Abstract
The International Cancer Genome Consortium (ICGC) aims to catalog genomic abnormalities in tumors from 50 different cancer types. Genome sequencing reveals hundreds to thousands of somatic mutations in each tumor, but only a minority drive tumor progression. We present the result of discussions within the ICGC on how to address the challenge of identifying mutations that contribute to oncogenesis, tumor maintenance or response to therapy, and recommend computational techniques to annotate somatic variants and predict their impact on cancer phenotype.
Introduction
Large-scale sequencing of cancer genomes often reveals many thousands of somatic missense (amino-acid changing) mutations in proteins. However, not all cancer mutations provide a selective (“driving”) advantage to cancer cells1,2. Many mutations are so-called “passengers” because their impact on protein function is either insignificant or the affected protein is not important for tumor progression. The important practical problem is to determine which mutations are likely drivers. Although the carcinogenicity of a particular mutation depends on concurrent genomic alterations in the cell, one can significantly reduce the number of potential driver candidates by determining the functional impact of each mutation. Thus, a key challenge is to distinguish between functional and non-functional mutations, and by extension between those that contribute to tumorigenesis (drivers) and those that do not (passengers) (see Box 1 for definitions).
Box 1. Definitions.
We define a functional variant as a genomic variant that affects the molecular function of a protein (as a gain, loss or switch of function). A non-functional variant does not significantly affect the molecular function of a protein. A driver variant confers a selective advantage to a particular tumor cell, while a passenger variant does not. It is important to distinguish between functional versus non-functional and driver versus passenger as they describe different concepts. For example, a mutation might dramatically affect the function of a protein without providing any selective advantage to the tumor (it is a functional passenger variant). Non-synonymous mutations are those that alter the amino acid sequence of a protein.
Cancer has been likened to an evolutionary process by which tumor cells gain a fitness advantage over their neighboring cells2. The process creates cells with altered abilities such as the circumvention of apoptosis and senescence, deregulated cell division, and failed responses to external cues such as contact-contact inhibition and ligand-mediated cell signaling3,4. Normal cells are reprogrammed by changes in the genome that are subsequently selected and clonally expanded. In a similar manner to the way germline mutations can leave behind patterns indicative of negative or positive selection over millions of years, somatic mutations that engender increases in tumor fitness also can leave telltale signs in the protein sequence. The analysis of a given protein can thus reveal a pattern of alterations that recurrently result in its loss of function, as in classic tumor suppressors, like TP53, RB1 or PTEN5.
Mutation events collected across several patient samples can also reveal signs of clustering in the peptide sequence or the three-dimensional protein structure that indicates a critical domain has been modulated. In the extreme case, the presence of the same amino acid change in the same position in different individuals can be a strong indicator of such gain of function or oncogenomic events, as is the case with the KRAS6 or BRAF7 oncogenes. Such patterns can be leveraged by informatics tools to predict if a particular mutational event induces a selectable phenotype.
We review the computational analyses that are commonly carried out after the detection of somatic mutations across a cohort of cancer samples to identify likely functional and likely driver mutations (Fig. 1). Our focus will be on single nucleotide variants (SNVs) and small indels (operationally defined here as variants shorter than 50 bp) that change the amino acid sequence or affect regulatory regions. The output of these analyses consists of prioritized lists of mutations, genes and pathways that may undergo follow-up experiments to demonstrate their actual role in cancer.
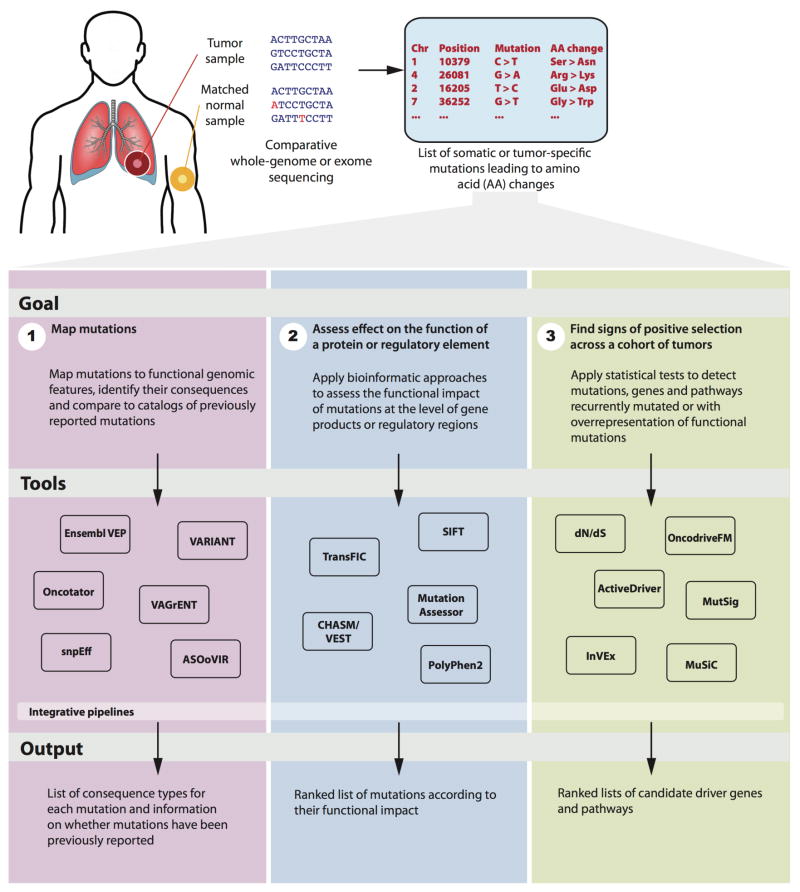
Figure 1.
Scheme depicting the three main approaches routinely employed in the analysis of cancer somatic mutations, as reviewed in this perspective. Although there are important relationships of precedence between elements from different approaches, they do not necessarily correspond to sequential steps. Tools employed in each of the approaches are shown in the middle. Integrative pipelines refer to tools that facilitate the use of methods across all approaches (e.g., IntOGen-mutations pipeline).
We divide the process of identifying functional and driver variants into three independent, but related, approaches (Fig. 1). The first consists of mapping mutations to annotated functional genomic features, identifying their consequences and determining if they have been previously reported. The second uses computational methods to predict the nature and magnitude of the functional impact of mutation in particular elements (e.g., proteins or regulatory regions). The third employs statistical methods to find signs of positive selection across the cohort. Figure 1 lists a subset of the computational tools employed in each of the approaches. In the sections that follow, we review the rationale and tools of each approach and conclude by presenting some of the unsolved challenges and future perspectives in the field.
Approach 1: Mutation mapping, annotation and comparison to known variants
The first step in determining the possible functional consequences of somatic mutations is to identify annotated genomic features that may be affected by them. Features that are more likely to encode genomic functions include protein-coding and non-coding transcripts, transcription factor binding sites and other potential regulatory regions. Less well-characterized features, such as highly conserved regions or regions of open chromatin, may also be of interest. There are a variety of software tools that infer the consequences of mutations, but frequently these use different terms and different definitions for the effect itself8–10 (Supplementary Table 1).
A large project such as the ICGC requires a common set of terms describing mutation consequences to facilitate the comparison of results among different groups. We have developed a standard set of ‘consequence terms’ drawn from the Sequence Ontology11 (see Supplementary Table 2). This list will be extended and updated as the project unfolds. Along with the Sequence Ontology term used to describe the effect of a mutation, we also identify a minimal set of ancillary information that annotation tools should provide for each relevant consequence term, such as coding DNA sequence (CDS), protein relative coordinates, and predicted amino acid substitutions. Several of these annotations will depend on the specific transcript the mutation falls within, and so we recommend that a transcript identifier always be included. Note that this caveat means that a single mutation can, and frequently will, be assigned multiple consequences on multiple transcripts.
We recommend using tools that can output mutation descriptions in the format defined by Human Genome Variation Society (HGVS) at all relevant levels (e.g. DNA-level for all mutations, and RNA and protein level descriptions where applicable). HGVS nomenclature provides a succinct and feature-centric format for variant descriptions, and some of the tools in Supplementary Table 1 (e.g. the Ensembl VEP) have options to produce output in this format. We propose a common ranking scheme for the term set that summarizes the effects of a mutation that falls in multiple genomic features, such as multiple transcripts (see Supplementary Table 2). In addition, the ranking may be used for prioritizing mutations for follow-up analysis.
When assigning consequence terms to variants, the source of all underlying annotations, such as gene models and regulatory elements, must be noted to clearly document the event. In the context of ICGC, we recommend using the GENCODE12 comprehensive set of gene models for all gene-associated annotations and identifying the specific release that was used. We advocate the use of GENCODE because of the detailed and frequently updated annotation of splice variants, pseudogenes and non-coding RNA loci, and the ready accessibility of all data for automated annotation via Ensembl and UCSC. Using the same gene models as the ENCODE project13 will also allow further integration of somatic mutation data and the wider set of ENCODE annotations.
Comparing the list of mutations to catalogues of known variants
An obvious step in determining the implication of detected variants is to identify those that have been observed previously in other cancers, that are involved in other diseases, or that exist as germline polymorphisms. The growing collection of somatic variants detected within the different ICGC projects is a useful source of information, as are databases such as dbSNP14, 1000 Genomes15, Catalogue of Somatic Mutations in Cancer (COSMIC)16 and databases of variants associated with hereditary diseases17,18. Several of the tools listed in Table 1 automatically report if the variant is already known. Since none of these sources are definitive, the ICGC recommends that, at a minimum, projects report matches to variants known in dbSNP, OMIM, 1000 Genomes and COSMIC along with the version number of the database. Although dbSNP has sometimes been used to filter for somatic mutations, historically it contained primarily germline variants. However, in newer releases, many somatic mutations including mutational hotspots are also present, for example in JAK2, KRAS and BRAF. Thus, although we recommend reporting matches in dbSNP we do not recommend using it to filter out somatic mutations.
Approach 2: Assessing the functional impact of mutations
For many variants, no further assessment can be made about their potential impact on cell operation. Nevertheless, for the specific subset of mutations that affect either protein coding sequences or known regulatory sites, one can make computational predictions about their potential effects. In this section we describe computational analyses that may shed light on the possible functions of these variants.
Mutations affecting protein coding sequence
A number of computational methods have been developed to differentiate “functional” or “disease-associated” non-synonymous mutations from “non-functional” or polymorphic variants19–24 (Supplementary Table 3). Some of these are specifically designed for cancer variants25–28. As a general rule, these approaches use evolutionary information (multiple sequence alignments), secondary and tertiary structure features, physico-chemical properties of amino acids, as well as information about the role of amino acid side chains in the 3D structure of proteins, such as protein surface placement in interaction sites.
Methods aimed at assessing the functional effect of non-synonymous mutations can be classified as “machine learning” and “direct”. Machine learning methods use relevant properties of the original and mutant residues (e.g., size, polarity), structural information (e.g., surface accessibility, hydrogen bonding), and/or evolutionary conservation and other features. These methods are then trained to distinguish between positive sets of disease-associated variants and negative control sets of presumably non-functional or passenger variants. In contrast, direct methods assess the effect of a mutation through a computed phenomenological score based on a particular theoretical model that does not require training sets.
Most of these computational approaches have been benchmarked on variants with pronounced phenotypic effects29 (e.g., functionally deleterious and Mendelian disease-associated variants) and appropriate negative control sets, reporting accuracies close to ~80%. Although not originally designed for this purpose, some of them have been widely employed to rank cancer somatic mutations for their likelihood to be drivers, without previously benchmarking their performance on this problem.
One of the main challenges to produce such benchmarking is the difficulty of collecting well-curated sets of driver and passenger mutations. A recent effort to circumvent this problem employed various datasets of likely driver and likely passenger mutations25. Under the assumption that each proxy dataset is incomplete in non-overlapping ways, this study compared the performance of three well-known methods and their impact scores transformed to account for the baseline tolerance across several datasets rather than on individual datasets25. In the future, when many more cancer genomes have been sequenced and we understand better the implication of genetic variants on cancer phenotype, it may be possible to collect gold standard datasets to perform more accurate validation.
Given the high-throughput nature of cancer genome projects, one important aspect to consider for tool selection is their computational efficiency when thousands of variants are analyzed. Precomputation of functional impact scores for all possible mutations in the human proteome is a useful remedy (as done by some tools presented in Supplementary Table 3). There is also at least one database (dbNSFP30) devoted to collecting and integrating such precomputed functional impact scores from different tools. In some cases it may be useful to visualize the location of mutations in protein 3D structure, if available, to further assess their potential role with respect to protein stability and/or function, for instance using MuPIT Interactive31 or the MutationAssessor web server22.
The output of any computational method should be interpreted as a ranked list of candidate driver variants based on the user-submitted mutations, with the vast majority not likely to be true positives. The purpose of this ranking is to prioritize mutations for further experimental testing. Using a combination of methods based on different theoretical principles (and hence independent error models) may help mitigate false positive and negative rates suffered by any one method alone, thus resulting in a cleaner list of candidates for experimental validation.
Mutations affecting regulatory sites
Only very recently has it become feasible to identify and characterize somatic noncoding mutations that affect putative regulatory sites. Predicting the functional effects of regulatory variants typically starts either by purely statistical approaches, such as the application of machine learning methods to learn motif models from the regulatory sequences, or by modeling the transcription factor (TF) to DNA binding biophysics aided by experimental data such as those obtained from micro-fluidics or protein binding experiments32,33. Both approaches result in predictions of binding sites for different TFs within regulatory sequences. There are several tools for making such predictions, such as The Meme Suite34, and the ENCODE project catalogues a number of relevant experimental data sets13. Furthermore, RegulomeDB provides an integrated approach to analyze regulatory variants35. It uses datasets from ENCODE13 and other sources and also uses motif models (eg. from JASPAR36).
When a somatic mutation falls within a TF binding site, it is possible to score its effect in multiple ways. Perhaps the simplest is to take the relevant binding site motif model36 and evaluate the score difference that the variant causes in that binding site’s match to the model. This is close in spirit to scores that are derived from multiple alignments, such as PFAM log E value37. However, the interpretation of this particular score is not straightforward because the actual binding probability of TF to DNA depends strongly on the factor concentration within the cell and the presence of other protein binding factors and may thus vary across cell types. Furthermore, it is not clear in general whether stronger or weaker predicted binding is better or worse for TF function, and clarifying this will require studying the particular promoter and gene in more detail.
Pleasance et al. (ref. 38) used a specific tool39 to address the functionality of mutations within promoters in a lung cancer cell line. Although somatic mutations did not differ significantly from the null expectation as a set, individual variants were predicted to have significant disruptive effects on potential binding motifs. More recently, systematic analyses integrating TF binding, histone marks, and other epigenomic data were used to identify pathways disrupted by Genome Wide Association Study (GWAS) at the regulatory level40.
In addition to promoters and enhancers, it is also important to consider possible effects of mutations in splicing, especially now that the connection between splicing and cancer is becoming increasingly clear (e.g., ref 41). Consequences of mutations in splicing regulatory elements are still difficult to predict but including additional experimental data, such as RNA-Seq, may lead to improvements in this area.
Given that the majority of somatic mutations reside in non-coding sequence, the need to computationally prioritize them for follow-up functional validation is clear. The recent discovery of melanoma driver mutations in the promoter sequence of telomerase reverse transcriptase (TERT) gene highlights the potential of regulatory variation to drive tumorigenesis43,44. As cancer genome projects are moving toward sequencing whole genomes, more non-coding driving mutations will likely be discovered. To facilitate such discoveries more computational method development to score regulatory variants is needed.
Approach 3: Finding signs of positive selection across a cohort
Independent of whether or not a functional consequence can be predicted for a given mutation, one can assess to what extent a given mutation has been observed at a higher frequency than expected. The rationale for assessing mutation frequency is that driver mutations provide an adaptive advantage to cancer cells (Box 1, e.g., BRAF V600E mutation found in melanoma7) and should thus be positively selected during the clonal evolution of tumors. Provided that similar selective pressures act on different patient tumors and that the same mutation is positively selected, one should be able to trace driver mutations by noting their higher frequency, a common trace of positive selection.
In principle, exploiting this fact to find driver genes is straightforward: it is simply a statistical comparison between the mutation rate observed in a gene versus what is expected under a neutral model. However, in practice this approach involves difficult choices with respect to the selection of appropriate models for neutral evolution. For example, germline variation should not be used to calibrate a null model for somatic mutation analysis26 because this reflects evolutionary pressures and mutation processes during species evolution rather than during the development of cancer. In addition, many cancers have defects in DNA repair processes that change the neutral mutation rate, which have different regional impacts38,45,46, and local mutation rate is variable depending on other factors such as replication timing47.
To accurately identify significantly mutated genes, gene-specific mutation rates should thus be computed. This can be done using synonymous mutations48 and/or mutations in introns and UTR sequences (eg. InVex)49; however, these approaches can only be effectively used in tumors with very high mutation rates. In other cases gene-specific mutation rates must be estimated taking into account factors known to affect mutation rate such as mutation context, replication timing and expression levels (eg. MuSiC50 and MutSig51).
Given the difficulties that are intrinsic to recurrence-based methods, new methods have been developed that try to infer signs of positive selection using alternative means. One such approach, OncodriveFM52, consists of detecting genes that exhibit a significant bias towards the accumulation of somatic mutations with high functional impact. This method employs well-known metrics of the functional impact of individual mutations (those in Supplementary Table 3) to detect genes and pathways with this functional impact bias52. Another novel approach, ActiveDriver53, involves the discovery of genes significantly enriched for somatic mutations that alter ‘active sites’ in proteins, such as signaling sites, regulatory domains or linear motifs, assuming that such active mutations are more likely to have a wide-spread downstream effect and lead to a phenotypic advantage for tumor cells53.
Supplementary Table 4 lists several statistical approaches recently developed to identify candidate driver genes with signs of positive selection in a cohort of tumors46,48–50,52–54. As some of these methods are based on different theoretical principles, we recommend applying multiple complementary methods and comparing their results.
Despite these recent advances, future methods will need to capture the high degree of inter-tumor heterogeneity, as different tumors may acquire the same hallmark of cancer by different means (known as analogous mutations55). This heterogeneity is clearly underestimated in the current driver/passenger model.
Challenges and future perspectives
Cancer genome sequencing is a rapidly expanding field, and consequently computational methods used to interpret these data are evolving. We have presented a review of classes of practical tools currently available for analysis of a subset of genetic variation data. Because of the rapid evolution of the field, we have purposely avoided recommending particular tools or methods. Instead we present general guidelines to assist in making educated choices of methods that can address particular research problems. A number of pipelines facilitate the user-friendly application of various tools presented here. For instance, CRAVAT56 maps mutations to their consequences on protein coding genes and it predicts their implication in cancer and disease using CHASM26 and VEST57. IntOGen-mutations58 provides a way to apply tools of the three approaches, including mapping mutations using Ensembl VEP8, reporting their functional impact on proteins (using MutationAssessor22, SIFT20, PolyPhen259 and TransFIC25) and identifying genes with signs of positive selection across a cohort using OncodriveFM52.
It is important to emphasize the limited capacity of these approaches to directly identify the causative mutations of tumor development. Rather, they are intended to prioritize candidates for follow-up experiments that may demonstrate their actual implication in the cancer phenotype. Reporting back the results of these rounds of validation experiments to the method’s authors could in principle help them improve their approaches. The current relative scarcity of established spaces for this information exchange should be specifically addressed as part of the development of this field. Furthermore, these validation experiments will contribute to expand the catalogs of well characterized driver and passenger mutations, thus creating appropriate datasets for the development of computational prediction tools.
There are three key challenges in the field of cancer mutation analysis (Box 2). The first is to improve the accuracy of prediction of the functional impact of a mutation. Because mutations do not occur in isolation, but coexist with other somatic alterations that work together to alter cellular processes, separate gene-by-gene analyses are error-prone. A promising direction is the integration of multiple sources of biological information60, and the use of pathway and network analyses in the interpretation of cancer genomes22,61,62.
Box 2. Current Challenges.
1. Assess the functional impact of sets of mutations
Most current methods cannot accurately predict changes in protein and cellular function because changes in tumor phenotype typically result from multiple genetic alterations.
2. Complement the identification of functional and driver mutations by the prediction of how mutations affect protein and cellular function
There is a need for methods that not only identify functional or driver mutations but also predict the likely cellular outcome resulting from mutations such as gain, loss or switch of function, and how mutations might affect cellular networks.
3. Apply predictive tools to biologically relevant questions such as drug resistance
The ideal method should not only predict the effect of multiple mutations in an integrative manner and how they affect protein and cellular outcome, but also tackle translational clinical challenges such as drug resistance.
The second challenge is to develop reliable computational methods for the classification of mutations by functional impact type: loss of function, gain of function or switch of function22,61,62. The computational classification of mutations by type as well as strength of impact will contribute to the more complete elucidation of functional alterations in a cancer genome. The rich information encoded in the 3D structure of proteins, which is not yet well utilized by current approaches, can be particularly useful for deducing both the functional type and cellular consequences of mutations.
Lastly, there is the practical challenge of identifying mutations that confer resistance or sensitivity to a particular form of therapy (see for example63,64). We look forward to the day when functional prediction methods support personalized therapeutics, in which the patient’s therapy is informed by analysis of the specific genetic alteration profile in an individual tumor. The development of better approaches for analysis of functional and driver mutations will help to facilitate this process and in so doing will support the future development of personalized cancer medicine.
Supplementary Material
Supplementary Table 2. Tools to annotate variants to genomic features that may be affected using Sequence Ontology Terms
Supplementary Table 3. Methods to assess the functional effect of nsSNVs that can be used in a high-throughput manner
Supplementary Table 4. Available tools to identify driver genes from a cohort of cancer patients
References
- 1.ICGC et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg R. a Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Futreal PA, et al. A census of human cancer genes. Nature Reviews Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nature reviews Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 7.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics (Oxford, England) 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w (1118)3; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina I, et al. VARIANT: Command Line, Web service and Web interface for fast and accurate functional characterization of variants found by Next-Generation Sequencing. Nucleic acids research. 2012;40:W54–8. doi: 10.1093/nar/gks572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoehndorf R, Kelso J, Herre H. The ontology of biological sequences. BMC Bioinformatics. 2009;10:377. doi: 10.1186/1471-2105-10-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Project G, Asia E, Africa S, Figs S, Tables S. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;135:0–9. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Research. 2010;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Medicine. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NHLBI Exome Sequencing Project (ESP) Exome Variant Server.
- 19.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Pérez A, López-Bigas N. Improving the Assessment of the Outcome of Nonsynonymous SNVs with a Consensus Deleteriousness Score, Condel. The American Journal of Human Genetics. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics (Oxford, England) 2009;25:1431–2. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Research. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Perez A, Deu-Pons J, Lopez-Bigas N. Improving the prediction of the functional impact of cancer mutations by baseline tolerance transformation. Genome medicine. 2012;4:89. doi: 10.1186/gm390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter H, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer research. 2009;69:6660–7. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminker JS, Zhang Y, Watanabe C, Zhang Z. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Research. 2007;35:W595–598. doi: 10.1093/nar/gkm405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capriotti E, Altman RB. A new disease-specific machine learning approach for the prediction of cancer-causing missense variants. Genomics. 2011;98:310–7. doi: 10.1016/j.ygeno.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Human mutation. 2011;32:358–68. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human non-synonymous SNPs and their functional predictions. Human mutation. 2011;32:894–9. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niknafs N, et al. MuPIT Interactive: Webserver for mapping variant positions to annotated, interactive 3D structures. Human Genetics. 2013 doi: 10.1007/s00439-013-1325-0. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science (New York, NY ) 2007;315:233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 33.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science (New York, NY ) 2009;324:1720–3. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic acids research. 2008;36:D102–6. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clifford RJ, Edmonson MN, Nguyen C, Buetow KH. Large-scale analysis of non-synonymous coding region single nucleotide polymorphisms. Bioinformatics (Oxford, England) 2004;20:1006–1014. doi: 10.1093/bioinformatics/bth029. [DOI] [PubMed] [Google Scholar]
- 38.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman MM, Birney E. An effective model for natural selection in promoters. Genome research. 2010;20:685–92. doi: 10.1101/gr.096719.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowper-Sal Lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nature genetics. 2012;44:1191–8. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature Genetics. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 42.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic acids research. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horn S, et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science (New York, NY ) 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 44.Huang FW, et al. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science (New York, NY ) 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nature genetics. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodis E, et al. A Landscape of Driver Mutations in Melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dees ND, et al. MuSiC: Identifying mutational significance in cancer genomes. Genome Research. 2012;22:1589–98. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013 doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic acids research. 2012;40:e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimand J, Bader GD. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Molecular Systems Biology. 2013;9:637. doi: 10.1038/msb.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science (New York, NY ) 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 55.Creixell P, Schoof EM, Erler JT, Linding R. Navigating cancer network attractors for tumor-specific therapy. Nature Biotechnology. 2012;30:842–848. doi: 10.1038/nbt.2345. [DOI] [PubMed] [Google Scholar]
- 56.Douville C, et al. CRAVAT: Cancer-Related Analysis of VAriants Toolit. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carter H, et al. Identifying Mendelian disease genes with the Variant Effect Scoring Tool. BMC Genomics. 2013;14(Supl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalez-Perez A, Perez-Llamas C, Santos A, Deu-Pons J, Lopez-Bigas N. IntOGen-mutations pipeline: To interpret catalogs of cancer somatic mutations. 2013 at < http://www.intogen.org/mutations/analysis>.
- 59.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masica DL, Karchin R. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer research. 2011;71:4550–61. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee W, Zhang Y, Mukhyala K, Lazarus RA, Zhang Z. Bi-Directional SIFT Predicts a Subset of Activating Mutations. PLOS one. 2009;4:e8311. doi: 10.1371/journal.pone.0008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ng S, et al. PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics (Oxford, England) 2012;28:i640–i646. doi: 10.1093/bioinformatics/bts402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science (New York, NY ) 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valencia A, Hidalgo M. Getting personalized cancer genome analysis into the clinic: the challenges in bioinformatics. Genome medicine. 2012;4:61. doi: 10.1186/gm362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makarov V, et al. AnnTools: a comprehensive and versatile annotation toolkit for genomic variants. Bioinformatics (Oxford, England) 2012;28:724–5. doi: 10.1093/bioinformatics/bts032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Habegger L, et al. VAT: a computational framework to functionally annotate variants in personal genomes within a cloud-computing environment. Bioinformatics (Oxford, England) 2012;28:2267–9. doi: 10.1093/bioinformatics/bts368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biology. 2007;8:R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong WC, et al. CHASM and SNVBox: toolkit for detecting biologically important single nucleotide mutations in cancer. Bioinformatics (Oxford, England) 2011;27:2147–8. doi: 10.1093/bioinformatics/btr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartl DL, Clark AG. Principles of Population Genetics. 4. Sinauer Associates, Inc; 2006. p. 545. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 2. Tools to annotate variants to genomic features that may be affected using Sequence Ontology Terms
Supplementary Table 3. Methods to assess the functional effect of nsSNVs that can be used in a high-throughput manner
Supplementary Table 4. Available tools to identify driver genes from a cohort of cancer patients