Abstract
Haem-dependent catalase is an antioxidant enzyme that degrades H2O2, producing H2O and O2, and is common in aerobes. Catalase is present in some strictly anaerobic methane-producing archaea (methanogens), but the importance of catalase to the antioxidant system of methanogens is poorly understood. We report here that a survey of the sequenced genomes of methanogens revealed that the majority of species lack genes encoding catalase. Moreover, Methanosarcina acetivorans is a methanogen capable of synthesizing haem and encodes haem-dependent catalase in its genome; yet, Methanosarcina acetivorans cells lack detectable catalase activity. However, inducible expression of the haem-dependent catalase from Escherichia coli (EcKatG) in the chromosome of Methanosarcina acetivorans resulted in a 100-fold increase in the endogenous catalase activity compared with uninduced cells. The increased catalase activity conferred a 10-fold increase in the resistance of EcKatG-induced cells to H2O2 compared with uninduced cells. The EcKatG-induced cells were also able to grow when exposed to levels of H2O2 that inhibited or killed uninduced cells. However, despite the significant increase in catalase activity, growth studies revealed that EcKatG-induced cells did not exhibit increased tolerance to O2 compared with uninduced cells. These results support the lack of catalase in the majority of methanogens, since methanogens are more likely to encounter O2 rather than high concentrations of H2O2 in the natural environment. Catalase appears to be a minor component of the antioxidant system in methanogens, even those that are aerotolerant, including Methanosarcina acetivorans. Importantly, the experimental approach used here demonstrated the feasibility of engineering beneficial traits, such as H2O2 tolerance, in methanogens.
Introduction
Methane-producing archaea (methanogens) are strictly anaerobic micro-organisms, which are only capable of growth by methanogenesis. Methanogenesis requires specific coenzymes and enzymes, many of which contain metal cofactors and function at low redox potentials (Thauer et al., 2008). Exposure of anaerobes to molecular oxygen (O2) results in autoxidation of cellular components, including flavoenzymes and metalloenzymes, leading to the production of reactive oxygen species (ROS), including superoxide (O2−) and H2O2. O2 and the ROS produced cause oxidative damage to enzymes, cofactors, coenzymes and general macromolecules, and may ultimately lead to cell death (Imlay, 2002). Thus, methanogens are sensitive to O2, and are only capable of growing and producing methane under anaerobic conditions. Nonetheless, methanogens are exposed transiently to O2, which would necessitate antioxidant and repair enzymes to facilitate O2 tolerance (Angel et al., 2011, 2012; Fetzer et al., 1993). Indeed, the majority of methanogen species can tolerate O2 exposure and there is evidence that some methanogens associated with termite gut can produce methane in the presence of low levels of O2 (Tholen et al., 2007). A detailed understanding of the molecular mechanisms underlying the oxidant tolerance of methanogens is lacking. Methanogens are of significant environmental and biotechnological importance, and an understanding of their antioxidant mechanisms may lead to the development of methods to enhance or inhibit methanogenesis.
Recent evidence suggests that strictly anaerobic bacteria and archaea contain antioxidant enzymes that differ from those found in aerobes and facultative organisms. For example, superoxide dismutase (SOD) and catalase are prevalent in aerobes and facultative microbes, but are found less frequently in strict anaerobes. It is hypothesized that anaerobes lack SOD and catalase because each enzyme produces O2 as an end product, which would serve to further propagate the production of ROS in anaerobes (Imlay, 2002). Rather, strict anaerobes contain enzymes such as superoxide reductase, peroxidase and rubrerythrin, which degrade O2− and H2O2 without producing O2 (Jenney et al., 1999; Lumppio et al., 2001). Nonetheless, there is evidence that catalase is part of the antioxidant system in some anaerobes. Catalase has been shown to be important in the tolerance to O2 and H2O2 by some sulfate-reducing bacteria, Bacteroides spp. and acetogens (Brioukhanov & Netrusov, 2004). There is limited evidence that indicates catalase contributes to the oxidant tolerance of methanogens. Methanobrevibacter arboriphilus, a methanogen that lacks cytochromes and is incapable of synthesizing haem, surprisingly possesses a haem-dependent catalase (Shima et al., 2001). Methanobrevibacter arboriphilus is thought to acquire haem from the external environment and convert apo-catalase to the active form. Supplementation of growth medium with haemin results in a substantial increase in Methanobrevibacter arboriphilus intracellular catalase activity, which increases the tolerance of this methanogen to H2O2 and O2 (Brioukhanov & Netrusov, 2012). Methanosarcina barkeri, a cytochrome-containing species that is capable of synthesizing haem, also exhibits intracellular catalase activity due to a haem-dependent catalase (Shima et al., 1999). Methanosarcina barkeri and Methanobrevibacter arboriphilus are both tolerant to O2 and millimolar levels of H2O2. The catalase gene in Methanosarcina barkeri is upregulated transcriptionally upon exposure of cells to sublethal concentrations of H2O2 (Brioukhanov & Netrusov, 2004). Recent evidence also revealed that methanogens are prevalent in aerated soils and once anaerobic conditions are restored, methanogenesis ensues (Angel et al., 2011). Among methanogens, members of the genera Methanosarcina and Methanocella dominate aerated soils, indicating these methanogens are extremely aerotolerant (Angel et al., 2012). Moreover, active transcription of the gene encoding a catalase (katE) was identified in samples of aerated soils, indicating that catalase is a potential component of the antioxidant system in aerotolerant Methanosarcina and Methanocella (Angel et al., 2011).
To ascertain the importance of catalase to the antioxidant system of methanogens, we used the cytochrome-containing species Methanosarcina acetivorans as a model, because it is aerotolerant, its metabolism has been extensively investigated and it has a robust genetic system (Ferry & Lessner, 2008; Guss et al., 2008; Horne & Lessner, 2013; Lessner et al., 2006). We have recently developed methods to assess the viability Methanosarcina acetivorans after challenge with oxidants. Importantly, the exogenous addition of catalase conferred a significant increase in the tolerance to H2O2, indicating catalase could be important in the oxidant tolerance of Methanosarcina acetivorans (Horne & Lessner, 2013). The goal of the present study was to examine the prevalence of catalase in methanogens, and to ascertain the importance of endogenous catalase in the H2O2 and O2 tolerance of Methanosarcina acetivorans. We also wanted to test the feasibility of engineering methanogen strains with increased oxidant tolerance. Therefore, we used a novel approach that employed the controlled expression of a bacterial catalase within Methanosarcina acetivorans. As the expression of the bacterial catalase could be tightly controlled, the approach used allowed for the specific assessment of the importance of catalase activity in the oxidant tolerance of Methanosarcina acetivorans.
Methods
Growth of Methanosarcina acetivorans.
Methanosarcina acetivorans strains were grown in high-salt (HS) medium supplemented with 125 mM methanol as a carbon and energy source, and 0.025 % Na2S as a reductant as described previously (Sowers et al., 1984). Growth was monitored spectrophotometrically as OD600 using a Genesys 10 Bio spectrophotometer (Thermo Scientific). The inducer tetracycline was added to a final concentration of 100 µg ml−1 where indicated.
Construction of an Escherichia coli KatG (EcKatG) expression strain of Methanosarcina acetivorans.
PCR was used to amplify katG from E. coli DH5α genomic DNA. The forward primer for the amplification contained the sequence for an NdeI restriction site (5′-GGTGGTCATATGAGCACGTCAGACGATATCCATAAC-3′), whilst the reverse primer contained a HindIII restriction site (5′-GGGGTAAGCTTTTACAGCAGGTCGAAACGGTCGAGG-3′). The PCR product was digested with NdeI and HindIII, and ligated with similarly digested pJK027A (Guss et al., 2008), generating pDL329. pDL329 contains E. coli katG fused to the PmcrB(tetO1) promoter in pJK027A. Methanosarcina acetivorans strain WWM73 was transformed with pDL329 and transformants selected as described previously (Guss et al., 2008). Successful integration of the plasmid into the chromosome of strain WWM73 was determined as described (Guss et al., 2008) and the resulting strain was named DJL20. Methanosarcina acetivorans strain DJL20 is capable of tetracycline-inducible expression of EcKatG.
Determination of catalase activity.
Cell lysates of Methanosarcina acetivorans strains were prepared by harvesting cells (50 ml) by centrifugation at 16 000 g at 4 °C. The cell pellets were frozen at −20 °C, thawed and resuspended in 0.5 ml of 50 mM Tris/HCl, pH 8.0. The cells were lysed by sonication and clarified lysates were obtained by centrifugation at 16 000 g at 4 °C. The catalase activity in cell lysates was determined spectrophotometrically (Beckman DU-7400) by monitoring the decrease in absorbance at 240 nm of 13 mM H2O2 in 50 mM Tris/HCl, pH 8.0. The amount of H2O2 consumed was determined using ϵ240 = 39.4 M−1 cm−1. One unit of activity was defined as 1 µmol H2O2 consumed min−1. The protein concentration of cell lysates was determined using the Bradford method (Bradford, 1976).
Oxidant challenge of Methanosarcina acetivorans.
The tolerance of Methanosarcina acetivorans cells to H2O2 was assessed using recently developed methods (Horne & Lessner, 2013). Specifically, cells of strain DJL20 were challenged with various concentrations of H2O2 for 1 h under anaerobic conditions and viability determined using the microtitre plate method (Horne & Lessner, 2013). To assess the ability of cells to grow when challenged with H2O2 or O2, Methanosarcina acetivorans strains were grown in 10 ml of HS medium, devoid of sulfide and resazurin to avoid the abiotic reduction of H2O2 and O2 by sulfide and the interference of oxidized resazurin with OD600 measurements. Mid-exponential-phase cultures were challenged by the direct addition of H2O2 or O2 to the culture tubes. Solutions of H2O2 were prepared freshly and 0.2 ml aliquots were added to mid-exponential-phase cells using a syringe and needle. Pure O2 was added to the desired percentage (v/v) of the headspace volume of each tube using a syringe and needle. To promote uniform O2 exposure, the tubes were incubated on their sides and then mixed by inverting them every hour.
Statistical analysis.
Viability determinations by the microtitre plate method were replicated independently a minimum of three times. Growth inhibition experiments and catalase activity assays were replicated three times. All data are presented as mean±sd. Plotting and calculation of the sd were performed in Microsoft Excel.
Results
Distribution of monofunctional catalase (KatE) and catalase peroxidase (KatG) in methanogens
Haem-containing monofunctional catalases (KatEs) have catalatic activity, but not peroxidatic activity, and are distributed widely within the three domains of life (Zamocky et al., 2008). Using E. coli KatE as a query, homologous proteins encoded in the genomes of several methanogens were identified (Table S1, available in Microbiology Online). Of the 57 sequenced methanogen genomes currently within the Integrated Microbial Genomes (IMG) system database (http://img.jgi.doe.gov), only 11 contain a putative KatE homologue, comprising ~19 % of the genomes. The presence of KatE is not restricted to certain methanogen orders or genera. However, all sequenced species within the order Methanococcales lack a putative KatE, whereas KatE is more prevalent in species of the orders Methanomicrobiales and Methanosarcinales. KatE from Methanosarcina barkeri and Methanobrevibacter arboriphilus have been characterized, with each demonstrated to have catalatic activity (Brioukhanov & Netrusov, 2004, 2012; Shima et al., 1999, 2001).
Haem-containing bifunctional catalases have catalatic activity and peroxidatic activity, and are also distributed within the three domains of life (Passardi et al., 2007). Of the 57 sequenced methanogen genomes within the IMG system database, only nine contain a putative KatG homologue, ~16 % of the genomes (Table S1). KatG appears restricted to species within the orders Methanobacteriales, Methanomicrobiales and Methanosarcinales. A KatG homologue from a methanogen has not been characterized experimentally. Only two methanogens possess a putative KatE and KatG, Methanolobus psychrophilus R15 and Methanosarcina acetivorans, both members of the Methanosarcinales.
Genome of Methanosarcina acetivorans encodes catalase, but cells lacked catalase activity
The genome of Methanosarcina acetivorans encodes a KatG (MaKatG; MA0972) homologue and a non-functional KatE (MA2081) homologue. Although the amino acid sequence of the putative KatE from Methanosarcina acetivorans is 88 % identical to characterized Methanosarcina barkeri KatE (Shima et al., 1999), sequencing of ma2081 confirms that the gene contains a frameshift causing a nonsense mutation that results in the synthesis of a truncated protein (173 of 496 predicted amino acids). Since half of the active-site residues identified in KatE (Díaz et al., 2012) are missing in the truncated protein (Fig. S1), Methanosarcina acetivorans likely does not possess a functional KatE, unlike Methanosarcina barkeri.
MaKatG is 77 % identical to Burkholderia pseudomallei KatG, for which the structure has been solved (Carpena et al., 2003), and 64 % identical to well-characterized EcKatG (Díaz et al., 2012). Moreover, the active-site residues are conserved in MaKatG (Fig. S2), indicating MaKatG may have catalatic and peroxidatic activities. MaKatG has not been detected in several proteomic analyses of Methanosarcina acetivorans (Lessner et al., 2006; Li et al., 2005a, b), suggesting that it may not be expressed or is expressed at a low level under non-stress conditions. The level of catalase activity in lysates derived from WT Methanosarcina acetivorans cells grown with methanol, harvested at mid-exponential phase or stationary phase, was below the detection limit. The lack of catalase activity suggests MaKatG is not expressed constitutively and is not induced upon entry into stationary phase. As catalase activity in Methanosarcina barkeri was shown to be induced by the addition of ROS (Brioukhanov et al., 2006), catalase activity was also measured in lysates from Methanosarcina acetivorans cells exposed to sublethal concentrations of H2O2 (1.5 mM) and O2 (5 %) for 1 or 4 h. The level of catalase activity in H2O2- or O2-challenged Methanosarcina acetivorans cells was also below the detection limit. Finally, to ascertain whether MaKatG encodes a functional catalase, MaKatG was expressed in E. coli and catalase activity measured in E. coli lysates. E. coli cells expressed soluble MaKatG, indicative of proper folding, but did not exhibit an increase in catalase activity (data not shown), suggesting MaKatG lacks catalase activity. Overall, these results suggest MaKatG is non-functional and therefore does not play a role in the antioxidant system in Methanosarcina acetivorans. However, we have demonstrated previously that exogenous addition of catalase does protect Methanosarcina acetivorans from H2O2 toxicity (Horne & Lessner, 2013), indicating catalase is a potential protective enzyme for Methanosarcina acetivorans. Therefore, we set out to construct a Methanosarcina acetivorans strain capable of inducible expression of recombinant catalase from a bacterium in order to determine whether endogenous catalase can protect Methanosarcina acetivorans from H2O2 and O2 toxicity, and whether it is possible to engineer a methanogen strain with increased oxidant tolerance.
Expression of recombinant EcKatG increased endogenous catalase activity in Methanosarcina acetivorans
To assess specifically the ability of KatG to protect Methanosarcina acetivorans from oxidants, the gene encoding EcKatG was fused to a tetracycline-inducible methanogen promoter (PmcrBTetO1) and the gene fusion (PmcrBTetO1–EckatG) was moved into the chromosome of Methanosarcina acetivorans strain WWM73 (Guss et al., 2008). The resulting strain, DJL20, exhibited similar growth rates and yields when grown with methanol in the presence or absence of tetracycline (data not shown), indicating induction of haem-dependent EcKatG is not inhibitory or detrimental to Methanosarcina acetivorans. Lysate derived from mid-exponential-phase cells of DJL20 grown in the presence of tetracycline (EcKatG-induced) exhibited a 100-fold increase in catalase activity [88±13 U (mg protein)−1], compared with activity in cells grown in the absence of tetracycline (uninduced), which was close to the detection limit [0.8±0.2 U (mg protein)−1]. The induced catalase activity [~90 U (mg protein)−1] in Methanosarcina acetivorans strain DJL20 was comparable to the intrinsic catalase activity [5–300 U (mg protein)−1] observed in other anaerobes (Brioukhanov & Netrusov, 2004). These results showed that EcKatG is expressed in an active form within Methanosarcina acetivorans, supporting the supposition that the machinery in Methanosarcina acetivorans recognizes EcKatG as a haemoprotein and facilitates the proper incorporation of haem into the active site of EcKatG. Importantly, to our knowledge this is the first example of the heterologous expression of a bacterial haem-dependent enzyme within a methanogen, indicating that methanogens (archaea) and bacteria likely contain similar mechanisms for haem incorporation.
Increase in endogenous catalase activity conferred increased resistance of Methanosarcina acetivorans to H2O2
The effect of increased catalase activity in strain DJL20 on the tolerance to H2O2 was assessed. Strain DJL20 was grown with methanol in the presence or absence of tetracycline to mid-exponential phase. Harvested cells were challenged with H2O2 for 1 h and viability was assessed using a recently developed microtitre assay (Horne & Lessner, 2013). EcKatG-induced cells exhibited no loss of viability when challenged with a concentration of H2O2 (6 mM) that is lethal to uninduced cells (Fig. 1a). These results demonstrate that expression of EcKatG within Methanosarcina acetivorans not only increased endogenous catalase activity, but that the cells were protected from the toxic effects of H2O2. Using the microtitre assay, the maximum dose of H2O2 resulting in a complete loss of viability of EcKatG-induced cells was 60 mM H2O2 (Fig. 1b), a 10-fold increase in the lethal H2O2 concentration over the uninduced cells. Thus, expression of EcKatG increased the tolerance of Methanosarcina acetivorans to high concentrations of H2O2.
Fig. 1.
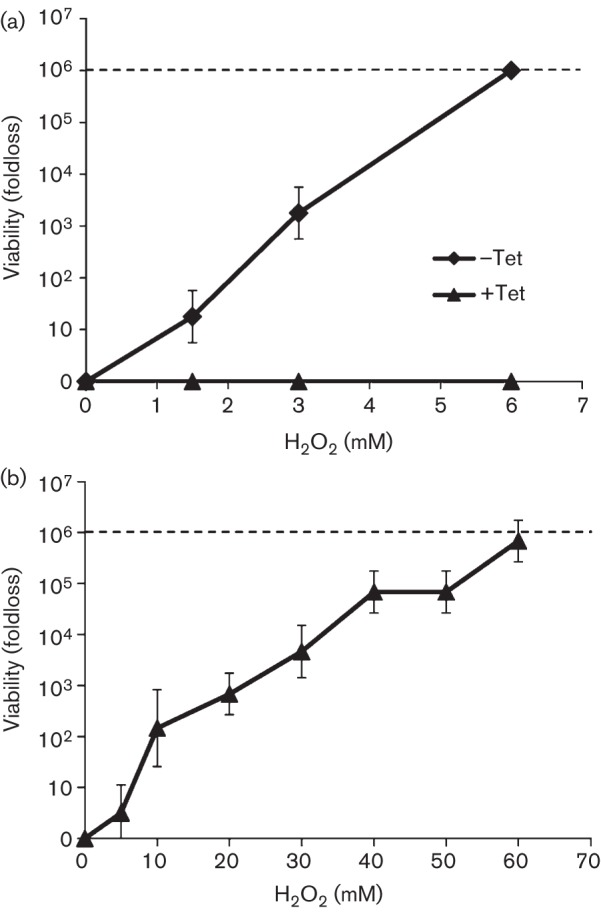
(a) Comparison of H2O2 tolerance of EcKatG-induced cells to uninduced cells of Methanosarcina acetivorans strain DJL20. Cells were grown in the absence (–Tet) or presence of tetracycline (+Tet), and challenged with 0, 1.5, 3.0 and 6.0 mM H2O2 for 1 h. (b) H2O2 tolerance of EcKatG-induced cells of strain DJL20 assessed by microtitre assay. In each graph the line depicts the highest fold loss that results in a complete absence of viable cells, and the data plotted are the mean±sd from three independent experiments.
The effect of H2O2 on actively growing cultures of Methanosarcina acetivorans was also examined. In order to test the intrinsic tolerance of strain DJL20 to H2O2, the cells were grown in medium devoid of sulfide, because we have shown that sulfide can protect Methanosarcina acetivorans from H2O2 (Horne & Lessner, 2013). Methanosarcina acetivorans exhibits a similar doubling time (~9 h) when grown with methanol in medium containing or lacking sulfide (data not shown). However, in medium devoid of sulfide, a longer doubling time was observed for EcKatG-induced cells (~18 h) compared with the uninduced cells (~9 h). The slower growth rate was specific to the absence of sulfide and induction of EcKatG, indicating that expression of a haem-dependent catalase in the absence of sulfide is inhibitory. Despite the slower growth rate, actively growing cultures of EcKatG-induced cells were more resistant to the addition of 1.5 or 3 mM H2O2 at mid-exponential phase, compared with the uninduced cells (Fig. 2). The uninduced cells immediately stopped growing upon the addition of 1.5 mM H2O2, a concentration which only resulted in a 10-fold loss of viability according to the microtitre assay (Fig. 1). The addition of 3 mM not only stopped growth of the uninduced cells, but resulted in cell lysis as evidenced by the decrease in OD600 (Fig. 2a). This result is consistent with the significant loss of viability of the uninduced cells when exposed to 3 mM H2O2 for 1 h as assessed by microtitre assay (Fig. 1). However, EcKatG-induced cells were able to overcome the addition of 1.5 mM H2O2 and continue to grow, albeit more slowly and with lower yield than the H2O2-free control (Fig. 2). The addition of 3 mM H2O2 did result in a cessation of growth of the EcKatG-induced cells; however, there was no apparent cell lysis, unlike the uninduced cells. Moreover, there were still a substantial number of viable cells remaining 40 h post-H2O2 addition, because growth was observed when fresh medium was inoculated with an aliquot of the 3 mM H2O2-exposed EcKatG-induced cultures (data not shown). These results demonstrated that expression of EcKatG can protect Methanosarcina acetivorans from acute H2O2 toxicity. To our knowledge this is the first example of engineered H2O2 resistance within a strict anaerobe, including methanogens.
Fig. 2.
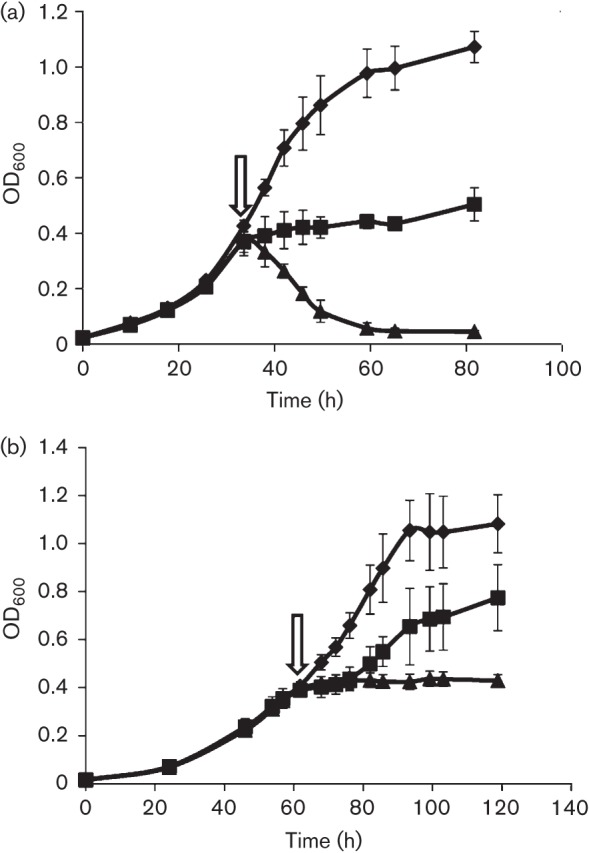
Effect of H2O2 on growth of EcKatG-induced cells compared to uninduced cells of Methanosarcina acetivorans strain DJL20. Mid-exponential-phase cultures of DJL20 grown in the (a) absence or (b) presence of tetracycline were challenged with 0 (diamonds), 1.5 (squares) or 3 mM (triangles) H2O2 at the time point indicated by the arrow. The data plotted are the mean±sd from three independent experiments.
Increase in endogenous catalase activity did not confer increased resistance of Methanosarcina acetivorans to O2
In the majority of natural environments, methanogens are exposed to O2, rather than high levels of H2O2. However, methanogens contain many enzymes, including low-potential flavoenzymes similar to those found in other strict anaerobes known to reduce O2 to H2O2 and/or O2− (Imlay, 2003). Therefore, catalase may serve a role in degrading endogenously produced H2O2 when cells are exposed to O2. Although the addition of atmospheric levels of O2 (20 %) results in a cessation of growth of Methanosarcina acetivorans, exposure to this O2 concentration for 1 h does not decrease viability (Horne & Lessner, 2013). Therefore, we investigated the effect of EcKatG expression on the ability of Methanosarcina acetivorans to grow in the presence of lower concentrations of O2, similar to studies with Methanobrevibacter arboriphilus (Brioukhanov & Netrusov, 2012). EcKatG-induced and uninduced cells of strain DJL20 were grown with methanol in medium devoid of sulfide. At mid-exponential phase, cells were challenged by the addition of 1 or 5 % O2 (v/v) to the headspace of the cultures (Fig. 3). Under these culture conditions the cells would be exposed to a maximum of ~190 µM O2, which is the maximum dissolved O2 concentration at 35 °C (Colt, 1984). Both the uninduced and EcKatG-induced cells were inhibited initially by the addition of 1 % O2, but were eventually able to resume growth. The addition of 5 % O2 to the headspace of uninduced and EcKatG-induced cells resulted in a cessation of growth. This result indicated that a significant increase in endogenous catalase activity did not confer an increase in the resistance of growing Methanosarcina acetivorans cells to O2. In contrast, increased endogenous catalase activity in Methanobrevibacter arboriphilus was observed to increase the ability of this methanogen to overcome the addition of O2 to growing cultures (Brioukhanov & Netrusov, 2012). However, a slight increase in the resistance of Methanosarcina acetivorans strain DJL20 to O2 by increased catalase activity could be masked by the slower growth rate of the EcKatG-induced cells in medium devoid of sulfide. Therefore, we attempted to determine the cause of the slower growth rate of the EcKatG-induced cells when grown in the absence of sulfide.
Fig. 3.
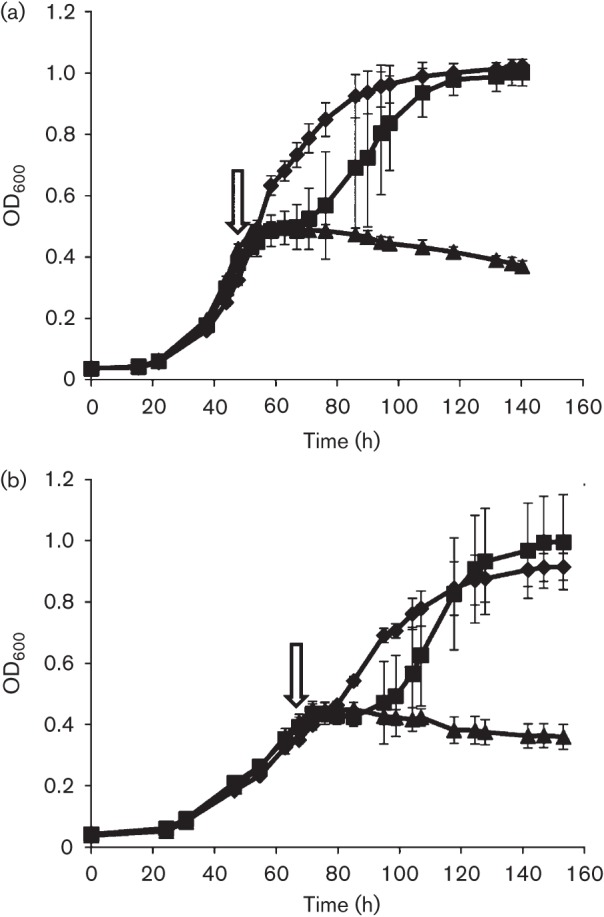
Effect of O2 on growth of EcKatG-induced cells compared to uninduced cells of Methanosarcina acetivorans strain DJL20. Mid-exponential-phase cultures of DJL20 grown in the (a) absence or (b) presence of tetracycline were not challenged with O2 (diamonds), or were challenged with 1 (squares) or 5 % (triangles) O2 at the time point indicated by the arrow. The data plotted are the mean±sd from three independent experiments.
Effect of the addition of exogenous haemin on growth and aerotolerance of Methanosarcina acetivorans
Methanosarcina acetivorans is capable of synthesizing haem necessary for incorporation into endogenous cytochromes, as well as recombinant EcKatG. However, in the absence of sulfide, expression of EcKatG could cause a depletion of haem, resulting in limitation of haem to metabolic enzymes, causing a slower growth rate. To determine whether haem was a limiting factor during growth of EcKatG-induced cells in the absence of sulfide, growth was monitored in medium supplemented with haemin. The addition of 30 µM haemin fully restored the growth rate of the EcKatG-induced cells with methanol in the absence of sulfide to that observed for EcKatG-induced cells grown in the presence of sulfide (Fig. 4). Although the molecular connection between sulfide and haem levels is not apparent, the ability of exogenous haemin to restore the normal growth rate indicated that EcKatG-induced Methanosarcina acetivorans cells had decreased levels or synthesis rates of haem when grown in the absence of sulfide.
Fig. 4.
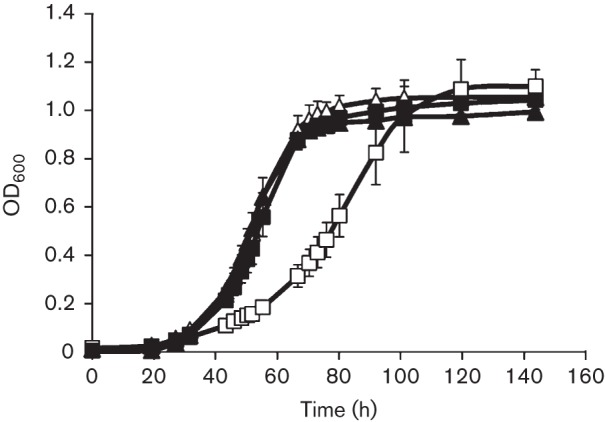
Growth of EcKatG-induced cells compared with uninduced cells of Methanosarcina acetivorans strain DJL20 in medium supplemented with haemin. Cultures of DJL20 were grown in the absence (triangles) or presence (squares) of tetracycline in medium lacking (open symbols) or containing (filled symbols) 30 µM haemin. The data plotted are the mean±sd from three independent experiments.
The exogenous addition of haemin was shown to affect positively the resistance of Methanobrevibacter arboriphilus to oxidants. Methanobrevibacter arboriphilus lacks cytochromes and is incapable of synthesizing haem. However, the addition of exogenous haemin results in an increase in endogenous catalase activity, due to conversion of the haem-dependent catalase (KatE) to the holo-form (Brioukhanov & Netrusov, 2012). The increased endogenous catalase activity was postulated to account for a significant increase in the resistance of Methanobrevibacter arboriphilus to both H2O2 and O2. Therefore, we assessed the effect of exogenous haemin on the endogenous catalase activity in EcKatG-induced and uninduced cells of Methanosarcina acetivorans strain DJL20. An increase in the endogenous catalase activity was not observed in either EcKatG-induced or uninduced cells when grown in the presence of sulfide and/or haemin, compared with cells grown in the absence of sulfide and/or haemin (data not shown). This result indicated that Methanosarcina acetivorans did not contain an endogenous haem-inducible catalase, unlike Methanobrevibacter arboriphilus, and that EcKatG is not limited for haem when expressed in Methanosarcina acetivorans cells grown in medium without sulfide or haemin. However, the exogenous addition of 30 µM haemin increased the resistance of both uninduced and EcKatG-induced cells of Methanosarcina acetivorans strain DJL20 to O2, albeit only slightly, as no inhibition was observed by the addition of 1 % O2 (Fig. 5) compared with cells grown in medium without haemin (Fig. 3). Haemin may cause a change in the expression of other antioxidant enzymes in Methanosarcina acetivorans, which could account for the increased resistance of strain DJL20 to O2. Haemin may also provide additional protection from O2 and/or H2O2 toxicity. For example, in a buffered solution, 30 µM haemin was able to decompose 3 mM H2O2 within 10 min (Fig. S3). Taken together, these data indicated that exogenous haemin can provide additional protection from oxidants and that the 100-fold increase in the endogenous catalase activity in Methanosarcina acetivorans did not increase the tolerance of growing cultures of Methanosarcina acetivorans to O2, at least under the experimental conditions tested here. This result is in contrast to the observed protection by increased haemin-dependent catalase activity in Methanobrevibacter arboriphilus, which was demonstrated to increase significantly the tolerance of this methanogen to O2, as well as H2O2 (Brioukhanov & Netrusov, 2012).
Fig. 5.
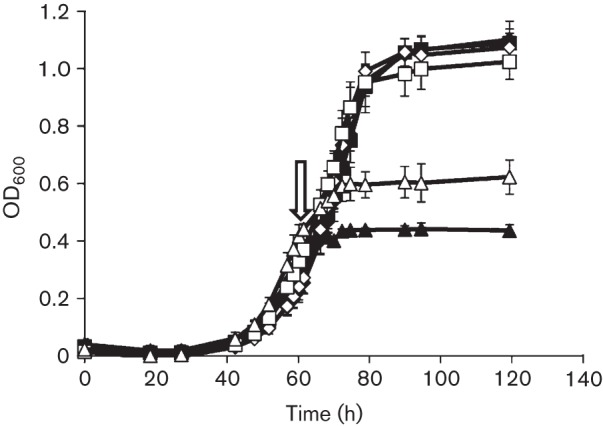
Effect of O2 on growth of EcKatG-induced cells compared to uninduced cells of Methanosarcina acetivorans strain DJL20 in medium supplemented with 30 µM haemin. Mid-exponential-phase cultures of DJL20 grown in the absence (open symbols) or presence (filled symbols) of tetracycline were not challenged with O2 (diamonds), or were challenged with 1 % (squares) or 5 % (triangles) O2 at the time point indicated by the arrow. The data plotted are the mean±sd from three independent experiments.
Discussion
The results from this study provide insight into the role of catalase in the antioxidant system of methanogens and the feasibility of engineering beneficial traits into methanogens. The limited number of methanogens that encode catalase suggests that it is not a core component of the antioxidant system in methanogens. Methanosarcina acetivorans encodes homologues of the haem-dependent catalases KatE and KatG, yet one contains a frameshift mutation (KatE) and the other does not appear to encode a functional catalase. In contrast, functional KatE in Methanosarcina barkeri is upregulated upon exposure of the cells to H2O2 and the O2−-generating chemical paraquat (Brioukhanov et al., 2006). Phylogenetic evidence suggests that catalase genes in methanogens were acquired by lateral gene transfer (Zámocký et al., 2012). The disparity in catalase activity between the two Methanosarcina species may be a result of environmental differences exerting varying selective pressure. For example, the genome of the marine species Methanosarcina acetivorans encodes hydrogenase, yet Methanosarcina acetivorans lacks detectable hydrogenase activity and does not consume or produce hydrogen during growth. In contrast, the freshwater species Methanosarcina barkeri possesses hydrogenase activity, and has the ability to consume and produce hydrogen. It is postulated that Methanosarcina acetivorans has lost the ability to use hydrogen because in the marine environment it would have to compete with sulfate reducers for hydrogen (Ferry & Lessner, 2008; Guss et al., 2009). Similarly, differences between freshwater and marine environments, such as the solubility of oxygen and microbial community composition, could alter the levels of O2 and ROS exposure to methanogens in marine and freshwater environments, and hence the selective pressure for utilizing specific antioxidant enzymes. It appears Methanosarcina acetivorans does not use the acquired katE or katG genes. It is unclear how widespread this phenomenon is in methanogens, because catalase activity has not been examined in the remaining methanogens that encode catalase (Table S1). Nonetheless, it should be noted that the presence of a gene encoding catalase in the genome of an individual species is not sufficient evidence to conclude that catalase plays a role in the antioxidant system of the organism.
To determine whether catalase provides an advantage to Methanosarcina acetivorans during exposure to O2 and H2O2, we employed a novel approach using the established Methanosarcina acetivorans genetic system. Heterologous expression of E. coli KatG in Methanosarcina acetivorans resulted in endogenous catalase activity within the range of activities observed for Methanosarcina barkeri and Methanobrevibacter arboriphilus. This increase in catalase activity improved the tolerance of Methanosarcina acetivorans to H2O2, but did not provide an advantage when cells were exposed to O2. Exposure of anaerobes to O2 results in the endogenous production of H2O2 and O2−; however, the rates of synthesis and levels of each ROS have not been determined in Methanosarcina acetivorans. H2O2 may not be the primary ROS produced during O2 exposure, which could explain the lack of additional tolerance by increased catalase activity. Alternatively, endogenously produced H2O2 may be scavenged sufficiently by a number of other antioxidant enzymes encoded in the genome of Methanosarcina acetivorans, including rubrerythrin (MA0639), peroxiredoxin (MA4103) and several iron–sulfur flavoproteins, which have been shown to scavenge H2O2 in other species (Cruz & Ferry, 2006; Lumppio et al., 2001). Methanosarcina acetivorans is more likely to come in contact with O2 rather than high concentrations of H2O2, and may have evolved H2O2-scavenging capabilities that do not involve catalase, even though catalase genes were acquired.
EcKatG is a haem-dependent enzyme and was active when expressed in Methanosarcina acetivorans cells grown in medium lacking haemin, revealing that haem synthesized in Methanosarcina acetivorans is inserted properly into EcKatG. However, when EcKatG was expressed in Methanosarcina acetivorans cells grown in medium lacking sulfide, a reduced growth rate was observed. Supplementation with haemin restored normal growth and did not induce additional catalase activity. The lack of catalase induction by haemin is similar to results seen with Methanosarcina barkeri (Brioukhanov & Netrusov, 2004). However, haemin induces catalase activity in Methanobrevibacter arboriphilus, a methanogen that is incapable of synthesizing haem. Addition of haem converts the apo-catalase to the active form (haem-containing) in Methanobrevibacter arboriphilus (Brioukhanov & Netrusov, 2012). Also, unlike Methanosarcina acetivorans, an increase in catalase activity in Methanobrevibacter arboriphilus correlated with increased tolerance of growing cultures to O2. A correlation with catalase activity and O2 tolerance has not been documented with Methanosarcina barkeri. Among methanogens, the use of catalase as a primary H2O2-scavenging enzyme may be unique to Methanobrevibacter arboriphilus. Interestingly, none of the sequenced Methanobrevibacter genomes within the IMG system database encode KatE or KatG, indicating that the use of KatE may be specific to Methanobrevibacter arboriphilus strains. However, since the levels of catalase activity in Methanobrevibacter arboriphilus are dependent on the concentration of haemin in the growth medium under the experimental conditions examined (Brioukhanov & Netrusov, 2012), it is difficult to distinguish protection solely due to catalase from that by haemin and possibly haemin-induced factors.
In summary, the results suggest catalase is not a key antioxidant enzyme in methanogens. Despite the fact that the majority of methanogens are tolerant to some O2, the key enzymes and factors that contribute to the observed aerotolerance are not clear. More detailed characterization is required to identify these enzymes and factors. Importantly, the recombinant approach described here could be used to identify and assess the importance of other enzymes (e.g. SOD, superoxide reductase, peroxidase) to the oxidant tolerance of methanogens. Finally, the results described here, along with previous studies (Lessner et al., 2010), highlight the ability to design methanogen strains with beneficial traits, which may aid in the development of methanogens as biological catalysts.
Acknowledgements
This work was supported in part by funding to D. J. L. from the National Institute of General Medical Sciences of the National Institutes of Health (grant no. P30 GM103450), National Science Foundation (grant no. MCB1121292), NASA Exobiology (grant no. NNX12AR60G) and the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000.
Abbreviations:
- ROS
reactive oxygen species
- SOD
superoxide dismutase
Footnotes
One supplementary table and three supplementary figures are available with the online version of this paper.
References
- Angel R., Matthies D., Conrad R. (2011). Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS ONE 6, e20453. 10.1371/journal.pone.0020453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel R., Claus P., Conrad R. (2012). Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J 6, 847–862. 10.1038/ismej.2011.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brioukhanov A. L., Netrusov A. I. (2004). Catalase and superoxide dismutase: distribution, properties, and physiological role in cells of strict anaerobes. Biochemistry (Mosc) 69, 949–962. 10.1023/B:BIRY.0000043537.04115.d9 [DOI] [PubMed] [Google Scholar]
- Brioukhanov A. L., Netrusov A. I. (2012). The positive effect of exogenous hemin on a resistance of strict anaerobic archaeon Methanobrevibacter arboriphilus to oxidative stresses. Curr Microbiol 65, 375–383. 10.1007/s00284-012-0168-6 [DOI] [PubMed] [Google Scholar]
- Brioukhanov A. L., Netrusov A. I., Eggen R. I. (2006). The catalase and superoxide dismutase genes are transcriptionally up-regulated upon oxidative stress in the strictly anaerobic archaeon Methanosarcina barkeri. Microbiology 152, 1671–1677. 10.1099/mic.0.28542-0 [DOI] [PubMed] [Google Scholar]
- Carpena X., Loprasert S., Mongkolsuk S., Switala J., Loewen P. C., Fita I. (2003). Catalase-peroxidase KatG of Burkholderia pseudomallei at 1.7 Å resolution. J Mol Biol 327, 475–489. 10.1016/S0022-2836(03)00122-0 [DOI] [PubMed] [Google Scholar]
- Colt J. (1984). Computation of Dissolved Gas Concentrations in Water as Functions of Temperature, Salinity, and Pressure. Bethesda, MD: American Fisheries Society. [Google Scholar]
- Cruz F., Ferry J. G. (2006). Interaction of iron-sulfur flavoprotein with oxygen and hydrogen peroxide. Biochim Biophys Acta 1760, 858–864. 10.1016/j.bbagen.2006.02.016 [DOI] [PubMed] [Google Scholar]
- Díaz A., Loewen P. C., Fita I., Carpena X. (2012). Thirty years of heme catalases structural biology. Arch Biochem Biophys 525, 102–110. 10.1016/j.abb.2011.12.011 [DOI] [PubMed] [Google Scholar]
- Ferry J. G., Lessner D. J. (2008). Methanogenesis in marine sediments. Ann N Y Acad Sci 1125, 147–157. 10.1196/annals.1419.007 [DOI] [PubMed] [Google Scholar]
- Fetzer S., Bak F., Conrad R. (1993). Sensitivity of methanogenic bacteria from paddy soil to oxygen and desiccation. FEMS Microbiol Ecol 12, 107–115. 10.1111/j.1574-6941.1993.tb00022.x [DOI] [Google Scholar]
- Guss A. M., Rother M., Zhang J. K., Kulkkarni G., Metcalf W. W. (2008). New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2, 193–203. 10.1155/2008/534081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss A. M., Kulkarni G., Metcalf W. W. (2009). Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri. J Bacteriol 191, 2826–2833. 10.1128/JB.00563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne A. J., Lessner D. J. (2013). Assessment of the oxidant tolerance of Methanosarcina acetivorans. FEMS Microbiol Lett 343, 13–19. 10.1111/1574-6968.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. (2002). How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv Microb Physiol 46, 111–153. 10.1016/S0065-2911(02)46003-1 [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2003). Pathways of oxidative damage. Annu Rev Microbiol 57, 395–418. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- Jenney F. E., Jr, Verhagen M. F., Cui X., Adams M. W. (1999). Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286, 306–309. 10.1126/science.286.5438.306 [DOI] [PubMed] [Google Scholar]
- Lessner D. J., Li L., Li Q., Rejtar T., Andreev V. P., Reichlen M., Hill K., Moran J. J., Karger B. L., Ferry J. G. (2006). An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc Natl Acad Sci U S A 103, 17921–17926. 10.1073/pnas.0608833103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessner D. J., Lhu L., Wahal C. S., Ferry J. G. (2010). An engineered methanogenic pathway derived from the domains Bacteria and Archaea . mBiol 1, e000243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Li L., Rejtar T., Karger B. L., Ferry J. G. (2005a). Methanosarcina acetivorans Part II: comparison of protein levels in acetate- and methanol-grown cells. J Proteome Res 4, 129–135. 10.1021/pr049831k [DOI] [PubMed] [Google Scholar]
- Li Q., Li L., Rejtar T., Karger B. L., Ferry J. G. (2005b). Methanosarcina acetivorans Part I: an expanded view of the biology of the cell. J Proteome Res 4, 112–128. 10.1021/pr049832c [DOI] [PubMed] [Google Scholar]
- Lumppio H. L., Shenvi N. V., Summers A. O., Voordouw G., Kurtz D. M., Jr (2001). Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J Bacteriol 183, 101–108. 10.1128/JB.183.1.101-108.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F., Zamocky M., Favet J., Jakopitsch C., Penel C., Obinger C., Dunand C. (2007). Phylogenetic distribution of catalase-peroxidases: are there patches of order in chaos? Gene 397, 101–113. 10.1016/j.gene.2007.04.016 [DOI] [PubMed] [Google Scholar]
- Shima S., Netrusov A., Sordel M., Wicke M., Hartmann G. C., Thauer R. K. (1999). Purification, characterization, and primary structure of a monofunctional catalase from Methanosarcina barkeri. Arch Microbiol 171, 317–323. 10.1007/s002030050716 [DOI] [PubMed] [Google Scholar]
- Shima S., Sordel-Klippert M., Brioukhanov A., Netrusov A., Linder D., Thauer R. K. (2001). Characterization of a heme-dependent catalase from Methanobrevibacter arboriphilus. Appl Environ Microbiol 67, 3041–3045. 10.1128/AEM.67.7.3041-3045.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Baron S. F., Ferry J. G. (1984). Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl Environ Microbiol 47, 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6, 579–591. 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- Tholen A., Pester M., Brune A. (2007). Simultaneous methanogenesis and oxygen reduction by Methanobrevibacter cuticularis at low oxygen fluxes. FEMS Microbiol Ecol 62, 303–312. 10.1111/j.1574-6941.2007.00390.x [DOI] [PubMed] [Google Scholar]
- Zamocky M., Furtmüller P. G., Obinger C. (2008). Evolution of catalases from bacteria to humans. Antioxid Redox Signal 10, 1527–1548. 10.1089/ars.2008.2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zámocký M., Gasselhuber B., Furtmüller P. G., Obinger C. (2012). Molecular evolution of hydrogen peroxide degrading enzymes. Arch Biochem Biophys 525, 131–144. 10.1016/j.abb.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]


