Abstract
Control over the simultaneous delivery of different functionalities and their synchronized intracellular activation can greatly benefit the fields of RNA and DNA biomedical nanotechnologies and allow for the production of nanoparticles and various switching devices with controllable functions. We present a system of multiple split functionalities embedded in the cognate pairs of RNA–DNA hybrids which are programmed to recognize each other, re-associate and form a DNA duplex while also releasing the split RNA fragments which upon association regain their original functions. Simultaneous activation of three different functionalities (RNAi, Förster resonance energy transfer and RNA aptamer) confirmed by multiple in vitro and cell culture experiments prove the concept. To automate the design process, a novel computational tool that differentiates between the thermodynamic stabilities of RNA–RNA, RNA–DNA and DNA–DNA duplexes was developed. Moreover, here we demonstrate that besides being easily produced by annealing synthetic RNAs and DNAs, the individual hybrids carrying longer RNAs can be produced by RNA polymerase II-dependent transcription of single-stranded DNA templates.
INTRODUCTION
We have developed a novel approach that separates functional nucleic acid strands and conditionally restores them to their original function (1). Conceptually, it resembles the widely used split-protein systems (2–4). To reveal the full potential of this technique, herein we propose to simultaneously split and restore multiple functionalities upon re-association of two cognate RNA–DNA hybrids (Figure 1). Besides the tighter control over synchronized activation, this novel approach may also help to resolve some problems associated with the clinical delivery of RNA-based therapies (5), including intravascular degradation (6) [will be significantly reduced for RNA–DNA hybrids (1)] and pharmacodynamics [fluorescent tags can be activated assisting in (Förster resonance energy transfer (FRET)) imaging of delivery and response (1)]. Moreover, additional chemical functionalities (targeting molecules, fluorescent tags, chemical analogs of nucleotides, etc.) can be introduced through direct modifications of the DNA strands in individual RNA–DNA hybrids thus, not interfering with the functions of the released RNA-based components. The new technique described here is anticipated to greatly benefit and expand the emerging fields of RNA and DNA nanotechnology (7–13).
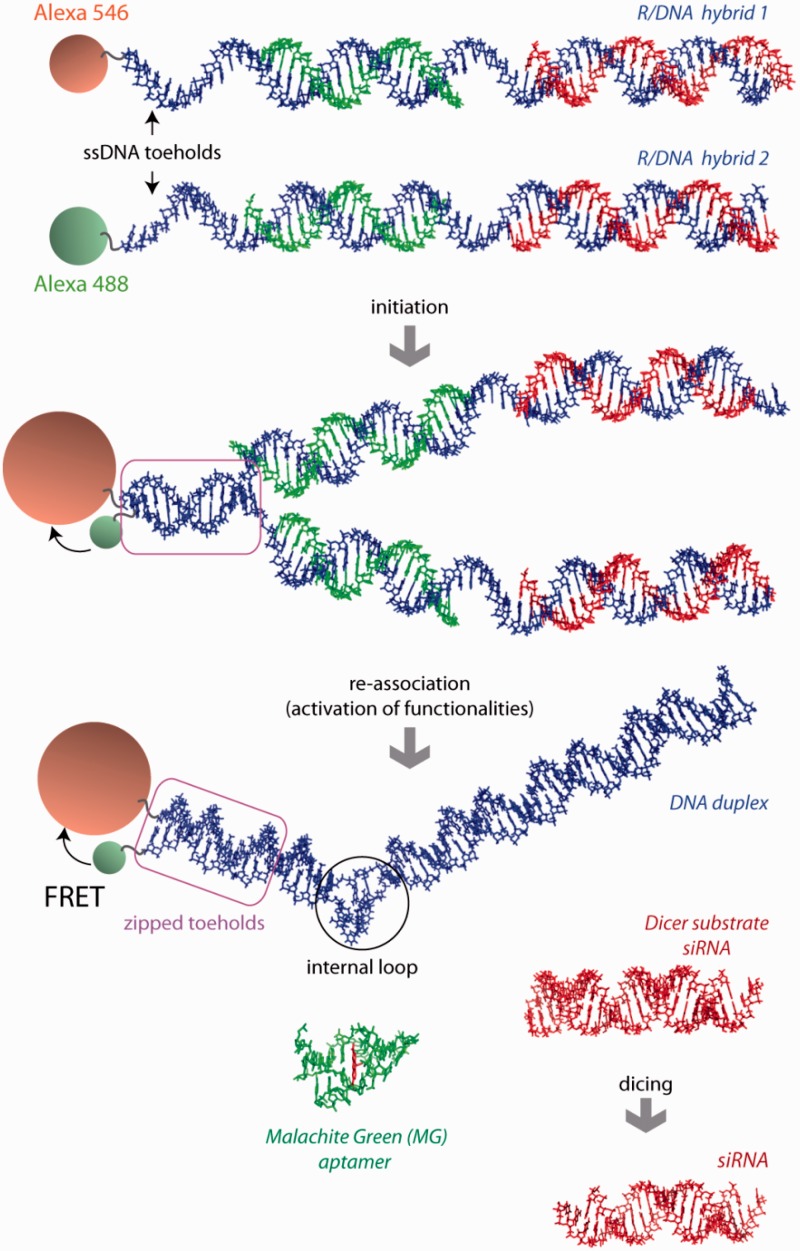
Figure 1.
Schematic representation of RNA–DNA hybrid re-association and release of multiple functionalities: FRET response, DS siRNA (in red) and MG RNA aptamer (in green). Three-dimensional (3D) structure of the two-stranded MG aptamer (in green) contains a bound dye (in red). PDB ID: 1f1t. Due to asymmetry of the MG aptamer, the resulting DNA duplex is also asymmetric and contains an internal loop.
MATERIALS AND METHODS
RNA and DNA sequences
All oligonucleotides were purchased from Integrated DNA Technologies, Inc. The RNA and DNA sequences are listed in the Supporting Information.
Hybrid RNA–DNA duplexes assemblies and native PAGE
The RNA and DNA oligonucleotides were mixed in hybridization buffer [89 mM Tris, 80 mM Boric Acid (pH 8.3), 10 mM magnesium acetate] at 100 nM final concentrations and incubated in a heat block at 95°C for 2 min followed by snap cool to room temperature and incubation for 30 min. Native polyacrylamide gel electrophoresis (PAGE) experiments were performed as described (14,15). Typically, assembly experiments reported were analyzed at 10°C on 7% (29:1) native polyacrylamide gels in the presence of 89 mM Tris-borate, pH 8.3, 2 mM Mg(OAc)2. A Hitachi FMBIO II Multi-View Imager was used to visualize SYBR Gold or Ethidium Bromide stained RNA–DNA hybrids.
Re-association and recombinant human Dicer assay
Hybrid RNA–DNA duplexes were prepared as described above to a final concentration of 3 μM. For re-association and dicing experiments, samples were incubated for 4 h at 37°C with or without recombinant human turbo dicer enzyme kit (Genlantis), containing an ultra-active form of human recombinant dicer enzyme, according to the manufacturer’s suggested protocol. Dicing reactions were analyzed on 2 mM Mg(OAc)2 native 7% PAGE (described above).
Release of Malachite Green aptamer
All fluorescent studies of Malachite Green (MG) aptamer release upon RNA–DNA hybrid re-association (at 100 nM final each) were carried out in hybridization buffer during the incubation at 37°C. Increase in MG emission signal (compared to free MG and MG in the presence of individual hybrids) confirms the release of RNA strands, their further assembly and formation of active MG aptamer. For all samples, the excitation was set at 425 nm.
FRET studies
To assess the re-association of RNA–DNA hybrids in vitro, FRET measurements were performed using a FluoroMax3 (Jobin-Yvon, Horiba). For all the experiments, the excitation wavelength was set at 460 nm, and the excitation and emission slit widths were set at 2 nm. In a first set of experiments, complementary DNAs were modified with Alexa488 or Alexa546. To follow the kinetics of recombination, an Alexa488 RNA–DNA hybrid containing sense RNA was first incubated for 2 min at 37°C and an Alexa543 RNA–DNA hybrid containing antisense RNA was then added in equimolar amounts as specified in the text. Upon excitation at 460 nm, the emissions at 520 and 570 nm were recorded simultaneously every 30 s to follow the process of re-association through FRET measurements. Static measurements were also performed after 3 h of co-incubation of equimolar amounts of the two fluorescently labeled hybrids. The decrease of Alexa488 fluorescence was fitted in Sigmaplot. A linear regression was applied to fit the data to a single exponential decay equation with three parameters as follows: y = y0 + ae-kt
Transfection of human breast cancer cells
For assaying the delivery of functional RNA–DNA hybrids, human breast cancer cell line MDA-MB-231 [with or without enhanced green fluorescent protein (eGFP)] was grown in D-MEM media (Gibco BRL) supplemented with 10% FBS and penicillin–streptomycin in a 5% CO2 incubator. All in vitro transfections in this project were performed using L2K purchased from Invitrogen. RNA–DNA hybrids were pre-incubated at 30°C with L2K. Prior to each transfection, the cell media was swapped with OPTI-MEM, and prepared hybrid/L2K (or control siRNA/L2K) complexes were added. The cells were incubated for 4 h followed by the media change (D-MEM, 10%FCS, 1% pen-strep) (16).
Interferon activation assay
Type I interferon (IFN) activity was measured using THP-1 cells engineered to express secreted alkaline phosphatase in response to type I IFN (Invivogen). THP-1 cells deficient for STING (stimulator of IFN genes) expression (Invivogen) were used as controls when examining DNA-dependent type IFN induction. THP-1 cells were cultivated in RPMI 1640 with 10% FBS, 10 mM HEPES, 1 mM pyruvate, penicillin–streptomycin and normocin (100 µg/ml). THP-1 cells were differentiated with 40 ng/ml phorbol 12-myristate 13-acetate (PMA) (Sigma) for 24 h and incubated for an additional 24 h in media lacking PMA prior to transfection. Nucleic acids were transfected using Lipofectamine LTX and PLUS or L2K reagents according to the manufacturer’s protocol (Invitrogen) at a final concentration of 10 nM. Culture supernatants were harvested 24 h post-transfection and assayed for alkaline phosphatase activity by incubating with the QUANTI-BLUE substrate (Invivogen) and measuring absorbance at 625 nm using a spectrophotometer.
Microscopy
To assess the re-association of R/DNA hybrids in cells, measurements were performed using a LSM 710 confocal microscope (Carl Zeiss) with a 63×, 1.4 NA magnification lens. MDA-MB-231 cells were plated in glass bottom petri dishes (Ibidi, Germany) and subjected to transfection with RNA–DNA hybrids as described above. In a first set of experiments, RNA–DNA hybrids individually modified with Alexa488 and Alexa546 were co-transfected into cells as described above. On the next day, the samples were fixed by incubation in 4% paraformaldehyde for 20 min at room temperature. Images of the cells were then taken to assess the appearance of FRET within the sample. For Alexa488 imaging, the 488 nm line of an Argon laser was used as excitation, and the emission was collected between 493 and 557 nm. For Alexa546 imaging, a DPSS 561 laser was used for excitation, and emission was collected between 566 and 680 nm. To evaluate the sensitized emission through FRET, images were taken exciting the sample with the 488 nm line and collecting emission between 566 and 680 nm. Because of spectral overlap, the FRET signal is contaminated by donor emission into the acceptor channel and by the excitation of acceptor molecules by the donor excitation wavelength. This bleed through was assessed through measurements performed with samples transfected with individual dyes and mathematically removed from the images of FRET. In another set of experiments, a DNA duplex containing one strand modified with Alexa 488 and another modified with Iowa Black FQ was used. This duplex was either transfected alone or co-transfected with an RNA–DNA hybrid able to recombine with the duplex. Alexa 488 fluorescence was monitored as described above. All images were taken with a pinhole adjusted to 1 airy unit.
Flow cytometry experiments
For statistical analysis with flow cytometry experiments, the MDA-MB-231 231 (with or without eGFP) cells grown in 12-well plates (10 × 104 cells per well) were lifted with cell dissociation buffer, washed twice with PBS and the level of expression of eGFP was determined by fluorescence-activated cell sorting (FACS) analysis on a FACScalibur flow cytometer (BD Bioscience). At least 20 000 events were collected and analyzed using the Cell quest software.
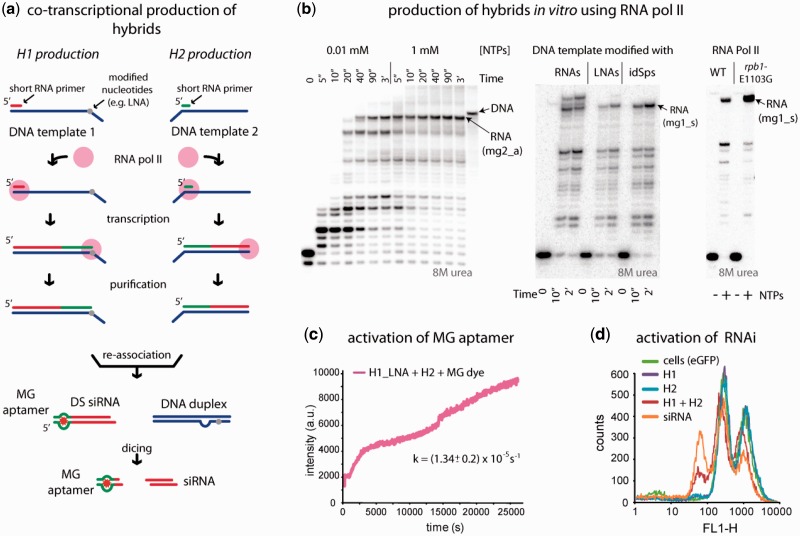
Co-transcriptional production of RNA–DNA hybrids
Wild-type (WT) and mutant Pol II variants were produced from the strains containing histidine-tagged Rpb3; WT RPB1 was deleted in the strain carrying rpb1-E1103G (17). For preliminary experiments performed in analytical scale, purified Pol II preparations were used. Reactions were done essentially as described in Kireeva et al. (18). In a typical experiment, the elongation complexes were formed with 10 µg Pol II immobilized on 25 µl Ni-NTA agarose beads (Qiagen) washed with transcription buffer (20 mM Tris–HCl, pH 7.9, 40 mM KCl, 5 mM MgCl2, 1 µM ZnCl2, 3 mM 2-mercaptoethanol). RNA primer was annealed to DNA template in transcription buffer at 1 µM final concentration, and 30 pmol of the short RNA–DNA hybrid was incubated for 10 min with immobilized Pol II. To remove the non-specifically bound RNA and DNA the immobilized elongation complexes were incubated in transcription buffer containing 300 mM KCl for 10 min, and then washed with regular transcription buffer. Transcription was started by addition of NTPs and stopped by addition of the gel loading solution (25 mM EDTA, 8 M urea). To analyze the release of RNA–DNA hybrids to supernatant, KCl was added to 300 mM final concentration after transcription was completed, the beads were precipitated by brief centrifugation, and the supernatant was removed. Transcription products were resolved in 20% denaturing polyacrylamide gel (19:1 acrylamide to methylene-bis-acrylamide ratio) in TBE and detected and quantified with Typhoon Phosphorimager (GE Healthcare).
To obtain extended RNA–DNA hybrids in the amounts sufficient for functional assays, WT and E1103G Pol II variants were immobilized on 1 ml Ni-NTA agarose cartridges (GE Healthcare) directly from whole cell lysates obtained from 0.5 l saturated yeast cultures as described in Kireeva et al. (19). The lysates were loaded to the cartridges using syringes attached to the cartridges (Supplementary Figure S8), and then the beads were washed consecutively with 10 ml each of the lysis buffer (150 mM Tris–acetate, pH 7.9, 50 mM potassium acetate, 5 mM MgCl2, 10 µM ZnCl2, 2 mM 2-mercaptoethanol, 0.5 mM EDTA) containing 1 M potassium acetate and with transcription buffer. The elongation complexes were formed by loading 1 ml of 1 µM primer/template hybrid in transcription buffer through the cartridge over 15–20 min, followed by washing with 5 ml transcription buffer containing 300 mM KCl, and 5 ml transcription buffer. Transcription was started by loading 1 ml of 1 mM NTPs through the cartridge. The resulting R/DNA hybrids were eluted with transcription buffer containing 300 mM KCl. Pol II was regenerated by washing with lysis buffer containing 1 M potassium acetate. The cartridges with immobilized Pol II were re-used for additional transcription rounds at least four times.
RESULTS
Rational design of hybrids and nomenclature
As a proof of principle, we designed several pairs of RNA–DNA hybrids, which upon re-association concurrently activate multiple functionalities including RNAi (targeting eGFP), a MG aptamer and a FRET signal. The design principle of the split functionalities is shown in Figure 1. For RNAi, we split either an asymmetric (25/27 nts) Dicer substrate short interfering RNA [DS siRNA (20)] or conventional (21/21 nts) siRNA. DS siRNA has to undergo the process of dicing (21,22) prior to loading into the RNA-induced silencing complex (RISC) required for RNAi (23–27). Dicer is an RNaseIII-like enzyme which is incapable of processing the RNA–DNA hybrids (Supplementary Figure S1) to make them loadable into the RISC (1,28). In addition, FRET dye pairs (Alexa488 and Alexa546) and the MG aptamer are also split between the same pair of RNA–DNA hybrids. Each of the DNA strands in the hybrids is decorated with a ssDNA toehold. The toeholds in the cognate hybrids are complementary to each other. When the cognate RNA–DNA hybrids are mixed together, hybridization of the toeholds initializes re-association of the DNA strands, displaces the RNAs from the RNA–DNA hybrid and induces re-association of the RNA duplexes. This results in FRET induction and release of the DS siRNA and MG aptamer. The complementary ssDNA toeholds in the hybrids are designed to avoid any stable secondary structures. The driving force for re-association after toehold zipping is the difference in free energies between the initial hybrids and the final RNA and DNA duplexes.
To aid in the prediction and analysis of the RNA–DNA hybrid constructs, we developed two computational approaches for predicting RNA–DNA interactions and one approach for designing RNA–DNA hybrids. The first interaction prediction method is a stochastic simulation algorithm. It provides the user with insights into the kinetic pathways involved in hybrid formation and hybrid re-association. It should be noted however, that this algorithm is relatively time consuming. It is in essence a ‘one bead per nucleotide’ method that keeps track of nucleotide base pairing as well as nucleotide–nucleotide distance constraints (minimum and maximum allowed distances as a means for determining entropic contributions). The system determines the next base pairing folding event in a probabilistic manner as a function of the maximum allowed spatial distance between any two nucleotides. The system is able to score RNA–RNA, DNA–DNA and RNA–DNA base pairing simultaneously. In addition, the program is a two-time-scale simulation system that balances regular and rare folding events. The program can simulate multi-pot assembly protocols and kinetic folding behaviors with possible differences in the starting states. An addition to StructureLab is used to visualize the folding and unfolding events (29). This approach is described in more detail in the Supporting Information.
A second relatively fast approach for multi-strand secondary structure prediction focuses on equilibrium thermodynamics as opposed to kinetics. The algorithm is deterministic and performs an innovative priority-queue based approach for the conformational search of nucleotide structures. The search is used for identifying the minimum free energy structure as well as for approximating probabilities of complex formation using a partition function approach. This methodology then can also be used for estimating concentrations of different RNA, DNA and hybrid complexes in solution. One advantage compared to existing multi-strand prediction approaches is that pseudo-knots of arbitrary complexity can be considered. More detailed information can be found in the Supporting Information.
The computational design of the presented RNA–DNA hybrids is straightforward, because the chosen siRNA sequence determines the majority of the reverse-complementary sequence of the DNA strands. The design of the DNA toeholds involves the design of de novo DNA sequences, such that the toehold regions are not frequently involved in stable secondary structure formation. This is accomplished using a fragment approach, detailed in the Supporting Information. The designs and structures generated by these algorithms are depicted in Supplementary Scheme S2.
In this work, the release and re-association of the split functionalities is kinetically studied through inclusion of an RNA aptamer, which behaves as a fluorescent reporter (30). We split the MG aptamer (31–34) into two domains and embedded these constructs in several non-overlapping positions within our cognate hybrids. In its reassembled form, this aptamer binds to the triphenylmethane dye, MG, with nanomolar affinity, which results in a substantial increase in photoemission (14,35) while also providing an accurate gauge of recombination rates in this study. Relative to the toehold, positions of the MG aptamer are varied within the different hybrids to determine whether the efficiency of RNA and DNA re-association and split functionality release is affected by the reporter location.
The naming of each hybrid will be used to illustrate the positioning of the functionality relative to the single-stranded toehold of the DNA strand. For example, H1(mg1_sDS) is a hybrid with one strand of the split MG aptamer located next to the toehold and the sense strand of DS siRNA embedded downstream. The cognate hybrid of H1(mg1_sDS) will be H2(mg2_aDS), which contains the second strand of the MG aptamer and the DS siRNA antisense strand. Hybrids consisting of two or three siRNA functionalities will be referred to in the same fashion, e.g. H1(mg1_sDSsDS) or H1(sDSsDSsDS). Sequences of the DNA and RNA tested in this work are listed in Supporting Information.
Hybrid re-association and release of split functionalities
To verify the simultaneous release of RNA-based functionalities programmed within RNA–DNA hybrids (schematically presented in Figure 2a), native PAGE was used. This commonly used technique allows visualizing the formation of re-association products—DNA duplex, MG aptamer and DS siRNA—based on their distinct mobilities. The results shown in Figure 2a (Supplementary Figure S2 for siRNA) demonstrate the conversion of cognate hybrids [H1(MG1_sDS) and H2(MG2_aDS)] into the more thermodynamically stable products after 3 h of incubation at 37°C. The additional higher order bands seen in the lane with two re-associated hybrids can be attributed to the formation of larger complexes due to the asymmetry of the DNA duplex. The MG aptamer duplex is asymmetric with one strand being six nucleotides longer. Therefore, the complementary DNA strands also vary in length, slowing DNA duplex formation after zipping of the ssDNA toeholds. This allows for multiple complementary DNA strands to anneal forming elongated duplexes. These results are consistent with the computational predictions (SI, computational approach). The computational approach also predicts (in addition to the expected siRNA and DNA duplexes) the existence of DNA quadruplexes (consisting of two copies of the sense DNA and two copies of the antisense DNA) in the case of the MG-aptamer binding DNA duplex. The corresponding higher-order band is indicated in Figure 2 and Supplementary Figures S1 and S2.
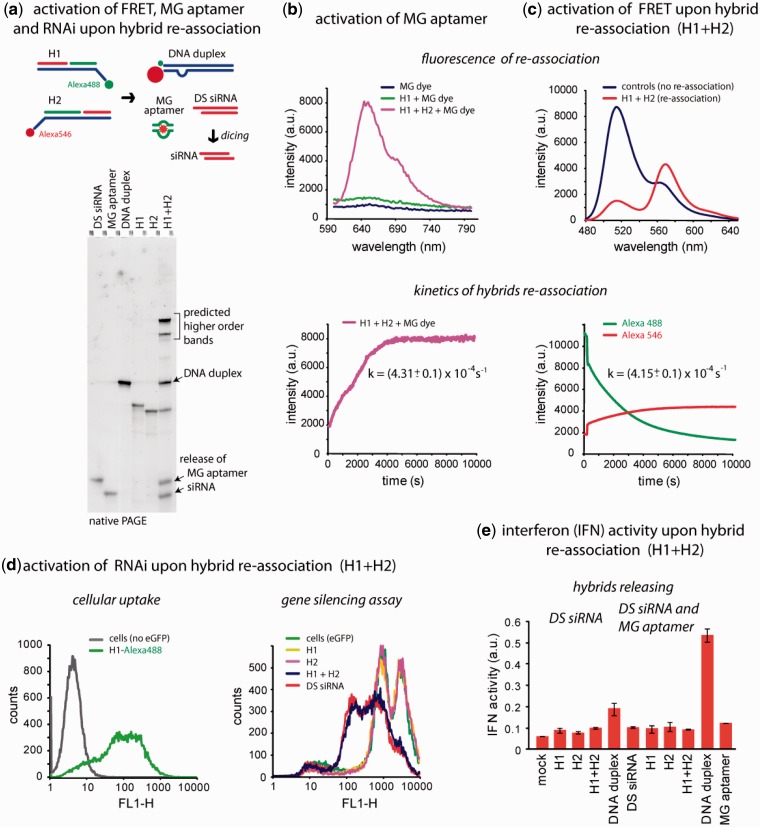
Figure 2.
Release of multiple functionalities (FRET, MG aptamer and DS siRNA) upon re-association of RNA–DNA hybrids. (a) Schematic of hybrid re-association and native PAGE demonstrating the release of DS siRNA and MG aptamer upon re-association of H1(mg1_sDS) and H2(mg2_aDS). Higher order bands on the gel (H1 + H2 lane) are in agreement with the computational predictions and can be attributed to asymmetry of the resulting DNA duplex (36). (b) Static and kinetics fluorescent experiments. Upper panel: activation of MG aptamer during the release. MG by itself is non-fluorescent (blue curve) and the presence of either one of the hybrids does not activate its fluorescence (green curve). However, re-association of two cognate hybrids leads to the release of individual MG aptamer strands, their assembly and further MG uptake leading to the significant increase of its fluorescence (magenta curve). Lower panel: kinetics time trace of the MG aptamer formation during hybrid re-association. (c) FRET activation. Upper panel: activation of FRET during re-association. Emission spectra of control DNA duplexes showing no FRET (blue curve) and re-associated hybrids with increased Alexa546 emission signal (red curve). Lower panel: FRET time traces during re-association of hybrids labeled with Alexa488 and Alexa546. (d) Cell culture experiments. Upper panel: cellular uptake of the fluorescently labeled hybrids. Lower panel: GFP knockdown assays. Three days after the transfection of cells, eGFP expression was statistically analyzed with flow cytometry experiments. As the control, DS siRNA duplexes against eGFP were used. (e) IFN activity was assessed using THP-1 IFN reporter cells that secrete alkaline phosphatase in response to type I IFN. Cells were transfected with hybrids, and culture supernatants were assayed for reporter activity after 24 h.
The fluorescence of the MG dye was monitored in the presence of the cognate hybrids. Prior to the acquisition of the emission spectrum, the cognate hybrids were incubated with the MG dye for 3 h at 37°C. The results show no emission of the MG dye when in the presence of individual hybrids; only hybrid re-association boosts the fluorescence. For the time trace experiments, the formation of the MG aptamer and subsequent fluorescence of the MG dye are tracked every 30 s. For the hybrids releasing the MG aptamer and one siRNA (Figure 2b), the complete re-association occurs after ∼90 min with the kMG aptamer release = (4.31 ± 0.1) × 10−4 s−1 (derived from the data fitting described in the Supporting Information). To confirm the release of MG aptamers irrespective of their position relative to the toehold and to estimate the rate of their release, we embedded aptamers at different positions in the hybrids (Supplementary Figure S3). Time trace experiments for these hybrids designed to release the MG aptamer together with two siRNAs show that the release rate is not significantly affected by the relative position of the MG aptamer. These results indirectly confirm the release of all RNA-based split functionalities regardless of their positions within the hybrids relative to the ssDNA toehold.
To visualize the delivery of the RNA–DNA hybrids and their re-association in solution as well as in cultured cells, the 3′-end of antisense-binding and the 5′-end of sense-binding DNA strands were fluorescently tagged with Alexa488 and Alexa546, respectively. These dyes are commonly used in FRET studies. When two fluorescently labeled hybrids are mixed, the double-stranded DNA (dsDNA) formation brings Alexa488 within the Förster distance (R0 = 6.31 nm) of Alexa546. As a result, when excited at 460 nm, the emission of Alexa546 increases while the signal of Alexa488 drops compared to the control DNA duplexes labeled with the single fluorescent dye, which are unable to re-associate (Figure 2c). Interestingly, the activation of FRET through DNA duplex formation reveals that the toehold zipping occurs at a rate comparable to the rate of the MG aptamer release [kFRET = (4.15 ± 0.1) × 10−4 s−1 and kMG aptamer release = (4.31 ± 0.1) × 10−4 s−1].
Intracellular re-association of hybrids
The ability of the cognate RNA–DNA hybrids to enter the cells and recombine releasing DS siRNAs was assessed with human breast cancer cells stably expressing eGFP (MDA-MB-231/eGFP) using flow cytometry experiments (Figure 2d). First, cells were co-transfected with only one hybrid at a time (H1 or H2) and 3 days after, the level of eGFP expression was analyzed with fluorescence microscopy and flow cytometry. All experiments were repeated at least three times. The results demonstrated no silencing in eGFP production caused by the individual hybrids. However, when cells were co-transfected with individually prepared complexes of L2K and individual cognate RNA–DNA hybrids (H1/L2K and H2/L2K), the level of silencing measured 3 days after was comparable to the silencing resulting from the transfections with control, pre-formed asymmetric DS siRNAs. The previously published results demonstrated that the individually pre-complexed with L2K hybrids were only able to re-associate within cells and not in medium (1). Interestingly, in the case of a conventional 21-mer siRNA release, individual hybrids containing antisense strands were able to trigger moderate silencing in eGFP production (Supplementary Figures S2c and S3c). These results are in agreement with previously published data (37,38).
While investigating whether these agents were cytotoxic or had immune stimulatory properties, we examined the type I IFN response (using a THP-1 IFN reporter cell line) following transfection of different nucleic acid duplexes. Transfection of individual hybrids or co-transfection of hybrids did not lead to significant IFN production, whereas direct transfection of dsDNA stimulated type I IFN (Figure 2e).
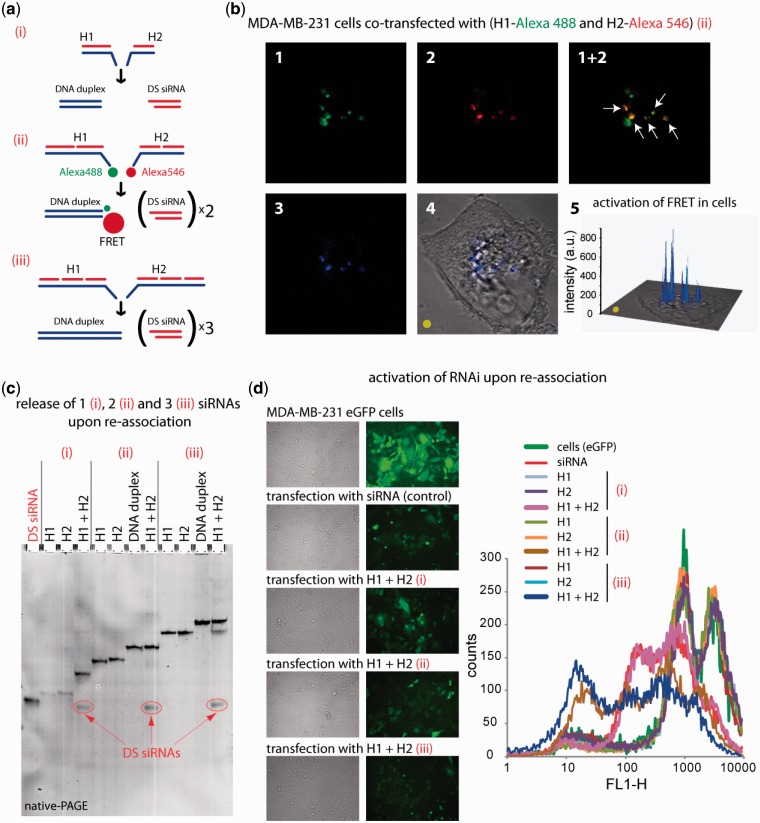
To visualize intracellular re-association, RNA–DNA hybrids labeled with Alexa488 and Alexa546 (Figure 3a) were co-transfected into MDA-MB-231 cells and examined by confocal microscopy the next day (Figure 3b). The overlap of the Alexa488 and Alexa546 fluorescence indicates that while a fraction of each dye is distributed in distinct endosomes, a significant amount of Alexa488 and Alexa546 are co-localized as evident from the appearance of the yellow signal. To further check whether FRET occurs within those endosomal compartments, the Alexa546 sensitized emission was imaged. The sample was excited at 488 nm, and the emission of Alexa546 was collected. The FRET signal [calculated as described previously (1)] remaining upon bleed through correction is presented in Figure 3b (4 and 5).
Figure 3.
RNA–DNA hybrid re-association with DS siRNA release and intracellular FRET tracking. (a) Schematic of different size hybrids re-association compared in these experiments [(i) H1(sDS) and H2(aDS); (ii) H1(sDS_sDS) and H2 (aDS_aDS); (iii) H1(sDS_sDS_sDS) and H2(aDS_aDS_aDS)]. (b) FRET experiments: cells were co-transfected with cognate hybrids labeled with Alexa 488 and Alexa 546, and images were taken on the next day. (c) Total SYBR Gold staining native PAGE demonstrating the release of DS siRNAs. Due to the full complementarities of the resulting DNA duplexes, there are no higher order bands observed on the gel. (d) GFP knockdown assays. Three days after the transfection of cells with auto-recognizing RNA–DNA hybrids programmed to release one [hybrids (i)], two [hybrids (ii)], and three [hybrids (iii)] DS siRNAs against eGFP, eGFP expression was observed by fluorescence microscopy and statistically analyzed with flow cytometry experiments. As the control, siRNA duplexes against eGFP were used. Image numbers in (b) correspond to: Alexa488 emission (1), Alexa546 emission (2), bleed-through corrected FRET image (3), differential interference contrast image with corrected FRET overlap (4), 3D chart representation of bleed-through corrected FRET image with the yellow dot indicating the correspondence (5).
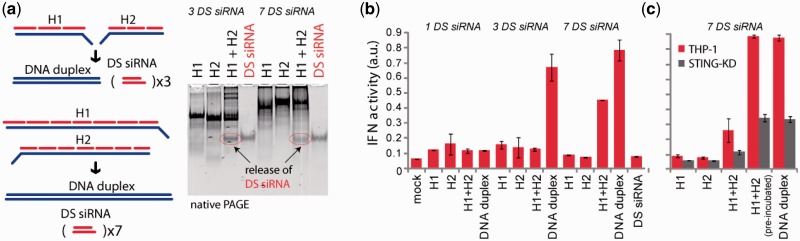
Figure 4.
Release of multiple DS siRNA targeting upon re-association of RNA–DNA hybrids. (a) Schematic of hybrid re-association and native PAGE demonstrating the release of three and seven DS siRNA. (b and c) IFN activity assessed using THP-1 IFN reporter cells that secrete alkaline phosphatase in response to type I IFN. Cells were transfected with hybrids, and cell culture supernatants were assayed for reporter activity after 24 h. (b) Cells were transfected with hybrids designed to release varying numbers of DS. (c) THP-1 and THP-1 deficient for STING (in gray) were transfected with hybrids releasing seven DS siRNAs.
Figure 5.
Co-transcriptional production of RNA-DNA hybrids by yeast RNA Pol II. (a) Schematics of R/DNA hybrid production. (b) Optimization of the full-length RNA synthesis. The RNA primers used for analytical transcription reactions were labeled at the 5′-end by phosphorylation with γ-[32P] ATP. RNA was annealed to DNA, and the elongation complexes were formed as described in Kireeva et al. (18). Transcription was initiated by addition of 1 mM NTPs unless indicated otherwise. Lower bands correspond to intermediate transcription stops. (c and d) Transcription products eluted from the Ni-NTA agarose cartridge containing immobilized Pol II were concentrated by ethanol precipitation, re-dissolved in standard buffer solution and MG2 aptamer (c) and DS siRNA (d) releases were monitored as described above. For (d), the lower relative efficiencies of the functionalities release can be explained by the lower concentrations of the initial hybrids and are consistent with published data (1).
It is anticipated, that the use of a higher number of DS siRNAs (and 21-mer siRNA) released upon hybrid re-association may potentially improve the silencing efficiency of the transfection experiments. To confirm this assumption, RNA–DNA hybrids designed to simultaneously release one, two and three DS siRNAs were tested (Figure 3c and d; Supplementary Figure S4). The results show the highest extent of silencing for hybrids releasing three DS siRNAs and the lowest for the release of only one DS siRNA. Interestingly, the silencing efficiencies for hybrids releasing two and three DS siRNAs are higher compared to control siRNAs.
To investigate the maximum number of DS siRNAs which can be released upon hybrids re-association, hybrids releasing one, three and seven DS siRNAs against previously tested glutathione S-transferase P1 (GSTP1) (1) were designed (Figure 4). Due to synthetic limitations of commercially available ssDNAs (maximum of 200 nts), the longest hybrids which we could obtain contained seven DS siRNAs. The successful release of DS siRNAs against GSTP1 was confirmed by native-PAGE (Figure 4a) and cell culture silencing experiments using A549 lung adenocarcinoma cells (Supplementary Figure S5).We also examined the type I IFN response for these longer hybrid systems. Hybrids releasing longer dsDNA substrates would be predicted to stimulate the type I IFN response, which has been shown previously to occur in response to dsDNA substrates larger than 60 bps (39). Importantly, addition of individual hybrids failed to induce type I IFN above background levels (Figure 4b). Furthermore, co-transfection of hybrids releasing fewer DS, and thus shorter dsDNA duplexes were not stimulatory for type I IFN induction (Figure 4b). However, simultaneous co-transfection of hybrids releasing seven DS siRNAs led to measurable type I IFN production (Figure 4b). Type I IFN induction was abrogated in cells deficient for STING, suggesting that intracellular re-association of hybrids led to DNA-dependent type I IFN production (Figure 4c). Induction of IFN was not as efficient as experiments where dsDNA was transfected into cells or where hybrids were pre-incubated together (and thus re-associated) prior to transfection (Figure 4c).
Optimization of hybrids design
The effects of introducing additional toeholds [total of two toeholds (2t) per each hybrid] and designing a completely complementary DNA duplex (through elongation of DNA strand for mg2_a), which was disrupted due to the asymmetry of MG aptamer, were also investigated (Supplementary Figure S6). These structural modifications aim to improve the kinetics of RNA–DNA hybrid re-association and thus enhance the release of RNA-based functionalities. The results indicate a slight improvement in the kinetics of MG aptamer release for hybrids with two toeholds (kMG aptamer release ∼4.8 × 10−4 s−1) and a higher increase in rates for elongated constructs (kMG aptamer release ∼ 6.6 × 10−4 s−1) compared to the ‘WT’ hybrids (kMG aptamer release ∼ 4.3 × 10−4 s−1). However, the silencing efficiency was not affected (data not shown). The results suggest the possibility to improve the release of functional aptamers through the rational design of RNA–DNA hybrids.
Co-transcriptional production of RNA–DNA hybrids
Previously, we developed protocols for co-transcriptional production of functional, chemically modified RNA nanoparticles using WT T7 RNA polymerase (9,10,40). However, this novel approach may not be suitable for functional RNA–DNA hybrid production (Figure 5a). The RNA–DNA hybrids characterized above contain short synthetic RNA oligonucleotides annealed to the synthetic single-strand DNA template; each RNA oligo encodes one separate split functionality. Extended RNA–DNA hybrids with a single long RNA molecule carrying several different split functionalities appear to be a promising alternative to short synthetic RNAs for applications in which coordinated delivery of the functionalities is essential. Pol II-directed synthesis of RNA–DNA hybrids containing ssDNA toeholds upstream from the RNA–DNA hybrid has been described previously (18). Pol II formed an elongation complex on the H2(mg2_aDS) template with the MG2 primer annealed to the single-stranded DNA oligo and reached the end of the template upon addition of NTPs (Figure 5b, left panel). The efficiency of the full-length runoff product formation was higher in 1 mM NTPs.
Next, we explored whether the downstream DNA toehold could be obtained by stopping transcription before RNA polymerase runs off the template by introducing two abasic sites (41); two RNA NMPs or two LNA NMPs into the template H1(mg1_sDS) 10 nts from its 5′-end. While the RNA bases stopped transcription inefficiently (Figure 5b, center panel; note the read-through after 2 min incubation with NTPs), abasic sites and the LNA modifications proved to be strong barriers to transcription elongation on a ssDNA template (Figure 5b, center panel). To ensure that the template modifications designed to stop transcription do not interfere with the DNA strand reannealing, the constructs carrying abasic sites and LNA NMPs were tested for siRNA release and MG aptamer activation (Supplementary Figure S7). Both constructs formed dsDNA and released the dsRNAs upon addition of H2 RNA–DNA hybrid as demonstrated by native PAGE (Supplementary Figure S7a) and were equally efficient in the release of siRNA against GFP (Supplementary Figure S7c). However, MG2 aptamer activation was significantly more efficient with H1(mg1_sDS) LNA than with a RNA–DNA hybrid containing abasic sites. Therefore, for larger scale enzymatic production of the extended RNA–DNA hybrid we chose the H1(mg1_sDS) LNA template. Hybridization of the RNA to DNA in the transcription product was confirmed by the high sensitivity of the transcription product to degradation by ribonuclease H (Supplementary Figure S8).
To further improve the homogeneity of the co-transcriptionally produced extended RNA–DNA hybrid, transcription pauses should be eliminated or decreased. To this end, we attempted to use a previously characterized fast mutant, rpb1-E1103G Pol II (17) in transcription reactions. The amount of the RNA product on H1(mg1_sDS) LNA template was significantly increased when transcription was done with the mutant Pol II (Figure 5b, right panel). However, when rpb1-E1103G Pol II transcribed the H2(mg2_aDS) DNA template, most of the accumulated RNA products were significantly longer than expected (data not shown), suggesting that the mutant Pol II might be switching from one single strand DNA template to another upon reaching the template end without releasing the template.
Enzymatic production of RNA–DNA hybrids for re-association assays was performed using rpb1-E1103G Pol II with the H1 template containing two LNA dNMPs. A WT yeast Pol II was used with the H2 template. Importantly, the Pol II elongation complexes containing 14-nt or longer RNA–DNA hybrid are unstable; the hybrid is quantitatively released from Pol II in 300 mM KCl (18). We exploited this property of Pol II and produced extended RNA–DNA hybrids free of Pol II and using transcription with Pol II immobilized on a Ni-NTA agarose cartridge. The procedure is described in detail in ‘Materials and Methods’ section and Supplementary Figure S8; it is different from conventional transcription by Pol II in solid phase (18) by the larger scale and immobilization of Pol II on a Ni-NTA agarose cartridge instead of loose Ni-NTA agarose suspension. Briefly, the primer hybridized to the template was passed through the Ni-NTA agarose cartridge containing immobilized Pol II. The resin was extensively washed with transcription buffer to remove non-specifically bound RNA and DNA and NTP solution was passed through the cartridge. The extended RNA–DNA hybrid was then eluted with the transcription buffer containing 300 mM KCl. The RNA–DNA hybrids obtained from the H1 and H2 templates were concentrated by ethanol precipitation, re-dissolved in hybridization buffer and subjected to functional tests for dsDNA and dsRNA re-association (Figure 5c and d; Supplementary Figure S9).
DISCUSSION
We demonstrated that the re-association of two hybrids leads to the simultaneous release of multiple functionalities with tunable efficiencies in vitro as well as inside mammalian cells. Using several different types of fluorescent reporters, the kinetics of RNA release and DNA re-association were found to be in accordance with one another. The enhanced silencing efficiencies observed with hybrids carrying multiple DS siRNA functionalities establishes how this novel technology provides an additional mechanism for delivering greater doses of nucleic acid functionalities while also limiting the possibility of degradation given its stable hybrid nature.
The successful assembly of a bipartite, asymmetric MG aptamer and the concomitant re-association of the DNA strands demonstrates how additional asymmetric RNA functionalities, such as miRNAs, can be built into this delivery system. The use of miRNAs would limit the off-target effects brought about by the unintended loading of the passenger strand into RISC. In contrast, by redesigning the hybrids to incorporate symmetric DNA strands, we were able to alter the rate of re-association illustrating how the structural and thus kinetic properties can be adjusted to meet varying cargo and delivery requirements. Additional improvements to the hybrid design can be developed and tested using the tunable, co-transcriptional production process we have developed. For example, aptamers can be built into the ends of the DNA strands opposite to the single-stranded toehold (Supplementary Figure S10). This provides an opportunity to test for cell-targeting capabilities via ligand-specific aptamers while also leaving the re-association capabilities of the RNA uncompromised. Our development of a novel RNA–DNA hybrid folding algorithm further facilitates this process of design manipulation by providing probabilistic determinations of thermodynamically stable secondary structures which may form during hybrid formation and subsequent re-association. The use of this predictive power vastly expands possibilities for modification and thus application of the hybrid technology.
Delivery of split functions in the context of two RNA–DNA hybrids has several important advantages such as the triggered release of multiple different RNAi inducers inside cells, simultaneous intracellular activation of other split functionalities (e.g. FRET), tight control over targeting specificity (e.g. hybrids can be decorated with cell surface specific recognition moieties such as aptamers; Supplementary Figure S10), real-time intracellular fluorescent tracking of the delivery and re-association of hybrids, introduction of additional functionalities without direct interference with RNAi processivity, increased retention time in biological fluids (1) and the possibility to fine-tune thermodynamics and kinetics of re-association by including chemical analogs (e.g. LNA) to the hybrid DNA strands. Importantly, to completely eliminate the possibility of gene silencing caused by individual hybrids, we recommend the use of asymmetric DS siRNAs (25/27-mers in length or longer). Their design can be assisted by newly developed software.
Notably, hybrids capable of releasing multiple DSs, and thus longer dsDNA byproducts, also stimulated a measureable type I IFN response in THP-1 cells. Previous studies have indicated that dsDNA longer than 60 bp is capable of inducing type I IFN through a STING-dependent mechanism (39). Here, we demonstrate that hybrids which can release up to three DSs do not induce significant IFN, even though the resulting dsDNA is long enough to trigger a response on its own. These data suggest that the kinetics of re-association of hybrids might be slower within cells, or that the dsDNA generated in cells upon re-association have relatively low concentrations locally. However, when hybrid lengths were increased to enable release of seven DSs, the resulting dsDNA was capable of inducing type I IFN production through a STING-dependent mechanism. These data suggest that there is a risk of immune stimulation as the number of DSs and corresponding DNA length are increased. Thus, in the future, hybrids could be designed which release a number of DSs that are sufficient for depletion of target genes, but fail to trigger host innate nucleic acid sensing pathways.
In this work we also demonstrated that RNA–DNA hybrids with RNA strands longer than 70 nt (which is a current sensible limit for chemical synthesis) are efficiently produced in vitro by Pol II. It opens new possibilities of combining multiple functionalities in just two RNA strands. This is, to our knowledge, the very first application of in vitro transcription by Pol II for preparative RNA production. It is important to mention, that in this setup both bacteriophage T7 RNA polymerase and Escherichia coli RNA polymerase could not extend the RNA primers to the desired full-length RNA–DNA hybrids (42). This unique ability of Pol II to extend the RNA primer to the end of the ssDNA template suggests that the primer-template complex might by itself serve as a therapeutic precursor. Provided that the pre-formed primer-template complexes are delivered to the nucleus, they would be converted to the full-length hybrids, subsequently releasing the functional RNAs and the byproduct DNAs. The advantage of using these hybrid precursors is that their high amounts can be chemically synthesized in vitro. To protect the ssDNA parts of the hybrid precursors against blood serum nucleases, phosphorothioate bonds can be introduced with further evaluation of Pol II activity in vitro. It is known that, for example, replication is not inhibited by 1–3 consecutive phosphorothioate bonds in the template DNA strand (43).
It is furthermore an advantage of the methodology, that the computational capabilities for structure prediction as well as for sequence design have been developed in parallel. The newly developed software aiming in design of hybrids will be available upon request and the website is currently under development. This adds further to the generality of the approach and simplifies the application of the RNA–DNA hybrid approach to novel cases. As previously shown in the case of DNA nanostructure design, computational approaches can significantly shorten and simplify the sequence design process (44).
The expression of long RNA–DNA hybrids from a vector is an exciting possibility to introduce various approaches of RNA nanotechnology to the expanding field of gene therapy (45). However, it will require significant modifications to the approach proposed in this work. The extended RNA–DNA hybrids (R-loops) are formed in mammalian cells, especially when RNA processing machinery is impaired (46). R-loops are frequently implicated in genomic instability and cancer (47). R-loops appear to have non-pathogenic biological functions, protecting CpG-containing DNA from methylation (48), promoting transcription termination (49), as well as recombination during immunoglobulin class-switch recombination (50). Extended RNA–DNA hybrids are stabilized by G-quadruplex structures in RNA and non-template DNA strands (51). It is possible to design a DNA sequence that will tend to form extended RNA–DNA hybrids while transcribed in the cell. However, production of two hybrids carrying split complementary RNAs appears to be challenging because the R-loops are preferentially formed when the transcript is G-rich, but not C-rich. Formation of ssDNA toeholds by transcription from a vector DNA poses additional challenges. For instance, an abaisic site or LNA introduced into the DNA vector before the transfection is likely to be repaired inside the cells; furthermore, these modifications will not survive DNA replication of viral-based vectors. Therefore, we currently focus mostly on possible therapeutic applications of RNA–DNA hybrids produced in vitro (by chemical synthesis or in vitro transcription).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [52–58].
ACKNOWLEDGEMENTS
The authors thank Lucyna Lubkowska for Pol II purification. Computational support by the NCI Advanced Biomedical Computing Center (ABCC) facility is highly appreciated. This work has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
FUNDING
This research was supported (in part) by the intramural research program of the NIH, National Cancer Institute, Center for Cancer Research. Funding for open access charge: NCI/CCR Intramural Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Afonin KA, Viard M, Martins AN, Lockett SJ, Maciag AE, Freed EO, Heldman E, Jaeger L, Blumenthal R, Shapiro BA. Activation of different split functionalities on re-association of RNA-DNA hybrids. Nat. Nanotechnol. 2013;8:296–304. doi: 10.1038/nnano.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassonnet P, Rolloy C, Neveu G, Vidalain PO, Chantier T, Pellet J, Jones L, Muller M, Demeret C, Gaud G, et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat. Methods. 2011;8:990–992. doi: 10.1038/nmeth.1773. [DOI] [PubMed] [Google Scholar]
- 4.Shekhawat SS, Ghosh I. Split-protein systems: beyond binary protein-protein interactions. Curr. Opin. Chem. Biol. 2011;15:789–797. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoerter JA, Krishnan V, Lionberger TA, Walter NG. siRNA-like double-stranded RNAs are specifically protected against degradation in human cell extract. PLoS One. 2011;6:e20359. doi: 10.1371/journal.pone.0020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinheiro AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L, Shapiro BA. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett. 2012;12:5192–5195. doi: 10.1021/nl302302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko SH, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Synergistic self-assembly of RNA and DNA molecules. Nat. Chem. 2010;2:1050–1055. doi: 10.1038/nchem.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 13.Afonin KA, Lindsay B, Shapiro BA. Engineered RNA Nanodesigns for Applications in RNA Nanotechnology. RNA Nanotech. 2013;1:1–15. doi: 10.2478/rnan-2013-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afonin KA, Danilov EO, Novikova IV, Leontis NB. TokenRNA: a new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. Chembiochem. 2008;9:1902–1905. doi: 10.1002/cbic.200800183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afonin KA, Leontis NB. Generating new specific RNA interaction interfaces using C-loops. J. Am. Chem. Soc. 2006;128:16131–16137. doi: 10.1021/ja064289h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T, Afonin KA, Viard M, Koyfman AY, Sparks S, Heldman E, Grinberg S, Linder C, Blumenthal RP, Shapiro BA. In silico, in vitro, and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Mol. Ther. Nucleic Acids. 2013;2:e80. doi: 10.1038/mtna.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kireeva ML, Komissarova N, Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J. Mol. Biol. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 19.Kireeva ML, Lubkowska L, Komissarova N, Kashlev M. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 2003;370:138–155. doi: 10.1016/S0076-6879(03)70012-3. [DOI] [PubMed] [Google Scholar]
- 20.Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 22.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 23.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 24.Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm. Res. 2011;28:2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]
- 25.Sashital DG, Doudna JA. Structural insights into RNA interference. Curr. Opin. Struct. Biol. 2010;20:90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 27.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro BA, Kasprzak W, Grunewald C, Aman J. Graphical exploratory data analysis of RNA secondary structure dynamics predicted by the massively parallel genetic algorithm. J. Mol. Graph. Model. 2006;25:514–531. doi: 10.1016/j.jmgm.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Leontis N, Afonin KA. RNA complexes featuring paranemic binding, methods of their production and sensors and analytical methods involving same. 2012 Patent application US8129516, Application number 12291697. [Google Scholar]
- 31.Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc. Natl Acad. Sci. USA. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolpashchikov DM. Binary malachite green aptamer for fluorescent detection of nucleic acids. J. Am. Chem. Soc. 2005;127:12442–12443. doi: 10.1021/ja0529788. [DOI] [PubMed] [Google Scholar]
- 33.Kolpashchikov DM, Stojanovic MN. Boolean control of aptamer binding states. J. Am. Chem. Soc. 2005;127:11348–11351. doi: 10.1021/ja051362f. [DOI] [PubMed] [Google Scholar]
- 34.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 35.Baugh C, Grate D, Wilson C. 2.8 A crystal structure of the malachite green aptamer. J. Mol. Biol. 2000;301:117–128. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 36.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J. Am. Chem. Soc. 2008;130:93–102. doi: 10.1021/ja071516m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hohjoh H. RNA interference (RNAi) induction with various types of synthetic oligonucleotide duplexes in cultured human cells. FEBS Lett. 2002;521:195–199. doi: 10.1016/s0014-5793(02)02860-0. [DOI] [PubMed] [Google Scholar]
- 39.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonin KA, Lin YP, Calkins ER, Jaeger L. Attenuation of loop-receptor interactions with pseudoknot formation. Nucleic Acids Res. 2012;40:2168–2180. doi: 10.1093/nar/gkr926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tornaletti S, Maeda LS, Hanawalt PC. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem. Res. Toxicol. 2006;19:1215–1220. doi: 10.1021/tx060103g. [DOI] [PubMed] [Google Scholar]
- 42.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J. Mol. Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 43.Thaler DS, Liu S, Tombline G. Extending the chemistry that supports genetic information transfer in vivo: phosphorothioate DNA, phosphorothioate RNA, 2′-O-methyl RNA, and methylphosphonate DNA. Proc. Natl Acad. Sci. USA. 1996;93:1352–1356. doi: 10.1073/pnas.93.3.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9:1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 46.Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 48.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ginno PA, Lim YW, Lott PL, Korf IF, Chedin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reaban ME, Griffin JA. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348:342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- 51.Wanrooij PH, Uhler JP, Shi Y, Westerlund F, Falkenberg M, Gustafsson CM. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012;40:10334–10344. doi: 10.1093/nar/gks802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 53.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu P, Nakano S, Sugimoto N. Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur. J. Biochem. 2002;269:2821–2830. doi: 10.1046/j.1432-1033.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- 55.Kasprzak W, Shapiro B. Stem Trace: an interactive visual tool for comparative RNA structure analysis. Bioinformatics. 1999;15:16–31. doi: 10.1093/bioinformatics/15.1.16. [DOI] [PubMed] [Google Scholar]
- 56.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 57.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W. Optimization and modifications of aptamers selected from live cancer cell lines. Chembiochem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]