10 cycles (30 seconds ON, 30 seconds OFF) on high at 4°C using the Bioruptor UCD-200 and water cooler (Diagenode, Denville, NJ) to a length of 200 bp and bigger. The sheared chromatin was centrifuged and the supernatant diluted. Approximately 300,000 cells were used per ChIP. Samples were incubated with 3 µg of anti-AhR antibody (H-211, sc-5579X, Santa Cruz Biotechnology) overnight at 4°C. After the bound chromatin was washed, the DNA-protein crosslinks were reversed using proteinase K and the AhR-enriched fraction of genomic DNA was purified. qRT-PCR was used to analyze ChIP DNA for enrichment at two different regions on the CYP1A1 and CYP1B1 promoters. Enrichment at the beta-actin promoter was used as a negative control. CYP1A1_1: forward primer: 5'-GCGCGAACCTCAGCTAGT-3', reverse primer: 5'-TTCCCGGGGTTACTGAGTC-3', UPL probe: #23; CYP1A1_2: forward primer: 5'-CCCCCTAGAGGGATGTCG-3', reverse primer: 5'-TGATTGGCAGAGCACAGAAA-3', UPL probe: #70; CYP1B1_1: 5'-GCAGAAACTTCAACCCGATAA-3', reverse primer: 5'-GAGCGGGGCGGAGAGT-3', UPL probe: #63; CYP1B1_2: 5'-TGTCAGGTGCCGTGAGAA-3', reverse primer: 5'-AGTCGGCTCCAGTCATATCC-3', UPL probe: #27; beta-actin: forward primer: 5'-CAAAGGCGAGGCTCTGTG-3', reverse primer: 5'-CCGAAAGTTGCCTTTTATGG-3', UPL probe: #77. 2.5 µl of diluted ChIP DNA was used as template for each real-time PCR reaction (20 µL reaction volume) with primer pairs at 0.3 µM and UPL probes at 0.2 µM. For all reactions PerfeCTa qPCR SuperMix Low ROX (Quanta Biosciences, Gaithersburg, MD) was used. The DNA polymerase was heat-activated at 95°C for 3 minutes followed by 45 cycles, denaturing at 95°C for 15 seconds, annealing and elongating at 60°C for 45 seconds. Data were collected and fold enrichment was calculated by subtracting the CT value of the ChIP DNA from the CT value of the input DNA fraction (adjusted for the dilution factor as described in the protocol). Relative enrichment = 2(CT(Input)- CT(ChIP)).
Statistical analysis
Significant differences between groups of data were determined using a t-test for all analyses. n = 3 was used for each data set unless otherwise noted. Statistical probability of p < 0.05 was considered significant.
Results
Hypoxia inhibits PCB 126 induced expression of CYP1A1
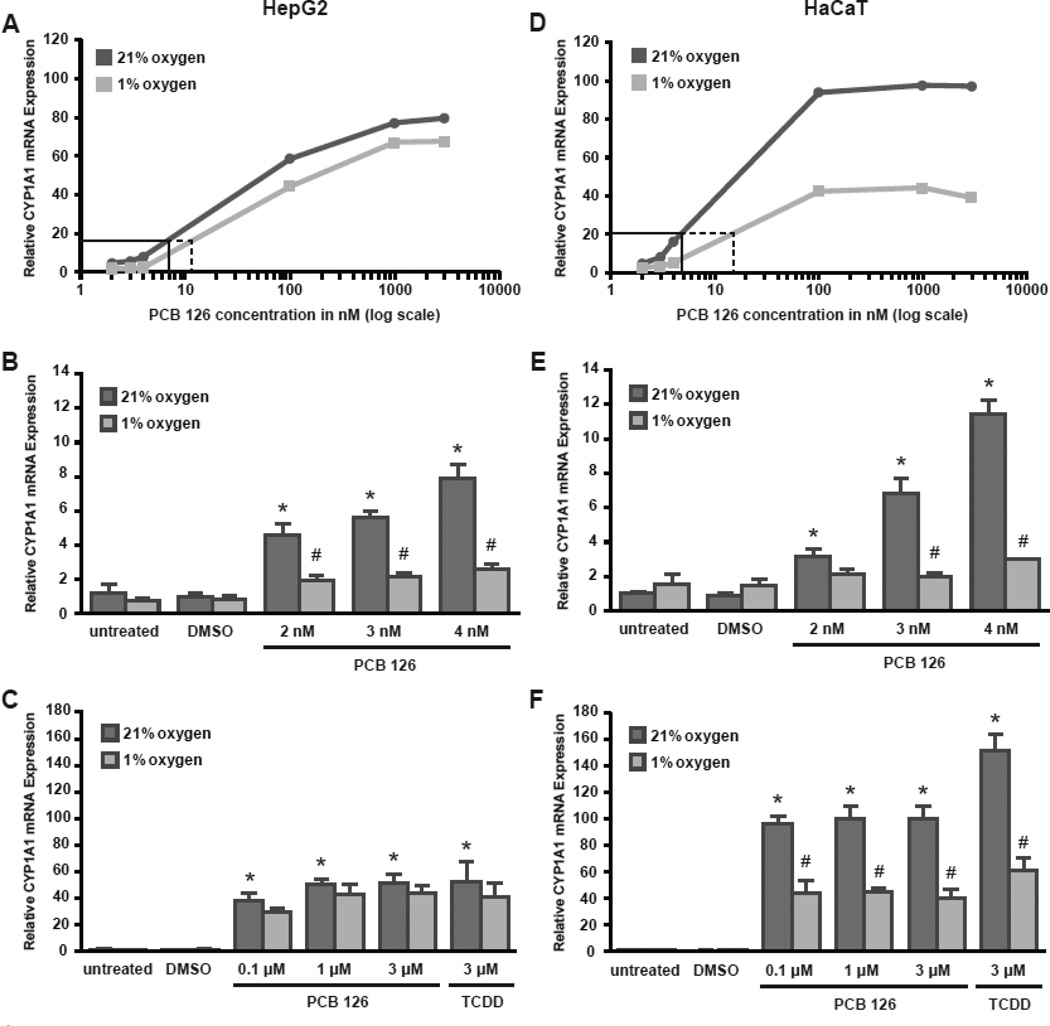
Hepatocellular carcinoma (HepG2) and keratinocyte (HaCaT) cell lines were used to assess the effect of hypoxia on AhR-dependent gene expression. Cell lines were treated with either 3 µM TCDD (positive control) or varying doses of the dioxin-like coplanar PCB 126 under normoxic or hypoxic conditions. The accumulation of CYP1A1 mRNA was then determined by qRT-PCR. TCDD and PCB 126 significantly induced expression of CYP1A1 mRNA in both HepG2 cells (Figure 1 B, C) and HaCaT cells (Figure 1 E, F). The low dose treatment groups showed a dose-response relationship with concentrations as low as 2 nM significantly inducing CYP1A1 mRNA expression. In the high dose treatment groups, HepG2 cells showed a lower overall response to PCB 126 treatment compared to HaCaT cells. Incubation in hypoxia for 8 hours prior to PCB 126 treatment significantly decreased CYP1A1 mRNA expression in both cell lines. To further assess the inhibitory effect of hypoxia, a dose-response curve was plotted for PCB 126 treated cells in normoxia and hypoxia, and EC20 values (the effective concentration to achieve 20% of the maximal response) were calculated (Figure 1 A, D). Both cell lines showed a shift of the dose-response curve to the right in hypoxic conditions compared to normoxia, thus increasing the EC20. The EC20 value for PCB 126 treated HepG2 cells increased from 7.1 nM in normal oxygen to 12.0 nM in hypoxia. In HaCaT cells the EC20 increased from 4.8 nM in normoxic conditions to 14.7 nM in hypoxia. To rule out that hypoxia had an effect on AhR or ARNT expression that could be causing the observed effects, mRNA levels of AhR and ARNT were measured in untreated cells in normoxia and hypoxia (Figure S1). No apparent differences in mRNA expression levels in normoxia or hypoxia were observed in either cell line. To extend these findings and to determine whether the observed CYP1A1 induction after PCB 126 treatment is AhR-specific, cell lines were treated with the AhR antagonist 6,2',4'-trimethoxyflavone (TMF) for 4 hours prior to treatment with PCB 126 (Figure S2 A, C). In both cell lines TMF did inhibit CYP1A1 mRNA expression after PCB 126 treatment compared to cultures treated with PCB 126 alone. Furthermore, cell lines were treated with the non-coplanar non-dioxin-like PCB 153 which is not a ligand for the AhR (Figure S2 B, D). PCB 153 did not induce CYP1A1 mRNA expression in either cell line, suggesting that induction of CYP1A1 is PCB 126 and TCDD-specific and likely mediated through AhR activation.
Figure 1. Oxygen concentration dependent induction of CYP1A1 mRNA expression.
HepG2 cells (left panel: A-C) and HaCaT cells (right panel: D-F) were incubated in normoxia (21% O2) or hypoxia (1% O2) for 8 hours prior to treatment with vehicle (DMSO), 2–4 nM PCB 126, 0.1–3 µM PCB 126 or 3 µM TCDD for 6 hours or 4 hours, respectively. CYP1A1 mRNA expression was determined by qRT-PCR. Analysis was performed with normalization to RPLP0 mRNA. (A, D) Dose-response curves for PCB 126 treated cells in normal oxygen and hypoxia. The EC20 is indicated by a solid black line in normoxic cells and a dashed line in hypoxic cells. (B, C and E, F) CYP1A1 mRNA expression levels of untreated and treated cells including standard error. RNA expression levels significantly (p < 0.05) greater than untreated controls are indicated by an asterisk (*). RNA expression levels significantly (p < 0.05) different between the PCB 126 treated samples in normoxia and hypoxia are indicated by a number sign (#). Error bars = SEM. n = 4 for PCB 126, n = 3 for TCDD.
As we observed significant inhibition of CYP1A1 mRNA expression in hypoxia, we performed western blots and enzyme activity assays to measure CYP1A1 protein levels and enzyme activity in different oxygen environments. We observed that after 24–48 hours of PCB 126 treatment in normoxia, CYP1A1 protein levels were increased compared to untreated controls in both cell lines (FIGURE 2 A, B). In contrast, cells incubated for 16 hours in hypoxia prior to PCB 126 treatment showed less protein expression under these conditions. To further investigate the effect of hypoxia on CYP1A1, we utilized the highly selective CYP1A1 luminogenic substrate Luciferin-1A1 to assess whether hypoxia negatively affects CYP1A1 enzyme activity (Figure 2 C, D). CYP1A1 enzyme activity was significantly increased in PCB 126 treated cells compared to untreated controls. PCB 126 treated cells in hypoxia showed significantly lower CYP1A1 enzyme activity compared to cells treated in normoxia. In summary, our findings suggest that hypoxia is an important modulator of PCB 126 induced CYP1A1 expression and function which might be due to hypoxic interference with AhR function.
Figure 2. CYP1A1 protein levels and enzyme activity are reduced in low oxygen environments.
Upper panel: HepG2 cells (A) and HaCaT cells (B) were subjected to normoxia or hypoxia for 16 hours prior to treatment with 3 µM PCB 126 for 48 hours or 24 hours, respecitively. Ten micrograms of whole cell lysate were western blotted for the presence of CYP1A1. Beta-tubulin was used as a loading control. Numbers over the lanes represent relative intensities of CYP1A1 compared to PCB 126 treated cells in normoxia. n.d. = not detectable, n.s. = non-specific band. Lower panel: HepG2 cells (C) and HaCaT cells (D) were incubated in normoxia or hypoxia for 16 hours before treatment with 3 µM PCB 126 for 24 hours. CYP1A1 activity was measured using a luminogenic CYP1A1 substrate and is depicted relative to untreated cells in normoxia. * = p < 0.005 vs. control. # = p < 0.005 normoxia vs. hypoxia. Error bars = SEM. n = 3.
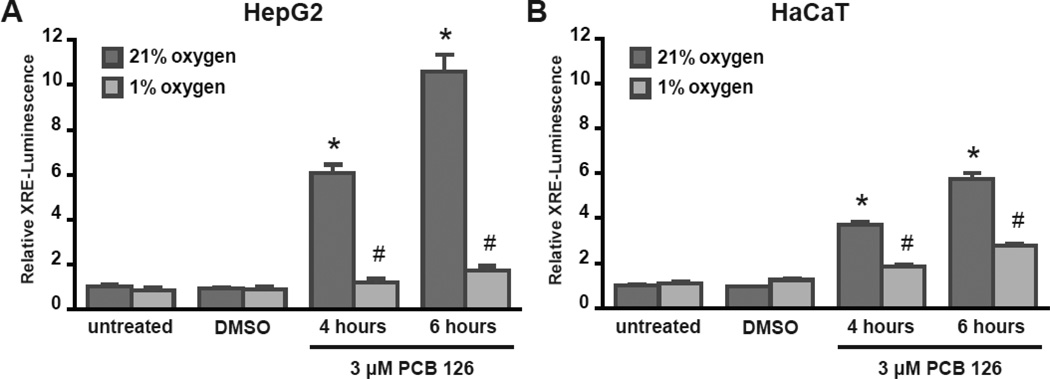
Hypoxia inhibits induction of XRE-luciferase reporter activity
To investigate the mechanism of hypoxic inhibition of gene expression after PCB 126 treatment and to assess AhR:ARNT function, HepG2 and HaCaT cells were transfected with a XRE-luciferase reporter construct containing 1.5 kb of the human CYP1A1 promoter (Morel and Barouki, 1998). Transfected cells were either cultured in normoxia or hypoxia for 16 hours prior to 4 or 6 hours of PCB 126 treatment and luminescence was measured (Figure 3). Consistent with our previous results, treatment with PCB 126 significantly induced expression of the XRE-luciferase reporter gene in normoxic conditions. However, hypoxia significantly inhibited reporter gene expression in PCB 126 treated cells suggesting that hypoxia perturbs the normal interaction of the AhR:ARNT heterodimer and subsequent activation of the CYP1A1 promoter. Cells similarly transfected with a HRE-luciferase reporter vector responded to hypoxia with a robust expression of the luciferase reporter gene upon hypoxia treatment, confirming the accuracy of hypoxic exposure (Figure S3). Next, we asked whether this hypoxic induction of a HRE-luciferase reporter could conversely be inhibited by pre-treatment with PCB 126. HepG2 and HaCaT cells transfected with a HRE-luciferase reporter showed strong luminescence under hypoxic conditions; however, treatment with PCB 126 for 4 hours prior to transferring cells to hypoxia inhibited HRE-luciferase reporter gene expression (Figure S3). Together, these data suggest that ARNT is a critical factor in both AhR and hypoxia signaling.
Figure 3. Hypoxia inhibits PCB 126 induced CYP1A1 promoter-luciferase reporter activity.
HepG2 cells (A) and HaCaT cells (B) were transfected with a XRE-luciferase (firefly) reporter vector and Renilla luciferase vector and then subjected to normoxia or hypoxia for 16 hours prior to treatment with 3 µM PCB 126 for 4 or 6 hours. Firefly luminescence was determined and normalized to Renilla luminescence, and is depicted relative to untreated cells in normoxia. * = p < 0.005 vs. control. # = p < 0.005 normoxia vs. hypoxia. Error bars = SEM. n = 3.
To further assess AhR:ARNT function and binding to a XRE sequence, an electrophoretic mobility shift assay (EMSA) was performed with nuclear protein extracts of untreated and PCB 126 treated HepG2 cells in normoxia and hypoxia (Figure S4). Compared to untreated cells in normoxia, binding to the radiolabeled XRE-probe was increased after 1 hour of PCB 126 treatment. In contrast, 8 hours of hypoxia pre-treatment, sufficient to induce HIF-1α expression prior to PCB 126 treatment, strongly decreased binding to the XRE-probe. In order to confirm the identity of the gel shift complex, cold and mutant competitors as well as an anti-ARNT antibody for a supershift reaction were used. Taken together, these data suggest that binding of the AhR:ARNT heterodimer to XRE sequences might be indirectly dependent on available oxygen concentrations.
ARNT overexpression relieves the cellular physiological effects of hypoxia on the AhR signaling pathway
The data described so far show that pre-treatment with hypoxic conditions was inhibitory to PCB 126 mediated AhR activation and target gene expression. Since hypoxia induces HIF-1α and leads to HIF-1α:ARNT dimerization and nuclear localization, we reasoned that ARNT bioavailability to the AhR would be restricted under hypoxic conditions and this could be the mechanism underlying the observed effects. To test this hypothesis, we overexpressed human ARNT in HepG2 cells along with the XRE-containing CYP1A1 promoter-luciferase reporter vector (Figure S5). Cells were subsequently exposed to PCB 126 with or without hypoxia pretreatment as described in previous experiments (Figure 4). Cells overexpressing ARNT showed an expectedly higher baseline of CYP1A1 promoter activity. Importantly, we found that ARNT overexpression was able to significantly rescue cells from the inhibitory effect of hypoxia on transactivation of the CYP1A1 promoter induced by PCB 126. Therefore, the mechanism of interference of the signaling crosstalk between the AhR and hypoxia pathways appears to be at least in part dependent on ARNT bioavailability. The responses of empty control vector transfected cells did not differ from cells transfected with the XRE-luciferase reporter vector alone.
Figure 4. ARNT overexpression relieves hypoxic inhibition of CYP1A1 promoter-luciferase reporter activity.
HepG2 cells were transfected with a XRE-luciferase (firefly) reporter vector, Renilla luciferase vector, and ARNT expression vector or empty control vector. Transfected cells were subjected to normoxia or hypoxia for 16 hours prior to treatment with 3 µM PCB 126 for 6 hours. Firefly luminescence was determined and is depicted relative to untreated cells in normoxia in the empty control vector group. * = p < 0.05 vs. control. # = p < 0.05 normoxia vs. hypoxia. ‡ = p < 0.05 empty vector vs. ARNT expression vector. Error bars = SEM. n = 3.
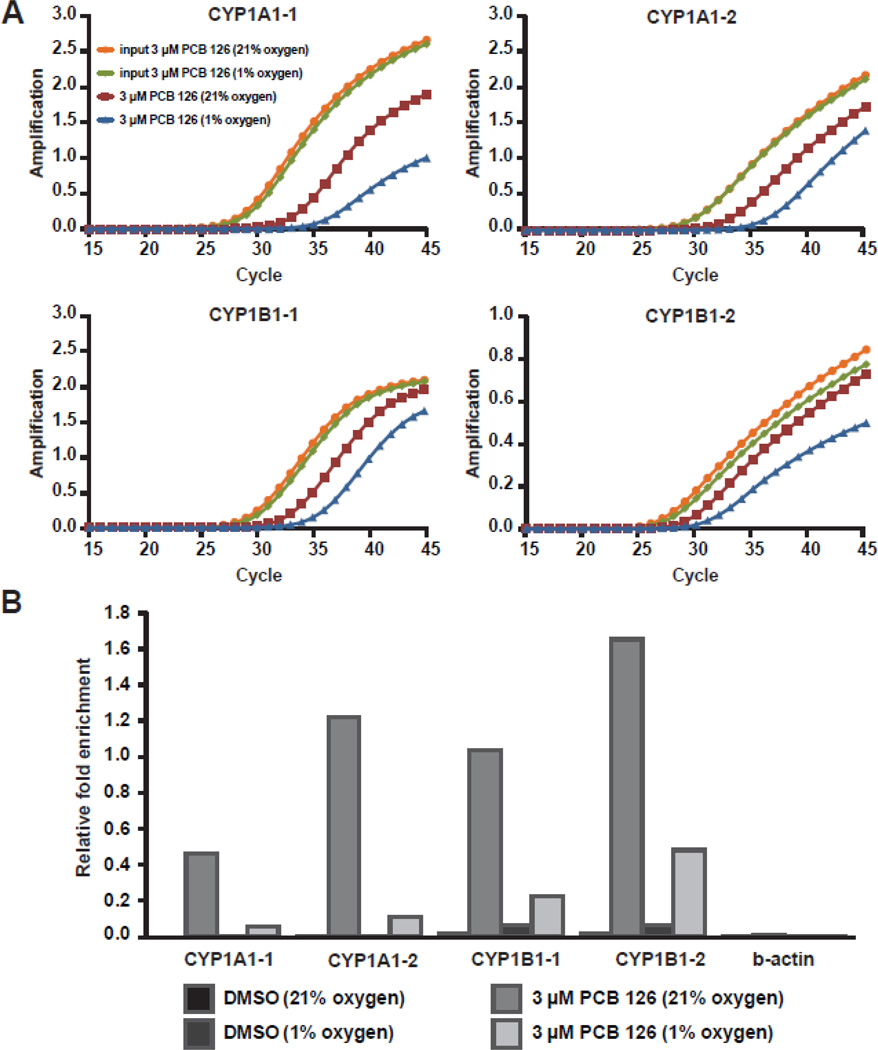
Differential binding of the AhR to CYP1 promoters
After ligand activation, the nuclear AhR:ARNT heterodimer induces transcription by binding to XRE sequences in AhR target genes. In order to determine whether hypoxia affects binding of the AhR to genomic XRE sequences in the CYP1A1 and CYP1B1 promoters, we performed chromatin immunoprecipitation (ChIP) with antibodies directed against human AhR (Figure 5). Our results show that the AhR was associated with multiple regions along the CYP1A1 and CYP1B1 promoters in HaCaT cells treated with PCB 126 under normal oxygen conditions. Enrichment at these loci was indeed decreased in PCB 126 treated cells cultured in hypoxia. DMSO-treated control cells showed only minimal enrichment of the AhR at the analyzed CYP1A1 and CYP1B1 promoter regions. Enrichment at the beta-actin promoter served as a negative control and showed no apparent binding of the AhR. These findings strongly suggest that binding of the AhR to chromatin at its cognate binding sites is at least in part dependent on oxygen.
Figure 5. Differential binding of the AhR to CYP1A1 and CYP1B1 promoters after PCB 126 treatment.
HaCaT cells were subjected to normoxia or hypoxia for 16 hours and subsequently treated with 3 µM PCB 126 for 1 hour. ChIP assays were performed using an anti-AhR antibody. Enriched DNA was analyzed using qRT-PCR primers designed to regions in the promoters of CYP1A1 and CYP1B1. Enrichment at the beta-actin promoter was used as a negative control. (A) Magnitude of the signal (delta Rn) vs. cycle number. (B) Enrichment levels calculated by subtracting CT values of ChIP DNA from input DNA in each sample.
Discussion
Both the AhR and HIF-1α are members of the class I bHLH/PAS (basic helix-loop-helix/PER-ARNT-SIM) protein family and form obligate heterodimers with their common binding partner ARNT, a member of the class II bHLH/PAS protein family, prior to actively transcribing their respective target genes (Kewley et al., 2004). Given the importance of ARNT in both the xenobiotic responsive AhR and hypoxia responsive HIF-1α pathways, it may be likely that ARNT is a central regulator of these pathways and can thus mediate crosstalk that can affect the regulation of gene expression. Although the influence of hypoxia, induced by chemical agents that induce HIF-1α transcriptional activity, on TCDD-induced AhR signaling has been studied, little is known about the effect of genuine hypoxia, that is low oxygen environments, on AhR signaling induced by PCBs. Previous studies that examined the role of ARNT in AhR-mediated cellular responses and the importance of ARNT in AhR and HIF-1α signaling have revealed variable results (Pollenz et al., 1999; Fleming et al., 2009; Nukaya et al., 2010). In the context of TCDD-mediated induction of the AhR, Pollenz et al. showed no competition for ARNT under hypoxic conditions that induced HIF-1α activity. In contrast, Fleming et al. suggested that in topminnow hepatocarcinoma cells, induction of the AhR by various AhR ligands, including PCB 126, was ARNT-dependent and HIF-1α might sequester ARNT under hypoxia-like conditions. Furthermore, Nukaya et al. showed that ARNT is required for AhR-mediated toxic responses in the mouse liver. Given these prior findings, we sought to analyze PCB 126 induced AhR signaling in different oxygen environments in human cell lines representative of target tissues for PCB toxicity in humans. Suitable tissues to assess the effects of hypoxia are the skin and the liver which both represent important target tissues of PCB toxicity. Importantly from a physiological perspective, the liver normally exhibits varying concentrations of oxygen in anatomically defined zones under physiological conditions and therefore is a useful model system to study PCB 126 induced AhR signaling under hypoxia (Oinonen and Lindros, 1998).
In the present study, we demonstrate that hypoxia significantly interfered with AhR functions after treatment with PCB 126 and that this hypoxic interference could negatively affect CYP1A1 mRNA expression, translating to less protein expression and enzyme activity. Our results show that CYP1A1 mRNA expression was significantly increased after exposure to PCB 126. Accumulation of CYP1A1 mRNA was significantly decreased in hypoxia compared to cells treated in a normal oxygen environment. We further observed an increase of the EC20 values calculated from dose-response curves in hypoxic conditions compared to normoxia. The observed reduction in CYP1A1 mRNA expression in hypoxia was likely due to inhibition of transcriptional activation of CYP1A1 since transactivation of a 5×XRE-containing CYP1A1 promoter-luciferase reporter vector was also inhibited by exposure to hypoxia followed by PCB 126 challenge. We further observed that CYP1A1 protein expression and enzyme activity induced by PCB 126 were significantly reduced in hypoxia compared to cells treated in 21% oxygen. To demonstrate that induction of CYP1A1 after PCB 126 exposure was mediated through the AhR, we exposed cells to PCB 126 in the absence or presence of the AhR antagonist TMF. Cells treated with TMF showed decreased CYP1A1 mRNA levels after PCB 126 treatment compared to cells treated with PCB 126 alone, indicating that the effects were likely mediated through the AhR. Furthermore, CYP1A1 was not induced by PCB 153, a non-coplanar non-dioxin-like PCB that does not activate the AhR. These data suggest that induction of CYP1A1 after PCB 126 treatment is likely mediated by the AhR and that hypoxic conditions perturb this normal transcriptional response.
Our data suggest that AhR function is indirectly dependent upon the environmental oxygen availability and that a hypoxic environment can therefore interfere with normal AhR signaling. Both the AhR and HIF-1α require ARNT as an obligate heterodimer prior to actively binding DNA and activating transcription of their respective target genes. We hypothesized that under hypoxic conditions HIF-1α binds to ARNT, thus limiting its bioavailability for AhR to induce a robust AhR transcriptional response after PCB 126 exposure. To further elucidate the mechanism of inhibition of the AhR transcriptional program by hypoxia and to assess the importance of the common factor ARNT, we first measured AhR transcriptional activity using the XRE-containing human CYP1A1 promoter driving a luciferase reporter. In both HepG2 and HaCaT cells PCB 126 exposure induced a time-dependent increase in XRE-dependent transcriptional activation of the CYP1A1 promoter reporter gene after PCB 126 exposure. Transcriptional responses were significantly decreased under hypoxic conditions that induced HIF-1α suggesting that the normal transcriptional function of the AhR:ARNT heterodimer is perturbed under these conditions. To assess the role of ARNT in the antagonistic relationship between the AhR and HIF-1α proteins, we overexpressed human ARNT along with the CYP1A1 promoter-luciferase reporter vector. Compared to cells transfected with an empty control vector that express only endogenous levels of ARNT, ARNT overexpressing cells showed increased ARNT protein levels. Importantly, overexpression of ARNT nearly completely relieved the inhibitory effect of hypoxia on CYP1A1 promoter activity in PCB 126 treated HepG2 cells, suggesting that ARNT sequestration by HIF-1α mediates the inhibitory effects observed under hypoxia. We interpret these data to mean that the increase in ARNT bioavailability enables AhR:ARNT heterodimer formation and AhR function even under hypoxic conditions when HIF-1α is also paired with ARNT. This finding clearly illustrates the crosstalk that exists at this important signaling node and provides a mechanism for the observed effects of hypoxia on PCB 126 mediated CYP1A1 transactivation. To more closely assess AhR:ARNT binding to target XRE sequences, EMSA experiments were performed. HepG2 cells showed increased gel shift complex formation after treatment with PCB 126 compared to untreated cells. However, the formation of gel shift complexes was decreased in PCB 126 treated cells in hypoxia compared to PCB 126 treated cells in normal oxygen. These data suggest that hypoxic induction of HIF-1α inhibits AhR:ARNT binding to a target XRE sequence in vitro. Further supporting evidence of inhibited AhR function and reduced AhR binding to target sequences in cells under hypoxic conditions was provided by chromatin immunoprecipitation experiments. HaCaT cells treated with PCB 126 in 21% oxygen showed strong enrichment of AhR binding at both the CYP1A1 and CYP1B1 target promoters. In contrast, AhR binding to these loci was blocked or reduced in low oxygen environments, further indicating a functional inhibitory crosstalk between the AhR and HIF-1α pathways. To test whether PCBs could conversely inhibit a robust hypoxic response, we utilized a hypoxia-inducible HRE-luciferase reporter and first pre-treated HepG2 and HaCaT cells with PCB 126 prior to exposure to hypoxia. HRE-luminescence was strongly induced in cells transfected with the HRE-luciferase reporter; however, cells pre-treated with PCB 126 for 4 hours showed a decreased luminescent signal. These data suggest that there is indeed an important interferential crosstalk between the AhR and HIF-1α signaling pathways and further suggest that ARNT is likely a critical factor in both AhR and hypoxia signaling.
Taken together, our findings suggest that AhR function is likely sensitive to environmental oxygen conditions and that hypoxia can therefore interfere with AhR signaling. In the presence of oxygen, the ligand-activated AhR binds to XRE target sequences inducing the expression of AhR target genes such as CYP1A1. However, HIF-1α induction prior to PCB 126 treatment squelches the normal AhR response to PCB 126, likely through an impaired formation of the AhR:ARNT heterodimer.
Conclusions
Due to the accumulation and long half-lives of PCBs, these organic contaminants pose a significant health risk to both humans and wildlife in the environment. As certain organs can experience varying concentrations of oxygen under normal physiologic conditions, it is critical to study the influence of oxygen concentrations on PCB-induced physiological processes in the human body.
The results in this study show that hypoxia interfered with AhR binding to genomic target gene sequences and that AhR target gene expression was perturbed in hypoxic environments. Our data implicate ARNT as a likely critical factor in the crosstalk between the AhR and HIF-1α signaling pathways and further suggest that AhR function is therefore indirectly sensitive to available oxygen concentrations through ARNT bioavailability. Conditions of reduced oxygen availability can significantly perturb the normal transcriptional response of the AhR after PCB 126 exposure which might ultimately affect the metabolism and toxicity of PCBs.
Supplementary Material
Highlights.
Significant crosstalk exists between AhR and HIF-1α signaling
Hypoxia perturbs PCB 126 induced AhR function and target gene expression
PCB 126 mediated activation of AhR activity inhibits HIF-1α signaling
AhR binding to CYP1A1 and CYP1B1 promoters is inhibited by hypoxia
ARNT overexpression relieves hypoxic inhibition of AhR function
Acknowledgements
This work was supported by National Institutes of Health (NIH) grant P42 ES013661-5099 (FED), P42 ES004940-5083 (BWF), and P50 ES006694. SUV received salary support from SRP Training Core P42 ES013661-5110. SUV further received a Superfund Research Program K.C. Donnelly Externship Administrative Supplement. PLS received salary support from NIH grant 2 T32 ES007091-31. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors would like to thank all the research staff in the Futscher lab for their valuable support and assistance during the K.C. Donnelly Externship training at The University of Arizona. Furthermore the authors would like to thank Dr. Weigel at the University of Iowa for providing space and reagents for the EMSA assays.
Abbreviations
- AhR
aryl hydrocarbon receptor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- bHLH/PAS
basic helix-loop-helix/PER-ARNT-SIM
- ChIP
chromatin immunoprecipitation
- CYP1A1
cytochrome P450 1A1
- DMSO
dimethyl sulfoxide
- EMSA
electrophoretic mobility shift assay
- HIF-1α
hypoxia-inducible factor-1α
- HRE
hypoxia response element
- PCB
polychlorinated biphenyl
- PCB 126
3,3',4,4',5-pentachlorobiphenyl
- PCB 153
2,2',4,4',5,5'-hexachlorobiphenyl
- qRT-PCR
quantitative real-time reverse transcription polymerase chain reaction
- RPLP0
ribosomal protein, large, P0
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TMF
6,2',4'-trimethoxyflavone
- XRE
xenobiotic response element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare that no conflicts of interest exist.
References
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb LS, Andersen ME, Broccardo CJ, Legare ME, Billings RE, Dean CE, Hanneman WH. Regional induction of CYP1A1 in rat liver following treatment with mixtures of PCB 126 and PCB 153. Toxicol. Pathol. 2004;32:467–473. doi: 10.1080/01926230490483306. [DOI] [PubMed] [Google Scholar]
- Crinnion WJ. Polychlorinated biphenyls: persistent pollutants with immunological, neurological, and endocrinological consequences. Altern. Med. Rev. 2011;16:5–13. [PubMed] [Google Scholar]
- Fleming CR, Billiard SM, Di Giulio RT. Hypoxia inhibits induction of aryl hydrocarbon receptor activity in topminnow hepatocarcinoma cells in an ARNT-dependent manner. Comp Biochem Physiol C Toxicol Pharmacol. 2009;150:383–389. doi: 10.1016/j.cbpc.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LG, DeCaprio AP, Nisbet ICT. PCB congener comparisons reveal exposure histories for residents of Anniston, Alabama, USA. Fresenius Environ. Bull. 2003;12:181–190. [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Kim JE, Sheen YY. Inhibition of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-stimulated Cyp1a1 promoter activity by hypoxic agents. Biochem. Pharmacol. 2000;59:1549–1556. doi: 10.1016/s0006-2952(00)00283-5. [DOI] [PubMed] [Google Scholar]
- Ludewig G, Lehmann L, Esch H, Robertson LW. Metabolic Activation of PCBs to Carcinogens in Vivo - A Review. Environ Toxicol Pharmacol. 2008;25:241–246. doi: 10.1016/j.etap.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel Y, Barouki R. Down-regulation of cytochrome P450 1A1 gene promoter by oxidative stress. Critical contribution of nuclear factor 1. J. Biol. Chem. 1998;273:26969–26976. doi: 10.1074/jbc.273.41.26969. [DOI] [PubMed] [Google Scholar]
- Nie M, Blankenship AL, Giesy JP. Interactions between aryl hydrocarbon receptor (AhR) and hypoxia signaling pathways. Environ Toxicol Pharmacol. 2001;10:17–27. doi: 10.1016/s1382-6689(01)00065-5. [DOI] [PubMed] [Google Scholar]
- Nukaya M, Walisser JA, Moran SM, Kennedy GD, Bradfield CA. Aryl hydrocarbon receptor nuclear translocator in hepatocytes is required for aryl hydrocarbon receptor-mediated adaptive and toxic responses in liver. Toxicol. Sci. 2010;118:554–563. doi: 10.1093/toxsci/kfq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinonen T, Lindros KO. Zonation of hepatic cytochrome P-450 expression and regulation. Biochem. J. 1998;329(Pt 1):17–35. doi: 10.1042/bj3290017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollenz RS, Davarinos NA, Shearer TP. Analysis of aryl hydrocarbon receptor-mediated signaling during physiological hypoxia reveals lack of competition for the aryl hydrocarbon nuclear translocator transcription factor. Mol. Pharmacol. 1999;56:1127–1137. doi: 10.1124/mol.56.6.1127. [DOI] [PubMed] [Google Scholar]
- Safe S. Toxicology, structure-function relationship, and human and environmental health impacts of polychlorinated biphenyls: progress and problems. Environ. Health Perspect. 1993;100:259–268. doi: 10.1289/ehp.93100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K, An WG, Melillo G, Blagosklonny MV, Costa M. Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis. 1999;20:1819–1823. doi: 10.1093/carcin/20.9.1819. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberhorn EM, Glauert HP, Robertson LW. Carcinogenicity of polyhalogenated biphenyls: PCBs and PBBs. Crit. Rev. Toxicol. 1990;20:440–496. doi: 10.3109/10408449009029331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.