Abstract
The human immunodeficiency virus type 1 (HIV-1) Vpr protein binds to the cellular uracil–DNA glycosylase UNG2 and induces its degradation through the assembly with the DDB1-CUL4 ubiquitin ligase complex. This interaction counteracts the antiviral activity exerted by UNG2 on HIV-1 gene transcription, as previously reported by us. In this work, we show that Vpr expression in the context of HIV-1 infection markedly decreases UNG2 expression in transformed or primary CD4+ T lymphocytes. We demonstrate for the first time that Vpr-UNG2 interaction significantly impairs the uracil excision activity of infected cells. The loss of uracil excision activity coincides with a significant accumulation of uracilated bases in the genome of infected cells without changes in cell division. Although UNG2 expression and uracil–DNA glycosylase activity are recovered after the peak of retroviral replication, the mutagenic effect of transient DNA uracilation in cycling cells should be taken into account. Therefore, the possible consequences of Vpr-mediated temporary depletion of endogenous nuclear UNG2 and subsequent alteration of the genomic integrity of infected cells need to be evaluated in the physiopathogenesis of HIV infection.
INTRODUCTION
Genome uracilation is generated either by misincorporation of deoxyuridine triphosphate (dUTP) during DNA polymerization or repair or by cytosine deamination either by spontaneous non-enzymatic processes (e.g. base alteration by chemicals or ionizing radiations) or through the action of a cytidine deaminase [reviewed in (1)]. The presence of uracil in DNA presents a potential threat for living organisms from yeast and bacteria to humans. When left unrepaired, uracil residues in U:G mismatches are 100% mutagenic. Owing to the DNA polymerase inability to discriminate between U and T in the template, unrepaired uracil bases result in the accumulation of G-to-A mutations on the complementary strand of DNA after the next round of replication.
Cytosine spontaneous deamination together with hydrolytic deamination is estimated to account for the accumulation of ∼100 mutations per genome per round of replication (2,3). Repair of uracil in DNA is ensured by the base excision repair (BER) pathway. The initial step is accomplished by a DNA glycosylase that catalyzes the hydrolysis of the N-glycosyl bond between uracil and the deoxyribose moiety. Then, an apyrimidinic/apurinic (AP) endonuclease creates a nick on the abasic site. Finally, the gap is repaired by the sequential action of DNA polymerase and DNA ligase activities (4). Five mammalian uracil–DNA glycosylases have been identified. Excision of uracil from U:A or U:G pairs in single- and double-stranded DNA is essentially supported by the nuclear uracil–DNA glycosylase UNG2. UNG1, an UNG2 isoform generated by the same unique UNG gene through the use of differentially regulated promoters and alternative splicing, is exclusively expressed in mitochondria and retains the same properties as UNG2 to ensure integrity of the mitochondrial genome (5). Besides UNG2, SMUG1 initially described as a single strand selective mono-functional UDG that excises uracil in U:A and U:G pairs (6), has recently been reported to exhibit a preferential activity towards double stranded genomic DNA in physiological conditions (7). SMUG1 also can remove some oxidized pyrimidines, suggesting a role in the repair of DNA oxidation damage (8,9). Finally, uracil from U:G can be removed by the thymine–DNA glycosylase (TDG) and the methyl-binding domain protein 4 (MBD4) that also excise thymine from T:G mismatches, preferentially in CpG sequences (3).
The function of the apparently redundant uracil–DNA glycosylases is tightly regulated and they are differentially expressed during the cell cycle (3,10). Indeed, UNG2 appears as the sole contributor to post-replicative repair of U:A lesions during S-phase through specific interaction with proliferating cell nuclear antigen and replication protein A at replication foci (11). Then, UNG2 is phosphorylated (11) and degraded by the proteasome to undetectable levels during the late S and G2 phases of the cell cycle. Conversely, SMUG1 and TDG are eliminated in cells entering the S-phase (11,12).
UNG2 function in maintaining genomic integrity is common to all cell types. However, its role is much more complex in activated B lymphocytes, in which UNG2 also facilitates mutagenic processing of AID-induced uracil in the switch (S) and V(D)J regions of immunoglobulin loci. Accordingly, UNG2 favors class-switch DNA recombination (CSR) and somatic hypermutation (SHM) and is critical for the maturation of the antibody response [for review see (2)]. UNG2 functional importance has specifically been highlighted by studies in Ung−/− mice and humans harboring UNG mutations. In both situations, absence of UNG2 expression is associated with a 5-fold increase in genomic mutation frequency (10), hyper-IgM syndrome and a significant perturbation of the acquired immune response caused by failure in class-switch recombination and altered somatic hypermutation (2,13,14). UNG2 deficiency also correlates with a global immunological imbalance with reduction of T-helper and NK-cells in spleen and deregulation of interferon γ, interleukin (IL)-2 and IL-6 levels (15). Finally, in aged mice, it results in an increased risk of developing follicular and diffuse large B-cell lymphoma (13).
A variety of viral proteins have the capacity to disturb DNA repair in the host cell. The mechanisms of such perturbation include transcriptional alteration of host genes coding for the DNA repair machinery, post-transcriptional modification of gene products and mislocalization and degradation or deregulation of host proteins that are associated with the DNA damage response resulting from their direct interaction with viral products [for review see (16)]. The regulatory Vpr protein is the main perturbator of the host cell DNA repair capacity in HIV-1-infected cells (17). Vpr is a virion-associated protein of an average length of 96 amino acids (∼15 kD) that was proposed to contribute to viral pathogenesis and disease progression in HIV-infected patients [reviewed in (17) and (18)]. In proliferating T-cells, Vpr enhances viral replication by 2- to 4- fold (19), and its expression is essential for efficient viral replication in non-dividing cells such as macrophages (20). This property has been attributed to the Vpr capacity to support the cytoplasmic–nuclear shuttling of the reverse transcription complex and viral genome nuclear import during early replicative events (21–24) and to stimulate HIV-1 gene transcription by interacting with the SP1, TFIIB and p300/CREB transcription factors (25–27). Because of its ability to interact with many cellular proteins, several functions have been ascribed to Vpr within the cell (28), including induction of apoptosis by promoting the formation of permeability transition pores in mitochondria (29–31), impairment of the host immune function for HIV-1 evasion (32) and blockade of the cell cycle in the G2 phase (33,34). This last function has been related to Vpr ability to activate the ataxia telangiectasia-mutated and Rad3-related (ATR) proteins (35–37) and to stimulate phosphorylation of the ATR substrates CHK1 and H2AX histone variant (36,38). Approaches based on yeast two-hybrid screening identified UNG2 as a Vpr-binding partner (39). The consequences of this interaction during HIV-1 replication have been widely debated. Vpr binding was initially proposed to induce viral packaging of UNG2 (40), thereby conferring to HIV-1 a replicative advantage by lowering the occurrence of mutations in the retroviral genome during reverse transcription (40–42). This model could not be confirmed using immortalized T-cells, in which UNG2 enzymatic activity was suppressed with the uracil glycosylase inhibitor (UGI) or in UNG2-defective B-lymphocytic cells as HIV-1 producer cells (43,44). Therefore, it was concluded that UNG2 is dispensable for HIV-1 replication. On the contrary, virion-associated UNG2 has been proposed to degrade APOBEC-3G-edited HIV-1 DNA, supporting an antiviral role for UNG2 (45).
Besides these studies, we and others have found that UNG2 cellular pool is actively depleted to undetectable levels in established cell lines overexpressing HIV-1 Vpr (44,46). Vpr-mediated UNG2 depletion results from accelerated natural turnover of the protein and requires both the direct interaction of the two proteins and the recruitment of DCAF1, DDB1 and CUL4 by Vpr (47–49). A similar mechanism is suggested to account for Vpr-induced proteasomal degradation of SMUG1 (44,49). In the past years, we questioned the requirement of Vpr-mediated UNG2 depletion in HIV-1-infected cells. We have demonstrated that UNG2 overexpression in an embryonic kidney carcinoma cell line is deleterious for LTR-driven HIV-1 gene transcription through an unknown mechanism independent of UNG2 enzymatic activity (46). We thus proposed that Vpr could act as a protective factor capable to counteract a detrimental effect exerted during the late steps of retroviral replication by actively degrading cellular UNG2 in infected cells (46). The recent study by Weil et al. (50) underlines that UNG2 can also behave as an HIV-1-host restriction factor counteracting early replication steps, but only in cells with high dUTP content. This model agrees with evidences indicating that a high uracil level in HIV reverse transcripts (>500 uracils per 10 kb HIV genome) prevents inappropriate strand transfer of HIV-1 DNA ends in the pre-integration complex, limits autointegration and favors chromosomal integration and viral replication in macrophages (51). Together with the study by Jones et al. (52) that reported the UNG2-dependent enhancement of early HIV-1 replication of R5- but not X4-dependent HIV-1 in primary lymphocytes, these results highlight a context-dependent role of UNG2 in HIV-1 replication.
In contrast to the studies that investigated the consequences of UNG2 viral packaging on HIV-1 infectivity, the outcome of Vpr-mediated UNG2 depletion in HIV-1 infected cells still remains unexplored. Therefore, the present study was undertaken to determine the consequences of Vpr expression on the global DNA repair capacity of HIV-infected cells. To this end, we designed and validated an in vitro enzymatic assay to specifically quantify U:A excision capacity in cell extracts. Using this assay, we demonstrate that Vpr expression in HIV-1 infected lymphoblastoid cell lines and in primary CD4+ T lymphocytes results in a transient decrease in the host cell uracil excision capacity. This drop in UNG2 activity is independent of Vpr cytostatic and pro-apoptotic properties. In chronically infected cells, it is accompanied by a significant increase of the uracil content in their genomic DNA. Although the uracil level decreases after full recovery of UNG2 protein levels, the consequences of such genomic alterations in cycling cells need to be taken into account.
MATERIALS AND METHODS
Reagents and antibodies
The following antibodies were used: rabbit polyclonal anti-UNG2 PU059 (53) (from Geir Slupphaug, Trondheim, Norway), which cross-reacts with human UNG1 (31 kDa) and UNG2 (37 kDa); anti-Vpr goat polyclonal antibody (sc-17493; Santa Cruz Biotechnology); anti-tubulin mouse monoclonal antibody T5168 and anti-rabbit IgG HRP-conjugated (A6154 Sigma); goat polyclonal anti-p24 (4999–9007; AbD Serotec); anti-p24 monoclonal antibody (clone 23-A5G4B9; NIH-AIDS Reagent Consortium); anti-goat IgG HRP-conjugated (Jackson ImmunoResearch); anti-mouse IgG HRP-conjugated (A10668) and Alexa488-conjugated (A11017) from Life Technologies; anti-SMUG1 antibody (ab15716; Abcam) that was reported to display neutralizing activity in uracil–DNA glycosylase assays. The UGI inhibitor was from New England Biolabs Inc.
Plasmids
The following plasmids encoding the viral molecular clones pNL4.3, pNL4.3-VprW54G, pNL4.3-VprR77Q, pNL4.3-VprR80A, pNL4.3ΔVpr (from W.C. Greene, Gladstone Institutes San Francisco, USA) were used to produce HIV-1, HIV-1 VprW54G and HIV-1 ΔVpr viruses. The pHR-Vpr construct was kindly provided by Vicente Planelles (University of Utah School of Medicine, Salt Lake City, USA). The pET28-His-TEV-UNG2 plasmid was obtained by PCR subcloning of UNG2 cDNA (aa 92–313) in the pET-His6-TEV plasmid (Addgene).
Cell culture
Immortalized H9 T-cells were maintained in RPMI 1640 medium. HEK 293T and MAGIC-5B cells (from M. Tatsumi, Tokyo, Japan) were maintained in DMEM medium. Complete media were supplemented with penicillin–streptomycin and 10% heat-inactivated fetal bovine serum (FBS) (Cambrex, France). Peripheral blood mononuclear cells were isolated from Buffy coats of healthy HIV-negative donors by Ficoll density gradient (Pharmacia, Piscataway, NJ). CD4+ T lymphocytes were isolated using a human CD4+ T cell enrichment kit (EasySep Stem Cell Technologies) and grown for 7 days in RPMI medium supplemented with 10% fetal calf serum, penicillin (100 U/ml) and streptomycin (100 pg/ml) at 37°C, 5% CO2, in a humid atmosphere under IL-2/ phytohemagglutinin stimulation.
Viral stock production
Viral stocks were produced by calcium phosphate transfection of HEK 293T cells. Briefly, cells were transfected with the proviral DNA constructs for 8 h, washed with pre-warmed DMEM/10% FBS. Viral supernatants were collected 48 h post-transfection, filtered and frozen in aliquots at −80°C. Viral stock titers were determined using the HIV-1 p24 enzyme-linked immunosorbent assay (ELISA) kit (Ingen, France).
Uracil–DNA glycosylase assays
The fluorescent probe PEG-U9 (5′-FAM-GCACUUAAGAAUUG-PEG-CAAUUGUUAAGUGC-3′DAB) was synthesized by Eurogentec. Uracil–DNA glycosylase activity was assayed in a reaction buffer containing 20 mM Tris-HCl pH 8.0, 50 mM KCl, 0.2 mM MgCl2 and 0.05% Brij-35. The mixture was incubated with 100 nM PEG-U9 for 20 min at 37°C. Fluorescence was recorded using a Tecan Infinite 200 fluorimeter.
Western blot experiments and analysis
Proteins were separated by SDS–PAGE (12.5% gels), transferred to nitrocellulose membranes (Millipore, France) and immunoblotted with the appropriate primary and horseradish peroxidase–conjugated secondary antibodies (Immunotech, France). Immune complexes were revealed by Enhanced Chemiluminescence (Supersignal Femto/Pico chemiluminescence susbtrate, Thermo Scientific) and images were acquired with the Gene Gnome imaging system (SynGene). Band intensities were quantified using the ImageJ software (NIH).
Stealth RNAi transfection
The Stealth RNAi sequence duplexes against UNG2 are as follows: sense, 5′-CGCGUUCGCUGCCUCCUCAGCUCCA-3′ and antisense, 5′-UGGAGCUGAGGAGGCAGCGAACGCG-3′. This target sequence corresponds to the 5′ untranslated region of UNG2 mRNA (positions 44–68 according to the NM_080911 entry). As a negative control, we used the Stealth RNAi scrambled control duplexes with high GC content (Invitrogen, France). Stealth RNAi duplexes (1 nM) were transfected using Lipofectamine 2000 following the manufacturer’s instructions (Invitrogen). To obtain optimal conditions for endogenous UNG2 expression, cells were maintained at a low density during the entire experiment. Forty-eight hours after transfection, cells were harvested and UNG expression analyzed in cell lysates.
Cell cycle analysis
Cells were collected, washed twice and resuspended in ice-cold PBS. Then, cells were fixed with 70% EtOH on ice for 1 h. Cells were pelleted at 1500 rpm for 5 min and resuspended in a solution containing 50 µg/ml propidium iodide, 0.1 mg/ml RNase A and 0.1% Triton X-100 in PBS. After 16 h at 4°C in the dark, the DNA content was determined by flow cytometry using an EPICS PROFILE XL4C cytofluorometer (Beckman Coulter) or a BD LSRFortessa cell analyzer (BD Biosciences). For cell cycle analysis of HIV-1-infected H9 T cells, a double-staining protocol to label intracellular HIV-1 antigens using the anti-p24 monoclonal antibody and the DNA content using propidium iodide was applied, as described by Zhou et al. (54).
Genome uracil content quantification
Genomic DNA was extracted from 3 × 107 cells using DNAzol (Invitrogen) according to the manufacturer’s recommendations. Pre-existing AP sites in DNA were protected by incubation with 200 mM methoxyamine at 37°C for 2 h. Then, 4 µg DNA was incubated with 0.2 U Escherichia coli-derived recombinant UDG (New England Biolabs) at 37°C for 15 min. AP sites, generated by excision of uracil residues contained in DNA, were titrated using the OxiSelect oxidative DNA Damage ELISA kit (Euromedex).
RESULTS
Design and validation of an in vitro uracil excision enzymatic assay
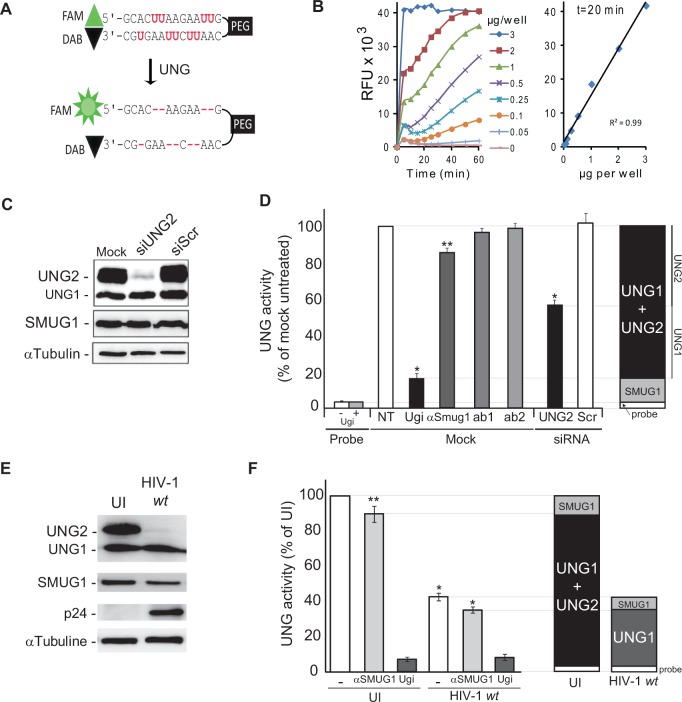
Our aim was to investigate the potential consequences of Vpr expression on the endogenous UNG2 protein levels and on the uracil excision capacity of HIV-1-infected cells. Thus, we developed and validated an in vitro assay to specifically detect U:A excision activity in host cell extracts. Based on previously reported data (55–57), we designed a fluorescent molecular beacon containing several uracil residues, flanked by self-complementary termini and carrying a FAM fluorophore and a DAB quencher at the 5′ and 3′ ends, respectively (Figure 1A, PEG-U9 probe). This molecule naturally forms a stem–loop structure that quenches the fluorophore as a result of the close proximity of the 5′-fluorophore and the 3′-quencher. Excision of PEG-U9 uracil bases disrupts the stem–loop structure and results in a fluorescent signal. Incubation of different amounts of total cell extracts from the HeLa-derived MAGIC-5B indicator cell line with 100 nM PEG-U9 in the appropriate reaction buffer for 20 min (Figure 1B) resulted in emission of fluorescence, while low background was observed in absence of cell extracts, attesting the specificity of the emitted signal. In our experimental conditions (incubation time of 20 min), this assay was linear with protein amounts ranging from 0 to 3 µg (Figure 1B). UNG1, UNG2 and SMUG1 can excise uracil residues in U:A pairs from double stranded DNA (3). To determine the respective contribution of each DNA glycosylase to the emitted signal, MAGIC-5B cell extracts were incubated with PEG-U9 in the presence of the UNG2/UNG1 inhibitor UGI (58) or after pre-incubation with anti-SMUG1 neutralizing antibodies (59). Fluorescence emission was reduced by >85% in the presence of UGI but only by 15% after pre-incubation with the anti-SMUG1 antibody. No inhibition was observed with isotype-matched irrelevant monoclonal antibodies (ab 1 and 2) (Figure 1D). To further validate the assay, we tested uracil–DNA glycosylase activity in MAGIC-5B cells in which expression of UNG2 was abolished by transfection of 1 nM siRNA specific for UNG2 (siUNG2). As monitored by immunoblot analysis (Figure 1C), UNG2 expression was strongly inhibited following siUNG2 transfection, whereas protein levels remained unchanged in cells transfected with the irrelevant siRNA (siScr). UNG1 and SMUG1 levels were unaffected. Following incubation with PEG-U9, fluorescence emission (and thus uracil excision activity) was reduced by 60% in siUNG2 cells in comparison with siScr cells. UNG1 and SMUG1 expression explained the residual enzymatic activity observed in siUNG2 cell lysates (Figure 1D). Thus, UNG2 is responsible for most uracil excision activity in MAGIC-5B cells that represents ∼40–60% of the overall DNA glycosylase activity. This observation is consistent with previous literature (59). Altogether, these experiments validate our in vitro enzymatic assay as a valuable tool to measure uracil–DNA glycosylase activity in extracts of HIV-1-infected cells.
Figure 1.
Vpr expression during HIV-1 replication decreases endogenous UNG2 levels in MAGIC-5B cells (A) Sequence and structure of the fluorescent PEG-U9 molecular beacon used as a substrate in UNG2 enzymatic assays. The oligonucleotide 5′ end is labeled with a FAM fluorophore and the 3′ end with a DAB quench. PEG: polyethylene glycol. Upon uracil excision, the fluorophore dissociates from the DAB quench, causing fluorescence emission. (B) Uracil DNA glycosylase activity in MAGIC-5B cells was determined by incubating different concentrations (0–3 µg proteins) of total cell lysate with the PEG-U9 molecular probe and then recording the fluorescence emission from 0 to 60 min after PEG-U9 addition (left panel). The correlation between fluorescence emission and protein concentration was linear during the 20 min incubation time (right panel). (C) UNG2 expression was assessed by western blotting using anti-UNG1/2 antibodies in MAGIC-5B cells transfected with 1 nM anti-UNG2 siRNA (siUNG2), irrelevant siRNA (siScr) or buffer alone (mock). Levels of SMUG1 and tubulin are shown as controls. (D) Uracil excision activity in total cell extracts from the experiment described in (C). Lysates were pre-incubated with the UNG1/UNG2 inhibitor UGI, anti-SMUG1 monoclonal antibodies or isotype-matched irrelevant monoclonal antibodies (ab1, ab2) to discriminate between the uracil excision activity as a result of SMUG1 or UNG1/UNG2. Values are the mean ± SD of three separate experiments performed in duplicate. Probe: background fluorescence. (E) Expression of UNG1, UNG2 and SMUG1 in UI and HIV-1 infected MAGIC-5B cells for 7 days was assessed by immunoblot analysis of total cell extracts. HIV-1 replication was monitored using the anti-p24 monoclonal antibody and equal loading was checked using an anti-Tubulin antibody. (F) Uracil excision activity in cell extracts from (E) was determined using the PEG-U9 uracilated fluorescent beacon. Activity attributable to UNG1/UNG2 or SMUG1 was discriminated by pre-incubating cell extracts with UGI or anti-SMUG1 antibodies as in D.
HIV-1 infection significantly reduces the uracil excision capacity of host cells
Next, endogenous UNG2 expression was analyzed over time in total extracts from HIV-1-infected MAGIC-5B cells (HIV-1 replication was demonstrated by intracellular p24 expression) (Figure 1E). UNG2 was significantly decreased at day 7 post-infection, and SMUG1 expression was significantly reduced in HIV-1-infected cells in comparison with uninfected (UI) cells. Uracil excision analysis using our enzymatic assay with the PEG-U9 probe showed that at day 7 post-infection, the uracil excision activity of HIV-1-infected cells was ∼40% of that of UI cells (Figure 1F). These data indicate that HIV-1-induced depletion of endogenous UNG2 results in a decrease of the host cell uracil excision capacity. To discriminate the respective contribution of UNG1, UNG2 and SMUG1 in the residual uracil–DNA glycosylase activity, fluorescence levels were recorded in the presence of UGI or after pre-incubation with anti-SMUG1 antibodies. The inhibitor further reduced the fluorescence emitted by HIV-1 infected-cell lysates to 10% of the value recorded in samples from UI cells, whereas the anti-SMUG1 antibodies only slightly decreased the residual uracil–DNA glycosylase activity in lysates from HIV-1-infected cells (Figure 1F). These results and those described in Figure 1D suggest that the residual uracil excision activity detected at day 7 post-infection can be attributed mostly to UNG1, the expression of which was barely affected at day 7 post-infection (Figure 1E), whereas SMUG1 depletion only accounts for a minor decrease in the uracil excision capacity of HIV-1-infected cells.
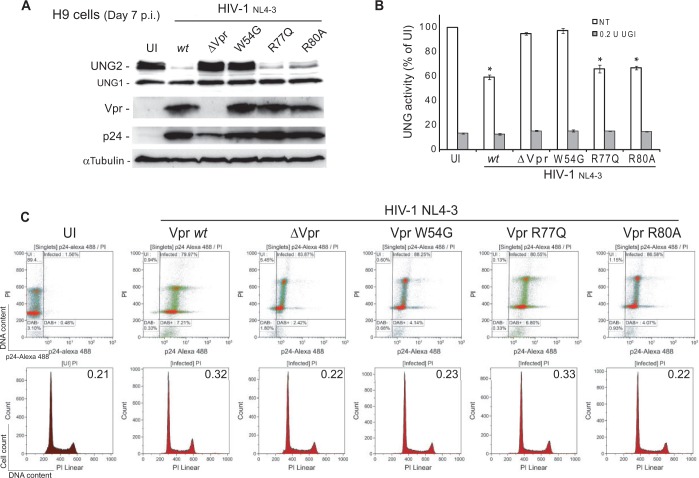
The decrease of uracil–DNA glycosylase activity in HIV-1-infected cells requires UNG2 binding to Vpr
Co-transfection experiments have suggested that HIV-1-induced UNG2 depletion requires Vpr interaction with UNG2 and Vpr-mediated recruitment of the ubiquitin–proteasome system (44,49). To determine whether Vpr is directly involved in the impairment of uracil excision capacity in HIV-1-infected cells, the H9 CD4+ T lymphoblastoid cell line was infected with wild type (wt) HIV-1 or with a HIV-1 mutant that does not express Vpr (HIV-1ΔVpr). At day 7 post-infection, the expression of endogenous UNG2 was decreased in cells infected with wt HIV-1 but not in cells infected with HIV-1ΔVpr, although p24 expression was still detected (albeit at lower level than in wt HIV-1-infected cells), demonstrating viral replication (Figure 2A). Despite the lower p24 protein level detected by western blotting in HIV-1ΔVpr cell extracts, cytometric analysis confirmed that the percentage of p24-positive cells was comparable in the absence or presence of Vpr (Figure 2C). The enzymatic assay with the PEG-U9 probe showed that, compared with UI cells, uracil–DNA glycosylase activity was reduced by ∼40% in wt HIV-1-infected H9 cell lysates, whereas it remained unchanged in HIV-1ΔVpr-infected lysates (Figure 2B). Addition of UGI to the reaction mixture further reduced the fluorescence to 20% of the values of UI cells in wt HIV-1 samples and also in HIV-1ΔVpr lysates (Figure 2B). Altogether, these data indicate that, when T lymphoblastoid cells are infected with wt HIV-1 that expresses the Vpr protein, UNG2 protein level is rapidly reduced and a significant loss of uracil–DNA glycosylase activity is observed. Previous studies showed that the W54 residue located at the beginning of the third α-helix of Vpr is required for direct interaction with UNG2 in yeast two-hybrid experiments (60). Moreover, the VprW54R mutant cannot recruit UNG2 into HIV-1 virions, although it is incorporated in viral particles as efficiently as wild-type Vpr (42). Ectopic expression of the VprW54R mutant was previously found unable to induce proteasomal degradation of UNG2 (44). To confirm the contribution of the W54 residue in Vpr binding to UNG2, we investigated the consequences of expressing the VprW54G mutant, which cannot interact with UNG2 in GST-pull down assays (Supplementary Figure S1) and yeast two hybrid experiments (60), by infecting H9 cells with HIV-1VprW54G for 7 days. Basal UNG2 expression persisted in infected cells that express high levels of VprW54G (Figure 2A) and the uracil–DNA glycosylase activity was comparable with that of UI cells (Figure 2B), despite efficient HIV-1 replication as attested by p24 levels.
Figure 2.
Vpr-UNG2 binding decreases uracil–DNA glycosylase activity in HIV-1-infected T cells. (A) Expression of UNG1 and UNG2 in H9 T lymphocytes infected with wild type HIV-1, HIV-1ΔVpr, HIV-1VprW54G, HIV-1VprR77Q or HIV-1VprR80A mutant was analyzed by immunoblotting with specific antibodies. Adequate Vpr expression was controlled. Cell infection was monitored using anti-p24 monoclonal antibodies and equal loading with an anti-tubulin antibody. UI cells are shown as controls. (B) Uracil excision activity in the corresponding total cell extracts was assessed by incubation with the PEG-U9 fluorescent molecular beacon and quantification of the fluorescence emission. UNG2 and UNG1 contribution was determined by pre-incubation with the UGI inhibitor. Values are the mean ± SD of duplicate experiments. Statistical significance was determined using the ANOVA test. (C) Cells from the experiment described in panel (A) were double stained with propidium iodide (DNA content) and anti-p24 antibodies (intracellular HIV-1 antigens). Cell cycle analysis was gated to HIV-1 positive cells. The G2M/G0G1 ratio is indicated for each condition.
Consequences of Vpr-induced UNG2 depletion on cell cycle progression and viability
UNG2 expression is finely regulated during cell cycle progression. As Vpr overexpression induces G2 arrest, we investigated the potential contribution of cell cycle perturbation in Vpr-induced UNG2 depletion. H9 cells were infected with HIV-1, HIV-1ΔVpr, HIV-1VprR77Q (which cannot induce cell apoptosis) (61) or with the HIV-1VprR80A mutant that cannot cause cell cycle arrest in G2 [this phenotype was validated by overexpression experiments (Supplementary Figure S2)] (60). After 7 days of infection, cells were stained with propidium iodide and the DNA content determined by flow cytometry only in p24-positive cells (∼80% of the cell population). Considering the total cell population, no significant modification in the cell cycle profiles of cells infected with HIV-1, HIV-1ΔVpr, HIV-1VprW54G, HIV-1VprR77Q or HIV-1VprR80A was observed compared with mock-infected cells (Figure 2C). However, as expected, a G2 arrest could be observed when cell cycle analysis was gated to the cells that express the highest p24 levels (data not shown). Thus, in our cell culture conditions, UNG2 depletion and loss of uracil–DNA glycosylase activity observed in HIV-1-infected cells cannot be ascribed to cell cycle arrest. This was confirmed by the observation that UNG2 expression was reduced also in HIV-1VprR80A-infected cells. In addition, flow cytometry analysis did not reveal any difference in the necrotic or apoptotic rate in cells with Vpr-induced inhibition of UNG2 expression as attested by Annexin V labeling and propidium iodide permeability assays (Supplementary Figure S3). Accordingly, Vpr-mediated UNG2 depletion in HIV-1 transfected cells is not a consequence of reduced cell viability.
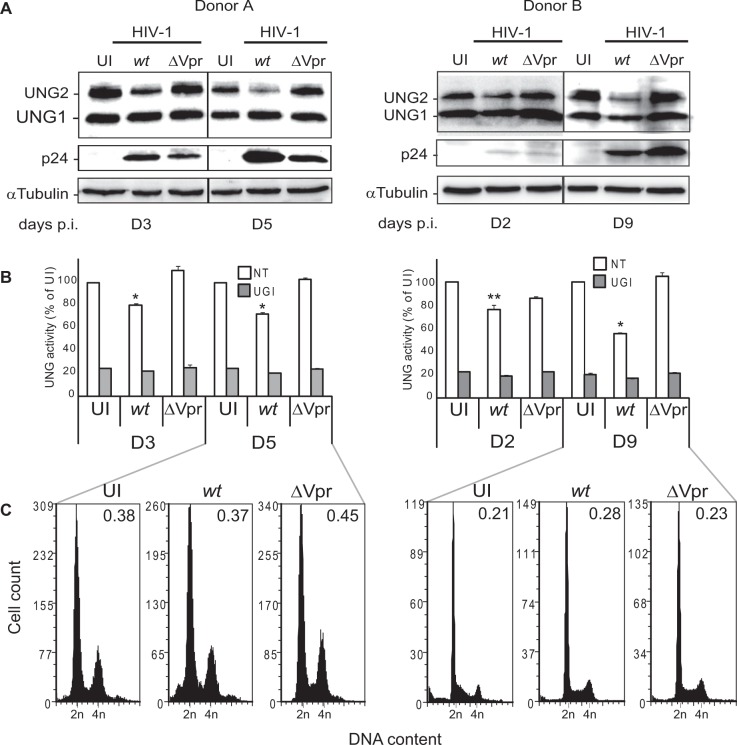
Uracil excision activity in chronically HIV-1 infected primary CD4+ T lymphocytes
To evaluate the possible physiological relevance of these results, we investigated the uracil excision capacity of HIV-1-infected primary CD4+ T lymphocytes. Primary CD4+ T cells, which were isolated from total peripheral blood lymphocytes of two healthy donors and negatively selected using coated magnetic beads (Supplementary Figure S4), were infected with wt HIV-1 or HIV-1ΔVpr at an MOI of 10. Cells were collected twice a week and UNG2 expression and uracil excision activity were determined in total cell extracts. UNG2 expression was significantly decreased at day 5 (donor A) and day 9 (donor B) post-infection (Figure 3A). As previously observed in T lymphoblastoid H9 cells, UNG2 reduction coincided with the peak of HIV-1 replication, as attested by p24 expression. Conversely, UNG2 level remained unaffected following infection with the HIV-1ΔVpr strain. Quantification of the enzymatic activity showed a significant loss of uracil–DNA glycosylase activity in cell extracts from CD4+ T cells infected with wt HIV-1 but not with the HIV-1ΔVpr strain (Figure 3B). Again, no significant difference in the cell cycle profile (particularly cell cycle arrest) was observed in these cultures (Figure 3C). Thus, UNG2 protein and uracil–DNA glycosylase activity are depleted in the absence of any cell cycle alteration in HIV-1-infected primary CD4+ T cells.
Figure 3.
Uracil DNA glycosylase activity during HIV-1 infection of primary CD4+ T lymphocytes. Primary CD4+ T lymphocytes were left uninfected (UI) or infected with wt HIV-1 or HIV-1ΔVpr (ΔVpr) and maintained in culture for 5 (donor A) or 9 days (donor B). (A) Expression of UNG1 and UNG2 was determined by immunoblotting at different time points following infection. HIV-1 infection was monitored using the anti-HIV-1 p24 antibody and equal loading with the anti-tubulin antibody. (B) Cell extracts were incubated with the PEG-U9 uracilated fluorescent molecular beacon and DNA glycosylase activity recorded as fluorescent emission. Values are the mean ± SD of triplicate measurements. (C) Cells collected at day 5 (donor A) or 9 (donor B) were stained with propidium iodide (DNA content) and cell cycle progression analyzed by flow cytometry.
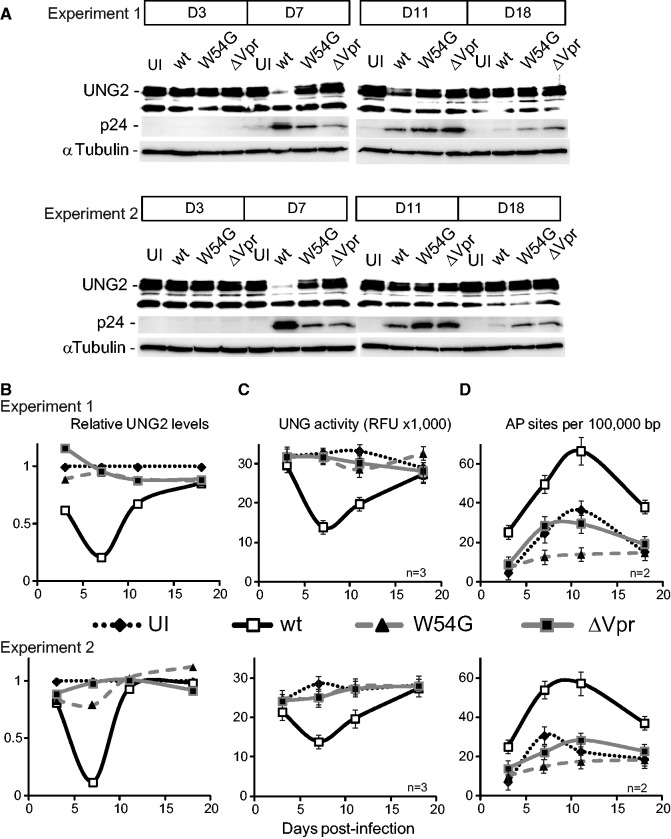
Chronic HIV-1 infection results in transient Vpr-mediated UNG2 depletion and increased uracil content in genomic DNA
Knock out of the Ung locus is associated with a 2.4- to 30-fold increase of uracilation in mice and yeast DNA (6,62). In humans, the study of cells from patients with biallelic germline UNG mutations also evidenced elevated genomic uracil levels (13). To investigate the consequences of Vpr-induced UNG2 depletion on genome integrity, we compared the genomic uracil content of HIV-1 (wt and mutants) infected and uninfected (UI) H9 cells cultured in similar conditions (two independent experiments). Genomic DNA was extracted from these cultures after 3, 7, 11 and 18 days in culture and then incubated in vitro with E. coli-derived recombinant uracil–DNA glycosylase. The AP sites generated by uracil excision were then revealed with a biotin-labeled aldehyde reactive probe and quantified using an ELISA assay (Figure 4). Genomic DNA extracted from cells after 7 days of infection with wt HIV-1 showed a drastic increase in uracil content, reaching 66 uracilated bases per 105 bases compared with UI cells (Figure 4D). At this time-point, both UNG2 protein levels (Figure 4A and B) and the global uracil DNA glycosylase activity (Figure 4C) were significantly decreased compared with UI cells (Figure 4). Analysis at later time-points revealed that the uracil content in DNA from HIV-1-infected cells started to decrease and returned almost to basal level at day 18 post-infection. No increase in uracil levels was observed in the genomic DNA from cells infected with the HIV-1VprW54G or HIV-1ΔVpr mutants compared with UI cells. In these cells, UNG2 expression was mostly unaffected and uracil DNA glycosylase activity unchanged over time. Cell cycle analysis throughout the experiment revealed no significant alteration in HIV-1-infected (wt and mutants) cells (Supplementary Figure S5A). Thus, the observed transient loss of UNG2 expression cannot result from cell cycle arrest or cell death. Moreover, the recovery of basal UNG2 expression, uracil excision activity and uracil level cannot be ascribed to the gradual emergence of less infected cell populations as attested by the constant increase of infected cells during the duration of the experiment (Supplementary Figure S5B). Therefore, following HIV-1 infection, Vpr expression and its interaction with UNG2 lead to a global reduction in uracil excision capacity of the host cells that correlates with a marked uracil accumulation in genomic DNA. Although UNG2 expression progressively returned to basal level as HIV-1 replication decreased, the presence of uracilated bases in the genomic DNA persisted for at least 18 days while cells were still normally cycling (Supplementary Figure S5A).
Figure 4.
Genomic uracil levels in HIV-1 infected lymphocytes. Cell extracts were prepared from uninfected H9 cells (UI) or infected with wt HIV-1, HIV-1ΔVpr (ΔVpr) or HIV-1 VprW54G (W54G) at different time-points. (A) UNG2 expression was monitored over time with anti-UNG2 antibodies and HIV-1 replication with anti-p24 antibodies. (B) The relative UNG2 expression was determined from (A) by densitometry scanning and normalized to α-tubulin levels. (C) Uracil excision activity over time was quantified in the corresponding cell extracts by using the fluorescent uracil–DNA glycosylase assay. (D) The variations in uracilation of genomic DNA from the corresponding samples (D) are plotted over time. For each experiment, n indicates the number of replicates.
DISCUSSION
Previous studies demonstrated that overexpression of HIV-1 Vpr results in cell depletion of the UNG2 and SMUG1 uracil DNA glycosylases (44,46,47,63). In the present study, we extended these observations and show that HIV-1 mediated Vpr expression results in UNG2 depletion and in a significant decrease of the global uracil DNA glycosylase activity in HIV-1-infected transformed and primary CD4+ T lymphocytes. This results in the accumulation of unrepaired uracil residues in the host cell genome. Our study provides first data on the effect of HIV-1 infection on the uracil excision machinery and the resulting DNA damage in infected T lymphocytes. Although uracilation of the host cell genome is transient and intense only at the peak of retroviral replication, its consequences need to be taken into account.
Here, we report that UNG2 expression in HIV-1-infected transformed and primary cells progressively decreases during HIV-1 replication and that this reduction is Vpr-dependent as indicated by the results obtained with the HIV-1ΔVpr mutant. This observation is consistent with the work by Wen et al. (49) who reported a dose-dependent depletion of UNG2 in Vpr-transfected epithelial cells. Moreover, the comparative analysis of cells infected with wild type HIV-1 or strains that lack Vpr or express a VprW54G mutant clearly shows that deregulation of UNG2 expression is mediated through Vpr-UNG2 binding. Indeed, the integrity of the UNG2-binding W54 residue in Vpr (localized between the second and third α-helix) is critical for UNG2 recruitment by the Vpr-CUL4-E3 ligase complex and its subsequent targeting to the proteasome (39,47,60).
HIV-1 ability to deplete the cellular pools of UNG2 and SMUG1 by enhancing the interaction between UNG2 and the CRL4 (DCAF1) complex is not unique among virus-encoded proteins [reviewed in (64)]. Indeed, human cytomegalovirus, adeno-associated virus type 2 and herpes simplex virus 1 have also developed strategies to eliminate, circumvent or exploit the DNA damage sensing and repair machinery of the host cell (65–67). Nevertheless, it is unclear why UNG2 is degraded during HIV-1 replication. Several hypotheses can be raised. We have previously reported that UNG2 overexpression leads to impairment of HIV-1 gene transcription through an unknown mechanism (46). In addition, uracilation of HIV-1 cDNA promotes the early phase of the HIV-1 life cycle by protecting the virus from autointegration (51). Finally, it cannot be excluded that accumulation of uracil bases in HIV-1 proviruses could have a protective effect by interfering with cytosolic sensors that can recognize the retroviral nucleic acids and activate antiviral innate immune responses, as suggested by others (68). Very recently, Weil et al. (50) demonstrated that UNG2 behaves as a restriction factor in cells with high dUTP levels and can prime viral DNA degradation. For all these reasons, HIV-1 may have acquired the capacity to control the host cell DNA repair machinery to maintain sufficient levels of retroviral DNA uracilation. Of note, UNG2 depletion in the host cell is tolerated for efficient HIV-1 replication, as described here. This observation corroborates previous studies reporting that production of fully infectious retroviral particles is achieved in cells lacking UNG2 activity (43).
Using an in vitro enzymatic assay developed in our laboratory, we found that the global uracil–DNA glycosylase activity in HIV-1-infected cells is also significantly decreased at the peak of retroviral replication. As observed for UNG2 reduced expression, this loss of activity strictly depends on the expression of a Vpr protein that can bind to UNG2. Moreover, our enzymatic assay combined with the use of UGI, an inhibitor of UNG1/UNG2 activity, or of anti-SMUG1 neutralizing antibodies allowed the quantification of the respective contribution of each uracil DNA glycosylase that can excise uracil in a U:A pair. This confirmed that the loss of uracil DNA glycosylase activity in HIV-1 infected cells is mainly because of UNG2 depletion and that the residual uracil DNA glycosylase activity detected in HIV-1-infected samples is consistent with unaffected UNG1 activity in the extracts. On the other hand, decreased SMUG1 expression moderately participates in the loss of the enzymatic activity.
In chronically infected cells, both UNG2 expression and uracil DNA glycosylase activity are significantly impaired at the peak of retroviral replication. Interestingly, a nearly total abolition was repeatedly observed in cultures containing only ∼60% of p24-positive cells. There is currently no explanation for such discrepancy. However, we speculate that in HIV-1-infected cultures, UNG2 depletion might also occur in uninfected cells in a Vpr-dependent manner. Moreover, in HIV-1-infected cultures, UNG2 expression and the associated enzymatic activity are progressively recovered as viral replication decreases. However, according to our findings, Vpr-induced down-regulation of the uracil excision machinery, although transient, results in a significant uracilation of the host cell genome that is apparently partially repaired in chronically infected cells after full recovery of UNG2 expression and activity. The values reported for genomic uracil here, showing a shift from 20 (uninfected) to 66 AP sites (HIV-1-infected H9 cells) per 105 bp are in good agreement with those reported by other groups. Specifically, inhibition of UNG2 enzymatic activity with UGI in HEK 293T cells induced 60–140 AP sites/105 bp from a basal level of 20–30 AP sites/105 bp in control cells (69). Accordingly, UGI overexpression in the human glioma U251 cell line induced a 3-fold increase in mutation frequency (70).
At the moment, the consequences of UNG2 reduction and increased genome uracilation on the outcome of HIV-1-infected cells and their possible significance in HIV-1 pathogenesis remain unknown. However, both the presence of uracil in DNA and the lack of UNG2 expression are toxic for cells (71,72). Spontaneous cytosine deamination is estimated at around 70–200 events per day in the human genome and this ratio needs to be cumulated with the enzymatic deamination generated by cytidine deaminase and replicative incorporation of dUMP in DNA (2). DNA glycosylases (mainly UNG2 and SMUG1) are involved in reducing mutations resulting from these events and also from oxidative stress. Decreased UNG2 expression through RNA interference results in accumulation of DNA damage, decreased cell proliferation, cell hypersensitivity to genotoxic stress (10,73,74) and increased cell apoptosis, especially in neuronal cells (75,76) leading to neurodegeneration (77,78). Accumulation of uracil in the human genome is associated with cancer [for review see (72)]. Recently, it was reported that in chronic myeloid leukemia cells in the chronic phase, UNG2 depletion through a BCR-ABL-kinase-dependent mechanism is involved in the accumulation of mutations in the cell genome (79). Although accumulation of uracil (∼3600 bases per genome) is observed in fibroblasts isolated from Ung−/− mice, SMUG1 compensates for the absence of UNG activity in these animals (80,81). As this is not the case in human cells (82), UNG2 depletion is expected to have a stronger pathophysiological impact.
Based on our experimental model, uracil accumulation does not result from a modification of the cell cycle, increased cell apoptosis or decreased cell viability. Genomic DNA analysis reveals a partial uracil reduction to almost basal levels after the peak of retroviral replication. Restoration of UNG2 expression may account for such repair. As the cell cycle was unaffected in our experimental conditions, possible multiple occurrences of base transitions during DNA replication cannot be ruled out. The presence of cells with a high level of unrepaired uracil the genome of HIV-1-infected patients still remains to be investigated. Increased levels of 7,8-dihydro-8-oxoguanine, an oxidative lesion, in DNA and marked reduction in DNA glycosylase activity for the repair of oxidative base lesions were detected in CD4+ T cells from patients with advanced HIV-1 infection (83). As these features are efficiently compensated by highly active antiretroviral therapy, HIV-1 replication directly accounts for these perturbations. Altogether, these data highlight the necessity to investigate the outcome of alterations of the uracil repair machinery in HIV-infected patients and to decipher the contribution of DNA repair enzymes in the pathogenesis of HIV-1 infection. The possible role of DNA repair imbalance in the predisposition to non-AIDS-defining malignancies, the occurrence of which is markedly higher in HIV-infected people than in the general population, needs to be investigated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by institutional funding from the Centre National de la Recherche Scientifique (CNRS); Universities of Montpellier [UM1 and UM2]; French National Agency of Research Against AIDS (ANRS) and Sidaction. ANRS grant (to D.F.). Funding for open access charge: Agence National de Recherche sur le SIDA (ANRS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Antoine Gross for helpful discussions. We are grateful to W.C. Greene and. V. Planelles for providing the HIV and Vpr expression plasmids and to M. Tatsumi for the MAGIC-5B cell line. We thank G. Sluppaugh for the gift of anti-UNG2 antibodies.
REFERENCES
- 1.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 2.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA—general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Visnes T, Doseth B, Pettersen HS, Hagen L, Sousa MM, Akbari M, Otterlei M, Kavli B, Slupphaug G, Krokan HE. Uracil in DNA and its processing by different DNA glycosylases. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009;364:563–568. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doseth B, Ekre C, Slupphaug G, Krokan HE, Kavli B. Strikingly different properties of uracil-DNA glycosylases UNG2 and SMUG1 may explain divergent roles in processing of genomic uracil. DNA Repair (Amst) 2012;11:587–593. doi: 10.1016/j.dnarep.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Boorstein RJ, Cummings A, Jr, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 9.Nagaria P, Svilar D, Brown AR, Wang XH, Sobol RW, Wyatt MD. SMUG1 but not UNG DNA glycosylase contributes to the cellular response to recovery from 5-fluorouracil induced replication stress. Mutat. Res. 2013;743–744:26–32. doi: 10.1016/j.mrfmmm.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An Q, Robins P, Lindahl T, Barnes DE. C –> T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagen L, Kavli B, Sousa MM, Torseth K, Liabakk NB, Sundheim O, Pena-Diaz J, Otterlei M, Horning O, Jensen ON, et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardeland U, Kunz C, Focke F, Szadkowski M, Schar P. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 2007;35:3859–3867. doi: 10.1093/nar/gkm337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kavli B, Andersen S, Otterlei M, Liabakk NB, Imai K, Fischer A, Durandy A, Krokan HE, Slupphaug G. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J. Exp. Med. 2005;201:2011–2021. doi: 10.1084/jem.20050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Noia JM, Williams GT, Chan DT, Buerstedde JM, Baldwin GS, Neuberger MS. Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 2007;204:3209–3219. doi: 10.1084/jem.20071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen S, Ericsson M, Dai HY, Pena-Diaz J, Slupphaug G, Nilsen H, Aarset H, Krokan HE. Monoclonal B-cell hyperplasia and leukocyte imbalance precede development of B-cell malignancies in uracil-DNA glycosylase deficient mice. DNA Repair (Amst) 2005;4:1432–1441. doi: 10.1016/j.dnarep.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhou PK, Sun Y, An J. Interaction between viral proteins and hosts and its disturbance in the cellular responses to ionising radiation. Int. J. Radiat. Biol. 2009;85:587–597. doi: 10.1080/09553000902954512. [DOI] [PubMed] [Google Scholar]
- 17.Fritz JV, Briant L, Mély Y, Bouaziz S, de Rocquigny H. HIV-1 viral protein r: from structure to function. Future Virol. 2010;5:607–625. [Google Scholar]
- 18.Sherman MP, Schubert U, Williams SA, de Noronha CM, Kreisberg JF, Henklein P, Greene WC. HIV-1 Vpr displays natural protein-transducing properties: implications for viral pathogenesis. Virology. 2002;302:95–105. doi: 10.1006/viro.2002.1576. [DOI] [PubMed] [Google Scholar]
- 19.Goh WC, Rogel ME, Kinsey M, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 20.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 21.Heinzinger NK, Bukinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl Acad. Sci. USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner DB. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J. Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J. Biol. Chem. 1998;273:13347–13352. doi: 10.1074/jbc.273.21.13347. [DOI] [PubMed] [Google Scholar]
- 24.Vodicka MA, Koepp DM, Silver PA, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agostini I, Navarro JM, Rey F, Bouhamdan M, Spire B, Vigne R, Sire J. The human immunodeficiency virus type 1 Vpr transactivator: cooperation with promoter-bound activator domains and binding to TFIIB. J. Mol. Biol. 1996;261:599–606. doi: 10.1006/jmbi.1996.0485. [DOI] [PubMed] [Google Scholar]
- 26.Kino T, Tsukamoto M, Chrousos G. Transcription factor TFIIH components enhance the GR coactivator activity but not the cell cycle-arresting activity of the human immunodeficiency virus type-1 protein Vpr. Biochem. Biophys. Res. Commun. 2002;298:17–23. doi: 10.1016/s0006-291x(02)02442-7. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Mukherjee S, Jia F, Narayan O, Zhao LJ. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J. Biol. Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 28.Planelles V, Benichou S. Vpr and its interactions with cellular proteins. Curr. Top. Microbiol. Immunol. 2009;339:177–200. doi: 10.1007/978-3-642-02175-6_9. [DOI] [PubMed] [Google Scholar]
- 29.Jacotot E, Ravagnan L, Loeffler M, Ferri KF, Vieira HL, Zamzami N, Costantini P, Druillennec S, Hoebeke J, Briand JP, et al. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J. Exp. Med. 2000;191:33–46. doi: 10.1084/jem.191.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacotot E, Costantini P, Laboureau E, Zamzami N, Susin SA, Kroemer G. Mitochondrial membrane permeabilization during the apoptotic process. Ann. N. Y. Acad. Sci. 1999;887:18–30. doi: 10.1111/j.1749-6632.1999.tb07919.x. [DOI] [PubMed] [Google Scholar]
- 31.Stewart SA, Poon B, Jowett JB, Chen IS. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J. Virol. 1997;71:5579–5592. doi: 10.1128/jvi.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams WV, Green DR, Weiner DB. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor kappa B. Nat. Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 33.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogel ME, Wu LI, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai M, Zimmerman ES, Planelles V, Chen J. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 2005;79:15443–15451. doi: 10.1128/JVI.79.24.15443-15451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmerman ES, Chen J, Andersen JL, Ardon O, Dehart JL, Blackett J, Choudhary SK, Camerini D, Nghiem P, Planelles V. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and gamma-H2AX focus formation. Mol. Cell. Biol. 2004;24:9286–9294. doi: 10.1128/MCB.24.21.9286-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakai-Murakami C, Shimura M, Kinomoto M, Takizawa Y, Tokunaga K, Taguchi T, Hoshino S, Miyagawa K, Sata T, Kurumizaka H, et al. HIV-1 Vpr induces ATM-dependent cellular signal with enhanced homologous recombination. Oncogene. 2007;26:477–486. doi: 10.1038/sj.onc.1209831. [DOI] [PubMed] [Google Scholar]
- 38.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 2003;278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 39.Bouhamdan M, Benichou S, Rey F, Navarro JM, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, et al. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J. Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J. Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Priet S, Gros N, Navarro JM, Boretto J, Canard B, Querat G, Sire J. HIV-1-associated uracil DNA glycosylase activity controls dUTP misincorporation in viral DNA and is essential to the HIV-1 life cycle. Mol. Cell. 2005;17:479–490. doi: 10.1016/j.molcel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J. Biol. Chem. 2004;279:28419–28425. doi: 10.1074/jbc.M403875200. [DOI] [PubMed] [Google Scholar]
- 43.Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 2005;79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 2007;282:11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- 46.Fenard D, Houzet L, Bernard E, Tupin A, Brun S, Mougel M, Devaux C, Chazal N, Briant L. Uracil DNA Glycosylase 2 negatively regulates HIV-1 LTR transcription. Nucleic Acids Res. 2009;37:6008–6018. doi: 10.1093/nar/gkp673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrofelbauer B, Hakata Y, Landau NR. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl Acad. Sci. USA. 2007;104:4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn J, Vu T, Novince Z, Guerrero-Santoro J, Rapic-Otrin V, Gronenborn AM. HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J. Biol. Chem. 2010;285:37333–37341. doi: 10.1074/jbc.M110.133181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen X, Casey Klockow L, Nekorchuk M, Sharifi HJ, de Noronha CM. The HIV1 protein Vpr acts to enhance constitutive DCAF1-dependent UNG2 turnover. PLoS One. 2012;7:e30939. doi: 10.1371/journal.pone.0030939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weil AF, Ghosh D, Zhou Y, Seiple L, McMahon MA, Spivak AM, Siliciano RF, Stivers JT. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl Acad. Sci. USA. 2013;110:E448–E457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan N, O'Day E, Wheeler LA, Engelman A, Lieberman J. HIV DNA is heavily uracilated, which protects it from autointegration. Proc. Natl Acad. Sci. USA. 2011;108:9244–9249. doi: 10.1073/pnas.1102943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones KL, Roche M, Gantier MP, Begum NA, Honjo T, Caradonna S, Williams BR, Mak J. X4 and R5 HIV-1 have distinct post-entry requirements for uracil DNA glycosylase during infection of primary cells. J. Biol. Chem. 2010;285:108603–186145. doi: 10.1074/jbc.M109.090126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 54.Zhou T, Dang Y, Baker JJ, Zhou J, Zheng YH. Evidence for Vpr-dependent HIV-1 replication in human CD4+ CEM.NKR T-cells. Retrovirology. 2012;9:93. doi: 10.1186/1742-4690-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krosky DJ, Bianchet MA, Seiple L, Chung S, Amzel LM, Stivers JT. Mimicking damaged DNA with a small molecule inhibitor of human UNG2. Nucleic Acids Res. 2006;34:5872–5879. doi: 10.1093/nar/gkl747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svilar D, Vens C, Sobol RW. Quantitative, real-time analysis of base excision repair activity in cell lysates utilizing lesion-specific molecular beacons. J. Vis. Exp. 2012;66:e4168. doi: 10.3791/4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang YL, Krosky DJ, Seiple L, Stivers JT. Uracil-directed ligand tethering: an efficient strategy for uracil DNA glycosylase (UNG) inhibitor development. J. Am. Chem. Soc. 2005;127:17412–17420. doi: 10.1021/ja055846n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cone R, Bonura T, Friedberg EC. Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2. Purification and preliminary characterization. J. Biol. Chem. 1980;255:10354–10358. [PubMed] [Google Scholar]
- 59.Visnes T, Akbari M, Hagen L, Slupphaug G, Krokan HE. The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair (Amst) 2008;7:1869–1881. doi: 10.1016/j.dnarep.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 60.Selig L, Benichou S, Rogel ME, Wu LI, Vodicka MA, Sire J, Benarous R, Emerman M. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajan D, Wildum S, Rucker E, Schindler M, Kirchhoff F. Effect of R77Q, R77A and R80A changes in Vpr on HIV-1 replication and CD4 T cell depletion in human lymphoid tissue ex vivo. AIDS. 2006;20:831–836. doi: 10.1097/01.aids.0000218546.31716.7f. [DOI] [PubMed] [Google Scholar]
- 62.Elder RT, Zhu X, Priet S, Chen M, Yu M, Navarro JM, Sire J, Zhao Y. A fission yeast homologue of the human uracil-DNA-glycosylase and their roles in causing DNA damage after overexpression. Biochem. Biophys. Res. Commun. 2003;306:693–700. doi: 10.1016/s0006-291x(03)01036-2. [DOI] [PubMed] [Google Scholar]
- 63.Langevin C, Maidou-Peindara P, Aas PA, Jacquot G, Otterlei M, Slupphaug G, Benichou S. HIV-1 Vpr modulates cellular expression of UNG2 via a negative transcriptional effect. J. Virol. 2009;83:10256–10263. doi: 10.1128/JVI.02654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15:119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Pyles RB, Thompson RL. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 1994;68:4963–4972. doi: 10.1128/jvi.68.8.4963-4972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Worrad DM, Caradonna S. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J. Virol. 1988;62:4774–4777. doi: 10.1128/jvi.62.12.4774-4777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prichard MN, Duke GM, Mocarski ES. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J. Virol. 1996;70:3018–3025. doi: 10.1128/jvi.70.5.3018-3025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo Y, Walla M, Wyatt MD. Uracil incorporation into genomic DNA does not predict toxicity caused by chemotherapeutic inhibition of thymidylate synthase. DNA Repair (Amst) 2008;7:162–169. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Radany EH, Dornfeld KJ, Sanderson RJ, Savage MK, Majumdar A, Seidman MM, Mosbaugh DW. Increased spontaneous mutation frequency in human cells expressing the phage PBS2-encoded inhibitor of uracil-DNA glycosylase. Mutat Res. 2000;461:41–58. doi: 10.1016/s0921-8777(00)00040-9. [DOI] [PubMed] [Google Scholar]
- 71.Hagen L, Pena-Diaz J, Kavli B, Otterlei M, Slupphaug G, Krokan HE. Genomic uracil and human disease. Exp. Cell Res. 2006;312:2666–2672. doi: 10.1016/j.yexcr.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 72.Sousa MM, Krokan HE, Slupphaug G. DNA-uracil and human pathology. Mol. Aspects Med. 2007;28:276–306. doi: 10.1016/j.mam.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 73.Studebaker AW, Ariza ME, Williams MV. Depletion of uracil-DNA glycosylase activity is associated with decreased cell proliferation. Biochem. Biophys. Res. Commun. 2005;334:509–515. doi: 10.1016/j.bbrc.2005.06.118. [DOI] [PubMed] [Google Scholar]
- 74.Pulukuri SM, Knost JA, Estes N, Rao JS. Small interfering RNA-directed knockdown of uracil DNA glycosylase induces apoptosis and sensitizes human prostate cancer cells to genotoxic stress. Mol. Cancer Res. 2009;7:1285–1293. doi: 10.1158/1541-7786.MCR-08-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kruman, Schwartz E, Kruman Y, Cutler RG, Zhu X, Greig NH, Mattson MP. Suppression of uracil-DNA glycosylase induces neuronal apoptosis. J. Biol. Chem. 2004;279:43952–43960. doi: 10.1074/jbc.M408025200. [DOI] [PubMed] [Google Scholar]
- 76.Kronenberg G, Harms C, Sobol RW, Cardozo-Pelaez F, Linhart H, Winter B, Balkaya M, Gertz K, Gay SB, Cox D, et al. Folate deficiency induces neurodegeneration and brain dysfunction in mice lacking uracil DNA glycosylase. J. Neurosci. 2008;28:7219–7230. doi: 10.1523/JNEUROSCI.0940-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones GJ, Barsby NL, Cohen EA, Holden J, Harris K, Dickie P, Jhamandas J, Power C. HIV-1 Vpr causes neuronal apoptosis and in vivo neurodegeneration. J. Neurosci. 2007;27:3703–3711. doi: 10.1523/JNEUROSCI.5522-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Power C, Hui E, Vivithanaporn P, Acharjee S, Polyak M. Delineating HIV-associated neurocognitive disorders using transgenic models: the neuropathogenic actions of Vpr. J. Neuroimmune Pharmacol. 2012;7:319–331. doi: 10.1007/s11481-011-9310-7. [DOI] [PubMed] [Google Scholar]
- 79.Slupianek A, Falinski R, Znojek P, Stoklosa T, Flis S, Doneddu V, Pytel D, Synowiec E, Blasiak J, Bellacosa A, et al. BCR-ABL1 kinase inhibits uracil DNA glycosylase UNG2 to enhance oxidative DNA damage and stimulate genomic instability. Leukemia. 2012;27:629–634. doi: 10.1038/leu.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 81.Andersen S, Heine T, Sneve R, Konig I, Krokan HE, Epe B, Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 82.Doseth B, Visnes T, Wallenius A, Ericsson I, Sarno A, Pettersen HS, Flatberg A, Catterall T, Slupphaug G, Krokan HE, et al. Uracil-DNA glycosylase in base excision repair and adaptive immunity: species differences between man and mouse. J. Biol. Chem. 2011;286:16669–16680. doi: 10.1074/jbc.M111.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aukrust P, Luna L, Ueland T, Johansen RF, Muller F, Froland SS, Seeberg EC, Bjoras M. Impaired base excision repair and accumulation of oxidative base lesions in CD4+ T cells of HIV-infected patients. Blood. 2005;105:4730–4735. doi: 10.1182/blood-2004-11-4272. [DOI] [PubMed] [Google Scholar]
- 84.DeHart JL, Zimmerman ES, Ardon O, Monteiro-Filho CM, Arganaraz ER, Planelles V. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol. J. 2007;4:57. doi: 10.1186/1743-422X-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rieger AM, Nelson KL, Konowalchuk JD, Barreda DR. Modified annexin V/propidium iodide apoptosis assay for accurate assessment of cell death. J. Vis. Exp. 2011;24 doi: 10.3791/2597. pii.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.