Abstract
Mortality from tobacco smoking remains the leading cause of preventable death in the world, yet current cessation therapies are only modestly successful, suggesting new molecular targets are needed. Genetic analysis of gene expression and behavior identified Chrna7 as potentially modulating nicotine place conditioning in the BXD panel of inbred mice. We used gene targeting and pharmacological tools to confirm the role of Chrna7 in nicotine CPP. To identify molecular events downstream of Chrna7 that may modulate nicotine preference, we performed microarray analysis of α7 KO and WT nucleus accumbens tissue, followed by confirmation with quantitative PCR and immunoblotting. In the BXD panel, we found a putative cis eQTL for Chrna7 in nucleus accumbens that correlated inversely to nicotine CPP. We observed that gain-of-function α7 mice did not display nicotine preference at any dose tested, while conversely, α7 KO mice showed nicotine place preference at a dose below that routinely required to produce preference. In B6 mice, the α7 nAChR-selective agonist, PHA-543613, dose-dependently blocked nicotine CPP, which was restored using the α7 nAChR-selective antagonist, MLA. Our genomic studies implicated an mRNA co-expression network regulated by Chrna7 in nucleus accumbens. Mice lacking Chrna7 demonstrate increased insulin signaling in the nucleus accumbens, which may modulate nicotine place preference. Our studies provide novel targets for future work on development of more effective therapeutic approaches to counteract the rewarding properties of nicotine for smoking cessation.
Keywords: nicotine, Chrna7, alpha7 nicotinic acetylcholine receptor, quantitative trait loci, conditioned place preference, BXD panel, insulin, behavioral genetics, genomics, reward
Introduction
Nicotine dependence as a result of tobacco use is the leading cause of preventable death in the world, contributing to 90% of lung cancer cases in the United States and 49,400 deaths from second-hand smoke per year (National Institutes of Health 2012). Several studies have demonstrated that genetic influences on nicotine addiction and dependence exist, with trait heritabilities between 46-84% (Swan et al. 1997; Kendler et al. 1999; Heath & Martin 1993; True et al. 1999). Although roughly 70% of adult smokers aspire to quit smoking, only 4-7% are successful without medication and only 25% of those using current cessation therapies are able to abstain from smoking for six months (American Cancer Society 2012). The inability to quit smoking is thought to be due to high rates of relapse from difficulty in managing cravings and withdrawal symptoms (National Institutes of Health 2008), thus, more effective treatments are needed.
It has been hypothesized that associative cues in smokers can maintain drug-seeking behavior and reinforcement, even in the absence of nicotine (Rose & Corrigall 1997; Caggiula et al. 2001). A conditioned stimulus choice test, conditioned place preference (CPP), has been used in animals since the early 1940s to model appetitive reward-like properties of drugs of abuse (Spragg 1940; Rossi NA 1976). Thus, using CPP to test an animal's drug-free response to contextual cues associated with nicotine may lead to discovery of neural pathways that play a role in nicotine reward and reinforcement (Bardo & Bevins 2000).
Behavioral genetics studies in animal models have been widely used to study the role of specific nicotinic subunits or other genes in nicotine's reward-like properties or other behaviors (Changeux 2010). Additionally, several human studies have associated the genetic variation within multiple nicotinic acetylcholine receptor (nAChr) genes with different nicotine dependence phenotypes (Ehringer et al. 2007; Bierut et al. 2007; Saccone et al. 2007; Zeiger et al. 2008; Amos et al. 2008; Berrettini et al. 2008; Hung et al. 2008; Saccone et al. 2010). However, few forward genetics approaches have been used to identify novel targets for intervention in nicotine behaviors. Modern genetic panels, together with high-density genotyping and use of expression genetics, have improved the prospects for using forward genetics to identify gene networks modulating complex traits such as nicotine dependence (Hitzemann et al. 2004; Carlborg et al. 2005; Broide et al. 2002).
To identify gene networks involved in nicotine dependence, we used a combination of behavioral genetics and pharmacological studies in mice, together with genetic analysis of gene expression. Our results implicate genetic variation in Chrna7 mRNA expression and its potential regulation of insulin signaling as modulators of nicotine conditioned place preference. These studies may have important implications for understanding and treating nicotine dependence in humans.
Methods and Materials
Mice
For all studies, male mice were housed 3-5 per cage and allowed at least a one-week acclimation period to the vivarium following shipment to Virginia Commonwealth University (VCU). Mice were maintained on a 12-hour light/dark cycle with ad libitum access to food and water. Adult mice were tested or had tissues harvested between 7-12 weeks of age during their light phase. C57BL/6J (B6, Stock No. 000664), DBA/2J (D2, Stock No. 000671), and BXD (B6 × D2) recombinant inbred mice were obtained from Jackson Laboratories (Bar Harbor, ME). Chrna7 knock-in, gain-of-function (α7 KI) mice (Chrna7 L250T +/−), bred from heterozygous breeding pairs, were obtained from Baylor College of Medicine (Houston, TX) (Broide et al. 2002). Chrna7 homozygous knock-out (α7 KO) mice (B6.129S7-Chrna7tm1Bay/J, Stock No. 003232) were either obtained from Jackson Laboratories or heterozygote breeding pairs were obtained from which WT and KO mice were bred and genotyped at VCU. Both α7 KI and α7 KO mice were backcrossed to the background strain, C57BL/6J, for an additional 8-10 generations and wild-type littermates (α7 WT) were used as controls. The animal facility was approved by the Association for Assessment and Accreditation of Laboratory Animal Care. Experiments were performed during the light cycle and approved by the Institutional Animal Care and Use Committee of VCU.
Drugs and Chemicals
(−)-Nicotine hydrogen tartrate salt and methyllycaconitine citrate (MLA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). PHA-543613 [N-[(3R)-1-azabicyclo[2.2.2]oct-3-yl]furo[2,3-c]pyridine-5-carboxamide] and cocaine hydrochloride were obtained from the Drug Supply Program of the National Institute on Drug Abuse (Rockville, MD). All drugs were dissolved in a vehicle of physiological saline (0.9% sodium chloride), filter sterilized, and administered at a volume of 0.1mL per 10g of mouse mass. Nicotine, PHA-543613, and MLA were administered subcutaneously (s.c.), while cocaine was given intraperitoneally (i.p.). All doses are expressed as the free base of the drug.
Place Conditioning Experiments
For all place conditioning experiments with BXD strains, α7 KO, KI, and WT mice, a five-day paradigm was performed as described previously (Kota et al. 2007). Each animal received cage enrichment, and on Wednesday, Thursday, and Friday of the week prior to place conditioning testing, the experimenter handled each mouse for approximately two minutes. The experimental apparatus (Med-Associates, St. Albans, VT, ENV3013) consisted of white and black chambers (20 × 20 × 20 cm each), which differed in floor texture (white mesh and black rod). The chambers were separated by a smaller grey chamber with a smooth PVC floor and partitions that allowed access to the black and white chambers. Briefly, on Day 1 (pre-conditioning day), mice were placed in the center chamber for 5 minutes, partitions were lifted, and mice were allowed to roam freely for 15 minutes. The times spent in the white and black chambers were used to establish baseline chamber preferences, if any. Mice were separated into vehicle and drug groups such that initial chamber biases in each group were approximately balanced. On days 2-4 (conditioning days), twice per day, mice were injected with vehicle or drug and subsequently paired with either the white or black chamber, where they were allowed to roam for 15 minutes. Vehicle-treated animals were paired with saline in both chambers and drug-treated animals received saline in one chamber and nicotine in the opposite chamber. Pairing of the drug with either the black or white chamber was randomized within the drug-treated group of mice. On day 5 (test day), mice did not receive an injection. They were placed into the center chamber for 5 minutes, the partitions were lifted, and they were allowed to roam freely for 15 minutes. Time spent in each chamber was recorded. Additional experimental details are provided in the Supplemental Information document.
Several studies evaluating the role of α7 nAChRs in nicotine conditioned place preference (CPP) were conducted. We tested nicotine CPP (0.1 and 0.5 mg/kg) in α7 KO, KI, and WT mice using the procedure described above. In separate studies, C57BL/6J mice were pretreated with either saline or PHA-543613 (4.0, 8.0 and 12.0 mg/kg) 15 min before nicotine administration (0.1 or 0.5 mg/kg). Finally, C57BL/6J mice were pretreated with either saline or MLA (10 mg/kg), 15 min later, treated with PHA-543613 (12 mg/kg), and another 15 min later, treated with nicotine (0.5 mg/kg).
A similar procedure was used for cocaine CPP as a positive control. We tested cocaine CPP (10 mg/kg) in α7 KO, KI and WT mice. Finally, C57BL/6J mice were pretreated with either saline or PHA-543613 (12 mg/kg), and 15 min later, received cocaine (10 mg/kg). Doses were chosen based on those previously shown to produce reliable CPP for nicotine (Walters et al. 2006; Grabus et al. 2006) and cocaine (Sora et al. 2001).
Behavioral Data Analysis and Statistics
For each drug-treated mouse, preference scores were calculated as time spent in the drug-paired side on test day minus time spent in the drug-paired side during baseline. For each saline-treated mouse, preference scores were calculated as the average of the white side on test day minus the white side during baseline and the black side on test day minus the black side during baseline. Strain means were then calculated. Since within-strain variability for place conditioning across the BXD panel was high for both nicotine and saline treatments, we performed a subtraction of BXD strain means: nicotine preference scores minus saline preference scores. This was done to normalize as well as remove possible confounds of the saline phenotype on the nicotine phenotypic measures. With one exception, none of the drugs used herein caused significant alterations in locomotor activity on test day at the doses tested (t-tests or one-way ANOVAs, where appropriate, with treatment as the factor, see Table S1)., Therefore, time spent in either chamber was not confounded by locomotor activity. BXD strains 27, 32, and 8 did show statistical differences by treatment (saline vs. nicotine) in locomotor activity on test day, however, these differences did not impact heritability calculations (Supplemental Table S2), or QTL mapping results (data not shown), thus, we could not justify removing these strains from our dataset. Statistical analysis of all behavioral studies was performed using one- or two-way analysis of variance (ANOVA), where appropriate. If a one-way ANOVA was significant, an appropriate post-hoc test was performed (Dunnett's for comparisons vs. control and Tukey's HSD for between-group comparisons when more than two groups existed). Post-hoc p-values of <0.05 were considered to be statistically significant.
QTL Mapping, Correlations, and Heritability Calculations
In order to identify putative genes underlying nicotine place conditioning, quantitative trait loci (QTL) mapping and genetic correlations were performed using BXD strain means for behaviors and expression data using GeneNetwork (Chesler et al. 2004) and R/qtl (Arends et al. 2010). Heritabilities for nicotine and saline place conditioning phenotypes were estimated using the intraclass correlation coefficient method at α=0.05 (Smith 1957; Lynch & Walsh 1998) using the ICC package in R (Wolak 2012).
Quantitative Reverse-Transcriptase PCR, Microarrays, Data Analysis, and Network Generation
For qRT-PCR experiments, single nucleus accumbens (NAc) samples from 9-week old, untreated C57BL/6J, DBA/2J, α7 KO, and WT mice were micro-dissected on ice and immediately flash-frozen in liquid nitrogen using a protocol described previously (Kerns et al. 2005). Samples were homogenized with a Polytron® (Kinematica AG) and extracted using a guanidine/phenol/chloroform method (STAT-60, Tel-Test, Inc.). Each RNA liquid layer was added to an RNeasy Mini Column (Qiagen) for cleanup and elution of total RNA. RNA quality and purity was determined using an Experion Automated Electrophoresis Station (Bio-Rad) and a Nanodrop 2000 (Thermo Scientific). 1μg of RNA was converted to cDNA using the iScript cDNA synthesis kit containing random hexamers (Bio-Rad) according to manufacturer's instructions. Primer sequences, Tm's, amplicon sizes, and cDNA dilutions used for each gene are listed in Table S10. Primer efficiencies were between 90-110% and each primer set resulted in only one PCR product. Data analysis was performed using the 2−[ΔΔCT] method (Heid et al. 1996). Statistical analysis of qRT-PCR data was performed using a Student's t-test between the two strains tested.
Affymetrix Mouse 430A 2.0 microarrays were performed on α7 KO and WT single NAc samples from individual animals (n=5/genotype). Sample collection and RNA extraction were performed as described above. All RNA RQI values were >9.0, 260/280 ratios were between 1.9-2.1 and 260/230 ratios were >2.0. Samples were randomized at all possible steps and 100ng of RNA input were used for each 16-hour IVT reaction. Remaining steps were performed according to the manufacturer's protocol. All microarrays passed each quality control measure and Pearson correlations of robust multi-array average signals between single chips were ≥ 0.996. Pairwise significance-scores (s-scores) between KO and WT chips were generated using the s-score package in R (Kerns et al. 2003). A one-class statistical analysis of microarrays against a mean=0 and 100 permutations, was performed in the MultiExperiment Viewer (MeV, Boston, MA). A delta of 0.314, a false-discovery rate of 9.8%, and s-scores ≥ |1.5| were used to identify significantly differentially regulated genes (Supplemental Table S9). Gene network construction was performed using Ingenuity Pathway Analysis (Ingenuity, Redwood City, CA), which uses Fisher's exact test to determine the probability of input genes belonging to the network by chance (Ingenuity Pathway Analysis 2005).
Immunoblotting
Nucleus accumbens from single untreated α7 KO or WT mice were microdissected and flash-frozen in liquid nitrogen as described above. Samples were triturated in 100uL of cold 1X LDS (Life Technologies, Grand Island, NY) containing 2X Halt protease and phosphatase inhibitor cocktail and 10mM EDTA (Thermo Fisher Scientific). Each sample was passed through a 28g syringe until brain tissue was no longer visible upon quick-spin centrifugation. Protein concentrations of whole sample homogenates were determined using the bicinchoninic acid assay (Thermo Fisher Scientific). Samples were balanced with 1X LDS, reduced with 50mM dithiothreitol and boiled for 10 minutes. For each antibody used herein, it was determined that 20μg of protein lie within the linear range of detection, thus 20μg of protein were loaded per lane on a 4-12% NuPage bis-tris gel (Life Technologies). Using 1X MOPS running buffer, electrophoresis was performed at 150V. The gel was transferred to a PVDF membrane at 10V for 24 hours using a freshly prepared transfer buffer containing 10% methanol. Coomassie staining of the gel and Ponceau staining of the membrane indicated efficient and even transfer. Prior to each primary antibody incubation, the membrane was blocked with 5% BSA in 1X wash buffer (TBS-T containing 0.3% Tween 20 and 1.5M NaCl) for 1 hour at room temperature. Primary and secondary antibody catalog numbers, dilutions, and incubation times are provided in Supplemental Table S11. Immunoblots were imaged on Kodak film using the chemiluminescent ECL prime reagent (GE Healthcare Life Sciences) and quantitated using ImageJ processing and analysis software (National Institutes of Health, Bethesda, MD). All proteins were normalized to the loading control, β-actin (ACTB). Statistical analysis of immunoblot data was performed using a Student's t-test between the two strains tested.
Results
Behavioral genetic analysis of nicotine place conditioning in BXD mice
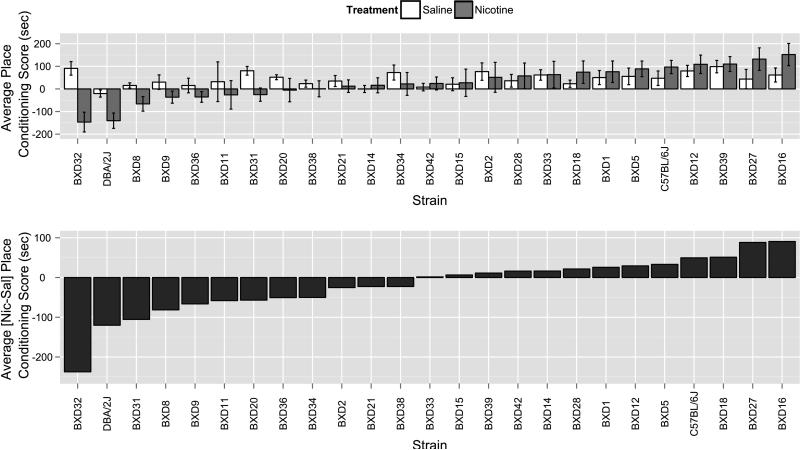
Following conditioning with either saline-saline or saline-nicotine and subsequent testing with no drug present, nicotine place preference scores (Figure 1a) were calculated across BXD strains tested (n=23 BXD strains, n=6-12 per treatment). Strains with positive scores are interpreted as having a preference for nicotine or saline, while negative scores indicate a place aversion. These scores followed a continuous distribution, indicative of a polygenic trait, and the nicotine scores were quite distinct from results seen with saline. The progenitor strains, C57BL/6J and DBA/2J, showed divergent phenotypes. Some BXD strains showed a trend for mild preference for the saline-treated side. The nicotine-saline subtracted phenotype (Figure 1b) in fact represents the nicotine response as the unsubtracted nicotine scores and subtracted scores were tightly genetically correlated (Pearson's r= 0.920, p=9.84E-14). The mean heritability for nicotine place preference was 18.7% (Table S2).
Figure 1. BXD strain distribution for place conditioning for 0.5mg/kg nicotine.
a) Following conditioning with either saline-saline (0.9%) or saline-nicotine (0.5mg/kg), nicotine place conditioning scores (black) on test day for BXD strains follow a continuous distribution, indicative of a quantitative trait. Progenitor strains, C57BL/6J and DBA/2J, show divergent phenotypes for this trait. Each point represents the mean ± SEM of n=6-12 mice per group. b) Transformed distribution, nicotine minus saline BXD strain means.
Genetic correlation analysis of nicotine place conditioning with traits within the GeneNetwork phenotype database (www.GeneNetwork.org) revealed that nicotine place preference significantly correlates inversely to the number of cholinergic neurons in multiple dorsal and ventral striatal sections (Table 1), suggesting a link between the nicotine place preference and cholinergic signaling in the striatum. However, nicotine preference scores also significantly correlated positively to a number of other phenotypes including exploratory behaviors (Table S3), suggesting a complex genetic architecture of the phenotype. Furthermore, no significant behavioral QTL was identified for the nicotine place conditioning phenotype (maximum LRS of 10.4 at 18.8 Mb on Chr X).
Table 1.
BXD place preference scores correlate inversely to the number of striatal cholinergic neurons.
| GeneNetwork ID | Phenotype | Authors | Spearma's rho | n Strains | p(rho) |
|---|---|---|---|---|---|
| 10106 | Central nervous system, morphology: Striatum cholinergic neurons, section 11 [n neurons/section] | Dains K, Hitzemann B, and Hitzemann R (1996) | −0.73567382 | 18 | 0.000268002 |
| 10107 | Central nervous system, morphology: Striatum cholinergic neurons, section 14 [n neurons/section] | −0.54437564 | 18 | 0.018084298 | |
| 10110 | Central nervous system, morphology: Striatum cholinergic neurons, section 22 [n neurons/section] | −0.64171408 | 18 | 0.003201896 |
Within multiple sections of the striatum, the number of cholinergic neurons (Dains et al. 1996) is significantly inversely correlated with nicotine place preference scores of BXD mice on day 5 of the CPP paradigm. All genetic correlations are Spearman correlations performed using GeneNetwork (GN ID = GeneNetwork Record ID, n = number of BXD strains used for correlations).
Basal Chrna7 mRNA expression in the NAc is inversely correlated with nicotine place conditioning in BXD mice
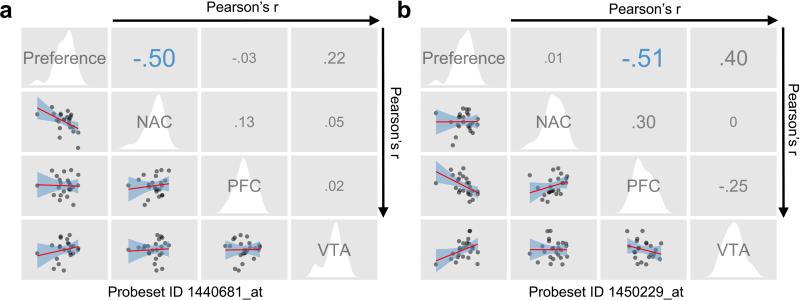
We previously generated basal genomic expression profiles across the mesolimbocortical dopamine pathway (NAc, PFC, and VTA) in many of the same BXD strains used for the studies here (Wolen et al. 2012). Therefore, to identify genes potentially functional in nicotine place conditioning we correlated these brain regional expression patterns to the transformed place conditioning behavioral data (nicotine-saline) across the BXD strains. This identified 2044, 2099, and 1253 probesets in NAc, PFC, and VTA, respectively, showing provisional correlations (p<0.05) (See Supplemental Tables S4-S6) with nicotine place conditioning. Further analysis of these expression correlates will be reported elsewhere, but for the purposes of this report, we focused the data analysis by surveying these results for nicotinic receptor-related genes that also contained putative cis expression QTL (eQTL). Such cis eQTL, resulting most frequently from genetic variation within or near the gene itself, have a larger effect size, and are primary genetic drivers of variation, while trans eQTL have secondary or tertiary effects (Schadt et al. 2003; Drake et al. 2006). We proposed that this might implicate specific nicotinic receptors not previously identified as modulating nicotine place conditioning. This analysis revealed only Chrna7 in NAc containing a putative cis eQTL and its expression significantly correlated with normalized nicotine place conditioning phenotype (see Table S7). Figure 2 depicts correlations between the normalized phenotype and Chrna7 mRNA expression (2a, probeset ID 1440681_at and 2b, probeset ID 1450229_at) in each of the three brain regions of the mesolimbic dopamine reward pathway (VCU BXD NA, VTA and PFC, RMA saline datasets). Although preference scores correlated significantly to Chrna7 mRNA expression in both NAc (Pearson's r= −0.50, p=9.04E-3, probeset 1446081_at) and PFC (Pearson's r= −0.51, p=9.07E-3, probeset 1450229_at), the putative cis eQTL only existed in the NAc for probeset 1446081_at, so we focused on this probeset for the remainder of analyses. Together, these data suggest that genetic variation in Chrna7 expression modifies nicotine place conditioning, with strains having low basal mRNA expression of Chrna7 in the NAc being more susceptible to the reward-like properties of nicotine.
Figure 2. The nicotine place conditioning phenotype is genetically correlated to Chrna7 basal mRNA expression in the nucleus accumbens, but not the prefrontal cortex, or ventral midbrain.
Conditioning scores (nicotine-saline) significantly correlate with basal Chrna7 expression (Chrna7 Probeset ID = 1440681_at, panel a) in the nucleus accumbens and prefrontal cortex (Chrna7 Probeset ID = 1450229_at, panel b), but not ventral midbrain (denoted as VTA). Correlation scattergrams (left of diagonal), univariate density plots (in white, along the diagonal), and Pearson's r values (right of diagonal) are displayed. For the correlation scattergrams, the linear fits are plotted in red with 50% confidence intervals in blue. Each point represents the mean for a BXD strain. A blue r value denotes a significant correlation, while grey r values are non-significant at an alpha = 0.05. All expression data are saline RMA values from the VCU BXD NA, PFC, and VMB Datasets, with probes containing SNPs between B6 and D2 genotypes removed.
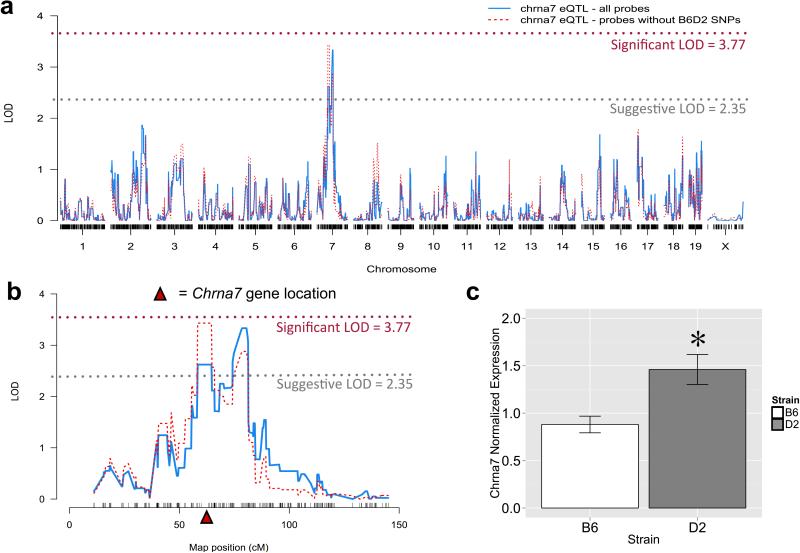
Figure 3a-b depicts a suggestive Chrna7 cis eQTL (probeset 1440681_at) present in the NAc across the BXD panel of mice. Following 2000 permutations, LOD scores of 2.35 and 3.77 denoted a suggestive or significant eQTL, respectively. These statistical limits are actually quite conservative since they reflect genome-wide corrections for multiple testing. Since two probes within this probeset have single nucleotide polymorphisms (SNPs) between B6 and D2 mice that may have influenced cRNA hybridization to the oligonucleotide microarray, causing a false cis eQTL, we excluded those probes and re-performed interval mapping, which confirmed the putative cis eQTL (Figure 3a-b). Microarray NAc data, showing higher Chrna7 transcript levels in D2 mice compared to B6 mice, were validated by qRT-PCR for Chrna7 from B6 and D2 NAc tissue samples (*p<0.001, t[12]=5.068, n=7/group, Student's t-test, Figure 3c). The Chrna7 gene is highly polymorphic between the B6 and D2 progenitor strains (285 SNPs, Supplemental File S8), thus expression differences may be attributed to one or more of these SNPs.
Figure 3. Genome-wide interval map for Chrna7 mRNA levels across the BXD RI panel.
a) A suggestive cis expression QTL (blue solid line) exists on chromosome 7 for Chrna7 mRNA levels in the nucleus accumbens (VCU BXD NA Dataset Saline RMA Values, Probeset ID 1440681_at). The cis eQTL remained after removal of the probes containing SNPs between B6 and D2 mice (red dotted line). The DBA/2J genotype for Chrna7 increases its expression. b) An enlargement of Chromosome 7 reveals two possible QTL peaks driving the mRNA expression of Chrna7, of which the proximal peak harbors Chrna7 (at 70.24Mb). C, qRT-PCR validation of microarray results. Basal mRNA expression of Chrna7 in the nucleus accumbens is significantly greater in D2 mice compared to B6 mice. Each point represents the mean ± SEM (* = p<0.01).
Knock-out of the α7 nAChR increases sensitivity to, while gain-of-function or agonism of the α7 nAChR, abolishes the nicotine CPP phenotype in mice
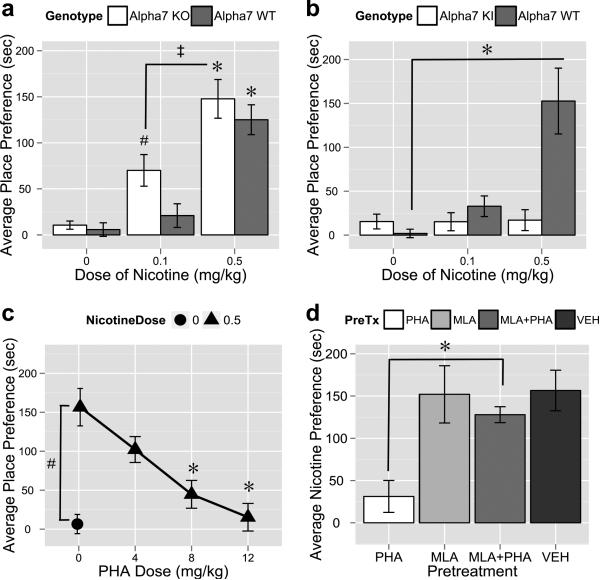
We employed a series of pharmacological and genetic manipulations in mice to confirm the involvement of the α7 nAChR in nicotine CPP as expected from the inverse correlation between basal Chrna7 mRNA expression and nicotine CPP. First, α7 KO mice demonstrated significant preference for 0.1mg/kg nicotine versus saline treatment (oneway ANOVA, followed by Dunnett's post-hoc vs. within-genotype saline group, F[2,20]=16.8854, #p<0.05, n=7-9/group). This dose of nicotine does not routinely produce place preference in C57BL/6J mice (Walters et al. 2006), suggesting an increase in sensitivity to the reward-like properties of nicotine in CPP. However, this difference in sensitivity between groups was not observed with 0.5 mg/kg of nicotine as both genotypes showed equal amounts of CPP (oneway ANOVAs, followed by Dunnett's post-hoc vs. within-genotype saline FKO[2,20]=16.8854, *pKO<0.01, nKO=7-8/group; FWT[2,21]=24.0645, *pWT<0.01, nWT=7-8/group, Figure 4a). Finally, the preference for 0.1mg/kg nicotine was significantly lower than preference for 0.5mg/kg nicotine in α7 KO mice (oneway ANOVA, followed by Tukey's HSD post-hoc to compare doses, FKO [2,20]=16.8854, ‡pKO<0.01, nKO=7-8/group). A twoway ANOVA evaluating nicotine doses (0, 0.1, and 0.5mg/kg) and mouse genotype (WT and KO) (F[5,41]=16.9029, n=7-9/group) revealed significant main effects of both dose (p<0.01) and genotype (p<0.05), but no significant interaction between dose and genotype (p=0.3371).
Figure 4. Deletion of the alpha7 nAChR results in increased sensitivity to nicotine place preference and knock-in or agonism of the alpha7 nAChR prevents nicotine place preference.
Panel a), a significant increase in place preference scores for 0.5mg/kg nicotine was observed in both alpha7 KO and WT mice. Alpha7 KO mice display place preference for 0.1mg/kg nicotine; preference for 0.1mg/kg nicotine was of significantly lower magnitude than preference for 0.5mg/kg nicotine. Panel b), Only wildtype (WT) mice, but not alpha7 knock-in (KI) mice, show nicotine place preference for 0.5mg/kg of nicotine. c), Place preference for 0.5 mg/kg nicotine was significantly higher than preference for saline in B6 mice. Pretreatment with PHA-543613 (PHA), dose-dependently blocked place preference for 0.5mg/kg nicotine at 8.0 and 12.0 mg/kg PHA. d), For CPP for 0.5mg/kg of nicotine, 12.0mg/kg PHA blocked preference; this was reversed by pretreatment with 10.0mg/kg of methyllycacontinine (MLA). MLA alone did not alter nicotine preference. Each point represents the mean ± SEM (*/‡ = p<0.01, # = p<0.05). Unless otherwise denoted, all symbols of significance denote comparisons for within-group vehicle treatment.
In contrast, gain-of-function, α7 KI mice did not develop preference to nicotine at either 0.1 or 0.5 mg/kg (oneway ANOVA, Tukey's HSD post-hoc vs. within-genotype saline control, FKI[2,29]=0.0080, pKI=0.9920, nKI=8-16/group), but WT littermates developed normal preference for 0.5 mg/kg nicotine (oneway ANOVA, Tukey's HSD post-hoc vs. within-genotype saline control, FWT[2,21]=12.1665, *pWT<0.01, nWT=8/group), Figure 4b). This follows the trend observed for low or no preference in BXD strains having high basal mRNA levels of Chrna7 (Figure 2). Finally, the α7-selective agonist, PHA-543613, was able to dose-dependently block preference in B6 mice for 0.5mg/kg nicotine (oneway ANOVA, followed by Dunnett's post-hoc vs. PHA0/Nic0.5, FB6[3,25]=10.4663, *p B6<0.01, n B6=6-8/group, Figure 4c), a dose that routinely produces place preference in this strain of mice (Walters et al. 2006). The highest dose of PHA-543613 (12.0 mg/kg) did not significantly alter preference scores in B6 mice on its own (data not shown, oneway ANOVA vs. vehicle, Tukey's HSD post-hoc, F B6 [1,14]=0.5321, p B6=0.4778, n B6=8/group). Furthermore, Figure 4d shows that blockade of nicotine preference by PHA-543613 can be reversed using 10.0 mg/kg of the α7-selective antagonist, MLA (oneway ANOVA, followed by Tukey's HSD post-hoc vs. PHA12/Nic0.5 alone, F B6 [1,13]=46.5249, *p B6<0.01, n B6=7-8/group). Also shown and previously reported, this dose of MLA was unable to block preference for 0.5mg/kg nicotine in B6 mice on its own (Walters et al. 2006).
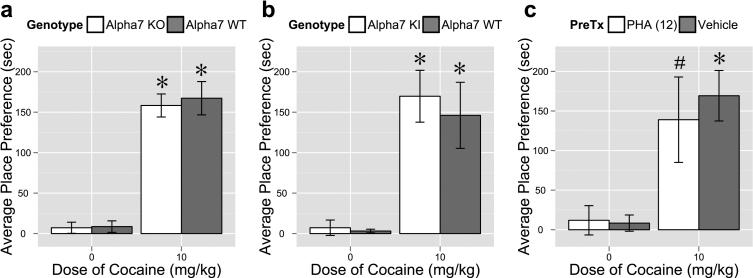
To control for possible learning deficits or enhancements in the gene-targeted mice and to determine if this effect was specific to nicotine, we tested cocaine CPP in α7 WT, KI, and KO mice. Mice of all three genotypes developed similar place preference to 10mg/kg cocaine (oneway ANOVAs, followed by Dunnett's post-hoc vs. within-genotype saline, FKO-a[1,13]=83.3130, *pKO-a<0.01, nKO-a=7-8/group, FWT-a[1,13]=47.3111, *pWT-a<0.01, nWT-a=7-8/group, FKI-b[1,16]=23.6439, *pKI-b<0.01, nKI-b=9/group, FWT-b[1,15]=13.8604, *pWT-b<0.01, nWT-b =8-9/group, Figure 5a-b), suggesting that not only are these mice responsive to pavlovian drug conditioning, but also that the α7 mechanism is not generalizable to another drug of abuse, cocaine. As an additional control to rule out Chrna7 involvement in place preference for cocaine, we attempted to block preference in C57BL/6J mice with PHA-543613, but saw no attenuation of cocaine place preference compared to control (oneway ANOVA, Dunnett's post-hoc vs. within-group vehicle, FB6[1,8]=23.0984, *pB6<0.01, nB6=5/group, oneway ANOVA, Dunnett's post-hoc vs. within-group PHA,, FB6[1,14]=4.9642, #pB6<0.05, nB6=8/group, Figure 5c).
Figure 5. Alpha7 knock-in and knock-out mice develop normal place preference to a 10mg/kg dose of cocaine; pre-treatment with PHA in C57BL/6J mice does not alter cocaine place preference.
Panels a and b, a significant increase in place preference scores for 10mg/kg cocaine compared to within-genotype saline treatment was observed for all genotypes tested. Panel c, C57BL/6J mice develop place preference to 10mg/kg cocaine. Pre-treatment with PHA did not block cocaine place preference in C57BL/6J mice. Each point represents the mean ± SEM (* = p<0.01, # = p<0.05).
Genetic interactions between Chrna7 and insulin-related genes in the NAc may contribute to preference for nicotine
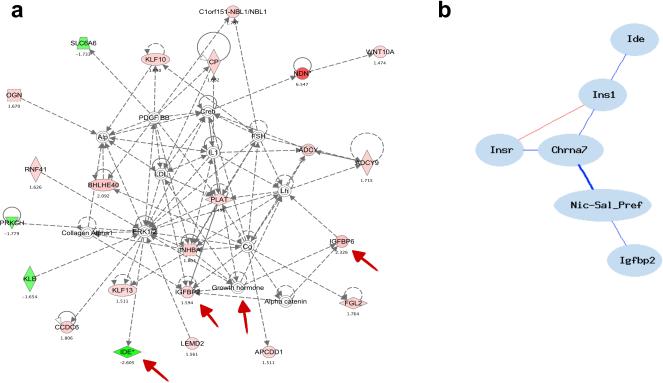
In order to explore possible mechanisms underlying the enhanced sensitivity to nicotine observed in α7 KO mice with the CPP test, contrasted to their WT counterparts, we performed microarray analysis on NAc samples from only α7 KO and WT mice. Following statistical analyses and filtering, differentially regulated genes revealed the top significant network (p=1.0E-49) as containing multiple genes involved in insulin signaling (Figure 6a). We observed that in α7 KO mice, expression of the insulin growth factor binding proteins, Igfbp2 and Igfbp6, were significantly increased, but Ide (insulin-degradation enzyme) expression, was significantly decreased in the NAc, compared to WT mice.
Figure 6. Knock-out of the α7 nAChR results in alterations to an insulin-related gene network.
a) Top-ranked biological network of genes differentially regulated in the NAc between α7 KO and WT mice (red = upregulated, green = downregulated, colorless = imputed gene, number below each gene = KO/WT s-score, red arrows denote insulin-related genes). b) Genetic correlation network performed using BXD mRNA expression data, displaying co-regulation of Chrna7 and multiple insulin-related genes in the NAc. (All correlations drawn are significant and used Pearson's r, red = positive, blue = negative. bold = r ≥ |0.5|, solid = |.41| ≥ r < |0.49|).
Finally, qRT-PCR performed from α7 KO and WT NAc samples confirmed significant down-regulation of Ide mRNA (p<0.05, t[7]= 2.957), up-regulation of Igfbp6 mRNA (p<0.05, t[7]=2.230), and a non-significant trend for differential regulation of Igfbp2 (p=0.11, t[7]=1.07, see Table 2). Furthermore, within the BXD panel of mice, we identified a co-expression network of Chrna7 and insulin-related genes in the NAc, which are significantly genetically correlated to each other as well as the transformed nicotine preference phenotype, suggesting novel and complex gene-gene interactions underlying place preference to nicotine (Figure 6b). The expression networks and qRT-PCR data suggest increased insulin signaling in both α7 KO and BXDs with low NAc levels of Chrna7 mRNA, thus perhaps suggesting a regulatory interaction.
Table 2.
Quantitative RT-PCR confirms differential insulin-related gene expression in the NAc of α7 KO and WT mice.
| Gene | Normalized Relative KO Expression | KO SEM | Normalized Relative WT Expression | WT SEM | p-value |
|---|---|---|---|---|---|
| Ide | 0.731 | 0.091 | 1.102 | 0.096 | 1.55E-02 |
| Igfbp2 | 1.768 | 0.249 | 1.166 | 0.166 | 1.08E-01 |
| Igfbp6 | 1.549 | 0.144 | 1.071 | 0.136 | 3.58E-02 |
| Ndn | 2.374 | 0.160 | 1.177 | 0.078 | 3.09E-03 |
Basal mRNA expression of Ide is significantly down-regulated in the NAc of KO mice compared to WT mice, while Igfbp6 is significantly up-regulated, and there is a trend toward up-regulation of Igfbp2. Each point represents the mean of ± SEM (nKO=4 and nWT=5). All genes were normalized to the housekeeping gene, Gapdh.
Knock-out of the α7 nAChR results in basal increases in insulin signaling in the nucleus accumbens
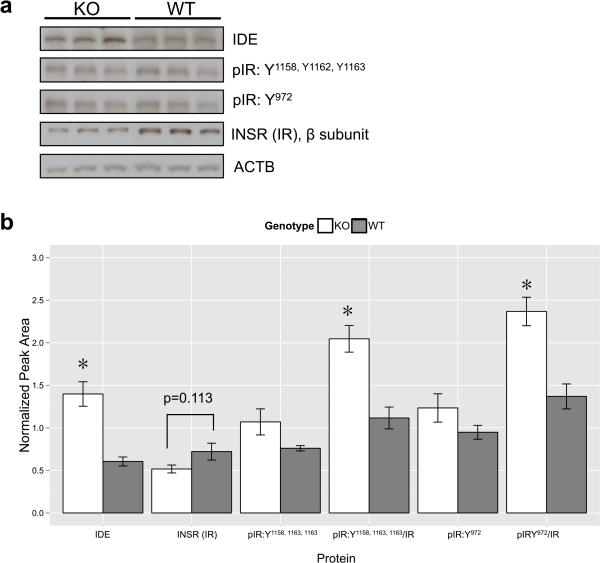
To confirm functional differences in insulin signaling in the nucleus accumbens of α7 KO and WT mice, we performed immunoblotting for proteins known to be involved in insulin signaling (Figure 7). Contrasted to our microarray data in which Ide mRNA was decreased in the NAc of α7 KO mice, we found that IDE protein was significantly upregulated compared to WT (*p<0.01, t[6]IDE=5.18). Additionally, we found a trend for decreased total insulin receptor levels, INSR-β (intracellular subunit) (p=0.113, t[6]INSR=1.87) and significantly increased phosphorylation of multiple sites of the INSR in α7 KO mice compared to WT mice (t[9]pIR(Y1158, Y1162, Y1163)/IR=4.60, t[9]pIR(Y932)/IR=4.49), Student's t-test, nKO=6, nWT=5.
Figure 7. Alpha7 knockout mice display differential basal expression of insulin-related proteins in the nucleus acumbens.
a) Representative immunoblots of nucleus accumbens samples from individual mice. IDE=insulin-degrading enzyme, pIR=phosphorylated insulin receptor (Y= tyrosine residue phosphorylated), INSR/IR=β subunit of the insulin receptor, ACTB=β-actin). b) Quantitation and statistical analyses of immunoblot samples (nKO=6, nWT=5) revealed that IDE protein levels as well the degree of phosphorylation of the total insulin receptor were significantly increased in alpha7 KO mice compared to WT mice, while total insulin receptor levels showed a trend for being decreased compared to WT mice. All proteins were normalized to ACTB (* = p<0.01).
Discussion
In this study, we combined behavioral and expression profiling across a genetic reference panel to identify genes underlying nicotine's reward-like behavioral response in mice. This implicated Chrna7 as a candidate gene. Interestingly, previous mouse studies have linked Chrna7 polymorphisms with strain differences in neuronal expression (Stitzel et al. 1996) and distribution of α7 nAChRs (Adams et al. 2001), as well as behaviors such as nicotine seizure sensitivity (Stitzel et al. 1998). However, until now, the role of Chrna7 in nicotine CPP had not been rigorously evaluated. Here, we verified the involvement of Chrna7 in nicotine CPP through the use of behavioral genetic studies in BXD mice, gene-targeted mice, and pharmacological tools. By extending our genomic analysis in α7 KO mice, we provide evidence that an insulin gene expression network in the NAc, regulated by Chrna7, may contribute to reward-like effects of nicotine CPP.
Using the BXD panel of mice, we found that basal Chrna7 mRNA expression in the NAc was significantly negatively correlated to place conditioning for nicotine, despite the lack of robustly significant behavioral QTL for nicotine place conditioning in the vicinity of Chrna7. This discrepancy might be accounted for by a number of factors, such as low heritability of nicotine preference coupled with the number of strains studied in these experiments, the influence of non-genetic factors influencing intra-strain and inter-strain variance in this phenotype, as well as the complexity of the behavioral assay. Furthermore, many genes or loci of small genetic effect underlying the place conditioning behavior, rather than one or few loci of large effect would likely mask detection of a significant behavioral QTL with the number of strains assayed herein. Studies with the α7 agonist, PHA-543613, dose-dependent attenuation of CPP and this was reversed by the α7 antagonist, MLA. α7 KI mice did not display nicotine CPP, but conversely, α7 KO mice were more sensitive to a low dose of nicotine, suggesting an increased perceived reward-like effect in these mice. Together, these behavioral data strongly implicate Chrna7 as a candidate gene modulating nicotine CPP reward-like phenotypes.
Previous studies using α7 KO mice or an α7 antagonist in rats have reported that in contrast to the β2-containing nAChR, the α7 nAChR is not required for nicotine CPP (Walters et al. 2006) or self-administration (Brunzell & McIntosh 2012), respectively. Additionally, it was reported that reductions in α7 nAChR activity due to antagonist administration into the NAc shell, as well as anterior cingulate cortex, dose-dependently increased the motivation of rats to self-administer nicotine (Brunzell & McIntosh 2012), suggesting that lower α7 nAChR activity may increase the intake of nicotine. By surveying the BXD panel and including a lower dose of nicotine in α7 KO mice, our studies have unmasked the first evidence that Chrna7 transcript levels in the NAc are both genetically regulated and may be an important factor in determining the magnitude of nicotine's reward-like effects as measured by CPP. Although CPP and self-administration were originally thought to be isomorphic models of drug reward, it is now commonly accepted that self-administration models reinforcement, and CPP models “reward” (and is influenced by other factors such as memory). Thus, these assays may measure overlapping as well as distinct components of drug-seeking behavior in animals (Bardo & Bevins 2000). Since the CPP test measures contextual cues associated with a drug's perceived “reward”, our data suggests that lower levels of Chrna7 mRNA in the NAc contributed to an altered neural response to cue-related behaviors, allowing nicotine-seeking behavior to persist.
Using microarray analysis, we discovered that gene-targeting of the α7 nAChR results in disruption of gene expression for an insulin-signaling network in the NAc (Fig. 6a). Independently, Chrna7 expression was genetically correlated with a network of insulin-related genes in the NAc across the BXD panel (Fig. 6b), strongly suggesting that our microarray results likely were not due to developmental compensation in α7 KO mice. Insulin-degrading enzyme, Ide, mRNA was significantly decreased in α7 KO mice, and previous rodent studies have demonstrated that functional disruption of this gene, results in hyperinsulinemia and glucose intolerance (Farris et al. 2003; Fakhrai-Rad et al. 2000). Additionally, Ide mRNA expression is altered in rats following nicotine treatment (Polesskaya et al. 2007), while an allele of the IDE gene is associated with plasma cotinine levels in both European and African American smokers (Hamidovic et al. 2012). Together with our studies here, these data implicate a possible role for Ide in nicotine addiction. Based on these studies, we predicted that in the NAc, α7 KO mice should have increased insulin signaling compared to WT mice.
Insulin and insulin growth factor 1 (IGF1) are produced peripherally and can readily cross the blood brain barrier via an active transport system to elicit endocrine signaling events. IGF1 is also secreted locally by neuronal cells (neurons, microglia, and astrocytes) and participates in paracrine signaling within the brain (Russo et al. 2005). Insulin and IGF1 signal through either of their receptors, INSR (Insulin receptor) or IGF1R (IGF1 receptor), or, through a hybrid receptor formed from dimerization of the INSR and IGF1R receptors (Russo et al. 2005). Activation of the receptor can trigger two canonical signaling pathways, PI3K-AKT and Ras-ERK. In rat NAc slices, insulin potentiates cocaine-induced DA release and this was reversed using a PI3K inhibitor (Schoffelmeer et al. 2011). Additionally, insulin receptor activation in rat striatal cultures causes increases in DAT mRNA expression and transporter function that resulted in increased dopamine reuptake (Patterson et al. 1998). Insulin regulation of DAT was blocked in cell culture with a PI3K inhibitor, providing further evidence for the PI3K-AKT pathway in insulin's modulation of dopamine uptake (Carvelli et al. 2002). These studies suggest a negative feedback loop in which insulin promotes both dopamine release and dopamine reuptake. Interestingly, α7 KO mice have been shown to have significantly higher levels of and longer persistence of nicotine-induced DA release in the NAc compared to WT mice (Besson et al. 2012). This enhancement of DA release within the brain's reward center in α7 KO mice could contribute to increased sensitivity to low dose nicotine observed in the CPP test.
Our studies also provide evidence of altered insulin signaling in α7 KO mice through multiple genes and proteins. Ide transcript levels in α7 KO mice are lower (Table 2) and IDE protein levels are higher compared to WT mice (Fig. 7b). Additionally, we found significantly higher activation of the INSR as well as a trend for decreased INSR protein levels in α7 KO mice (Fig. 7). Furthermore, our microarray experiment revealed that α7 KO mice, compared to WT mice, had higher NAc transcript levels of genes for two insulin growth factor binding proteins, Igfbp6 and Igfbp2, which regulate IGF1 bioavailability.
Taken together, our studies provide evidence that Chrna7 modulates nicotine reward. Our genomic and molecular studies suggest that Chrna7 influences an expression network of insulin-signaling genes and their proteins which may alter downstream dopamine signaling and lead to altered nicotine reward-like behaviors. These studies are the first to elucidate the genetic interplay between Chrna7 and insulin signaling in the NAc and a role for such interactions in nicotine behavior. Future studies will directly test these relationships between nicotine CPP, Chrna7, and the insulin signaling pathway in the NAc. These genetic and protein interactions have implications for uncovering new therapeutic targets for nicotine cessation pharmacotherapies.
Supplementary Material
Acknowledgements
This research was supported by the National Institutes of Health – National Institute on Drug Abuse: R01 DA12610 (MID), R01 DA032246 (MID and MFM), and T32 DA007027 (WLD). JH conceived and carried out the molecular genetics studies, performed statistical analyses, and drafted the paper. MFM and MID conceived and designed the BXD behavioral studies and assisted in drafting the manuscript. PPM carried out the cocaine CPP studies. MDB generously donated the α7 KI mice. All authors read and approved the final manuscript. None of the authors have reported biomedical financial interests or potential conflicts of interest. We thank Tie Han, Cindy Evans, and Lisa Merritt for technical assistance with the BXD behavioral studies.
References
- Adams CE, et al. Alpha7-nicotinic receptor expression and the anatomical organization of hippocampal interneurons. Brain Res. 2001;922(2):180–190. doi: 10.1016/s0006-8993(01)03115-8. [DOI] [PubMed] [Google Scholar]
- American Cancer Society . American Cancer Society: Guide to Quitting Smoking. Atlanta, GA: 2012. [Google Scholar]
- Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature Publishing Group. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends D, et al. Bioinformatics. Oxford, England: 2010. R/qtl: high throughput Multiple QTL mapping. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardo MT, Bevins RA. Conditioned place preference: what does it add to our preclinical understanding of drug reward? Psychopharmacology. 2000;153(1):31–43. doi: 10.1007/s002130000569. [DOI] [PubMed] [Google Scholar]
- Berrettini W, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychiatry. 2008;13(4):368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson M, et al. Alpha7-nicotinic receptors modulate nicotine-induced reinforcement and extracellular dopamine outflow in the mesolimbic system in mice. Psychopharmacology. 2012;220(1):1–14. doi: 10.1007/s00213-011-2422-1. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broide R, et al. Increased sensitivity to nicotine-induced seizures in mice expressing the L250T alpha 7 nicotinic acetylcholine receptor mutation. Molecular Pharmacology. 2002;61(3):695–705. doi: 10.1124/mol.61.3.695. [DOI] [PubMed] [Google Scholar]
- Brunzell DH, McIntosh JM. Alpha7 nicotinic acetylcholine receptors modulate motivation to self-administer nicotine: implications for smoking and schizophrenia. Neuropsychopharmacology. 2012;37(5):1134–1143. doi: 10.1038/npp.2011.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiula AR, et al. Cue dependency of nicotine self-administration and smoking. Pharmacol Biochem Behav. 2001;70(4):515–530. doi: 10.1016/s0091-3057(01)00676-1. [DOI] [PubMed] [Google Scholar]
- Carlborg O, et al. Methodological aspects of the genetic dissection of gene expression. Bioinformatics. 2005;21(10):2383–2393. doi: 10.1093/bioinformatics/bti241. [DOI] [PubMed] [Google Scholar]
- Carvelli L, et al. PI3-kinase regulation of dopamine uptake. J Neurochem. 2002;81(4):859–869. doi: 10.1046/j.1471-4159.2002.00892.x. [DOI] [PubMed] [Google Scholar]
- Changeux J-P. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nat Rev Neurosci. 2010;11(6):389–401. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, et al. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nature neuroscience. 2004;7(5):485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Dains K, Hitzemann B, Hitzemann R. Genetics, neuroleptic response and the organization of cholinergic neurons in the mouse striatum. The Journal of pharmacology and experimental therapeutics. 1996;279(3):1430–1438. [PubMed] [Google Scholar]
- Drake TA, Schadt EE, Lusis AJ. Integrating genetic and gene expression data: application to cardiovascular and metabolic traits in mice. Mammalian genome: official journal of the International Mammalian Genome Society. 2006;17(6):466–479. doi: 10.1007/s00335-005-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(5):596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Fakhrai-Rad H, et al. Insulin-degrading enzyme identified as a candidate diabetes susceptibility gene in GK rats. Hum Mol Genet. 2000;9(14):2149–2158. doi: 10.1093/hmg/9.14.2149. [DOI] [PubMed] [Google Scholar]
- Farris W, et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid b-protein, and the b-amyloid precursor protein intracellular domain in vivo. 2003;100(7):4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabus SD, et al. Nicotine place preference in the mouse: influences of prior handling, dose and strain and attenuation by nicotinic receptor antagonists. Psychopharmacology. 2006;184(3-4):456–463. doi: 10.1007/s00213-006-0305-7. [DOI] [PubMed] [Google Scholar]
- Hamidovic A, et al. Gene-centric analysis of serum cotinine levels in African and European American populations. Neuropsychopharmacology. 2012;37(4):968–974. doi: 10.1038/npp.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Martin NG. Genetic models for the natural history of smoking: evidence for a genetic influence on smoking persistence. Addict Behav. 1993;18(1):19–34. doi: 10.1016/0306-4603(93)90005-t. [DOI] [PubMed] [Google Scholar]
- Heid CA, et al. Real time quantitative PCR. Genome research. 1996;6(10):986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, et al. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcoholism, clinical and experimental research. 2004;28(10):1437–1448. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Ingenuity Pathway Analysis IPA Network Generation Algorithm. 2005:1–26. [Google Scholar]
- Kendler KS, et al. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29(2):299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- Kerns RT, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. Journal of Neuroscience. 2005;25(9):2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Zhang L, Miles MF. Application of the S-score algorithm for analysis of oligonucleotide microarrays. Methods. 2003;31(4):274–281. doi: 10.1016/s1046-2023(03)00156-7. [DOI] [PubMed] [Google Scholar]
- Kota D, et al. Nicotine dependence and reward differ between adolescent and adult male mice. The Journal of pharmacology and experimental therapeutics. 2007;322(1):399–407. doi: 10.1124/jpet.107.121616. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. 1st ed. Sinauer Associates; 1998. [Google Scholar]
- National Institutes of Health . Tobacco Addiction - June, 2012: A research update from the National Institute on Drug Abuse. Bethesda, MD: 2012. [Google Scholar]
- National Institutes of Health . Tobacco and Nicotine Research An Update from the National Institute on Drug Abuse – August 2008. Bethesda, MD: 2008. [Google Scholar]
- Patterson TA, et al. Food deprivation decreases mRNA and activity of the rat dopamine transporter. Neuroendocrinology. 1998;68(1):11–20. doi: 10.1159/000054345. [DOI] [PubMed] [Google Scholar]
- Polesskaya OO, et al. Nicotine causes age-dependent changes in gene expression in the adolescent female rat brain. Neurotoxicol Teratol. 2007;29(1):126–140. doi: 10.1016/j.ntt.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Rose JE, Corrigall WA. Nicotine self-administration in animals and humans: similarities and differences. Psychopharmacology. 1997;130(1):28–40. doi: 10.1007/s002130050209. [DOI] [PubMed] [Google Scholar]
- Rossi NA RL. Affective states associated with morphine injections. Physiol Psychol. 1976;4:269–274. [Google Scholar]
- Russo VC, et al. The insulin-like growth factor system and its pleiotropic functions in brain. Endocr. Rev. 2005;26(7):916–943. doi: 10.1210/er.2004-0024. [DOI] [PubMed] [Google Scholar]
- Saccone NL, et al. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav. 2010;9(7):741–750. doi: 10.1111/j.1601-183X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422(6929):297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schoffelmeer ANM, et al. Insulin modulates cocaine-sensitive monoamine transporter function and impulsive behavior. Journal of Neuroscience. 2011;31(4):1284–1291. doi: 10.1523/JNEUROSCI.3779-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. ON THE ESTIMATION OF INTRACLASS CORRELATION. Annals of Human Genetics. 1957;21(4):363–373. doi: 10.1111/j.1469-1809.1972.tb00291.x. [DOI] [PubMed] [Google Scholar]
- Sora I, et al. Molecular mechanisms of cocaine reward: combined dopamine and serotonin transporter knockouts eliminate cocaine place preference. Proc Natl Acad Sci USA. 2001;98(9):5300–5305. doi: 10.1073/pnas.091039298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spragg SDS. Morphine Addiction in Chimpanzees. Comparative Psychology Monographs. 1940;15:1–132. [Google Scholar]
- Stitzel JA, Blanchette JM, Collins AC. Sensitivity to the seizure-inducing effects of nicotine is associated with strain-specific variants of the alpha 5 and alpha 7 nicotinic receptor subunit genes. The Journal of pharmacology and experimental therapeutics. 1998;284(3):1104–1111. [PubMed] [Google Scholar]
- Stitzel JA, Farnham DA, Collins AC. Linkage of strain-specific nicotinic receptor alpha 7 subunit restriction fragment length polymorphisms with levels of alpha-bungarotoxin binding in brain. Brain Res Mol Brain Res. 1996;43(1-2):30–40. doi: 10.1016/s0169-328x(96)00149-0. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58(2):182–190. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- True WR, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56(7):655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Walters CL, et al. The β2 but not α7 subunit of the nicotinic acetylcholine receptor is required for nicotine-conditioned place preference in mice. Psychopharmacology. 2006;184(3-4):339–344. doi: 10.1007/s00213-005-0295-x. [DOI] [PubMed] [Google Scholar]
- Wolak M. Functions facilitating the estimation of the Intraclass Correlation Coefficient. 2012:1–9. [Google Scholar]
- Wolen AR, et al. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PloS one. 2012;7(4):e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger JS, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17(5):724–734. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.