Abstract
Transcription is apparently risky business. Its intrinsic mutagenic potential must be kept in check by networks of DNA repair factors that monitor the transcription process to repair DNA lesions that could otherwise compromise transcriptional fidelity and genome integrity. Intriguingly, recent studies point to an even more direct function of DNA repair complexes as co-activators of transcription and the unexpected role of “scheduled” DNA damage/repair at gene promoters. Paradoxically, spontaneous DNA double-strand breaks also induce ectopic transcription that is essential for repair. Thus, transcription, DNA damage and repair may be more physically and functionally intertwined than previously appreciated.
Introduction
Accurate processing of genetic information by transcription is vital for development and survival of the organism. Execution of these gene expression programs in a stage- and cell type-specific manner requires the coordinated assembly of the transcription apparatus at select gene promoters (Lemon and Tjian, 2000). Transcriptional activation involves the initial recognition of key regulatory DNA elements at promoters by sequence-specific DNA-binding activators and the core transcription machinery, along with the recruitment of essential cofactors (Fong et al., 2012; Lemon and Tjian, 2000; Naar et al., 2001; Roeder, 2005). Within this large protein ensemble termed the preinitiation complex (PIC), a series of enzymatic reactions and extensive remodeling of protein-DNA and protein-protein transactions must occur before transcription commences (He et al., 2013).
The highly choreographed cascade of events leading to gene activation provides numerous points of regulation and fine tuning. This remarkable flexibility, a necessary property to ensure target specificity and transcriptional fidelity, also makes transcription particularly sensitive to perturbations in the genome including DNA damage. Chromosomal DNA is under relentless attack from both endogenous byproducts of cellular metabolism (e.g. reactive oxygen species) and exogenous sources of environmental stress (e.g. ultraviolet light). These genotoxic agents create DNA breaks and adducts that, if left unresolved, can be detrimental to both DNA replication and transcription, and, ultimately, cell function and survival (Hoeijmakers, 2001). These genomic insults may be especially pertinent to stem cells where the consequences of unrepaired DNA damage can be profound. Mutations acquired by stem cells become amplified through self-renewal and, at the same time, propagated to progenitor cells that can differentiate to form a substantial part of a tissue, or in the case of embryonic stem cells, an entire organism (Mandal et al., 2011). In fact, proliferation of these damaged cells often manifests itself in diseases such as premature aging, developmental disorders and cancer (Diderich et al., 2011; Iyama and Wilson, 2013).
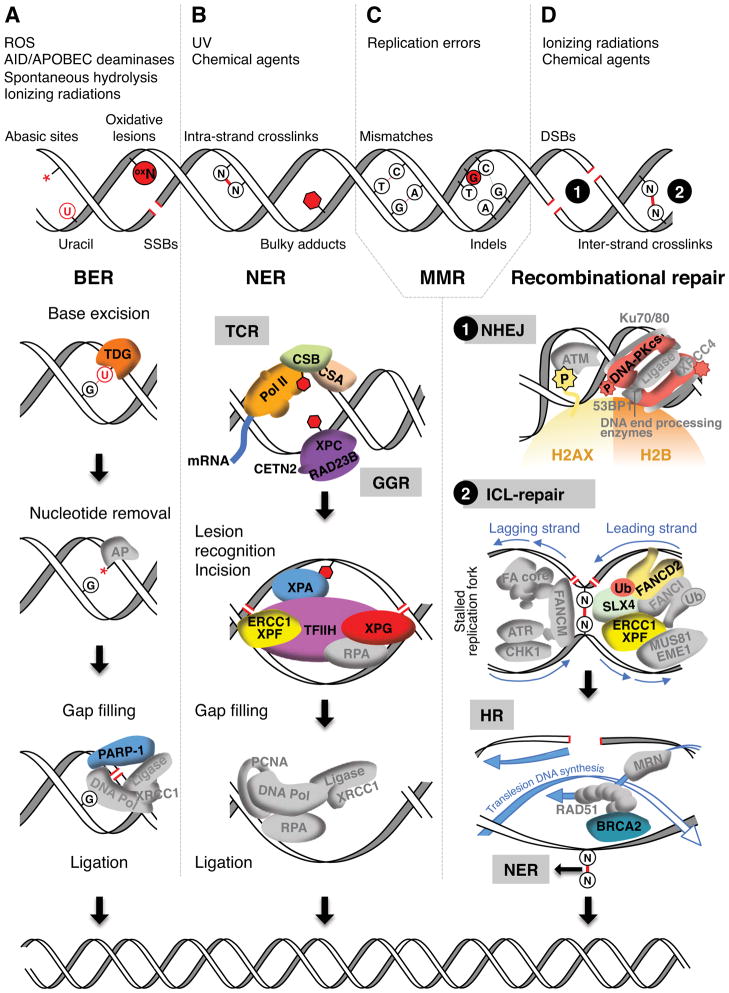
Considering the frequency at which DNA damage occurs (~104 events per day (Lindahl and Barnes, 2000)) and the broad spectrum of damage incurred by a cell, it is remarkable that the overwhelming majority of these offending DNA lesions are repaired with impressive accuracy and efficiency. Indeed, cells have evolved multiple, often overlapping, mechanisms to sense and demarcate the DNA lesions, while their repair is carried out by one (or more) of the four major pathways: base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR) and recombinational repair (Figure 1 and see reviews (Iyama and Wilson, 2013; Sancar et al., 2004)).
Figure 1. Major DNA repair pathways in mammals.
Exogenous and endogenous genotoxic agents (top) generate a variety of DNA damage, such as single and double strand breaks (SSBs, DSBs), insertions and deletions (indels). Lesions are detected and repaired by four major DNA repair pathways: base excision repair (BER, A), nucleotide excision repair (NER, B), mismatch repair (MMR, C) and recombinational repair (D). Mechanisms of BER, NER and recombinational repair are depicted. For each pathway, factors discussed in this review are in colors.
A. Removal of uracil from DNA by BER. Thymine DNA glycosylase (TDG) removes the nitrogenous base and generates an abasic site (*). Apurinic/apyrimidinic (AP) endonuclease finalizes the nucleotide removal and creates a nick in the sugar-phosphate backbone. Poly ADP-ribose polymerase 1 (PARP-1) senses the SSB and recruits DNA polymerase and ligase to fill in the gap.
B. Removal of large bulky adducts by NER. Transcribing RNA polymerase II (Pol II) stalls at DNA lesions and triggers transcription-coupled NER (TCR). TCR is initiated by the recruitment of Cockayne syndrome proteins A and B (CSA, CSB) to the arrested polymerase. DNA damage on the non-transcribed regions of the genome is repaired by global genome NER (GGR) instead. DNA lesion is recognized by the repair complex comprising Xeroderma pigmentosum C (XPC), RAD23B and Centrin 2 (CETN2). Completion of TCR and GGR requires the recruitment of downstream NER factors (XPA, RPA, TFIIH, ERCC1-XPF, XPG). ERCC1-XPF and XPG endonucleases incise the damaged strand a few bases 5′ and 3′ to the DNA lesion, respectively. The gap is filled in by DNA polymerase and sealed by ligase.
D. Removal of DSBs and inter-strand cross-links (ICL) by recombinational repair. 1. Repair of DSBs by non-homologous end joining (NHEJ). DSB sites are marked by Ataxia telangiectasia mutated (ATM) kinase-mediated phosphorylation of histone H2A variant X (γ-H2AX) (Burma et al., 2001). Ku proteins direct the binding of the catalytic subunits of the DNA-dependent protein kinase (DNA-PKcs) to the exposed DNA ends. Autophosphorylation of DNA-PKcs facilitates DNA-ends processing and resealing.
2. Repair of ICLs by Fanconi Anemia (FA), homologous recombination (HR) and NER pathways (see review (Deans and West, 2011)). During S-phase, converging replication forks stall at ICLs and are sensed by FANCM protein, which recruits downstream FA proteins (FA core) and initiates ATR (ATM- and Rad3-related)-CHK1 checkpoint response. The FA core complex ubiquitinates FANCD2 and FANCI. This facilitates the recruitment of endonucleases (SLX4, XPF/ERCC1, MUS81/EME1) and resection of the lesion from one of the two cross-linked strands. Translesion DNA synthesis proceeds through the uncut strand, generating the template for the homologous recombination machinery (MRN, BRCA2, RAD51) to complete DNA replication across the nicked DNA strand. NER removes the remaining adducts.
Given that transcription and DNA repair both involve intimate transactions with DNA, it is perhaps not surprising that these two processes are often coupled and, as we shall discuss in this review, perhaps also interdependent and cross functional. It is well known that there is preferential repair of the transcribed DNA strand in expressed genes by transcription-coupled repair (TCR), a sub-pathway of NER (Hanawalt and Spivak, 2008; Mellon et al., 1987). Central to this repair process is the recruitment of Cockayne Syndrome B (CSB), TFIIH, and Xeroderma pigmentosum G (XPG) to RNA polymerase II arrested at damaged site (Saxowsky and Doetsch, 2006; Svejstrup, 2002). In addition, all three factors have well-defined roles in transcription (Citterio et al., 2000; Ito et al., 2007; Schaeffer et al., 1993) while XPG and TFIIH are also key players in global genome repair (GGR), the other branch of NER, thus highlighting the interconnected nature of transcription and DNA repair (Kamileri et al., 2012a). In this review, we discuss recent findings suggesting a more complex, unanticipated interplay between transcription and DNA repair beyond TCR. A growing list of proteins and protein complexes that were long thought to function exclusively in DNA repair are revealing themselves to be involved in transcription as well. In catastrophic events like double strand breaks (DSBs), damage-induced ectopic transcription at such lesion appears to be an essential initiating event of the repair process. On the other hand, there is also accumulating evidence pointing to a role of eliciting DNA damage at gene promoters that can influence transcriptional activation. This extensive two-way crosstalk between transcription and DNA repair may arise in part from the fact that transcription is an inherently mutagenic process.
DNA repair factors that double as transcription factors
Mutations in NER factors CSB, XPG, and the helicase subunits of TFIIH (XPB and XPD) generate complex multi-symptom phenotypes that cannot be readily explained by defects in DNA repair alone. The realization that the same set of proteins also participates in transcriptional control resolved this long standing puzzle by pointing to a transcriptional component in disease etiology (Compe and Egly, 2012; Kamileri et al., 2012a). Since then, more factors in the NER and other repair pathways have also been implicated in transcriptional activation (Bradsher et al., 2002; Fong et al., 2012; Iben et al., 2002; Le May et al., 2010a). By taking full advantage of their DNA nuclease, glycosylase, or helicase and related ATPase activities, repair proteins facilitate the remodeling of the epigenetic landscape and topology of chromatin at gene promoters. Perhaps more surprisingly, they can also recruit cofactors and/or interface with the transcription apparatus by acting as classical transcriptional activators and co-activators (Table 1 and Figure 2).
Table 1.
DNA repair factors involved in repair and transcription
| Protein (Official Name) | Repair Pathway | Enzymatic Activity | Function in DNA Repair | Function in Transcription |
|---|---|---|---|---|
| CSB (ERCC6) | TCR (NER) | DNA-dependent ATPase | Initiates TC-NER at stalled Pol II |
|
| DNA-PKcs | NHEJ BER |
Protein kinase |
|
|
| FANCD2 | ICL | [ND] | Initiates ICL repair | Activates transcription of TAp63 and promotes senescence of tumorigenic cells |
| FANCP (SLX4) | ICL | [ND] | Acts as a scaffold for multiple structure- specific endonucleases | Cooperates with FANCD2 in transcriptional activation of TAp63 |
| TFIIH | TCR (NER) GGR (NER) |
|
|
|
| PARP-1 | NHEJ HR BER NER |
DNA-dependent poly(ADP- ribosyl)transferase | Interacts physically and functionally with components in NHEJ, HR, BER and NER pathways |
|
| TDG | BER | DNA glycosylase | Excises damaged nitrogenous bases |
|
| XPC-RAD23B-CETN2 | GGR (NER) BER |
[ND] |
|
|
| XPF (ERCC4) | TC-NER GG-NER ICL |
Structure-specific endonuclease | Incises the damaged strand 5′ to the DNA lesion |
|
| XPG (ERCC5) | TCR (NER) GGR (NER) |
Structure-specific endonuclease | Incises the damaged strand 3′ to the DNA lesion |
|
Definitions are as follows: BER, base excision repair; GG-NER, global genome nucleotide excision repair; HR, homologous recombination; ICL, inter-strand cross-link repair; NER, nucleotide excision repair; NHEJ, non-homologous end joining; NR, nuclear receptor; TC-NER, transcription-coupled nucleotide excision repair [ND], not determined
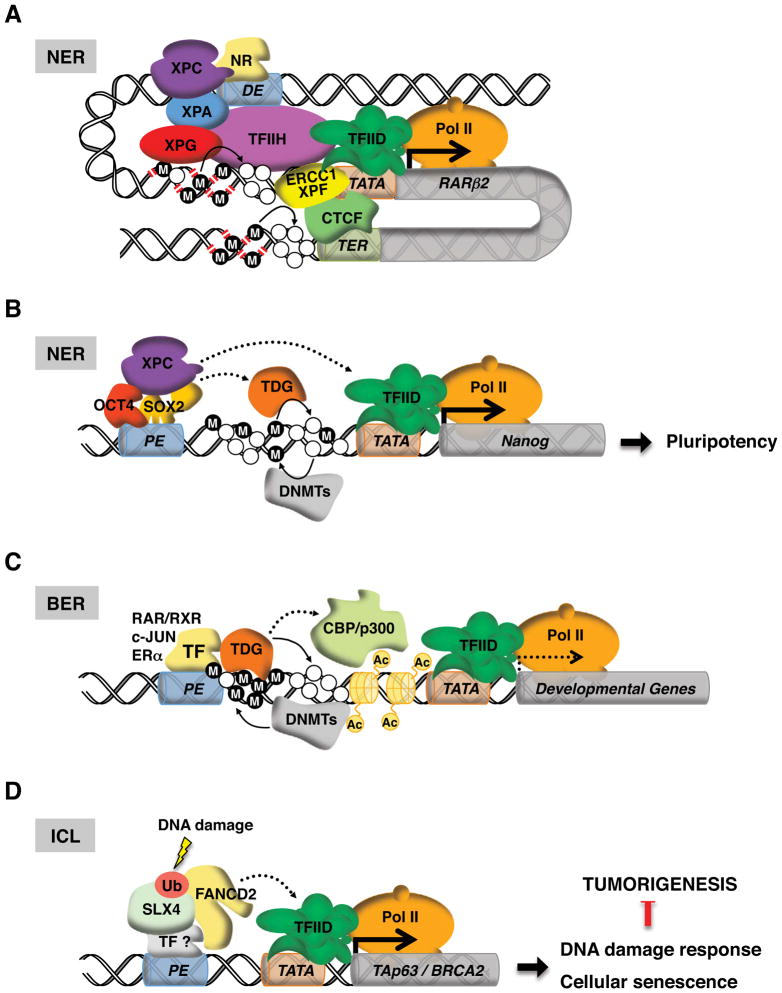
Figure 2. Proteins classically ascribed to DNA repair also participate in transcriptional control.
A–B. Nucleotide excision repair (NER). A. XPC potentiates transcriptional activation of nuclear receptors (NR) by nucleating the assembly of the entire NER machinery at the promoter (TATA) of responsive genes (RARβ2, retinoic acid receptor β2 gene). XPG and ERCC1-XPF endonucleases create DNA nicks, enabling DNA demethylation (open circles represent demethylated cytosines; filled circles denote 5-methylcytosines) at gene promoter and terminator (TER), CTCF recruitment and looping between proximal and distal regulatory elements (DE, distal enhancer). B. In embryonic stem cells, the XPC complex functions as a transcriptional co-activator for stem cell-specific transcription factors OCT4 and SOX2 to maintain pluripotency. The mechanism by which the XPC complex stimulates transcription remains to be elucidated (dashed arrow). XPC can potentially regulate transcription by stimulating TDG-mediated DNA demethylation at gene regulatory regions. Cytosines can be converted back to 5-methylcytosines by DNA methyltransferases (DNMTs).
B. Base excision repair (BER). Thymine DNA glycosylase (TDG) bridges CBP/p300 histone acetyltransferase to sequence specific transcription factors (RAR/RXR, c-JUN, ERα) and participates in active DNA demethylation at promoters (TATA) of transcriptionally-poised, developmentally-regulated genes. Re-methylation of DNA is carried out by DNMTs.
C. Inter-strand cross-link repair (ICL). Upon DNA damage, mono-ubiquitinated Fanconi anemia protein D2 (FANCD2) and its repair partner SLX4/FANCP bind and activate gene promoters that are implicated in tumor suppression and cellular senescence (e.g., TAp63, BRCA2). The mechanism by which FANC proteins activate transcription is unclear (dashed arrow).
Nucleotide excision repair
XPC is a member of the Xeroderma pigmentosum family that is dispensable for TCR but essential for initiating GGR (Venema et al., 1990). If XPC functions purely as a sensor for bulky DNA lesions, scanning for damage by XPC ought to be transient (Camenisch et al., 2009), and its interaction with DNA should display neither sequence nor positional preference. Instead, in HeLa cells and human fibroblasts, XPC was found to bind hormone-inducible gene promoters (e.g. RARβ2) and nucleate the assembly of an entire NER complex in a transcription-dependent but DNA damage-independent manner, thus arguing for an expanded role of NER factors in gene regulation (Le May et al., 2010b) (Figure 2A). Indeed, depletion of XPC disrupted NER complex assembly at RARβ2 and significantly attenuated its transcriptional response to retinoic acid induction. Optimal expression of RARβ2 requires DNA breaks elicited by endonucleases XPG and XPF, which, by a yet unknown mechanism, enable CTCF recruitment and gene looping (Le May et al., 2012). These coordinated events are accompanied by active DNA demethylation that could depend on the DNA damage-inducible protein GADD45A, although its involvement remains controversial (Barreto et al., 2007; Jin et al., 2008; Schmitz et al., 2009). Consistent with these observations, the ERCC1-XPF complex has also been shown to regulate transcription initiation of genes associated with growth in mice (Kamileri et al., 2012b).
It is worth noting that the absence of NER factors blunted but did not abolish transcriptional activation of only a subset of genes (Le May et al., 2010b). The intrinsic affinity of XPC for DNA (Krasikova et al., 2013) alone cannot explain how XPC is directed to this subset of hormone-inducible and transcriptionally-poised gene promoters. Instead, we favor a model where XPC is recruited to promoters by gene-specific activators, or potentially through recognition of DNA bends and distortions induced by binding of nuclear hormone receptors to their cognate response elements (Nardulli and Shapiro, 1993; Robinson et al., 1998; Sugasawa et al., 2001). Assembly of the NER complex at gene promoters can be further stabilized by interactions with core components of the PIC (Kamileri et al., 2012b; Yokoi et al., 2000). Therefore, DNA repair factors may act as “facilitators” of gene activation by creating a favorable structural and epigenetic configuration for transcription in a gene- and possibly cell type-specific manner.
Additional evidence of cell type-specific transcriptional regulation by NER factors came from a recent study using an unbiased in vitro reconstituted system to biochemically screen for factors that are required for transcription of the stem cell pluripotency gene Nanog (Chambers et al., 2003; Mitsui et al., 2003). The use of a highly integrated in vitro transcription assay uncovered an activity enriched in an embryonic stem (ES) cell nuclear extract that is essential for the key stem cell-specific activators OCT4 and SOX2 to activate Nanog (Fong et al., 2011). This newly detected activity turned out surprisingly to be the XPC-RAD23B-Centrin 2 complex (referred to as XPC hereafter) (Araki et al., 2001) (Figure 2B). Although this result mirrors to some extent what is known about the role of XPC and other NER factors in general transcription, how XPC mediates OCT4/SOX2-specific activated transcription in ES cells seemed to differ fundamentally from its action in differentiated cell types in several respects. First, XPC can directly potentiate the transcription of Nanog in the absence of other XP proteins (except TFIIH that is required to form an active prototypic PIC), suggesting that the assembly of a NER complex initiated by XPC and the subsequent DNA breaks induced by XPF and XPG are dispensable for Nanog activation at least in vitro. Furthermore, XPC occupies distal enhancers of a remarkably high number of OCT4/SOX2-target genes in ES cells (~70%) (Fong et al., 2011). It also became clear that XPC is not merely a passive partner of OCT4 and SOX2 because disruption of the XPC complex compromises ES cell transcriptional responses, self-renewal and somatic cell reprogramming. Instead, a wealth of evidence suggests that XPC acts as a functionally important stem cell selective co-activator for OCT4 and SOX2 wherein XPC is recruited to enhancers likely via a direct interaction with the activators (Fong et al., 2011). Coopting a DNA repair factor by activators to drive stem cell-specific transcription may also provide the added benefits of protecting the integrity of genes essential for self-renewal and pluripotency from DNA damage (Etchegaray and Mostoslavsky, 2011).
Base excision repair
Active DNA demethylation is thought to play a key role in transcriptional control and resetting of epigenetic memory during embryonic development and cellular reprogramming (Wu and Zhang, 2010). Although there are multiple mechanisms by which 5-methylcytosine (5mC) is removed in mammals, a common end point to some of these processes appears to be excision of the deaminated and/or oxidized derivatives of 5mC by the BER enzyme thymine DNA glycosylase (TDG) (Cortazar et al., 2007; Franchini et al., 2012). However, how prevalent and where TDG-mediated active DNA demethylation occurs in the mammalian genome is not well understood. Four recent reports suggest that it is extensive (Cortazar et al., 2011; Cortellino et al., 2011; Shen et al., 2013; Song et al., 2013). Dynamic DNA demethylation occurs preferentially at promoters of silent and developmentally poised genes, and distal enhancers of active genes in mouse ES cells (Shen et al., 2013; Song et al., 2013). Given the pervasive nature of DNA methylation/demethylation in the genome, it was surprising that TDG knockdown in mouse ES cells only affected the expression of a handful of genes. Apparently, active DNA demethylation at distal enhancers is dispensable for ongoing transcription. Presumably, the binding of regulatory factors (i.e. activators) is not particularly perturbed by the loss of regulated 5mC/C turnover at enhancers (Shen et al., 2013). On the other hand, the ability to dynamically control DNA methylation homeostasis at promoters of developmentally- and transcriptionally-poised genes by TDG appears to be essential for the rapid and complete transcriptional response to inducers like retinoic acid (Cortazar et al., 2011; Cortellino et al., 2011) (Figure 2C). Therefore, TDG depletion evidently compromised primarily the reactivation of signal-dependent and developmentally-poised genes during ES cell differentiation and embryogenesis. This may also explain why TDG inactivation is embryonically lethal, a rather unusual phenotype given that many related DNA glycosylases are dispensable for embryogenesis (Cortazar et al., 2007).
Independent of its catalytic activity, TDG can also potentiate transcription by acting as a scaffold to bridge the transcriptional co-activator CBP/p300 to transcription factors like c-JUN (Chevray and Nathans, 1992), RAR/RXR (Cortellino et al., 2011; Um et al., 1998) and estrogen receptor α (ERα) (Chen et al., 2003) to facilitate histone modification (Figure 2C). Interestingly, the NER factor XPC has been shown to stimulate the DNA glycosylase activity of TDG via a direct interaction in vitro (Shimizu et al., 2010). This raises the intriguing possibility that XPC could also regulate transcription by coordinating with TDG and BER in active DNA demethylation at gene regulatory regions (Figure 2B). Recently, it has been shown that the most dynamically regulated and differentially methylated regions (DMRs) across different cell types are overrepresented by enhancers and transcription factor binding sites that display tissue or cell type-specific regulation (Ziller et al., 2013). In light of this finding, it is tempting to speculate that NER and BER factors could contribute to this process by cooperating with cell type-specific transcription factors.
Recombinational repair
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome characterized by developmental abnormalities, bone marrow failure and increased predisposition to squamous cell carcinoma of the skin (D’Andrea and Grompe, 1997). In response to DNA damage that creates inter-strand cross-links, mono-ubiquitination of Fanconi anemia (FA) protein FANCD2 by the FA core complex facilitates the binding of FANCD2 to chromatin (Montes de Oca et al., 2005) and initiates the repair process by recruiting SLX4/FANCP and other downstream FA proteins (Yamamoto et al., 2011). However, a recent study uncovered an unanticipated role for the mono-ubiquitinated form of FANCD2 (FANCD2-Ub) in the transcriptional activation of the tumor suppressor gene TAp63 (Park et al., 2013). It is unclear how FANCD2-Ub regulates transcription, but its recruitment to the regulatory region of the TAp63 promoter is dependent on SLX4, presumably through the recognition of the ubiquitin moiety on FANCD2 by the ubiquitin binding domains in SLX4. Genome-wide analysis of FANCD2-Ub binding sites identified many DNA damage-dependent or -enhanced targets including BRCA2, a key player in homologous recombination repair (Roy et al., 2011). This suggests that FANCD2-Ub and SLX4 may cooperate to generate a coordinated response to DNA damage by repairing the lesion and perhaps simultaneously acting as transcription factors to establish a gene expression program that promotes cellular senescence of damaged and potentially tumorigenic cells (Figure 2D). In fact, using the same DNA damage-induced posttranslational modification (i.e. mono-ubiquitination) on FANCD2 as a trigger for both transcriptional activation and DNA repair provides a potentially elegant solution to synchronizing the two processes. It is worth noting that XPC is both ubiquitinated and sumoylated in response to UV-induced DNA damage (Sugasawa et al., 2005; Wang et al., 2005). It would therefore be of interest to examine the functional consequences of posttranslational modifications on XPC in transcription and DNA repair and the putative crosstalk between the two processes.
Transcription facilitates DNA damage
As vital as transcription is, it is not entirely an innocuous process. Transcription inevitably exposes the DNA template to attacks by genotoxic agents and generates potentially harmful DNA structures that are prone to mutagenesis and recombination (Figure 3A). Oddly enough, gene activation sometimes also requires transient and localized DNA damage at promoters that must be repaired (Figure 3B). Therefore, involvement of DNA repair factors in transcriptional control might originate as an adaptive measure by cells to preserve genetic information that evolved to take on additional roles in transcriptional regulation, thus further blurring the line between transcription and DNA repair factors, and, to a certain extent, the processes they mediate.
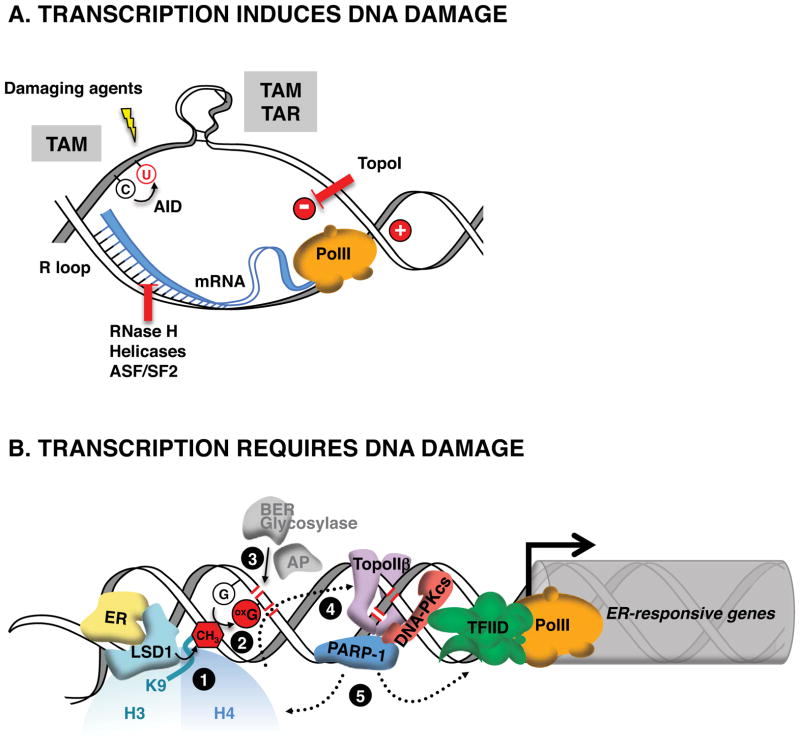
Figure 3. Transcription and DNA repair intersect.
A. Transcription is a mutagenic process. R loops form when nascent messenger RNAs (mRNAs) hybridize back to their template. Negative (−) and positive (+) supercoiling accumulate behind and ahead of elongating RNA polymerase II (Pol II), and stabilize R loops. The displaced single-stranded DNA is highly susceptible to chemical modifications (DNA damaging agents and deamination by activation-induced cytidine deaminase (AID)), and to the formation of recombinogenic secondary structures that are prone to transcription-associated mutagenesis (TAM) and recombination (TAR). Topoisomerase I (TopoI), RNase H, helicases and splicing factors (ASF/SF2) can prevent or disrupt the formation of mutagenic R loop structures.
B.
“Scheduled” DNA damage promotes transcriptional activation. Upon ligand binding, estrogen receptor (ER) activates histone H3 demethylase LSD1 at responsive genes
 . The demethylation reaction releases reactive oxygen species that converts nearby guanines (G) into 8-oxo-guanines (oxG)
. The demethylation reaction releases reactive oxygen species that converts nearby guanines (G) into 8-oxo-guanines (oxG)
 . oxG removal by base excision repair (BER) creates DNA nicks
. oxG removal by base excision repair (BER) creates DNA nicks
 that facilitate entrance of the endonuclease topoisomerase IIβ (TopoIIβ)
that facilitate entrance of the endonuclease topoisomerase IIβ (TopoIIβ)
 . TopoIIβ-induced double strand breaks recruit PARP-1 and DNA-PKcs repair enzymes, which induce a permissive chromatin architecture for transcription initiation
. TopoIIβ-induced double strand breaks recruit PARP-1 and DNA-PKcs repair enzymes, which induce a permissive chromatin architecture for transcription initiation
 .
.
Transcription-associated mutagenesis and recombination
The very act of transcription requires the separation of the two strands of a DNA double helix by RNA polymerase II (RNAPII). As RNAPII transverses the DNA, negatively supercoiled DNA accumulated behind the advancing polymerase can cause strand opening and expose single-stranded (ss) regions to chemical modifications and genotoxic agents (Rahmouni and Wells, 1992). For example, deamination of cytosine by spontaneous hydrolysis or by activation-induced cytidine deaminase (AID)/APOBEC: a family of deaminases that preferentially target ssDNA regions that, if left unresolved by BER, could lead to C-to-T substitutions (Conticello, 2008; Frederico et al., 1990). In addition to nucleotide substitutions, ssDNA regions are prone to recombination as well as expansion and contraction of tri-nucleotide repeats. These likely arise as a result of attempts by transcription and replication enzymes to negotiate local secondary structures (e.g. hairpins and loops) formed at DNA sequence repeats (see review (Kim and Jinks-Robertson, 2012)). It therefore comes as no surprise that high levels of transcription are often associated with increased spontaneous rates of mutagenesis and recombination events in phenomena known as transcriptional-associated mutation (TAM) and transcription-associated recombination (TAR) (Aguilera, 2002; Lippert et al., 2011; Mischo et al., 2011; Takahashi et al., 2011).
The good and bad of R loops
R loops are three-stranded nucleic acid structures containing an RNA-DNA duplex and a displaced, single stranded DNA (Reaban et al., 1994). They occur naturally during lagging strand DNA synthesis when DNA primase synthesizes a short RNA oligomer on ssDNA, and during transcription in the form of a transient hybrid between the newly synthesized messenger RNA (mRNA) and the template DNA at the catalytic center of RNAPII, where the length of the RNA-DNA hybrid can dictate stability and processivity of the transcription complex (Bochkareva et al., 2012; Nudler, 2012). The recombinogenic potential of R loop is highlighted in immunoglobulin (Ig) class switch recombination (CSR) at the Ig heavy chain locus, a process that is critical to diversifying the antibody repertoire during an immune response (Lieber, 2010). Transcription through the G-rich switch (S) regions generates R loops that can exceed 1 kilobase in size (Yu et al., 2003). This enables AID-dependent modification of the displaced ssDNA. Deamination of cytosine to uracil by AID is thought to provoke base excision by the BER factor uracil DNA glycosylase, followed by recruitment of endonucleases to create single- and double-strand breaks within the S region. Class switching as a result of forming new S-S junctions is then achieved by the non-homologous end joining (NHEJ) recombinational repair mechanism. R loops are therefore functionally important regulatory structures that appear to impact a wide array of cellular processes (Aguilera and Garcia-Muse, 2012).
R loops are also sources of TAM and TAR (Figure 3A). In addition to an increased vulnerability of ssDNA regions to attacks by genotoxic agents and nucleases, the RNA-DNA hybrid in an R loop can potentially prime error-prone DNA synthesis (Aguilera and Garcia-Muse, 2012). As in the case of CSR, ectopic R loops can generate DNA strand breaks by nucleases and promote downstream recombination events (Li and Manley, 2006). Indeed, defects in the molecular pathways limiting R loop genesis and clearance have been linked to human diseases (Lin and Wilson, 2012; Yuce and West, 2013). Fortunately, redundant mechanisms exist to suppress ectopic R loop structures induced by transcription. DNA strand opening due to torsional strain generated by transcription (or replication) is counteracted by Topoisomerase I (TopoI) that removes DNA supercoils. In fact, it has recently been shown that transcription of exceptionally long genes (>200 kilobases) is particularly sensitive to TopoI inactivation (King et al., 2013). A key step in R loop formation is the invasion and hybridization of the nascent mRNA to the template strand. It has been shown that coating of mRNA by splicing factor ASF/SF2 precludes RNA-DNA hybridization by packaging nascent mRNA into ribonucleoprotein complexes (Li and Manley, 2005). RNA-DNA hybrids can also be removed through unwinding by helicases or digestion of the RNA moiety by RNase H (Aguilera and Garcia-Muse, 2012). Co-transcriptional R loops are therefore rare but highly disruptive structures that pose major threats to genome stability when one of these fail-safe mechanisms is compromised (Gan et al., 2011; Wahba et al., 2013).
Transcriptional activation may require “scheduled” DNA damage
Cells have developed multiple mechanisms to minimize the mutagenic potential of DNA damage. In the case of DSBs in gene bodies, Polycomb repressive complexes (PRCs) accumulate at break sites and are thought to mediate transcriptional silencing as an adaptive response to reduce interference between repair and the transcription apparatus (Vissers et al., 2012). Transcriptional inhibition has also been observed in BER of oxidative DNA damage sites that do not arrest transcribing RNAPII (and therefore do not evoke TCR), likely as a preventive measure against miscoding by RNAPII (Khobta and Epe, 2012). While global damage on DNA is generally associated with gene silencing, targeted DSB formation and localized DNA base damage have been implicated in transcriptional activation by nuclear hormone receptors (Ju et al., 2006; Lin et al., 2009; Perillo et al., 2008). Eliciting a “scheduled” DNA base damage appears to be a common first step in hormone-dependent activation. Liganded androgen receptor recruits AID to deaminate cytosine (Lin et al., 2009) while estrogen receptor activates resident lysine-specific demethylase 1 (LSD1) to demethylate histone 3 lysine 9, an oxidative process that releases hydrogen peroxide and converts nearby guanines into 8-oxo-guanines (Perillo et al., 2008). Their ensuing repair by DNA glycosylases is thought to generate transient DNA nicks that act as entry points for DNA endonucleases like Topoisomerase IIβ (TopoIIβ) (Ju et al., 2006; Perillo et al., 2008) or LINE-1 repeat encoded ORF2 endonuclease (Lin et al., 2009). The resulting DSBs are thought to relax DNA strands and facilitate the recruitment of other damage sensing and DNA repair enzymes (e.g. PARP-1 and DNA-PK) to collectively induce a permissive chromatin architecture for transcriptional activation (Ju et al., 2006). These observations are reminiscent of the proposed role of DNA breaks induced by NER factors XPG and XPF in chromatin looping (Le May et al., 2012). Perhaps relaxation of DNA strands via DSBs or nicks permits chromosome bending thereby facilitating enhancer-promoter communication and dynamic gene transcription (Figure 3B).
It is thus evident that DNA damage can have opposite effects on transcription. We presume that there must exist a mechanism that enables cells to distinguish between these “scheduled” DNA damage events linked to gene activation from those that arise spontaneously and cause undesirable consequences. After all, both processes could generate damage-induced assembly of DNA repair complexes that are, for all intents and purposes, highly similar if not identical to each other. Perhaps the transient nature of transcription-induced lesions like DSBs by TopoIIβ prevents them from eliciting a persistent DNA damage response (DDR) (Ciccia and Elledge, 2010). It is also likely that transcription can limit the amplification of a DDR by suppressing the accumulation and spreading of the phosphorylated form of histone variant H2AX (γ-H2AX) that would normally mark DSB sites and promote the retention of repair factors (Iacovoni et al., 2010).
DNA damage induces “unscheduled” transcription
Efficient repair of DSBs by recombinational mechanisms (i.e. homologous recombination (HR) or NHEJ) requires the coordinated action of factors that sense, mark and process the lesions before the repair machinery can be recruited to the damage sites. Although there is a wealth of information on protein factors involved in the initial phase of DSB repair, two recent studies point to an unanticipated role of small non-coding RNAs (ncRNAs) in repair that is conserved from plants to humans (Francia et al., 2012; Wei et al., 2012). These ~21 nucleotide short ncRNAs, termed DSB-induced small RNAs (diRNAs), are derived from ectopic transcription induced at DSB sites and processed by machinery in the microRNA biogenesis pathway. They are proposed to guide downstream DSB repair proteins to damaged sites to facilitate the repair process (Francia et al., 2012). It would be of interest to examine whether diRNAs are also involved in the repair of physiological DSBs created in V(D)J recombination and CSR during an immune response (Lieber, 2010).
It is worth noting that in Arabidopsis thaliana, plant-specific RNA polymerases IV and V are able to generate transcripts at and near DSB sites in both sense and antisense strands. In human cells, RNAPII can also initiate transcription from both strands but is conspicuously excluded from regions immediately surrounding the DSB sites (Wei et al., 2012). This is likely due to the mutual antagonism between γ-H2AX accumulation at DSB sites and RNAPII transcription (Iacovoni et al., 2010). This also underscores a fundamental difference in how RNAPIV/V and RNAPII can or cannot navigate DSB sites. Since the experimentally-induced DSBs were created at a single defined position within the protein coding region of an integrated reporter (Wei et al., 2012), a more immediate question is what drives ectopic initiation when regulatory sequences critical for the assembly of a typical PIC are likely absent at DSB sites. Therefore, an alternative PIC recruitment mechanism must exist. Whether or not this DSB-induced PIC is identical to the “canonical” PIC assembled at gene promoter remains to be elucidated. However, given the recent appreciation that the make-up of a functional PIC is more flexible than previously thought (D’Alessio et al., 2009; Goodrich and Tjian, 2010; Muller et al., 2010), and that many DNA repair factors can function as transcription factors, it is conceivable that in this context DNA repair factors could directly recruit RNA polymerase to the damaged sites and initiate transcription. Thus, damage-induced PICs could be compositionally and perhaps functionally distinct from prototypical PICs.
Conclusion
In this review, we have examined various ways in which DNA repair factors can impact gene activation. But they all share one common thread: DNA repair factors are often required for transcription of developmentally-regulated and activator-dependent genes, but less so for constitutive housekeeping transcription. Based on these observations, DNA repair factors likely facilitate de novo PIC assembly at gene promoters, a rate-limiting step for gene activation (Lemon and Tjian, 2000; Michel and Cramer, 2013), by facilitating chromatin remodeling and enhancer-promoter communication.
Development of a multicellular organism from a zygote is critically dependent on a highly controlled process of cellular proliferation and differentiation. Proper execution of these processes is largely regulated at the transcriptional level (Levine and Tjian, 2003). The reverse is also true; reprogramming of cells with restricted developmental potential back to a pluripotent state requires dynamic changes in chromatin organization, histone/DNA modifications and reactivation of a complex transcriptome (Apostolou et al., 2013; Phillips-Cremins et al., 2013; Polo et al., 2012; Wei et al., 2013; Zhang et al., 2013). The expanded roles of DNA repair factors in transcriptional control may explain why inactivation of factors in the NER, HR, NHEJ and Fanconi anemia (FA) repair pathways poses such strong barriers to somatic cell reprogramming by OCT4, SOX2, KLF4 and c-MYC (Fong et al., 2011; Gonzalez et al., 2013; Molina-Estevez et al., 2013; Muller et al., 2012; Takahashi and Yamanaka, 2006). On the other hand, rapid ectopic induction of transcription of a large number of genes by these factors during the initial phase of reprogramming could promote TAM and TAR due to a surge in transcriptional load and replication stress (Helmrich et al., 2012), thus activating a DNA damage response that is prohibitive to the conversion process (Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009). This is consistent with the observation that there is a dramatic increase in frequency of DSBs, as marked by accumulation of γ-H2AX and FANCD2 foci, in partially reprogrammed cells (Gonzalez et al., 2013; Muller et al., 2012). Taken together, these finding suggest that a major threat to the intrinsic ability of somatic cells to reprogram could indeed come from within.
Requirement of DNA repair factors in stem cell pluripotency and in development may not be just a preventive mechanism to safeguard genome integrity but also a proactive mechanism to ensure that their respective transcriptional programs are robust yet dynamic enough to change in response to developmental cues. The “symbiotic” relationship between transcription and DNA repair may originate from the fact that integrity of one is dependent on the other.
Acknowledgments
The authors wish to thank M. Botchan, J-M. Egly, D. Rio and C. Inouye for critical reading of the manuscript. C. Cattoglio is a postdoctoral fellow of California Institute for Regenerative Medicine (CIRM training program TG2-01164).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera A. The connection between transcription and genomic instability. The EMBO journal. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell stem cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. The Journal of biological chemistry. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. The EMBO journal. 2012;31:630–639. doi: 10.1038/emboj.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM. CSB is a component of RNA pol I transcription. Molecular cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. The Journal of biological chemistry. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, Naegeli H. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. The EMBO journal. 2009;28:2387–2399. doi: 10.1038/emboj.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen D, Lucey MJ, Phoenix F, Lopez-Garcia J, Hart SM, Losson R, Buluwela L, Coombes RC, Chambon P, Schar P, Ali S. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. The Journal of biological chemistry. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Molecular and cellular biology. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome biology. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA repair. 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio JA, Wright KJ, Tjian R. Shifting players and paradigms in cell-specific transcription. Molecular cell. 2009;36:924–931. doi: 10.1016/j.molcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea AD, Grompe M. Molecular biology of Fanconi anemia: implications for diagnosis and therapy. Blood. 1997;90:1725–1736. [PubMed] [Google Scholar]
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderich K, Alanazi M, Hoeijmakers JH. Premature aging and cancer in nucleotide excision repair-disorders. DNA repair. 2011;10:772–780. doi: 10.1016/j.dnarep.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Mostoslavsky R. eNERgizing pluripotent gene transcription. Cell stem cell. 2011;9:285–286. doi: 10.1016/j.stem.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Fong YW, Cattoglio C, Yamaguchi T, Tjian R. Transcriptional regulation by coactivators in embryonic stem cells. Trends in cell biology. 2012;22:292–298. doi: 10.1016/j.tcb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147:120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annual review of genetics. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes & development. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Georgieva D, Vanoli F, Shi ZD, Stadtfeld M, Ludwig T, Jasin M, Huangfu D. Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state. Cell reports. 2013;3:651–660. doi: 10.1016/j.celrep.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nature reviews. 2010;11:549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nature structural & molecular biology. 2012;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. The EMBO journal. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben S, Tschochner H, Bier M, Hoogstraten D, Hozak P, Egly JM, Grummt I. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Molecular cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Iyama T, Wilson DM., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA repair. 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS genetics. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science (New York, N Y. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 2012a;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Kamileri I, Karakasilioti I, Sideri A, Kosteas T, Tatarakis A, Talianidis I, Garinis GA. Defective transcription initiation causes postnatal growth failure in a mouse model of nucleotide excision repair (NER) progeria. Proceedings of the National Academy of Sciences of the United States of America. 2012b;109:2995–3000. doi: 10.1073/pnas.1114941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khobta A, Epe B. Interactions between DNA damage, repair, and transcription. Mutation research. 2012;736:5–14. doi: 10.1016/j.mrfmmm.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nature reviews. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Yandava CN, Mabb AM, Hsiao JS, Huang HS, Pearson BL, Calabrese JM, Starmer J, Parker JS, Magnuson T, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasikova YS, Rechkunova NI, Maltseva EA, Pestryakov PE, Petruseva IO, Sugasawa K, Chen X, Min JH, Lavrik OI. Comparative analysis of interaction of human and yeast DNA damage recognition complexes with damaged DNA in nucleotide excision repair. The Journal of biological chemistry. 2013;288:10936–10947. doi: 10.1074/jbc.M112.444026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Egly JM, Coin F. True lies: the double life of the nucleotide excision repair factors in transcription and DNA repair. Journal of nucleic acids. 2010a;2010 doi: 10.4061/2010/616342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Fradin D, Iltis I, Bougneres P, Egly JM. XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Molecular cell. 2012;47:622–632. doi: 10.1016/j.molcel.2012.05.050. [DOI] [PubMed] [Google Scholar]
- Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Molecular cell. 2010b;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes & development. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes & development. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wilson JH. Nucleotide excision repair, mismatch repair, and R-loops modulate convergent transcription-induced cell death and repeat instability. PloS one. 2012;7:e46807. doi: 10.1371/journal.pone.0046807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harbor symposia on quantitative biology. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O’Shea SH, Jinks-Robertson S. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Blanpain C, Rossi DJ. DNA damage response in adult stem cells: pathways and consequences. Nat Rev Mol Cell Biol. 2011;12:198–202. doi: 10.1038/nrm3060. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Michel M, Cramer P. Transitions for regulating early transcription. Cell. 2013;153:943–944. doi: 10.1016/j.cell.2013.04.050. [DOI] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Molecular cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Molina-Estevez FJ, Lozano ML, Navarro S, Torres Y, Grabundzija I, Ivics Z, Samper E, Bueren JA, Guenechea G. Impaired Cell Reprogramming in Non-Homologous End Joining Deficient Cells. Stem cells (Dayton, Ohio) 2013 doi: 10.1002/stem.1406. [DOI] [PubMed] [Google Scholar]
- Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, Houghtaling S, Grompe M, D’Andrea AD. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- Muller F, Zaucker A, Tora L. Developmental regulation of transcription initiation: more than just changing the actors. Current opinion in genetics & development. 2010;20:533–540. doi: 10.1016/j.gde.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Muller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K, Moreau LA, Schambach A, Park IH, London WB, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annual review of biochemistry. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Nardulli AM, Shapiro DJ. DNA bending by nuclear receptors. Receptor. 1993;3:247–255. [PubMed] [Google Scholar]
- Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Kim H, Kim JM, Primack B, Vidal-Cardenas S, Xu Y, Price BD, Mills AA, D’Andrea AD. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Molecular cell. 2013;50:908–918. doi: 10.1016/j.molcel.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science (New York, N Y. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmouni AR, Wells RD. Direct evidence for the effect of transcription on local DNA supercoiling in vivo. Journal of molecular biology. 1992;223:131–144. doi: 10.1016/0022-2836(92)90721-u. [DOI] [PubMed] [Google Scholar]
- Reaban ME, Lebowitz J, Griffin JA. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. The Journal of biological chemistry. 1994;269:21850–21857. [PubMed] [Google Scholar]
- Robinson CE, Wu X, Morris DC, Gimble JM. DNA bending is induced by binding of the peroxisome proliferator-activated receptor gamma 2 heterodimer to its response element in the murine lipoprotein lipase promoter. Biochemical and biophysical research communications. 1998;244:671–677. doi: 10.1006/bbrc.1998.8305. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS letters. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual review of biochemistry. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chemical reviews. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science (New York, NY. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Molecular cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Uchimura Y, Dohmae N, Saitoh H, Hanaoka F, Sugasawa K. Stimulation of DNA Glycosylase Activities by XPC Protein Complex: Roles of Protein-Protein Interactions. Journal of nucleic acids. 2010;2010 doi: 10.4061/2010/805698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes & development. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. The Journal of biological chemistry. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, van Hoffen A, Natarajan AT, van Zeeland AA, Mullenders LH. The residual repair capacity of xeroderma pigmentosum complementation group C fibroblasts is highly specific for transcriptionally active DNA. Nucleic acids research. 1990;18:443–448. doi: 10.1093/nar/18.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers JH, van Lohuizen M, Citterio E. The emerging role of Polycomb repressors in the response to DNA damage. Journal of cell science. 2012;125:3939–3948. doi: 10.1242/jcs.107375. [DOI] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic acids research. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell stem cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. The Journal of biological chemistry. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature immunology. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yuce O, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Molecular and cellular biology. 2013;33:406–417. doi: 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jiao W, Sun L, Fan J, Chen M, Wang H, Xu X, Shen A, Li T, Niu B, et al. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell stem cell. 2013;13:30–35. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]