Abstract
Clinical topoisomerase I (Top1) and II (Top2) inhibitors trap topoisomerases on DNA, thereby inducing protein-linked DNA breaks. Cancer cells resist the drugs by removing topoisomerase-DNA complexes, and repairing the drug-induced DNA double-strand breaks (DSBs) by homologous recombination (HR) and non-homologous end-joining (NHEJ). Because numerous enzymes and cofactors are involved in the removal of the topoisomerase-DNA complexes and DSB repair, it has been challenging to comprehensively analyze the relative contribution of multiple genetic pathways in vertebrate cells. Comprehending the relative contribution of individual repair factors would give insights into the lesions induced by the inhibitors and genetic determinants of response. Ultimately, this information would be useful to target specific pathways to augment the therapeutic activity of topoisomerase inhibitors. To this end, we put together 48 isogenic DT40 mutant cells deficient in DNA repair and generated one cell line deficient in autophagy (ATG5). Sensitivity profiles were established for three clinically relevant Top1 inhibitors (camptothecin and the indenoisoquinolines LMP400 and LMP776) and three topoisomerase II inhibitors (etoposide, doxorubicin and ICRF-193). Highly significant correlations were found among Top1 inhibitors as well as Top2 inhibitors, while the profiles of Top1 inhibitors were different from those of Top2 inhibitors. Most distinct repair pathways between Top1 and Top2 inhibitors include NHEJ, TDP1, TDP2, PARP1 and Fanconi Anemia genes whereas HR appears relevant especially for Top1 and to a lesser extent for Top2 inhibitors. We also found and discuss differential pathways among Top1 inhibitors and Top2 inhibitors.
Keywords: topoisomerase I, topoisomerase II, camptothecin, etoposide, synthetic lethal
Introduction
DNA topoisomerases are the target of widely used anticancer and antibacterial drugs (1–3). Topoisomerase I (Top1) releases DNA torsional stress during replication and transcription by cleaving one strand of duplex DNA and generating Top1-DNA cleavage complexes (Top1cc) that allow the untwisting of supercoiled DNA. Top1 inhibitors kill cycling malignant cells by trapping Top1cc, leading to their conversion to DNA double-strand breaks (DSBs) (4) when replication forks encounter the single-strand breaks (SSBs). Although there are two major repair pathways: homologous recombination (HR) and non-homologous end joining (NHEJ) for DSB repair, studies in yeast demonstrate that the DSBs induced by Top1 inhibitors are repaired by HR during S phase (5). Other repair pathways for Top1cc are excision of the 3′-tyrosyl-DNA bond by tyrosyl-DNA-phosphodiesterase 1 (TDP1) (6–8), and endonuclease cleavage and elimination of the DNA strand attached to the Top1 by Mus81-Eme1 (9), XPF-ERCC1 (10) and CtIP (11). Two camptothecin (CPT) derivatives, topotecan and irinotecan are approved by the FDA and across the world as anticancer agents. However, because of their limitations (chemically instability, limited drug accumulation in drug efflux overexpressing cells and reversible trapping of Top1cc) (12), non-camptothecin Top1 inhibitors, such as the indenoisoquinolines [LMP400 (NSC 743400) and LMP776 (NSC 725776)] are in clinical trial (13, 14). We previously reported that 1 μM CPT generates similar amounts of Top1cc as 1μM of LMP400 or LMP776 (14).
Although topoisomerases II (Top2α and β), like Top1, relaxes DNA supercoiling by cleaving the DNA backbone and forming transient tyrosyl-DNA cleavage complex intermediates (Top2cc), they differ from Top1 in several ways (1, 15–17). They cleave both strands of one DNA duplex simultaneously to allow another duplex to pass through Top2-linked DSB. This enables Top2 not only to relax supercoiling but also to disentangle DNA knots and catenanes at the end of DNA replication. Thus, Top2 generates DSBs in cycling cells especially during mitosis phase, in which both HR and NHEJ are available for repair (18). Additional notable differences between Top2 and Top1 include the covalent linkage of Top2 at the 5′- rather than 3′-DNA ends of the breaks, and ATP requirement for catalysis. Top2 inhibitors are divided into two classes, catalytic inhibitors and poisons that trap Top2cc (1, 3, 19). A classical Top2 catalytic inhibitor is ICRF-193 (20), which blocks ATP hydrolysis and inhibits reopening of the ATPase domain in Top2, thereby trapping the crossing DNA inside the enzyme. Top2 poisons, on the other hand, inhibit the religation of Top2cc. They are widely used clinically and belong to 2 subclasses, non-intercalating and intercalating drugs. Non-intercalating drugs primarily represented by etoposide (VP-16) are highly specific for Top2cc (4, 21). Intercalators, such as doxorubicin, on the other hand, not only trap Top2cc but also kill cells by intercalation and generation of oxygen radicals (22). The removal of Top2cc primarily involves tyrosyl-DNA-phosphodiesterase 2 (TDP2), which was discovered recently as the prevalent cellular 5′-tyrosyl phosphodiesterase responsible for excising Top2cc (23). Accordingly, knocking out TDP2 in human (23) or chicken DT40 cells (24) increases the cytotoxicity of etoposide.
Thus, various repair pathways related to numerous genes are required for cellular tolerance to topoisomerase inhibitors (25–28). However, it is hard to tell which genes are more important than others due to a lack of comprehensive system to compare the contribution of genes to drug sensitivity in vertebrate cells. Furthermore, it is not certain whether intercalators and non-intercalating Top2 inhibitors, or CPT and non-CPT Top1 inhibitors induce similar DNA lesions and are repaired by different pathways. The comprehensive analysis of DNA repair gene mutants would allow the characterization of the drug-induced DNA lesions, and identify genes that could be targeted to augment the selectivity and overcome resistance to topoisomerase inhibitors.
To this end, we organized a panel of isogenic DNA repair mutant chicken DT40 cell lines. Indeed, DT40 cells provide the largest collection of isogenic DNA repair mutant clones in vertebrate cells, due to high gene targeting efficiency and stable karyotype of the chicken B lymphocyte line (29). Furthermore, since DT40 cells lack G1/S checkpoint due to impaired p53(30) and proliferate at an extremely high rate (≈ 8 hours doubling-time), DT40 and malignant cancer cells may share comparable characteristics in cellular responses to chemotherapeutic agents. Taking advantage of the panel of repair-deficient DT40 cells, we recently demonstrated the importance of poly(ADP-ribose) polymerase (PARP) trapping for the anticancer activity of clinical PARP inhibitors (31). We also showed the relevance of such an approach with a small DT40 mutant library to screen for genotoxic agents (32).
In the present study, we profiled and compared the sensitivity of forty-nine DT40 mutant cells to three Top1 inhibitors (CPT and the two indenoisoquinolines in clinical trial, LMP400, and LMP776) and three Top2 inhibitors (the non-intercalator etoposide, the intercalator doxorubicin, and ICRF-193, a Top2 catalytic inhibitor). In addition, we generated a novel knockout cell line (ATG5−/− cells) to examine the effect of autophagy in comparison with DNA repair.
Materials and Methods
Cell lines and cell culture
The DT40 cell lines used in this study were obtained from the Laboratory of Radiation Genetics, Graduate School of Medicine in Kyoto University (Kyoto, Japan) in 2011–2012. All the mutant cell lines except for ATG5−/− cell line were previously authenticated by Southern blotting and/or RT-PCR and/or Western blotting (see the references of Supplementary Table 1). The gene disruption of ATG5 in ATG5−/− cells was authenticated in this study by Southern blotting (Supplementary Fig. 1). DT40 cells were cultured at 37°C with 5% CO2 in RPMI-1640 medium (11875-093, Invitrogen, Carlsbad, CA) supplemented with 1% chicken serum (16110-082, Invitrogen, Carlsbad, CA), 10−5 M β-mercaptoethanol (M-3148, Sigma-Aldrich, St. Louis, MO), penicillin-streptomycin (15140-122, Invitrogen), and 10% fetal bovine serum (100-106, Gemini Bio-Products, West Sacramento, CA).
Drug preparations
CPT, LMP400 (NSC 743400) and LMP776 (NSC 725776) were obtained from the Drug Synthesis and Chemistry Branch, National Cancer Institute (Bethesda, MD, USA). Drug stock solutions were made in DMSO at 10 μM for CPT and 100 μM for LMP400 and LMP776. Etoposide (E1383, Sigma-Aldrich) and ICRF-193 (I4659, Sigma-Aldrich) were dissolved in DMSO at 1 mM. Doxorubicin (D1515, Sigma-Aldrich) was dissolved in distilled water at 100 μM. Paclitaxel (Taxol, T1912, Sigma-Aldrich) was dissolved in DMSO at 1 μM. All stock solutions were stored at −20ºC in dark. We diluted the stock solutions with culture medium. Maximum concentrations were 40 nM for CPT, 240 nM for LMP400, 120 nM for LMP776, 800 nM for etoposide, 1,600 nM for ICRF-193, 50 nM for doxorubicin, 10 nM for paclitaxel. Due to the hyper-resistance of NHEJ mutants (KU70, LIGIV, and DNA-PK deficient cells) to CPT, we used 320 nM CPT as a maximum concentration. We prepared 5 different concentrations by 1:2 serial dilution.
Measurement of cellular sensitivity
Two hundred DT40 cells in 20 μl of culture medium were seeded into 384-well white plates (#6007680 Perkin Elmer Life Sciences), and then added 20 μl of culture medium containing drugs. Most outer wells were not used to avoid error associated with an evaporation issue while PBS or culture medium was added in the most outer wells. Plates were incubated at 37ºC for 72 hours, allowing untreated wild-type cells to divide 9 times. Cell survival was determined using the ATPlite 1-step kit (Perkin Elmer Life Sciences). In brief, 20 μl ATPlite solution was directly added to each well of 384-well white plates. Five minutes after adding the ATPlite solution, luminescence was measured by Envision 2104 Multilabel Reader (PerkinElmer). All procedures were performed in triplicate.
Evaluation of relative cellular sensitivity
One 384-plate allowed us to examine sensitivity to two kinds of drugs in 7 different cell lines at once. Wild-type cells were always included in each plate. To evaluate the relative cellular sensitivity of each mutant to wild-type cells, sensitivity curves were drawn by setting the survival of untreated cells as 100%, under conditions where cell number per well was linearly correlated with fluorescence signal until 100,000 cells per well (Supplementary Fig. 2). IC90 values (inhibition concentration 90%; IC90) for each drug and cell line were determined as the crossing points between the 10% viability line and survival curve connecting average points for each drug concentration (see Supplementary Fig. 3A–B). The IC90 of each mutant was divided by the IC90 of wild-type cells that were cultured on the same plate, and then the quotient was converted into logarithmic scale (base 2). Each score was plot on the same bar graph.
Flow cytometry
Cells were fixed with 70% ethanol, stained with propidium iodide, and then analyzed.
Statistical analyses
Pearson’s correlation analysis was used to examine the correlation between drugs.
Generation of the ATG5−/− DT40 mutant cells
To generate ATG5−/− DT40 mutant cells, we constructed ATG5-targeting vectors to replace exon 3 with puromycin and blasticidin resistance gene cassettes (puro and bsr) (Supplementary Fig. 1). The primers used to amplify the left arm including the probe sequence from DT40 genomic DNA were 5′-GGGATGGCAGTGTCTCTTATTTACTTCAAG-3′ and 5′-TGAGGTCATTCACATGAATGAGAACGGTTT-3′. The amplified PCR product was digested with KpnI restriction enzyme, and the 5′ fragment was used as the probe for Southern blot while the 3′ fragment was used for the left arm. The primers used to amplify the right arm were 5′-GACAGAGGGCCAGAGCACCATCTGATCAGT-3′and 5′-CCTGGCACCACCTTCTCAATGCATTTGAGA-3′. We transfected linearized ATG5-targeting vectors sequentially by electroporation. The genomic DNA of transfectants was digested with XbaI, and targeted clones were confirmed by Southern blot analysis. The sizes of the hybridizing fragments of the wild-type locus and the loci targeted by ATG5-puro or ATG5-bsr are 8.0 kb, 5.8 kb and 4.6 kb respectively.
Results
Experimental design and validation
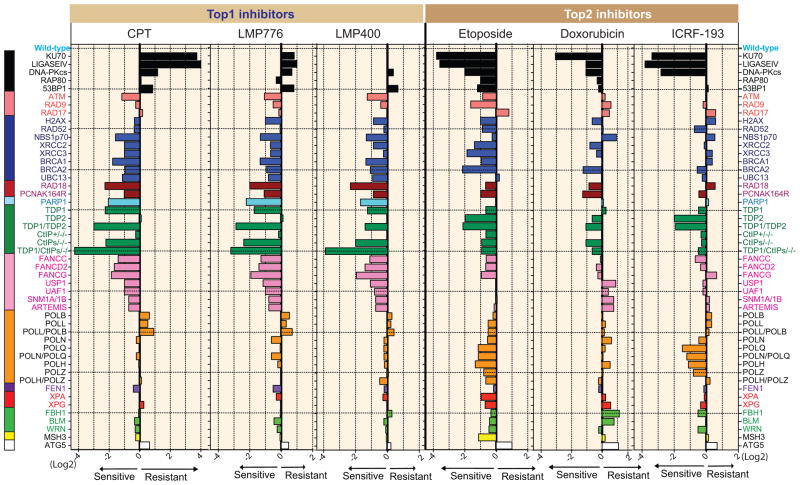
The isogenic DT40 cell lines used for drug profiling are listed in Supplementary Table 1, and color-coded in Figure 1. Differential sensitivity curves of wild-type, BRCA1−/− and KU70−/− cells used to generate the plots in Figure 1 are shown in supplementary figure 3A–B. We also validated the hypersensitivity of BRCA1−/− cells to CPT and etoposide and the hypersensitivity of KU70−/− to etoposide by flow cytometry analyses (Supplementary Fig. 3C–D). BRCA1−/− cells showed profound deleterious alteration at concentrations of CPT (10 nM) and etoposide (200 nM) where the wild-type cell were not apparently affected. On the other hand, the flow cytometry analyses of the KU70−/− cells showed such alteration to CPT but not to etoposide, consistent with the ATPlite assays. IC90 (inhibitory concentration required to inhibit 90% cell survival) were then determined for each drug in each cell line, which enabled the results to be plotted (see Fig. 1) after calculating the fold difference in IC90 for each mutant compared to the wild-type (31). Values were normalized to wild-type cells (0 value in Log scale). Bars to the left indicate hypersensitivity and to the right resistance compared to wild-type cells. Numbers at the bottom (X-axis) correspond to IC90 Log2 ratios. The selectivity of our bioassay for genotoxic agents was confirmed with paclitaxel (Taxol), a specific mitotic inhibitor devoid of DNA damaging effects (33), which showed no significant difference compared to wild-type cells (Supplementary Fig. 4).
Figure 1.
Sensitivity profiles of the indicated topoisomerase inhibitors in the selected DNA repair-deficient DT40 panel. Negative (the left side) and positive (the right side) scores indicate that the indicated cells are sensitive and resistant to the drug, respectively. The bars are colored according to the main DNA repair function of deficient gene(s): black, non-homologous end joining (NHEJ); salmon, damage checkpoint; blue, homologous recombination (HR); brown, translesion synthesis (TLS); aqua, PARP1; green, removal of Top1 or Top2 covalent complexes; pink, Fanconi anemia (FA) pathway; orange, DNA polymerases and translesion synthesis; purple, FEN1 (flap endonuclease); red, nucleotide excision repair (NER); light green, DNA helicases; yellow, MSH3 (mismatch repair); white, ATG5 (autophagy). Wild-type cells are shown at the top with a reference value of 0. The average IC90 with standard deviations (n = 7~19) of each drug in wild type cells was 20.2 ± 4.6 (nM) for CPT, 35.2 ± 7.3 (nM) for LMP776, 89.5 ± 21.2 (nM) for LMP400, 374.3 ± 97.5 (nM) for Etoposide, 16.4 ± 1.7 (nM) for doxorubicin, and 497.3 ± 166.2 (nM) for ICRF-193.
Distinct repair profiles for Top1 versus Top2 inhibitors
Figure 1 shows the response patterns for each of the 6 drugs tested. The three Top1 inhibitors are on the left and the three Top2 inhibitors on the right. Testing the overall response patterns pairwise (Table 1) using Pearson’s correlation coefficient analysis showed highly significant correlation among the three Top1 inhibitors with 0.80–0.91 Pearson’s correlation values (p < 10−12). Likewise, the Pearson’s correlation values were highly significant (0.54–0.76) among the three Top2 inhibitors (p < 10−4). By contrast, the activity profiles of the Top1 inhibitors were greatly different from those of Top2 inhibitors without significant correlation across the two drug classes (Table 1).
Table 1.
Similarities across Top1 inhibitors and Top2 inhibitors and dissimilarities between Top1 and Top2 inhibitors. Pearson’s correlation coefficients were computed for all four tested topoisomerase inhibitors across the DT40 DNA repair mutant panel as shown in Figure 1. For n = 50, a Pierson coefficient < 0.451 corresponds to a significance probability P < 0.001, two-tailed test). Paclitaxel is shown as negative control (see Supplementary Fig. 4).
| CPT | 1 | ||||||
| LMP776 | 0.91 | 1 | |||||
| LMP400 | 0.80 | 0.92 | 1 | ||||
| Etoposide | −0.37 | −0.15 | −0.05 | 1 | |||
| Doxorubicin | −0.16 | 0.06 | 0.18 | 0.73 | 1 | ||
| ICRF-193 | −0.48 | −0.26 | −0.19 | 0.76 | 0.54 | 1 | |
| Paclitaxel | 0.02 | 0.02 | 0.02 | −0.004 | −0.19 | 0.02 | 1 |
| CPT | LMP776 | LMP400 | Etoposide | Doxorubicin | ICRF-193 | Paclitaxel | |
The genes contributing to the differential profiles of Top2 and Top1 inhibitors include KU70, LIGASE IV, DNA-PKcs (all part of NHEJ; black bars, top of Fig. 1), PARP1, TDP1, TDP2 and Fanconi Anemia genes (pink bars; Fig. 1). These results indicate that our bioassay can clearly distinguish Top1 inhibitors from Top2 inhibitors according to the characters of DNA lesions. The results for the NHEJ, TDP1, TDP2 and CtIP mutant cells are consistent with independent reports (8, 11, 24, 34). Common genes conferring similar sensitization to both Top1 and Top2 inhibitors include ATM, H2AX, and HR pathway genes (XRCC2, XRCC3, BRCA1, BRCA2).
Differential contribution of NHEJ to Top1cc and Top2cc repair
While Top1 and Top2 inhibitors were both more active in HR-deficient cells, one of the most striking difference was their opposite effects in NHEJ-deficient cells. Indeed, KU70 knockout cells (and to a lesser extend LIGASE IV or DNA-PKcs knockout cells) were more than 10-fold resistant to CPT while they were more than 10-fold hypersensitive to etoposide (black bars at the top of Fig. 1). Doxorubicin and ICRF-193 behave comparably to etoposide with respect to NHEJ, reinforcing the finding that NHEJ is specifically required for the repair of DSBs induced by Top2 inhibitors (34, 35).
Overall, the profiling patterns of the three Top1 inhibitors were very similar to each other (Fig. 1 and Table 1). Hypersensitivity to HR (blue), RAD18, PARP1, TDP1, CtIP and Fanconi Anemia (pink) deficient/mutant cells, underlines the importance of these genes for Top1cc repair [reviewed in (28)]. Previous studies indicated that NHEJ has a toxic effect on HR by competing DNA ends, and thereby the loss of NHEJ increases the cellular tolerance to CPT (34). Interestingly, mutants in core NHEJ factors (KU70, LIGASE IV, and DNA-PKcs shown with black bars) displayed different responses to the three Top1 inhibitors. While all NHEJ mutants were resistant to CPT and weakly resistant to LMP776, there was neither sensitivity nor resistance to LMP400. These results suggest that targeted Top1 inhibitors can damage chromosomal DNA in a slightly different manner such that accessibility of NHEJ factors to the DSBs may differ for each Top1 inhibitor.
Unexpected function of TDP2 and NHEJ for ICRF-193-induced lesions and complex effects of doxorubicin
Closer analysis of the profiling patterns among Top2 inhibitors showed that the etoposide dependency for HR-related genes (blue) was more restricted for doxorubicin and ICRF-193. Moreover, the hypersensitivity of TDP2-deficient cells to ICRF-193 was unexpected because ICRF-193 has been reported to generate Top2 topological complexes (closed protein clamps) rather than Top2cc (36). Thus, the hypersensitivity of TDP2-deficient and NHEJ-deficient cells to ICRF-193 revealed that ICRF-193 can act as a Top2 poison like etoposide, and/or that TDP2 is a cofactor of NHEJ for the removal of Top2cc topological complexes.
Another notable finding is that doxorubicin, which unambiguously generates Top2cc (37, 38) did not sensitize TDP2 deficient cells as well as etoposide and ICRF-193, and showed relatively low correlation with etoposide and ICRF-193 (Fig. 1 and Table 1), indicating additional cell killing mechanisms of doxorubicin other than Top2cc targeting.
The autophagy-related gene, ATG5, contributes to cell death by Top2 inhibitors
To test the involvement of autophagy in cellular responses to topoisomerases inhibitors, we generated ATG5-deficient cells (Supplementary Fig. 1), as ATG5 is a key player in autophagic cell death (39). Indeed, overexpression of ATG5 in HeLa cells induces more cell death in the presence of etoposide and doxorubicin (40). ATG5−/− cells showed consistent mild resistance to Top2 inhibitors and to a lesser extent to Top1 inhibitors, which agrees with the fact that ATG5 is required for autophagic cell death upon cellular stress by etoposide and doxorubicin (40).
Targeting genes to induce synthetic lethality for topoisomerase inhibitors
From our results of Top1 inhibitors, the importance of HR genes, RAD18, PARP1, TDP1, phosphorylation of Ser332 of CtIP, Fanconi Anemia genes were comparably high. Hence, all these genes can induce synthetic lethality for Top1 inhibitors. Notably, the dual mutant of TDP1 and CtIP (8) was the most sensitive mutant in this panel to all Top1 inhibitors, suggesting that targeting both genes can further augment the cytotoxicity of Top1 inhibitors. As for etoposide, NHEJ core components can be the best candidate followed by TDP2 and HR-related genes.
Discussion
Usefulness of the DT40 repair panel profiling for categorizing DNA-damaging and genotoxic agents
In this study, we describe a comprehensive genetic analysis of multiple DNA repair pathways in response to topoisomerase inhibitors in 50 cell lines constituting a DT40 repair panel (DR panel). This is the first report comparing quantitatively and in parallel the significance of as many as 48 repair and one autophagy genes for drug sensitivity in a panel of vertebrate cells. The profiles of topoisomerase inhibitors targeting Top1 vs. Top2 were highly correlated among each other. On the other hand, the profiles to Top1 inhibitors were very different from those of Top2 inhibitors. Extension of the DR panels to other classes of DNA-targeted drugs will probably further reveal its value by showing differences between drugs with different targets and mechanism of action. Using a subset of the DR panel, we recently discovered that clinical PARP inhibitors such as olaparib and niraparib target PARP-DNA complexes by trapping them on DNA and that not only HR but also replication bypass and Fanconi pathways are critical for survival of cells treated with these PARP inhibitors (31). Because the DR panel includes multiple repair pathways, it could be used as a sensitive tool to screen potential genotoxic agents (32). Indeed, one would expect genotoxics to be selectively more cytotoxic in at least a subset of DNA repair-deficient cell lines. This point is supported by our demonstration that the microtubule inhibitor, paclitaxel (Taxol) showed no significant hypersensitivity in any of the DR panel lines.
Differences among topoisomerase inhibitors from the same group
Our comprehensive profiling revealed previously unappreciated differences within drug classes. Indeed, for the Top1 inhibitors, the sensitization conferred by NHEJ genes was different for CPT and the two indenoisoquinolines. Biochemical studies show that the indenoisoquinolines induce Top1cc at different genomic sites compared to CPT, and tend to induce persistent Top1cc that can target transcription in addition to replication complexes (14, 41), suggesting the accessibility of the NHEJ machinery to DSB ends might be different depending on the types of lesion induced by the indenoisoquinoline Top1 inhibitors.
For the Top2 inhibitors, it is unexpected that etoposide and ICRF-193 are more highly correlated with each other than etoposide and doxorubicin since etoposide and doxorubicin are both categorized as Top2 poisons whereas ICRF-193 is categorized as a catalytic Top2 inhibitor (1, 3, 17, 20, 36). Considering that TDP2-deficient cells were weakly sensitivity to doxorubicin but hypersensitive to etoposide, the contribution of Top2cc for cell killing appears higher for etoposide than doxorubicin. This result is supported by the fact that doxorubicin is not only a Top2cc-targeting drug but also a powerful DNA intercalator and inducer of oxygen radicals (22). On the other hand, the fact that ICRF-193 was significantly more cytotoxic in TDP2 deficient cells suggests the formation of Top2cc and induction of DNA breaks by ICRF-193, which is also consistent with the critical involvement of NHEJ for ICRF-193-treated cells.
Determination of targeting genes for synthetic lethality
The DR panel profiling enabled us to list up the priority genes to be targeted for synthetic lethality. For instance, the PARP1-deficient cells are among the most sensitive mutants to the three Top1 inhibitors, confirming the synergistic combination of Top1 and catalytic PARP inhibitors such as veliparib (31, 42). Likewise, inhibiting of HR, TDP1, RAD18 as well as CtIP-BRCA1 binding will synergize the cytotoxicity of Top1 inhibitors. On the other hand, the sensitivity of PARP1 deficient cells was equivalent to those of wild-type to the three Top2 inhibitors, suggesting that PARP1 inhibitors would not induce synthetic lethality with Top2 inhibitors. The profiles indicates that NHEJ genes especially KU70 gene are the prior targets for Top2 inhibitors. Since many key genes involved in DNA damage and repair responses are often mutated in human cancer, if the information about the defective genes in each tumor cells is available in advance, we can expect more effective cell killing effect by choosing the anti-cancer drug that induces the DNA damage repaired by the defective gene. The data presented in this study thus provide a rational approach to measure the importance of individual repair pathways and genes as targets in chemotherapy.
Supplementary Material
Acknowledgments
We thank Dr. Minoru Takata and Dr. Ishiai (Kyoto University, Japan) for sharing FANCC, FANCD2, FANCG and SNM1A/1B mutant DT40 cell lines, and Dr. Keith Caldecott (University of Sussex, UK) for kindly providing TDP2 mutant DT40 cell line.
Grant Support
J. Murai is a recipient of a fellowship from the Japan Society for the Promotion of Science (JSPS) and the Kyoto University Foundation. Y. Pommier and J. Murai were supported by the Intramural Program of the National Cancer Institute, Center for Cancer Research (Z01 BC 006150). Y. Maede, H. Shimizu and S. Takeda were supported by the JSPS Core-to-Core Program.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–33. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–50. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long BH, Stringfellow DA. Inhibitors of topoisomerase II: structure-activity relationships and mechanism of action of podophyllin congeners. Adv Enzyme Regul. 1988;27:223–56. doi: 10.1016/0065-2571(88)90019-2. [DOI] [PubMed] [Google Scholar]
- 5.Nitiss J, Wang JC. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci U S A. 1988;85:7501–5. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11534–9. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–5. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 8.Murai J, Huang SY, Das BB, Dexheimer TS, Takeda S, Pommier Y. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J Biol Chem. 2012;287:12848–57. doi: 10.1074/jbc.M111.333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regairaz M, Zhang YW, Fu H, Agama KK, Tata N, Agrawal S, et al. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. The Journal of cell biology. 2011;195:739–49. doi: 10.1083/jcb.201104003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic acids research. 2011;39:3607–20. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pommier Y, Cushman M. The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther. 2009;8:1008–14. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antony S, Agama KK, Miao ZH, Takagi K, Wright MH, Robles AI, et al. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67:10397–405. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 15.Deweese JE, Osheroff MA, Osheroff N. DNA Topology and Topoisomerases. Biochem Mol Biol Educ. 2009;37:2–10. doi: 10.1002/bmb.20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–37. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–53. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 18.Wohlbold L, Fisher RP. Behind the wheel and under the hood: functions of cyclin-dependent kinases in response to DNA damage. DNA repair. 2009;8:1018–24. doi: 10.1016/j.dnarep.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andoh T, Ishida R. Catalytic inhibitors of DNA topoisomerase II. Biochim Biophys Acta. 1998;1400:155–71. doi: 10.1016/s0167-4781(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 21.Ross W, Rowe T, Glisson B, Yalowich J, Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984;44:5857–60. [PubMed] [Google Scholar]
- 22.Doroshow JH. Anthraycyclines and anthracenediones. In: Chabner BA, Longo DL, editors. Cancer chemotherapy and biotherapy. 2. Philadelphia: Lippincott-Raven; 1996. pp. 409–34. [Google Scholar]
- 23.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–8. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 24.Zeng Z, Sharma A, Ju L, Murai J, Umans L, Vermeire L, et al. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic acids research. 2012 doi: 10.1093/nar/gks622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng C, Brown JA, You D, Brown JM. Multiple Endonucleases Function to Repair Covalent Topoisomerase I Complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci U S A. 2002;99:13669–74. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik M, Nitiss JL. DNA repair functions that control sensitivity to topoisomerase-targeting drugs. Eukaryot Cell. 2004;3:82–90. doi: 10.1128/EC.3.1.82-90.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–88. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 30.Takao N, Kato H, Mori R, Morrison C, Sonada E, Sun X, et al. Disruption of ATM in p53-null cells causes multiple functional abnormalities in cellular response to ionizing radiation. Oncogene. 1999;18:7002–9. doi: 10.1038/sj.onc.1203172. [DOI] [PubMed] [Google Scholar]
- 31.Murai J, Huang S-yN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji K, Kogame T, Choi K, Wang X, Lee J, Taniguchi Y, et al. A novel approach using DNA-repair-deficient chicken DT40 cell lines for screening and characterizing the genotoxicity of environmental contaminants. Environ Health Perspect. 2009;117:1737–44. doi: 10.1289/ehp.0900842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orr GA, Verdier-Pinard P, McDaid H, Horwitz SB. Mechanisms of Taxol resistance related to microtubules. Oncogene. 2003;22:7280–95. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adachi N, Suzuki H, Iiizumi S, Koyama H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: implications for the repair of topoisomerase II-mediated DNA damage. J Biol Chem. 2003;278:35897–902. doi: 10.1074/jbc.M306500200. [DOI] [PubMed] [Google Scholar]
- 35.Caldecott KW. Tyrosyl DNA phosphodiesterase 2, an enzyme fit for purpose. Nat Struct Mol Biol. 2012;19:1212–3. doi: 10.1038/nsmb.2455. [DOI] [PubMed] [Google Scholar]
- 36.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA. 1994;91:1781–5. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross WE, Glaubiger DL, Kohn KW. Protein-associated DNA breaks in cells treated with adriamycin or ellipticine. Biochim Biophys Acta. 1978;519:23–30. doi: 10.1016/0005-2787(78)90059-x. [DOI] [PubMed] [Google Scholar]
- 38.Tewey KM, Chen GL, Nelson EM, Liu LF. Intercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984;259:9182–7. [PubMed] [Google Scholar]
- 39.Zalckvar E, Yosef N, Reef S, Ber Y, Rubinstein AD, Mor I, et al. A systems level strategy for analyzing the cell death network: implication in exploring the apoptosis/autophagy connection. Cell death and differentiation. 2010;17:1244–53. doi: 10.1038/cdd.2010.7. [DOI] [PubMed] [Google Scholar]
- 40.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nature cell biology. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 41.Antony S, Kohlhagen G, Agama K, Jayaraman M, Cao S, Durrani FA, et al. Cellular topoisomerase I inhibition and antiproliferative activity by MJ-III-65 (NSC 706744), an indenoisoquinoline topoisomerase I poison. Mol Pharmacol. 2005;67:523–30. doi: 10.1124/mol.104.003889. [DOI] [PubMed] [Google Scholar]
- 42.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic acids research. 2011;39:3607–20. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.