Abstract
Metabolic diseases and comorbidities represent an ever-growing epidemic where multiple cell types impact tissue homeostasis. Here, the link between the metabolic and gene regulatory networks was studied through experimental and computational analysis. Integrating gene regulation data with a human metabolic network prompted the establishment of an open-sourced web portal, IDARE (Integrated Data Nodes of Regulation), for visualizing various gene-related data in context of metabolic pathways. Motivated by increasing availability of deep sequencing studies, we obtained ChIP-seq data from widely studied human umbilical vein endothelial cells. Interestingly, we found that association of metabolic genes with multiple transcription factors (TFs) enriched disease-associated genes. To demonstrate further extensions enabled by examining these networks together, constraint-based modeling was applied to data from human preadipocyte differentiation. In parallel, data on gene expression, genome-wide ChIP-seq profiles for peroxisome proliferator-activated receptor (PPAR) γ, CCAAT/enhancer binding protein (CEBP) α, liver X receptor (LXR) and H3K4me3 and microRNA target identification for miR-27a, miR-29a and miR-222 were collected. Disease-relevant key nodes, including mitochondrial glycerol-3-phosphate acyltransferase (GPAM), were exposed from metabolic pathways predicted to change activity by focusing on association with multiple regulators. In both cell types, our analysis reveals the convergence of microRNAs and TFs within the branched chain amino acid (BCAA) metabolic pathway, possibly providing an explanation for its downregulation in obese and diabetic conditions.
INTRODUCTION
Several diseases caused by dysfunction in metabolism have become prevalent in human populations worldwide. Among these, cardiovascular disease (CVD) represents the leading cause of death worldwide. Obesity is a major risk factor for CVD, in particular when accompanied with insulin resistance, hypertension and altered blood lipid profiles (1). These in combination are referred to as the metabolic syndrome that also confers risk to develop diabetes and cancer (1).
High-quality genome-scale metabolic reconstructions are now available that represent the entire network of metabolic reactions a given organism is known to exhibit (2,3). Metabolic fluxes within the network adapt according to enzyme activity, substrate, cofactor, energy, metabolite and product availability as well as posttranslational regulation (4,5). Current technologies allow the characterization of global phenotypes on the transcriptome level through deep sequencing of RNA and DNA molecules. However, global measurements of proteome activity or metabolic fluxes remain a bottleneck. To address the latter limitation, it is possible to leverage the ability of mathematical models to integrate various data types to reveal central changes in metabolism. These mathematical representations allow the computation of physiological states. For estimating reaction activities, a method was proposed (6) where the expression levels serve as a soft-constraint to favor consistent solutions in concordance with the mass conservation in the metabolic network.
Alterations in the expression status are an initial step for a metabolic shift and can serve as a predictor of the metabolic activity cells are able to sustain. For this reason, the regulator molecules actuating this shift represent candidate therapeutic targets. In adipocytes, two transcription factors (TFs), peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer binding protein α (CEBPα), have been shown to be the key regulators: they are required to initiate terminal differentiation and are sufficient to convert other cell types to adipocytes (7), manifested through their genome-wide binding profile that positions them as master regulators of the adipocyte expression profile (8–10). Several antidiabetic drugs have been developed that activate PPARγ (11). The widely used CVD drugs statins on the other hand impact cholesterol levels through genes regulated by the signal-responsive TFs sterol-regulatory element binding factors (SREBFs) and liver X receptors (LXRs) (12). It is highly likely that interactions among TFs could play a role in disease, yet less is known so far how their targets overlap. Recent studies have also placed attention on the role of noncoding RNA regulators known as microRNAs (miRNAs) during adipocyte differentiation of cell culture and in vivo models (13,14), identifying counteracting regulators such as the miR-27 family and let-7 (15–18). We have recently identified several miRNAs as primary PPARγ target genes in mouse adipocytes (19), yet it remains unclear to what extent these different regulators converge to control the metabolic phenotype and whether identifying their convergence points could improve therapeutic interventions.
The Encyclopedia of DNA Elements (ENCODE) project has built an extensive list of functional elements in the human genome, including regulatory elements bound by TFs that control gene activity (20). Human umbilical vein endothelial cells (HUVECs) belong to the panel of ENCODE cell types with most data available and are also widely used as a model cell line in CVD research. Here, we hypothesized that observing the regulation of metabolic genes via multiple regulators (epigenetic, transcriptional and posttranscriptional) could indicate a plausible high relevance in a disease context. Moreover, to delineate the metabolic activity shifts affected by these key nodes, such an integrative analysis could become informative coupled with mathematical modeling of reaction activities. To allow data sources of gene regulation [such as ENCODE (20)] and metabolic network models (2,3) to be explored in an integrative manner, we used a tripartite graph representation and developed an interactive web portal, Integrated Data Nodes of Regulation (IDARE, http://systemsbiology.uni.lu/idare.html, see User Guide in Supplementary Material), that can be used to visualize global or tissue-specific data. This integrative experimental and computational analysis allowed us to address the connectivity between the human regulatory and metabolic networks.
Using just the overlap of TF-gene associations and the metabolic network, we observed a strong enrichment of disease-associated nodes among genes that show TF binding in multiple HUVEC ChIP-seq studies, including the nitric oxide synthase (NOS) gene family. We collected further experimental data on TFs and miRNAs in adipocytes differentiated from Simpson–Golabi–Behmel syndrome (SGBS) preadipocyte cell line, an established model for human adipogenesis (21). Interestingly, each of the previously characterized dyslipidemia genes LDLR (LDL receptor) (22), LPIN1 (lipin 1) (23) and LPL (lipoprotein lipase) (24) that belong to the triacylglycerol synthesis and release pathway are highlighted as shared TF- and miRNA-associated genes. Moreover, the cell fate determining TFs were observed to form a multi-TF feed-forward loop with binding sites nearby genes from the cholesterol synthesis and fatty acid activation pathways. Finally, the convergence of miRNAs and TFs highlight the branched-chain amino acid (BCAA) metabolism as a key nonlipid pathway for which altered regulation by the factors studied here may provide an explanation for its association with obesity and diabetes.
MATERIALS AND METHODS
Cell culture and differentiation
The human preadipocyte cell line isolated from a Simpson-Golabi-Behmel syndrome patient (SGBS) have previously been shown to be in many ways identical to differentiated primary adipocytes from healthy donors but maintain their differentiation capacity longer than other isolated cells (21), therefore representing an optimal model system for high-throughput analysis. The SGBS cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/Nutrient Mix F12 (Gibco, Paisley, UK) containing 8 mg/l biotin, 4 mg/l pantothenate, 0.1 mg/ml streptomycin and 100 U/ml penicillin (OF medium) supplemented with 10% FBS in a humidified 95% air/5% CO2 incubator. The cells were seeded into culture medium flasks or plates, which were coated with a solution of 10 µl/ml fibronectin and 0.05% gelatine in phosphate-buffered saline (PBS). Confluent cells were cultured in serum-free OF medium for 2 days followed by stimulation to differentiate with OF media supplemented with 0.01 mg/ml human transferrin, 200 nM T3, 100 nM cortisol, 20 nM insulin, 500 µM IBMX (all from Sigma-Aldrich) and 100 nM rosiglitazone (Cayman Chemical, Ann Arbor, USA). After day 4, the differentiating SGBS cells were kept in OF media supplemented with 0.01 mg/ml human transferrin, 100 nM cortisol and 20 nM insulin. SGBS cells differentiate within 10–12 days as determined by microscopic analysis (Oil red O staining). At this time point, the cells are filled with small-sized lipid droplets and are most responsive, whereas at later time points (20 days), the lipid droplets fuse and cells are less active (Dr Martin Wabitsch, personal communication). RNA samples were collected at 0, 4, 8 and 12 h and on days 1, 3 and 12 of differentiation and chromatin samples from day 0 (H3K4me3) and day 10 (TFs and H3K4me3). To find LXR-responsive genes, the day 10 differentiated SGBS cells were stimulated with 1 µM T0901317 for 4 h (synthetic agonist for LXRs), while control cells received DMSO (final concentration 0.1%).
miRNA transfection
MiRNA mimics for miR-27a, miR-29a and miR-222 (Thermo Scientific Dharmacon, Colorado, USA) or a scrambled double-stranded siRNA sequence as control (siCtrl) (Eurogentec, Liège, Belgium) were introduced into 4 days differentiated SGBS adipocytes using Lipofectamine RNAiMAX reagent (Invitrogen, Halle, Belgium) according to manufacturer’s instructions. Shortly, miRNA mimics or siRNAs were mixed with Lipofectamine RNAiMAX reagent, incubated for 20 min and diluted with plain DMEM-F12 medium to a final concentration of 100 nM. The first differentiation medium was replaced by the transfection mixture and incubated for 2 h before changing to the second differentiation medium (see above). Twenty-four hours after transfection, the cells were collected for RNA extraction.
RNA extraction and real-time quantitative polymerase chain reaction
Total RNA was extracted using TriSure (Bioline, London, UK). One milliliter of TRIsure was added per a confluent six-well to lyse the cells. RNA was extracted with 200 μl of chloroform and precipitated from the aqueous phase with 400 μl of isopropanol by incubating at −20°C overnight. cDNA was synthesized by using 1 μg of total RNA, 0.5 mM dNTPs, 2.5 μM oligo-dT18 primer, 1 U/μl RiboLock RNase Inhibitor (Fermentas, Vilnius, Lithuania) and 10 U/μl M-MuLV Reverse Transcriptase (Fermentas) for 1 h at 37°C. The reaction was stopped by 10-min incubation at 70°C. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed with Applied Biosystems 7500 Fast Real-Time PCR System using Absolute Blue qPCR SYBR Green Low ROX Mix reagent (Thermo Fisher Scientific, Surrey, UK). Five microliters of cDNA template was used with 1 μl of gene-specific primer pairs (10 μM) and 10 μl of the qPCR SYBR mixture in a final reaction volume of 20 µl. The PCR reaction started with 15 min at 95°C to activate the polymerase. The PCR cycling conditions were as follows: 40 cycles, of which each was composed of 15 s at 95°C, 15 s at 55°C and 30 s at 72°C. Fold inductions were calculated using the formula 2−(ΔΔCt), where ΔΔCt is [Ct(target mRNA) − Ct(RPL13A)]differentiated − [Ct(target mRNA) − Ct(RPL13A)] and the Ct is the cycle at which the threshold is crossed. PCR product quality was monitored using post-PCR melt curve analysis. The primer sequences are provided in Supplementary Table S1.
miRNA assays
The miRNA detection was performed by using TaqMan MicroRNA Reverse Transcription Kit with TaqMan MicroRNA Assays (Applied Biosystems). The miRNA cDNA synthesis and miRNA real-time PCR were done following manufacturer’s instructions and by using an Applied Biosystems 7500 Fast Real-Time PCR System. Relative expression levels in the undifferentiated and the 5-day differentiated adipocytes were calculated using the formula 2−(ΔΔCt), where ΔΔCt is [Ct(target miRNA) − Ct(U6)]differentiated − [Ct(target miRNA) − Ct(U6)]undifferentiated and the Ct is the cycle at which the threshold is crossed.
Microarray profiling
Total RNA in triplicates from the differentiation time series, LXR agonist stimulation and the miRNA transfections were processed according to the manufacturer instructions to prepare cDNA that was hybridized on microarrays (for the time series and ligand stimulation, the array hybridizations were performed on Illumina HT-12 v3 arrays at the Turku Centre for Biotechnology, Microarray and sequencing facility; for the miRNA transfections, on Illumina HT-12 v4 arrays at DNA Vision, Charleroi, Belgium). The raw data files were processed and quality controlled using the R/Bioconductor lumi package. Raw and normalized expression values are available via GEO (GSE41578). Genes that had a detection P <0.05 were selected for statistical analysis performed using the limma package. The F-test was used to assess significance of overall dynamic response over the differentiation and a two-tailed t-test to compare specific time points to day 0 undifferentiated cells (Benjamini–Hochberg adjusted P <0.01 was considered significant). For the miRNA transfections, statistical analysis was based on t-test significance comparing mean expression levels on miRNA transfection to a scrambled siRNA control transfection, and similarly the LXR agonist-treated cells were compared with solvent-treated cells. The expression profiles of metabolic genes or TFs were clustered for visualization using self-organizing maps [GEDI software (25)] and AutoSOME (26) as instructed in the tool documentation. Enriched pathways from the human metabolic reconstruction (2) were determined using a hypergeometric test testing for overrepresentation. Genes from Gene Ontology categories with similar gene numbers as Recon1 (1040) were obtained using the GO Online SQL Environment (http://www.berkeleybop.org/goose), as of 12 August 2013: cell projection (747), envelope (630), locomotion (775) and receptor activity (464). The number of probes detected in the array is indicated in brackets for each category from a total of 12 756 detected probes.
miRNA array profiling
Total RNA samples from time points day 0, day 1, day 3 and day 12 of the differentiation time series used for mRNA array analysis (see above) were also used to profile miRNAs using miChip arrays (v.11.0) arrays (27) at the EMBL Genomics Core facility at Heidelberg. The raw signal values from total RNA array hybridizations were median normalized and then further normalized to the respective signals from day 0 samples. Only probes corresponding to mature human miRNAs were included in the analysis.
Chromatin immunoprecipitation
Nuclear proteins were cross-linked to DNA by adding formaldehyde directly to the medium to a final concentration of 1% for 8 min at room temperature. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M and incubating for 5 min at room temperature on a rocking platform. The medium was removed and the cells were washed twice with ice-cold PBS. The cells were then collected in lysis buffer [1% sodium dodecyl sulphate (SDS), 10 mM EDTA, protease inhibitors, 50 mM Tris–HCl, pH 8.1] and the lysates were sonicated by a Bioruptor UCD-200 (Diagenode, Liege, Belgium) to result in DNA fragments of 200–500 bp in length. Cellular debris was removed by centrifugation and the lysates were diluted 1:10 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, protease inhibitors, 16.7 mM Tris–HCl, pH 8.1). Chromatin solutions were incubated overnight at 4°C with rotation with antibodies against H3K4me3 (4 µl per immunoprecipitation (IP) of 17-614, Millipore, Billerica, MA, USA), PPARγ (mixture of 0.5 µl per IP of sc-7196x, Santa Cruz Biotechnologies, Santa Cruz, CA, USA and 5 µl per IP of 101700, Cayman, Ann Arbor, MI USA), CEBPα (5 µl per IP of sc-61, Santa Cruz Biotechnologies) and LXRα (5 µl per IP, kind gift from Eckardt Treuter, Karolinska Institute, Stockholm, Sweden). The LXR antibody recognizes also LXRβ that maintains a constant low level of expression during differentiation. The immuno complexes were collected with 20 µl of MagnaChIP protein A beads (Millipore) for 1 h at 4°C with rotation. Nonspecific background was removed by incubating the MagnaChIP protein A beads overnight at 4°C with rotation in the presence of bovine serum albumin (250 µg/ml). The beads were washed sequentially for 3 min by rotation with 1 ml of the following buffers: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 8.1), high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris–HCl, pH 8.1) and LiCl wash buffer (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1). Finally, the beads were washed twice with 1 ml of TE buffer (1 mM EDTA, 10 mM Tris–HCl, pH 8.1). The immuno complexes were then eluted by adding 500 µl of elution buffer (25 mM Tris–HCl, pH 7.5, 10 mM EDTA, 0.5% SDS) and incubating for 30 min on rotation. The cross-linking was reversed and the remaining proteins were digested by adding 2.5 µl of proteinase K (Fermentas) to a final concentration of 80 µg/ml and incubating overnight at 65°C. The DNA was recovered by phenol/chloroform/isoamyl alcohol (25:24:1) extractions and precipitated with 0.1 volume of 3 M sodium acetate, pH 5.2, and 2 volumes of ethanol using glycogen as carrier. Immunoprecipitated chromatin DNA was then used as a template for real-time quantitative PCR or for library preparation and sequencing (performed at EMBL Core facility).
PCR of chromatin templates
Real-time quantitative PCR of ChIP templates was performed using chromatin-region–specific primers in a total volume of 20 µl with Applied Biosystems 7500 Fast Real-Time PCR System using Absolute Blue qPCR SYBR Green Low ROX Mix reagent (Thermo Fisher Scientific, Surrey, UK). The PCR cycling conditions were preincubation for 15 min at 95°C, 40 cycles of 15 s at 95°C, 15 s at 55°C and 30 s at 72°C and a final elongation for 10 min at 72°C. Relative association of chromatin-bound protein was calculated using the formula 2−(ΔCt)*100, where ΔCt is Ct(output) − Ct(IgG control), output is the DNA immunoprecipitated with TF-specific antibodies and IgG control is the DNA from immunoprecipitations using nonspecific control antibody. The primer sequences are provided in Supplementary Table S1.
Discretization of array expression values and constraint-based model
Metabolic changes resulting from human SGBS preadipocyte cell differentiation were qualitatively predicted from gene expression data using an implementation of the constraint-based method by Shlomi et al. (6). Constraint-based modeling is a widely used mathematical approach for the description and analysis of metabolic networks. It relies on the stoichiometric structure and does not require detailed kinetic parameters. By assuming steady state for the intracellular metabolites, the respective dynamic balance equations can be simplified to easy to handle linear equations. Besides the law of mass conservation, other constraints might be included, e.g. enzyme capacities, irreversibility information or measured uptake and secretion rates, as well as optimality considerations, to further constrain the possible solution space, i.e. the possible flux values, which can be realized within the given network structure. Recent efforts, like the aforementioned applied method of Shlomi et al., focus on the generation of context-specific and thus more predictive models via the additional integration of omics data. A consistent version of the generic human metabolic model Recon1 (2) served thereby as modeling platform on which own microarray data were overlaid as soft-constraints based on gene-protein-reaction associations to allow the prediction of network activity distributions. LPIN1 was missing and owing to its central role in adipocytes, was added to the model and assigned to the triacylglycerol pathway. Continuous log2-normalized expression values were first discretized into three categories: lowly expressed (−1), moderately expressed (0) and highly expressed (1) genes, based on mean expression ± 0.5*standard deviation cutoffs. These values were mapped to the reactions contained in Recon1 and used as input for the metabolic reaction activity prediction.
Heptamer enrichment analysis and miRNA target identification
To identify heptamer motifs whose frequency is significantly different in the 3′-untranslated regions (3′-UTRs) of downregulated transcripts, relative to their frequency in the entire set of 3′-UTRs, we considered all RefSeq transcripts for which a corresponding probe set was significantly downregulated (P < 0.01, log2-fold change <−0.3) 24 h after miRNA mimic transfection. The 3′-UTR sequences of the RefSeq transcripts were downloaded from the UCSC genome browser (14.05.2012) and the human miRNA sequences were obtained from miRBase release 18 (http://www.mirbase.org/). The enrichment analysis was performed using a Bayesian model originally introduced for comparing miRNA frequencies between samples (28,29) that have been successfully applied to determine motif enrichments of small RNAs (30,31).
The transcripts for the list of putative target transcripts of miR-27a, miR-29a and miR-222 were selected based on two criteria: (i) at least one probe set corresponding to the transcript was significantly downregulated on the respective miRNA transfection and, (ii) the transcripts 3′-UTR contained at least one hit for any of the possible heptameric reverse complements for the corresponding seed sequences of miR-27a (CUGUGAA or ACUGUGA), miR-29a (GGUGCUA, AUGGUGC or UGGUGCU) or miR-222 (AUGUAGC), respectively.
ChIP-seq analysis
The HUVEC H3K4me3 peak data available from ENCODE (wgEncodeUwHistoneHuvecH3k4me3StdPkRep1) was overlapped with transcription start site (TSS) coordinates from Refseq to limit the analysis to active genes in HUVECs. Gene to disease associations were obtained from DisGeNET (32). A list of endothelial-relevant disease-associated genes was compiled by combining genes associated with CVDs, vascular diseases, coronary artery diseases, cerebrovascular disorders, peripheral arterial occlusive disease and pulmonary arterial hypertension. ENCODE data from untreated HUVEC cells was retrieved as peak coordinate files (UTA cMYC, SYDH GATA2, SYDH MAX, SYDH cJUN and SYDH cFOS). Other public data were obtained from the SRA database as .sra files (SRR576805 ETS1 from VEGFA stimulation, SRR351351 MEF2C from statin stimulation, SRR390745 p65 from TNF stimulation, SRX096362 FLI1 representing an endothelial cell developmental TF, SRR518265 HIF1A from hypoxia and PPARG samples SRX032890 and SRX019521, each with their respective control samples) that were converted to fastq files using sratools v.2.1.7. (data from own experiments were already in fastq format). Raw reads were first quality controlled using the FASTQ software v.0.10.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). A deviation from the expected GC-content was observed in the input sample of SGBS cells and this sample was replaced in the downstream analysis by a new input obtained from similarly differentiated cells. All reads that were detected as read artifacts or that had low-quality base pair calling were removed and read stacks collapsed using the FASTX software v.0.0.13 (minimum quality score of phred 10 across the read length was required) (http://hannonlab.cshl.edu/fastx_toolkit/index.html). The reads that passed the quality control steps were aligned to the hg19 human genome using the Bowtie (33) software v0.1.25 (one mismatch allowed, maximum three locations in the genome from which the highest quality match was reported). The mapping capability with these settings was tested by aligning all 36-mers of the hg19 fasta genome available via UCSC and was determined to be 0.88 and used in the subsequent peak-calling step.
SGBS histone data were analyzed using the EpiChIP software v.0.9.7 (34), where the H3K4me3 signal was quantified from −750 to +1250 region centered at Refseq TSS coordinates. This region was detected to have the highest signal by window analysis. TF peak detection from SGBS was performed using the Quest software (35) v.2.4. run in the advanced mode with default settings applied except for the mappable genome fraction (set to 0.88) and enrichment (ChIP enrichment set to 15 and ChIP to background enrichment to 2.5). Fastq files and signal tracks from SGBS cells can be accessed via NCBI GEO (GSE41578). The final peak lists were filtered to remove peaks with log10Qvalue <3. We chose to apply two cutoffs to detect both low-occupancy (enrichment >15) and high-occupancy (enrichment >30) binding sites. In the text, the complete list of low- and high-occupancy genomic regions (Supplementary Table S2) has been analyzed unless otherwise specified. The public HUVEC ChIP-seq data were processed with default settings (enrichment 30), which corresponds to settings used in their respective publications (information was available for three of five studies via GEO). To assess what biological pathways could be most affected by the given TF, a genomic region enrichment test was performed using the GREAT software (binomial P-value, false discovery rate (FDR) 1%) (36). The same software was used to obtain the peak to gene association files for analyzing TF convergence on shared targets. The complete gene association and enrichment term lists for the SGBS TFs can be found in Supplementary Table S3
Gene metanodes and IDARE web portal
Gene Metanodes (Figure 1) showing gene-related data were generated with Matlab®. Recon1 metabolic gene Entrez IDs were used for data mappings. Based on homogeneous (HUVEC TFs) or heterogeneous (SGBS) data we customized the metanode visual appearance. The open sourced IDARE web portal and its HUVEC and SGBS instances are built using HTML5 standards and javascript libraries jQuery, highchart.js, bootstrap and cytoscapeweb. The release contains a configuration-based python workflow responsible for building graph objects from Recon1 SIF and XGMML pathways into javascript object notation files (full description on the User Guide provided in the Supplementary Material). In addition, gene metanode image files, annotations from hg19 and time course expression data are integrated along with reaction and metabolite relationships. The images and graph object files are then deployed to the appropriate directories according to the instance workflow configurations. This architecture is extendable and allows for easy inclusion of other data sets like HUVEC as well as custom pathways. IDARE instances can be deployed locally or on web and cloud servers.
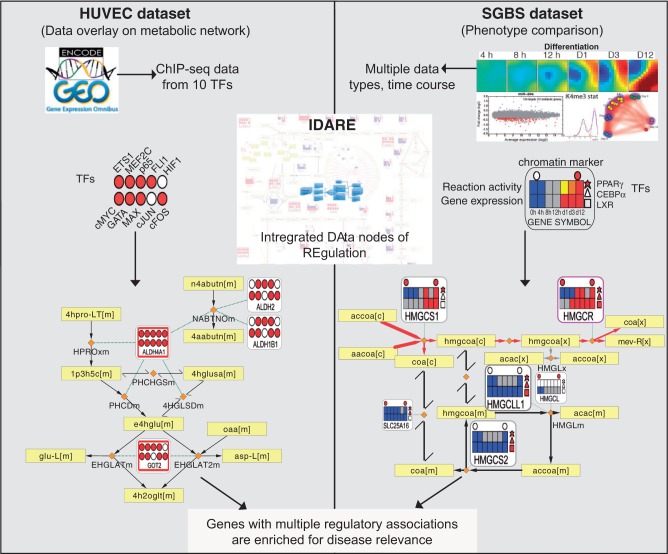
Figure 1.
Conceptual overview of the analysis performed that links the regulatory and metabolic networks for exposing disease-relevant genes. A summary of the data sets and data integration approach is shown. By integrating regulatory network components with metabolic pathways, gene metanodes shown from the IDARE webportal provide a simple and intuitive means to analyze relationships between the metabolic and regulatory networks. HUVEC data set: Overlaying TF-binding data indicated in filled color circles with the metabolic network reaction backbone can be used to examine the co-occurrence of transcriptional regulators that is informative of likely disease association. Yellow rectangular nodes represent metabolites and small orange diamonds represent reactions in the metabolic network. SGBS data set: Often the biological question involves comparison of phenotypic states. Constraint-based modeling methods, established in context of metabolic reconstructions, can be used as exemplified with data from adipocyte differentiation. Those pathways that are predicted to shift to an active state in adipocytes compared with preadipocytes are indicated by the red edge color, yellow and black color correspond to active or inactive reactions in both states. The heatmaps show for each differentiation time point the gene expression (below) and predicted reaction activity (above) where blue indicates low/absent gene expression or inactive reaction, gray, moderate expression or undetermined reaction and red stands for highly expressed gene and active reaction, respectively. The regulators can be added as different shapes. Here, circles above represent the presence (red) or absence (white) of the H3K4me3 marker for active transcription and the three different polygons to the right represent PPARγ (star), CEBPα (triangle) and LXR (square) peak associations (red, present; white, not present).
RESULTS
Integrating gene regulation data with a human metabolic reconstruction
A metabolic network typically consists of metabolite and reaction nodes. To make such models actionable in context of disease pathways or drug target identification, it becomes useful to integrate the regulation of genes that catalyze the reactions within the model. We included versatile nodes (referred to as gene metanodes) associated to reactions, which can represent any gene-centric data collected from different experiments (Figure 1). TF binding data indicated in filled circles in the metanode can be overlaid with the metabolic network that includes rectangular nodes representing metabolites and diamonds representing reactions. Exemplified using HUVEC data, the visualization can be used to examine the co-occurrence of transcriptional regulators. The display options are flexible including presenting the regulators as different shapes, as shown in SGBS. Moreover, the edges forming the network backbone can be colored to represent reaction activity obtained using constraint-based modeling methods. The metanode heatmaps show for each differentiation time point the gene expression (below) and predicted reaction activity (above). By integrating regulatory network components with metabolic pathways, gene metanodes shown from the IDARE web portal (see Supplementary Data—IDARE User Guide for further details) provide a simple and intuitive means to analyze relationships between the metabolic and regulatory networks.
To illustrate the concept and to test whether such data could be informative to highlight key parts of the large metabolic reconstruction networks, we collected ChIP-seq data from HUVECs that represent the most studied primary cell type among ENCODE data sets. Using the H3K4me3 chromatin mark to focus on active loci, we associated Recon1 genes to TF peaks from 10 studies, 5 collected from NCBI GEO database and 5 available from ENCODE (see ‘Materials and Methods’ section for details). Disease information was collected from the DisGeNET database (32), focusing on endothelial-relevant disease. Table 1 shows those genes that are associated with eight or more TFs while the complete TF and disease association result is shown in Supplementary Table S4. Interestingly, expressed genes associated with between seven and nine TFs are >2-fold enriched in vascular disease-relevant genes, a result that points to the prominent link between high TF-mediated gene expression regulation and disease. The hypergeometric P-values for the different number of TF associations to Recon1 genes are shown in Figure 2A, comparing vascular disease-relevant associations to all disease associations (the observed increasing trend in significance and enrichment apply also when analysis is not restricted to metabolic genes, data not shown).
Table 1.
Endothelial disease relevant genes exposed by association to multiple TFs in HUVEC data
| Symbol | Pathway | Entrez GeneID | Number of disease | Number of TFs |
|---|---|---|---|---|
| NOS3 | Arginine and proline metabolism | 4846 | 140 | 8 |
| PTGS2 | Eicosanoid metabolism | 5743 | 97 | 8 |
| HMOX1 | Heme degradation | 3162 | 56 | 8 |
| ABCA1 | Transport, extracellular; transport, golgi apparatus | 19 | 20 | 9 |
| PTGS1 | Eicosanoid metabolism | 5742 | 12 | 8 |
| LIPG | Triacylglycerol synthesis | 9388 | 9 | 10 |
| ADA | Nucleotides; purine catabolism | 100 | 9 | 9 |
| PDE4D | Nucleotides | 5144 | 9 | 8 |
| PDE4B | Nucleotides | 5142 | 8 | 9 |
| ABCC4 | Transport, extracellular | 10 257 | 7 | 9 |
| PDE3A | Nucleotides | 5139 | 7 | 8 |
| PIK3CG | Inositol phosphate metabolism | 5294 | 7 | 8 |
| SLC12A2 | Transport, extracellular | 6558 | 6 | 9 |
| PAFAH2 | Glycerophospholipid metabolism | 5051 | 4 | 9 |
| GCLM | Glutathione metabolism | 2730 | 3 | 10 |
| MTHFD1L | Folate metabolism | 25 902 | 3 | 10 |
| GALNT2 | O-glycan biosynthesis | 2590 | 3 | 9 |
| PPAP2B | Triacylglycerol synthesis | 8613 | 1 | 10 |
| NNMT | NAD metabolism | 4837 | 1 | 10 |
The 19 Recon1 metabolic genes annotated with endothelial-relevant disease terms in the DisGeNET database (32) and having an active TSS mark (H3K4me3) with putative binding of at least 8 TFs from ChIP-Seq HUVEC studies (see ‘Materials and Methods’ section for details) are presented.
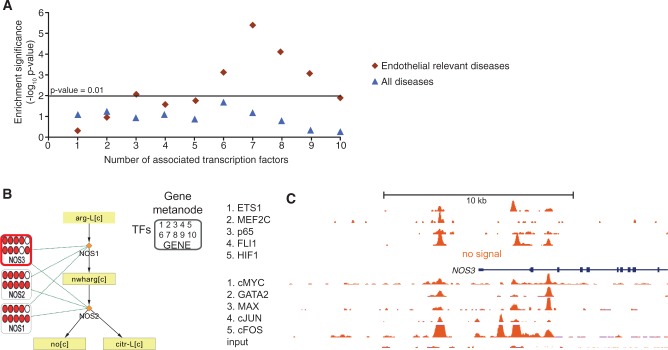
Figure 2.
Gene metanodes reveal frequent TF association to disease-relevant genes exemplified by nitric oxide synthases in HUVEC data. The result of disease enrichment tests for genes associated with 1–10 TFs are shown (A), where the horizontal line corresponds to hypergeometric P <0.01. The endothelial relevant diseases (diamonds) is compared with all diseases (triangles). (B) The three enzymes encoding nitric oxide synthases (NOS1, NOS2 and NOS3) and the two reactions that convert l-arginine to NO for regulating vascular dilation are shown. Data related to genes are displayed in gene metanodes superimposed on the metabolite-reaction network. Peak associations from 10 ChIP-seq studies in HUVECs (ETS1, MEF2C, p65, FLI1, HIF1α available via NCBI SRA and cMYC, GATA2, MAX, cJUN, cFOS and input available via ENCODE) are displayed in the indicated order where color indicates TF association and the respective TF signal tracks are shown in C. The value range 1–100 is used in the first five tracks and 1–78 in the ENCODE tracks.
The gene with most disease associations overall, the endothelial nitric oxide synthase, NOS3 (also known as ENOS), is visualized in Figure 2B. The release of NO is a key paracrine signal in the vascular system that is essential for the regulation of blood flow and pressure (37). The two other nitric oxide synthases (NOS1 and NOS2) can catalyze the same reaction to convert l-arginine to NO. Interestingly, each of these gene metanodes show multiple TF associations (Figure 2B). The respective genomic region around NOS3 TSS with TF signal from the 10 ChIP-seq experiments is shown in Figure 2C. The other multi-TF–associated nodes in the proline-arginine metabolic pathway shown in Supplementary Figure S1 include ALDH4A1 that participates in multiple amino acid pathways and is known to cause the autosomal recessive disorder known as type II hyperprolinemia (38) and MTAP from the coronary artery disease genome-wide association study (GWAS) reported locus on chromosome 9 (39). Encouraged by these findings, we next evaluated whether the same principle generalizes in human adipocytes that represent a key cell type in obesity and metabolic disease. However, a more limited set of available genome-wide regulator profiles motivated the collection of experimental data and in parallel using mathematical predictions based on the metabolic reconstruction to expose relevant pathways as described in more detail below.
Predicted metabolic activity changes during adipocyte differentiation
To outline plausible metabolic activity changes during adipocyte differentiation using the SGBS cell line, which represents an established human adipocyte cellular model isolated from a SGBS patient (21), we leveraged the constraint-based modeling approach (see ‘Materials and Methods’ for details) to predict the dynamic activity changes of metabolic reactions (6). Based on a time-course measurement of the transcriptome of differentiating SGBS preadipocytes a dynamic shift is evident, in particular, in the expression levels of metabolic genes among which 18%, 2-fold more when compared with other gene categories with similar numbers of genes or even with all detected genes (Supplementary Figures S2 and S3) (25,26), are differentially regulated. An overall trend of increasing levels from day 1 onward results in 219 upregulated metabolic genes by day 12 of differentiation, compared with 98 downregulated genes (Supplementary Table S5).
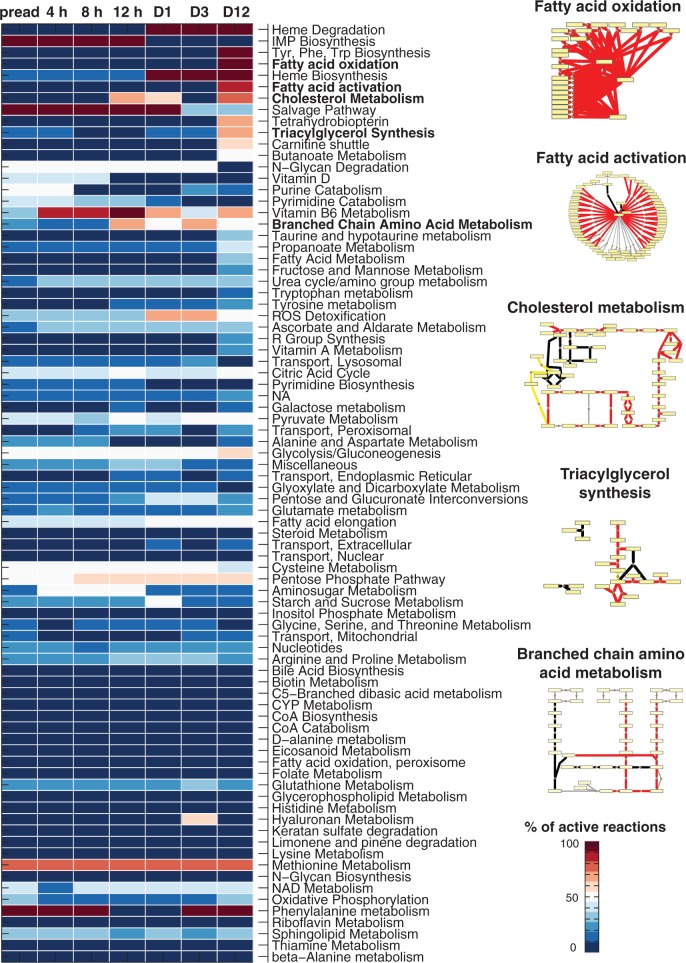
As gene expression levels alone are insufficient to describe the metabolic adaptation that occurs during terminal differentiation, we used them as soft-constraints to predict reaction activity for Recon1 (2,6). The predicted pathway activity changes are shown in Figure 3 and the complete prediction results for all seven differentiation time points and the 2469 consistent reactions contained in Recon1 are provided in Supplementary Table S6 (‘Materials and Methods’ section). Consistent with the mRNA level changes, a much higher number of reactions were predicted active in adipocytes than in preadipocytes (556 compared with 290, respectively) with 259 reactions predicted to become active during differentiation. Five pathways with highest predicted activation between preadipocytes and adipocytes were cholesterol metabolism (76% reactions predicted to change), fatty acid activation (64%) and oxidation (93%), triacylglycerol synthesis (60%) and branched chain amino acid (BCAA: valine, leucine, isoleucine) metabolism (50%) (for metabolites and enzymes involved refer to Supplementary Table S7), highly involved in lipid metabolism and metabolic diseases, suggesting the ability of the approach to recapitulate adipocyte characteristics. On a metabolite level, these pathways converge at acetyl-CoA, which can produce intermediates to be converted to fatty acids or to be consumed in the energy-producing mitochondrial oxidation. Pathways excluded from further analysis contained reactions predicted undetermined in one of the two phenotypic states, concretely, heme biosynthesis with 10 reactions, all predicted active in adipocytes, but nine of them undetermined in preadipocytes; heme degradation with only two reactions, both predicted active in adipocytes but undetermined in preadipocytes and the biosynthesis of tyrosine, phenylalanine and tryptophan, which contains only one reaction (O2 + l-Phenylalanine + Tetrahydrobiopterin -> Tetrahydrobiopterin-4a-carbinolamine + l-Tyrosine), predicted active in adipocytes and inactive in preadipocytes.
Figure 3.
Predicted metabolic activity changes during adipocyte differentiation and reaction backbones of selected pathways. A heatmap representing the predicted percentage of active reactions for each differentiation time point (columns) and metabolic pathway (rows) is shown for the Recon1 pathways (2). Blue color corresponds to no active reactions and red to complete activation. Notice that reactions can change between active and either inactive or undetermined. The pathways are shown in a descending order of activity considering the difference between preadipocytes (‘preadip’) and adipocytes (‘day 12’) and secondly alphabetically. Pathways highlighted (cholesterol metabolism, fatty acid activation and oxidation, triacylglycerol synthesis and BCAA metabolism) represent those with highest predicted activation with high prediction confidence score. The metabolic pathway backbones of these pathways are shown based on the stoichiometric information in the generic human metabolic model Recon1 (2). These pathways are predicted to shift to an active state in adipocytes compared with preadipocytes as indicated by the red edge color, yellow and black colors correspond to active or inactive reactions in both states, respectively. Yellow rectangular nodes represent metabolites and small orange diamonds represent reactions.
More than 300 metabolic reactions are predicted to change from preadipocytes to adipocytes (Supplementary Table S6). In agreement with an increased gene expression for the majority of metabolic genes, most reaction changes predicted are activations, especially those in lipid metabolism pathways. The BCAA metabolism pathway appeared as the most affected nonlipid pathway based on the predictions.
Genome-wide measurements of transcriptional, posttranscriptional and chromatin-level regulation in adipocytes
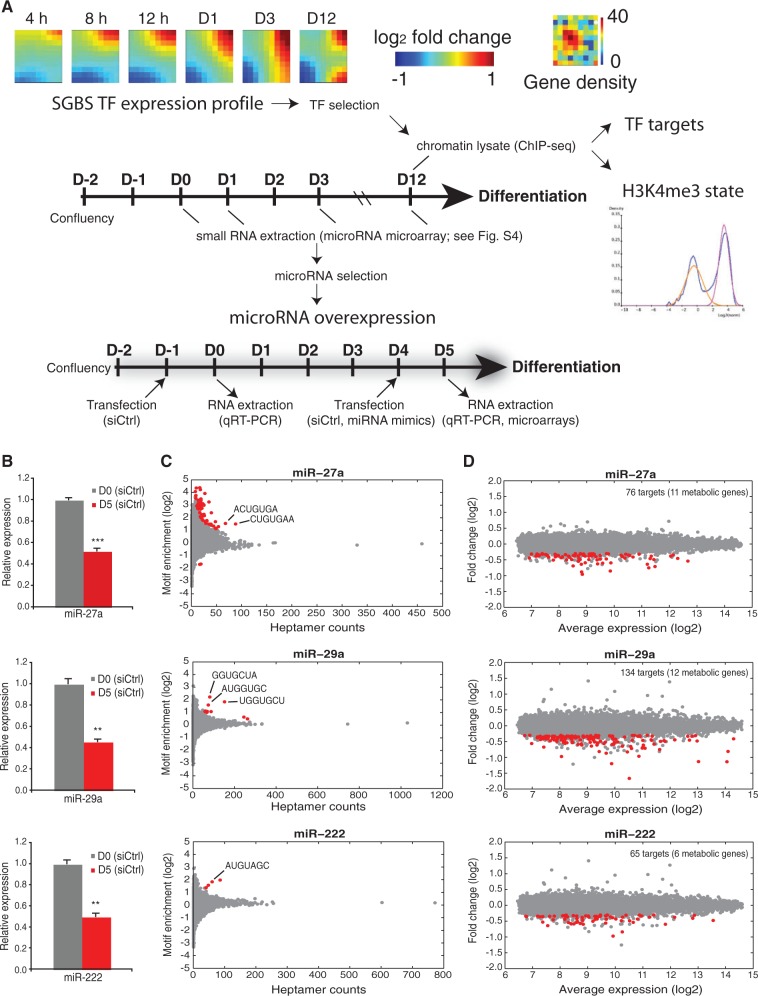
We performed a diverse set of genome-wide measurements from SGBS adipocytes, summarized in Figure 4A. We first focused on identifying the most highly induced TFs and their putative targets. In agreement with metabolic gene expression changes, a highly dynamic TF expression profile is also observed (Figure 4A). Among the highest upregulated TFs (see Supplementary Table S8), PPARG and CEBPA have been previously associated with a key role in adipocyte differentiation and global regulation of metabolic genes (8,40,41). The most upregulated TF gene was the signal responsive LXRA (also known as NR1H3), an established regulator of cholesterol reverse transport (42) for which the genome-wide occupancy has not yet been studied in adipocytes. We obtained the genome-wide binding profiles of PPARγ, CEBPα and LXRα to reveal their possible interplay in SGBS adipocytes, by collecting ChIP samples for sequencing (ChIP-seq). We also included published primary adipocyte (day 9) (10) and SGBS PPARγ (day 20) (41) data sets retrieved from the Sequence Read Archive database for comparison. The ChIP-seq reads that satisfied quality criteria were aligned to the hg19 human genome (see ‘Materials and Methods’ section for details). Statistics about initial and final read numbers are indicated in Supplementary Table S2.
Figure 4.
Selection of regulatory molecules for further study. (A) The TF expression profile used to select the highest upregulated TFs during adipogenesis is visualized using GEDI maps (25) that cluster TF genes with similar expression profiles. The number of genes in each cluster is displayed in the Gene density panel beside. For the identification of their genome-wide targets, ChIP-seq samples were collected as indicated on day 12, including lysates used to assess the H3K4me3 chromatin marker status using TSS activity status mixture modeling. The schematic representation of the experimental procedure indicates the sample collection intervals for microarrays, miRNA microarrays and ChIP-seq. For the identification of primary target mRNAs of the selected miRNAs, a similar differentiation procedure was used, accompanied by transfections with miRNA mimics for miR-27a, miR-29a or miR-222 or scrambled siRNA as a control at day 4. Samples were collected 24 h after transfection for microarray and RT-qPCR analysis. (B) RT-qPCR analysis of the relative expression values of the endogenous miR-27a, miR-29a and miR-222 from undifferentiated and 5-day differentiated SGBS cells. The measured expression values were normalized to U47 snRNA and are shown relative to undifferentiated cells, value of which was set to 1 (gray bars). Data points indicate the mean expression values of triplicate experiments and the error bars represent SD. Student’s t-test was performed to determine the significance of downregulation on differentiation (*P < 0.05; **P < 0.01; ***P < 0.001). (C) Enrichment analysis of heptamer motifs in the 3′-UTR sequences of significantly downregulated transcripts. The count of all possible heptamer motifs (each represented by a circle) and their log2-enrichment within the 3′-UTRs of downregulated transcripts are depicted on the x-axis and y-axis, respectively. The significantly enriched heptamers are marked in red. The most enriched abundant heptamers are corresponding to the reverse complement sequences of the overexpressed miRNA seeds as indicated (see ‘Materials and Methods’ section for details). (D) MA-plot depicting the log2-expression levels (x-axis) of all transcripts in cells transfected with indicated miRNA mimics and the log2-fold change relative to cells transfected with siCtrl. The significantly downregulated transcripts (unadjusted P < 0.01, log2-fold change < −0.3) containing at least one putative binding site for the respective miRNA are marked in red. The total number of putative miRNA targets is indicated (with metabolic target genes in brackets).
In addition to regulation of the transcriptional output, the measured expression changes of metabolic genes could also be controlled at the posttranscriptional level. In particular, miRNAs have emerged as important regulatory molecules and regulation of a number of miRNAs have been described as important for successful adipogenesis and lipid accumulation (43). We hypothesized that different miRNAs that become down-regulated during adipogenesis might contribute to allow the observed upregulation of metabolic genes to take place, serving as gatekeepers in preadipocytes. To investigate this possibility, we performed microarray profiling to detect differentially regulated miRNAs on days 0, 1, 3 and 12 of differentiating SGBS cells, revealing several candidate miRNAs for further analysis (Supplementary Figure S4). We focused on miRNA clusters with several members repressed already at early stages of differentiation, which lead to the selection of miR-27a that has previously been studied in mouse models, and two miRNAs whose role in adipocytes has not been characterized, miR-29a and miR-222 for further experiments. Their downregulation was validated by RT-qPCR to occur already by day 5 of differentiation (Figure 4B). To identify candidate target genes, we performed a miRNA mimic transfection (corresponding to a specific overexpression of each miRNA) at day 4 of differentiation and analyzed by microarrays the mRNA profiles at 24 h posttransfection and compared these with the cells similarly transfected with a scrambled control siRNA. An analysis for enriched heptamer motifs in 3′-UTRs of the downregulated mRNAs from the microarray analysis at day 5 reveals enrichment of motifs complementary to respective miRNA seed sequences (Figure 4C and D), suggesting that the observed mRNA downregulation could be due to their direct targeting by the miRNA.
Finally, we used the H3K4me3 chromatin modification, indicative of the transcriptional potential of the associated TSS, to evaluate changes in chromatin state of metabolic genes. We collected ChIP samples from undifferentiated and differentiated SGBS cells for sequencing and analyzed the ChIP-seq data obtained and public data from primary preadipocytes and adipocytes (10,41).
Predicted target gene profiles and their overlap
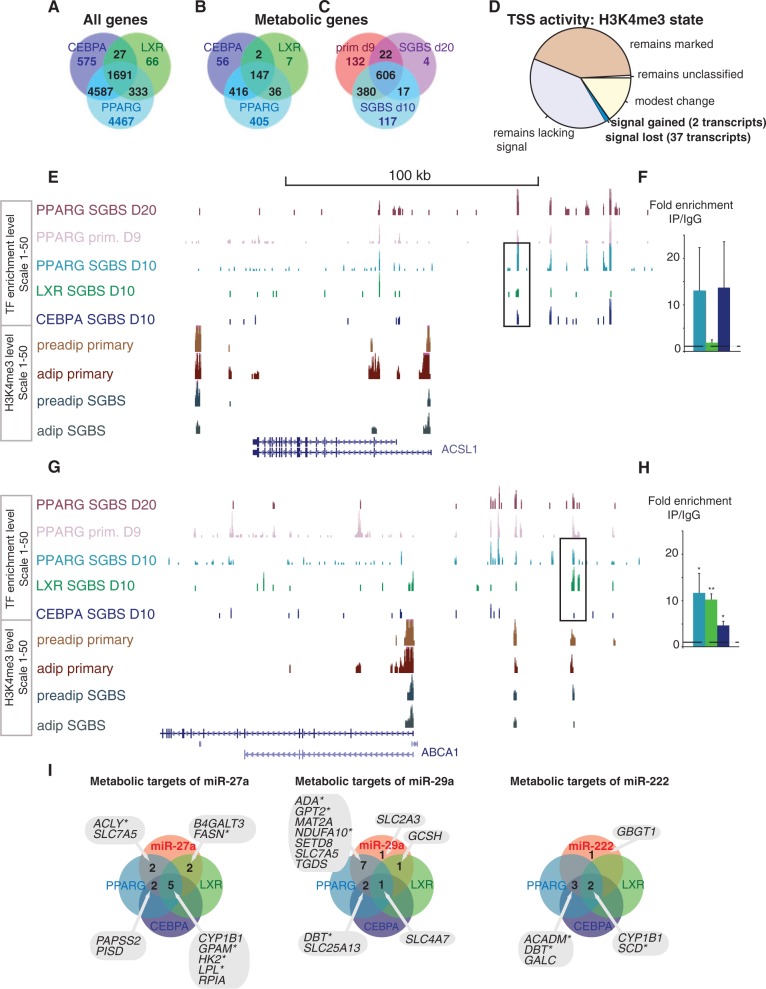
The comparison of all genes (in A) or metabolic genes (in B) associated to each TF is shown in Figure 5. Figure 5C shows the intersection of metabolic genes associated with PPARγ in the three independent data sets (1278). This first genome-wide mapping of LXR binding in human adipocytes revealed 2117 associated putative target genes. For CEBPα, we obtained 6880 putative target genes, while for PPARγ, the 11 078 putative target associations kept reflect peak associations that were found in our data set and observed in at least one of the two public data sets (10,41). From these genes, 1691 were associated with peaks from all three TFs (Figure 5A, Supplementary Table S3), with 147 common metabolic putative target genes.
Figure 5.
Overlaps in the genome-wide target profiles of PPARγ, CEBPα, LXR and the miRNAs −27a, −29a and −222 from SGBS adipocytes. Venn diagrams comparing putative target genes for PPARγ, CEBPα and LXR (in A and B) or between different studies that profile PPARγ binding (B) as obtained using the GREAT tool (36) are shown. Among all genes (A), 1691 genes are associated with all TFs, representing a large fraction from each individual TF peak-gene association list. Metabolic genes (C) show a similar highly overlapping target association profile. (C) In total, 606 PPARγ target associations to metabolic genes are supported by ChIP-seq data from day 10 and day 20 differentiated SGBS cells (41) and day 9 differentiated primary adipocytes (44), with an additional 419 metabolic genes supported by at least two data sets. The pie chart (D) illustrates how many genes gain or lose the H3K4me3 signal, showing that most genes retain their activity marker status. (E) The ChIP-seq signal tracks from PPARγ studies in SGBS cells (41) and primary adipocytes (44), CEBPα and LXR from SGBS adipocytes and H3K4me3 from primary and SGBS cells are displayed at the ACSL1 locus that shows high-occupancy binding of PPARγ, CEBPα and LXR. The ChIP-qPCR validation comparing enrichment with specific antibody to IgG unspecific control for the PPARγ and CEBPα occupied region indicated is shown in (F). (G) Similarly as above, the TF signal tracks show multiple peaks at the ABCA1 locus including the LXR response elements that show significant enrichment also with PPARγ and CEBPα antibodies as validated using ChIP-qPCR in (H). The enrichment values are shown relative to the enrichment of IgG and indicate the mean enrichment values of triplicate experiments and the error bars represent SEM. One sample t-test was performed to determine the significance of TF enrichment compared with IgG (*P < 0.05; **P < 0.01). (I) Venn analysis of metabolic target genes of the tested miRNAs and their targeting by TFs. The lists of metabolic genes targeted by the individual miRNAs and by the TFs PPARγ, CEBPα or LXR are overlapped to identify the metabolic genes under combinatorial multilevel regulation. The genes significantly changing during SGBS differentiation are indicated with a star.
To evaluate TSS activity changes, we analyzed the H3K4me3 ChIP-seq data using a mixture model method (34) that separates histone marker labeled gene TSS from those lacking the marker (Supplementary Figure S5) based on their read count distribution estimates. According to this analysis, most genes did not completely switch their TSS activation state during differentiation in SGBS cells (15 263 active; 19 311 inactive; 470 unclassified), while among the transcripts with altered TSS activity, the H3K4me3 marker mostly decreased: 1223 metabolic gene TSS are labeled with H3K4me3 in both preadipocytes and adipocytes, 1125 are inactive, TSS activity decreased for 37 transcripts (corresponding to 25 gene loci) while only 2 gained the activity marker (Figure 5D and Supplementary Table S9). As indicated, there was considerable agreement between primary adipocyte (10) and SGBS data (Supplementary Table S9).
The large intersection of TF-associated genes with the metabolic genes from Recon1 (1069 out of 1496 genes, Figure 5B) suggests a high contribution of these 3 TFs to the metabolic changes observed on terminal adipocyte differentiation. Interestingly, the metabolic pathways with most changes (those highlighted in Figure 3) show extensive TF binding. The acyl-CoA synthetase long-chain family member 1 (ACSL1) gene of the fatty acid activation pathway was among the top genes associated with high-occupancy binding sites for all three TFs (Figure 5E). The co-localized binding seen in Figure 5E was rather exceptional; genome-wide overlap in peak regions by all three TFs occurred only at 223 peak locations. At gene ontology term level (Supplementary Table S3), the TFs overlap in regulation of cholesterol efflux. Several genomic regions each bound by one or several of the TFs studied concentrated in the vicinity of ATP-binding cassette, sub-family A member 1 (ABCA1) a key gene in cholesterol reverse transport (Figure 5G). Interestingly, ACSL1 is required for oleate- and linoleate-mediated inhibition of cholesterol efflux through ABCA1 in macrophages indicating cross-talk between the pathways (45). ChIP-qPCR validation from independent immunoprecipitations is shown for the two regions indicated (Figure 5F and H), and the CDH1 and FABP4 loci that served as a negative and positive control region, respectively, are shown in Supplementary Figure S6.
Next we identified target genes for each miRNA based on repression in the array experiment and a seed match analysis (putative targets are indicated in red in Figure 4 and listed in Supplementary Table S10), ranging from 65 (miR-222) to 134 (miR-29a) significant target calls per miRNA. Figure 5I shows the metabolic target genes of each miRNA overlapped with TF target association lists (those genes that are significantly regulated during differentiation are indicated with a star).
Interestingly, those putative miR-27a and miR-222 targets that are overlapping with more than one TF are in fact mainly associated with high-occupancy binding, namely PISD, CYP1B1, the mitochondrial glycerol-3-phosphate acyltransferase (GPAM), hexokinase 2 (HK2), LPL, RPIA, the C-4 to C-12 straight chain acyl-CoA dehydrogenase (ACADM) and stearoyl-CoA delta-9-desaturase (SCD), suggesting that key metabolic genes are under tight combinatorial transcriptional and posttranscriptional regulation. Interestingly, the DisGeNET resource reports a disease association for each of these genes, only HK2 is not supported by this data source but in light of recent literature is implicated in cancer (46,47). All but two putative miRNA targets were also associated with TF-mediated regulation. Moreover, as previously reported (17,16), we could confirm PPARG among the miR-27a targets in adipocytes. There is also overlap between the miRNAs: according to our analysis miR-27a and miR-222 both target CYP1B1, miR-27a and miR-29a target the amino acid transporter solute carrier family 7 member 5 (SLC7A5) while miR-29a and miR-222 target dihydrolipoamide branched chain transacylase E2 (DBT).
In conclusion, the putative shared TF and miRNA target genes that our data integration revealed were ACADM, DBT, GPAM, HK2, LPL and SLC7A5. Taken together, by collecting a diverse and comprehensive set of regulatory data on adipocyte differentiation we were able to show that enzymes crucial or rate limiting for lipid metabolism were often associated with miRNAs and high occupancy binding by multiple TFs, suggesting tight combinatorial effects of TF upregulation and miRNA downregulation driving metabolic changes in adipogenesis.
Cell fate determining TFs engage signal-dependent TFs in a feed-forward motif
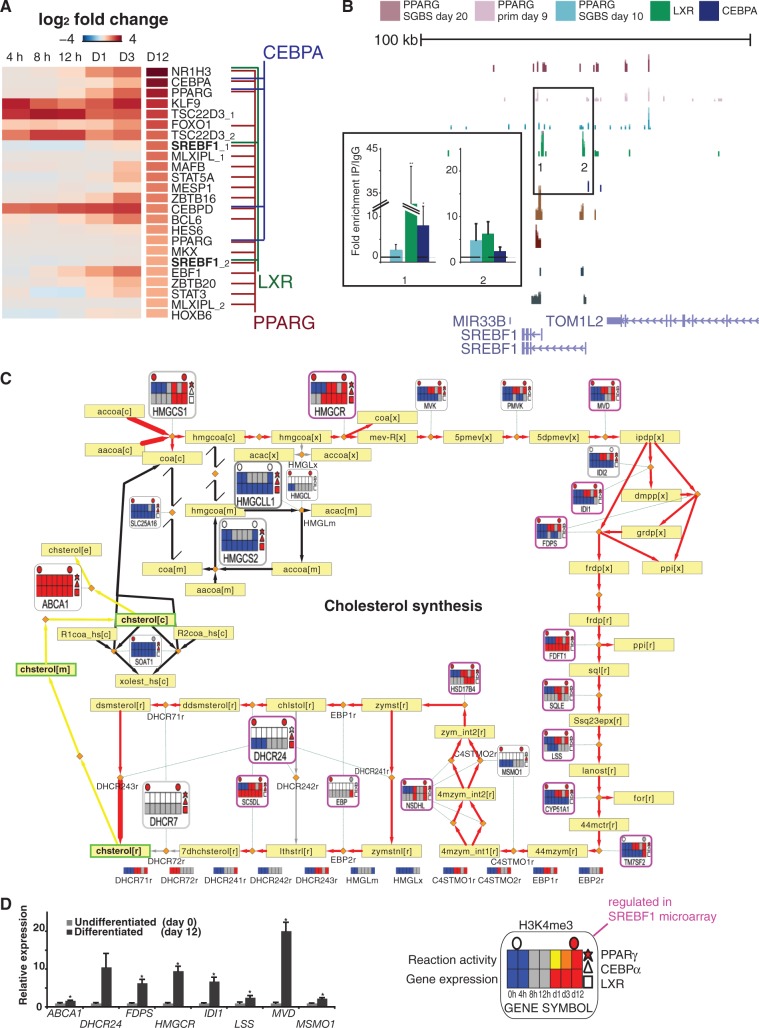
Defects in adipocyte differentiation represent an important early event in obesity and related metabolic dysfunction. To address the role of cell fate master regulators interfacing the metabolic and regulatory networks, heatmaps showing the top-ranked upregulated TFs according to differential expression between day 12 and day 0 are shown in Figure 6A. Focusing on target gene associations to high occupancy binding by the three TFs (lines connecting the studied TFs to upregulated genes), the prominent role for PPARγ in regulating other TFs in adipogenesis becomes apparent. Notably, each TF studied here binds its own regulatory region, while CEBPα and LXR show few high-occupancy interactions to the other upregulated TFs, in contrast to PPARγ. In fact, the only other TF association is the LXR binding to SREBF1.
Figure 6.
Integrated analysis of the regulation of the cholesterol synthesis pathway. (A) The average logarithmic fold change values from 4, 8 and 12 h and days 1, 3 and 12 of differentiation displayed as a heatmap and sorted based on day 12 values identify LXRA (NR1H3), CEBPA and PPARG as the most upregulated TF genes. Association to high-occupancy ChIP-seq peaks of PPARγ, CEBPα and LXR in SGBS cells are indicated with colored lines identifying autoregulation of each TF, regulation of SREBF1 by LXR and PPARγ, of CEBPD and PPARG by CEBPα and regulation of majority of TF genes shown by PPARγ. (B) The ChIP-seq signal tracks as in Figure 4 are shown at the SREBF1 locus. Regions with high enrichment for one or several TFs were selected for validation by ChIP-qPCR (numbered in the figure). The enrichment values using antibodies against all three TFs are shown relative to the enrichment of IgG and indicate the mean enrichment values of triplicate experiments and the error bars represent SEM. One sample t-test was performed to determine the significance of TF enrichment compared with IgG (*P < 0.05; **P < 0.01). (C) The metabolic pathway of cholesterol synthesis is shown with several omics data overlayed extending the metanode features presented in Figure 2. The pathway starts with the condensation of acetyl-CoA (accoa[c]) and acetoacetyl-CoA (aacoa[c]) to form 3-hydroxy-3-methylglutaryl-CoA (hmgcoa[c]) catalyzed by HMG-CoA synthase encoded by the gene HMGCS1. The end point metabolite is cholesterol (chsterol[r][m][c][e], r—endoplasmic reticulum, m—mitochondria, c—cytosol, e—extracellular). The start and end reactions are indicated by a thicker arrow and genes discussed further in the text are shown as larger metanodes for clarity. For details of heatmap, node and edge descriptions, see Figure 1, and for a complete list of metabolite names, Supplementary Table S7. Here, regulation in the SREBF1 microarray (48) is indicated by purple lining of the node. The reaction activity heatmap is blank if the gene is associated to multiple reactions with different predicted activity and in those cases the respective reaction activity heatmaps can be found below the pathway with the reaction naming matching those shown on the pathway. (D) RT-qPCR validation of the relative expression values of selected genes from the cholesterol synthesis pathway during SGBS differentiation. The measured expression values are shown normalized to RPL13A mRNA and relative to undifferentiated cells (set to 1). Data points indicate mean expression values of triplicate experiments and the error bars represent SEM. Student’s t-test was performed to determine the significance of upregulation (*P < 0.05).
We confirmed the binding of LXR to the prominent LXR peaks in the SREBF1 locus by ChIP-qPCR in adipocytes (enrichment was also observed for CEBPα), while the prominent PPARγ peak further upstream is supported by all three ChIP-seq studies (Figure 6B). According to our data, the cell fate regulating TFs form two closely connected feed-forward motifs to these two signal responsive TFs, both known to play key roles in cholesterol metabolism, with PPARγ associated to SREBF1 through both LXR and CEBPα (Figure 6A and Supplementary Figure S7).
As a representative of a ligand-responsive candidate drug target TF, we examined the high-occupancy LXR binding sites and confirmed binding to 11 previously reported LXR targets (Supplementary Table S3 in bold). Notably, all these genes were also upregulated during differentiation. To test the ligand-responsiveness of genes in the loci occupying the LXR binding sites (<500 kb from the TSS), we performed a microarray with the LXR agonist T0901317 (Supplementary Table S11) from a 4-h ligand stimulation of differentiated SGBS cells and could validate 39 additionally ligand-responsive genes at this early time point (Supplementary Table S12). Among known target genes, MYLIP1, SREBF1, ABCA1 and ABCG1 were significantly ligand responsive at this time point, while the upregulation of stearoyl-CoA delta-9-desaturase (SCD) was modest and did not pass the multiple testing correction (unadjusted P < 0.05). New putative target associations to ADH1B, TMEM17 and WSB1 were supported by the array data and high-occupancy binding sites. However, the majority of responsive genes were associated with less-prominent LXR binding under the unstimulated condition represented by the ChIP-seq experiment, including ELOVL6, LIPA, SCARB1, HSD17B7 and LPIN1 that function in lipid metabolism. Therefore, ligand stimulation greatly impacts LXR-mediated regulation of the metabolic network, in agreement with observations from mouse liver (49).
The interaction of PPARγ, LXR and SREBF1 has been studied before on selected target genes (50), and therefore our focus here was how they interact on pathway level. First, to confirm that SREBF1-mediated regulation converges with the pathways observed to change most during adipogenesis (Figure 3 and Supplementary Figure S2), we obtained a list of SREBF1-regulated genes observed in human myocytes (48). Cholesterol metabolism was the most significant pathway (hypergeometric P = 3.15e-06, Supplementary Table S13). The TF feed-forward circuits additionally engage the acyl-CoA synthetase enzyme genes, namely ACSL3, −4, −5 (Supplementary Figure S8) and the previously highlighted ACSL1 (Figure 5), in fatty acid activation that fuels triglyceride synthesis.
The integrative pathway map of cholesterol metabolism is shown in Figure 6C. Red edges represent predicted reaction activation from the preadipocyte state and black edges represent reactions predicted inactive. A detailed description of all the components can be found in the figure legend. In short, filled shapes indicate the putative binding of a TF (polygons on the right) or the presence of the active TSS marker (circles above) from ChIP-seq data. The central heatmaps display the discretized gene expression (below) and the reaction activity during differentiation as predicted by the mathematical model (above).
Based on the microarray data, all genes participating in the active branch of the cholesterol synthesis pathway, except CYP51A1, show an increased expression on differentiation. RT-qPCR validation for eight induced genes is displayed in Figure 6D. Genes involved in the inactive branch, namely 3-hydroxy-3-methylglutaryl-CoA (HMGC)-lyase-like-1, HMGC-synthase-2, SLC25A16 and SOAT1, show a constant low level of expression. To place the candidate regulatory motifs including SREBF1 (Supplementary Figure S7) in context of the ChIP-seq data, we used these integrative pathway maps to check which expression profiles potentially reflect more complex dynamics that can be achieved through feed-forward motifs by identifying those that could not be explained by simple direct binding by the three most up-regulated TFs selected for ChIP-seq.
Binding sites for at least one of the three TFs were associated to most genes in the pathway (81%). PPARγ has high-occupancy binding sites in the vicinity of HMGC-synthase 1 and mevalonate kinase (MVK) indicating that it may have a predominant role in regulating these enzymes at key upstream reactions of the synthesis pathway (Figure 6C, upper left corner). All TFs bound nearby the HMGC-reductase (HMGCR) gene locus encoding the rate limiting enzyme and gene loci encoding enzymes of the initial steps of the alternative ketogenesis pathway (mitochondrial) that start by producing HMGC from acetyl-CoA and acetoacetyl-CoA, namely HMGC-synthase 2 and HMGC-lyase-like-1. Concordant regulation by SREBF1 observed in myocytes concentrates on HMGCR, and, in particular, on the central and terminal parts of the pathway, including all genes starting from MVD. Among these, five were not bound by the other TFs in the ChIP-seq data, suggesting that their upregulation occurs indirectly via the regulation of SREBF1. These include farnesyl diphosphate synthase (FDPS) that has been reported to synthesize isoprenoid natural ligands for PPARγ (51), constituting a putative metabolite positive feedback loop. Interestingly, nodes associated to multiple TFs reappear at the end of the synthesis pathway (lower left corner) at the dehydrocholesterol reductases (DHCR)-7 and −24 loci, the latter being a known LXR target gene (52) in addition to ABCA1. However, only ABCA1 is significantly up-regulated in the microarray after 4 h of ligand stimulation (Supplementary Table S11 and S12), which suggests that more complex dynamics may be used to control the terminal step of the pathway at the DHCR24 locus.
In summary, a tight regulatory circuit between TFs necessary for adipocyte differentiation and those implicated in proper cholesterol homeostasis was observed and associated with the major lipid synthesis pathways. More generally, analyzing TF binding in context of the metabolic network allows formulating testable hypothesis about the regulatory mechanism.
Convergence of miRNAs and TFs exposes further disease-relevant nodes
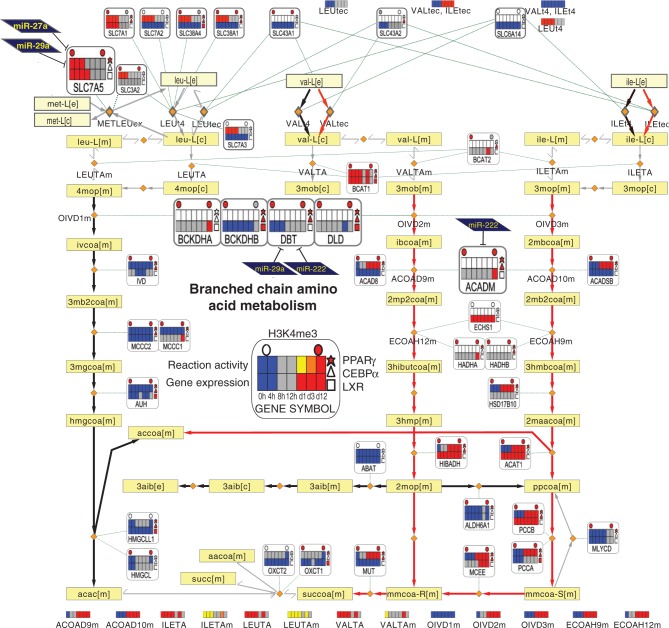
Our data also revealed several convergence points of miRNAs and TFs on metabolic genes (Supplementary Figure 5I). Figure 7 shows the BCAA catabolism pathway that represents the main nonlipid pathway highlighted here. The early steps include several genes that are upregulated during differentiation, two of which are potentially targeted by all three TFs studied, the branched chain keto acid dehydrogenase E1 beta (BCKDHB) and the dihydrolipoamide dehydrogenase (DLD), while DBT is a putative target of PPARγ, CEBPα, miR-222 and miR-29a. Together with BCKDHA, these genes in fact encode for the large protein complex called branched-chain alpha-keto acid dehydrogenase. According to our results, this enzyme complex is the key regulatory point under the control of multiple regulators and an interesting target for further analysis. Moreover, the cytosolic branched chain aminotransferase 1 (BCAT1) that catalyzes the first steps of the BCAA turnover in cytosol is associated with all 3 TFs. The upregulated genes downstream include propionyl CoA carboxylase beta (PCCB) and methylmalonyl CoA epimerase (MCEE) whose genomic loci are occupied by both PPARγ and CEBPα and PCCA that is associated to all three TFs. Thus, similarly to cholesterol synthesis pathway, the TFs studied converge especially at the initial and terminal steps of the pathway, with the added complexity of posttranscriptional regulators at the large main enzyme complex. As described earlier, the SLC7A5 gene that functions in BCAA transport is targeted by two miRNAs (miR-27a and miR-29a). The BCAA pathway from HUVECs is shown in Supplementary Figure S9. Among the miRNA target profiles available from HUVECs, a study of miR-663 targets reported regulation of SLC7A5 (54), and moreover, 9 out of 10 TFs in HUVECs were associated with this gene. As in adipocytes, the BCAT1 gene is associated with multiple TFs in HUVECs revealing key similarities between the regulator profiles and multi-regulator nodes of these cell types.
Figure 7.
Integrated metabolic pathway of BCAA metabolism. The BCAA (valine, leucine and isoleucine) metabolism pathway is shown. The metanode and edge representation is identical to Figure 6. While leucine degradation is predicted inactive in both preadipocytes and adipocytes (left part), the degradation of both valine and isoleucine is predicted to become active in adipocytes (red edges). The respective end products acetyl-CoA and succinyl-CoA can be fed into the TCA cycle. The important intermediates malonyl-CoA and acetoacetate link to lipogenesis or ketone body formation, respectively. This pathway has similarities with FAO, sharing the enzyme ACADM and the metabolite propionyl-CoA (ppcoa[m]). The metanodes indicate that two nodes are associated with both TFs and miRNAs: The component of the large multienzyme complex, DBT, is associated with PPARγ, CEBPα, miR-29a and miR-222, while among BCAA transporters contained in Recon1, SLC7A5 is associated to PPARγ, miR-27a and miR-29a. The genes discussed further in the text are shown as larger metanodes for clarity.
To visually assess the convergence of the studied TFs and miRNAs on the other highly activated metabolic pathways, we extended gene metanode metabolic maps to two additional pathways (fatty acid oxidation in Supplementary Figure S8 and triglyceride synthesis in Figure 8). All five maps, as well as the remaining 94 pathways in Recon1, and associated data can be interactively explored in our IDARE web portal (http://systemsbiology.uni.lu/idare.html).
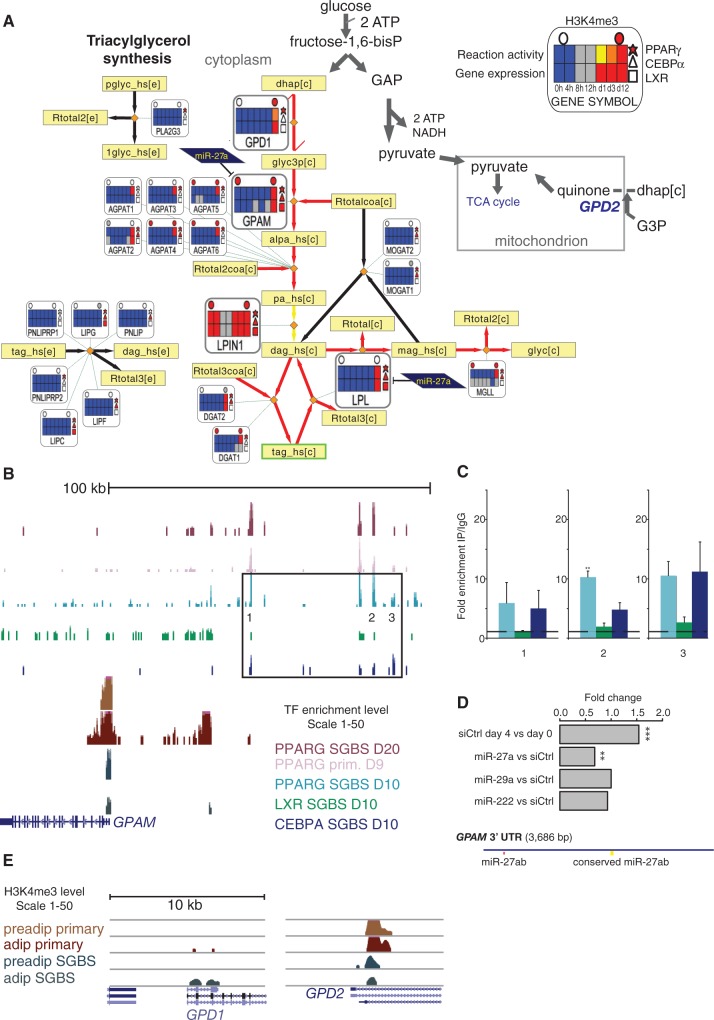
Figure 8.
Integrated metabolic pathway of triacylglycerol synthesis. (A) The triacylglycerol synthesis pathway from Recon1 (2) is shown. The metanode and edge representation is identical to Figure 6. This pathway represents the synthesis of triacylglycerol (tag_hs[c]) from glycerol-3-phosphate (glyc3p[c]), which can be generated from the reduction of dihydroxyacetone phosphate (dhap[c]) by Glycerol-3-phosphate dehydrogenase (GPD1) (55). The GPD1 and GPD2 genes encode enzymes that function in an opposing manner in the conversion between DHAP and G3P. The mitochondrial GPD2 functions in a catabolic pathway that fuels the TCA cycle, whereas GPD1 plays a role in triglyceride synthesis. The initial and committing step in glycerolipid biosynthesis is catalyzed by GPAM. GPAM, LPIN1 and LPL are all associated with all three TFs and in addition GPAM and LPL are targeted by miR-27a. The synthesis of both triacylglycerol and glycerol is predicted to shift to active in adipocytes. The genes discussed further in the text are shown as larger metanodes for clarity. (B) The ChIP-seq signal tracks as in Figure 5 are shown at the GPAM locus. Regions with high enrichment for one or several TFs were selected for validation by ChIP-qPCR (numbered in the figure). Each region was tested for enrichment using antibodies against all three TFs and IgG as a control as is shown in adjacent plots (C). The enrichment values are shown relative to the enrichment of IgG and indicate the mean enrichment values of triplicate experiments and the error bars represent SEM. One sample t-test was performed to determine the significance of TF enrichment compared with IgG (*P < 0.05; **P < 0.01). (D) The GPAM 3′UTR is shown with miRNA target predictions from TargetScan. Two binding sites for miR-27a can be seen, one of which is conserved and previously validated in mouse liver (56). Fold change values from miRNA mimic transfections are displayed from the microarray data. Two sample t-test was performed to determine the significance of silencing compared with siCtrl transfection (**adjusted P < 0.01; ***adjusted P < 0.001). (E) Signal tracks at the vicinity of their TSS regions of GPD1 and GPD2 show the H3K4me3 ChIP-seq signal from primary preadipocytes and adipocytes (10) compared with SGBS preadipocytes and adipocytes. At the GPD1 locus, the H3K4me3 signal increases in SGBS and primary adipocytes, whereas a decrease in signal is observed at the GPD2 TSS in SGBS cells.
MiR-27a is known to engage in the main TF circuitry through the inhibition of PPARγ (15–17). The triglyceride pathway was identified to contain multiple shared target associations, as shown in Figure 8. The regulatory associations from TFs and miRNAs converge along the pathway at three key enzymes: (i) GPAM that catalyzes the initial and committing step in glycerolipid biosynthesis, playing a pivotal role in the regulation of cellular triacylglycerol and phospholipid levels (57), (ii) LPIN1 that catalyzes the penultimate step in triglyceride synthesis including the dephosphorylation of phosphatidic acid to yield diacylglycerol (23) and (iii) LPL that catalyzes the release of fatty acids from triglycerides (24) [extracellular LPL facilitates fatty acid import, whereas also intracellular activity has been observed that could serve in fatty acid export (58)]. The enzymes from reactions directly connected to these highly regulated gene nodes are also upregulated and associated with PPARγ (and some CEBPα) binding, including the AGPAT gene family members (directly downstream GPAM), the triacylglycerol synthesizing DGAT1 and DGAT2, and the lipase MGLL, supporting a tight transcriptional regulation of triglyceride synthesis spread across the pathway. We selected the GPAM locus for further validation experiments, as it was the first enzyme targeted by both TFs and miR-27a. We could confirm binding to several prominent peaks upstream of the GPAM locus (Figure 8B and C). Furthermore, miRNA motif analysis of the 3′UTR revealed two miR-27a binding sites, one of which corresponds to a conserved site that has been shown functional in mice (56). In agreement, transfection with miR-27a mimic, but not that of mir-29a or mir-222, significantly decreased GPAM mRNA levels (Figure 8D).
Finally, we also examined the H3K4me3 data in context of the pathways identified to change most. A reciprocal change in the TSS activity affecting carbohydrate and lipid metabolism was observed for two genes encoding enzymes involved in glycerol metabolism: the glycerol-3-phosphate dehydrogenases GPD1 and GPD2. Glycerol-3-phosphate (G3P) can be synthesized from glucose via an intermediate step that forms dihydroxyacetone phosphate (DHAP). This metabolite and NADH are converted to G3P by GPD1, and G3P can subsequently be converted to lipids (55). GPD1 increased H3K4me3 levels in differentiated cells, also in primary adipocytes (Figure 8E), matching its expression profile with a 7.9-fold increase in transcription (Supplementary Table S5). GPD2 in turn can convert G3P to quinone to fuel mitochondrial oxidation; in agreement with a shift to lipogenic metabolism its TSS activity was repressed (Figure 8E) and a low level of expression maintained.
Altogether, we obtained four novel genome-wide target gene profiles associating the TF LXRα and the miRNAs −27a, −29a and −222 with their likely target genes in human adipocytes to support the analysis performed using public data of TF binding for additional 12 TFs from adipocyte and endothelial cells. Such data on regulatory factors are highly relevant to describe their role in different cell types and potentially in driving disease. However, often the volume of data from genome-wide assays hinders tangible conclusions to be drawn. We provide here an easily accessible tool to analyze links between the metabolic and regulatory networks, to identify different regulatory mechanisms at the pathway level and for the discovery of key nodes as demonstrated here using the convergence of regulators to highlight such genes in the two cell types.
DISCUSSION
Despite the growing amount of data collected from gene expression studies, a common framework or a model to capture the key systems properties is often lacking. Here, we collect a comprehensive data set from own experiments and public data focusing on two key cell types implicated in the pathogenesis of metabolic syndrome, namely endothelial cells and adipocytes. To study in an integrated manner how components of transcriptional and posttranscriptional regulation impact the expression of metabolic genes, we introduce gene metanodes and the web portal IDARE (Integrated Data nodes of Regulation) for interactive data exploration of various data types within the metabolic network context.
The endothelium-derived relaxing factor NO and the catalyzing enzymes nitric oxide synthases that generate NO from the amino acid l-arginine represent a key discovery in CVD research. Together with genes implicated in hereditary monogenic disease (ALDH4A1) (38) or in recent GWAS studies (MTAP) (39), these enzymes from the arginine-proline metabolic pathway represent those associated with most TF binding in our analysis of 10 ChIP-seq studies from HUVEC cells.
TFs represent key factors to establish a cellular phenotype; however, they do not function in isolation. For a more comprehensive view, the analysis was extended from TFs to include the evaluation of chromatin marker levels and miRNAs. Three most upregulated TFs (PPARγ, CEBPα and LXRα) and prominent members of miRNA families (miR-27a, miR-29a and miR-222) downregulated on adipocyte differentiation were selected for genome-wide analysis. We validated the combinatorial binding in ChIP-qPCR and repression of mRNA using miRNA mimics for GPAM, representing a gene associated with TF and miRNA binding at a committing step in glycerolipid biosynthesis. Supporting the relevance of maintaining tight regulation of its expression level, Brockmöller et al. (59) reported a significant association between lowered GPAM mRNA levels and poor survival in breast cancer, a misregulation that in context of our results could be linked to the increased expression levels of miR-27a reported in invasive breast cancers (60). These results are highly supportive of the relevance of regulatory associations following the approach used here. However, mammalian regulatory regions may overlap and span across gene boundaries resulting inevitably in some false target gene associations. As further possible caveats, target mRNA dynamics impact detection of the miRNA regulatory effect and ChIP-seq signal only implies TF binding that constitutes a necessary, but not sufficient, condition to alter mRNA synthesis.
Despite possible inaccuracies in representing true regulatory interactions, the integrated analysis on metabolic pathway regulation clearly implicated the dyslipidemia loci LPL (24) and LPIN1 (23) as well as LDLR (22) (the latter is missing from Recon1), each associated with multiple TFs and miRNAs. Furthermore, the GPD1 locus that we identify among genes with increased TSS marker levels has recently been described to cause infantile hypertriglyceridemia (61). Thus, our analysis holds promise to identify key regulatory nodes that are important in different diseases by using microarray and sequencing data that are readily available from multiple tissues.
Our data extends data collected on PPARγ and CEBPα in human adipocytes (10,40,41,44), while for LXR and miR-27a, our genome-wide data on target genes are the first reported in human adipocytes and can be compared with data obtained from liver (49,56) and foam cells (62). PPARγ and CEBPα represent cell fate determining TFs widely studied in context of adipocytes. However, their interplay with signal-dependent TFs is less well understood, including LXRα that increased at expression level most during differentiation. The first glimpse to the LXR genome-wide binding profile through ChIP-seq showed binding in a few hundred to few thousand regions (high- versus low-occupancy cutoff), in agreement with a similar number of binding sites reported from unstimulated mouse liver cells (49). Most strikingly, among all upregulated TFs, only SREBF1 was associated with TFs other than PPARγ, being bound by LXR and CEBPα. The cholesterol synthesis and fatty acid activation pathways were each associated with this putative multi-TF feed-forward circuit. It is intriguing that precisely these pathways have been reported to contribute to generation of endogenous PPARγ ligands (51,63), potentially providing a metabolite positive feedback to substantiate transcriptional autoactivation required for cell differentiation.
On activating signal, our microarray using an LXR agonist revealed regulation of several genes that in our ChIP-seq data were initially associated with low occupancy binding. Similar study in mouse liver (49) showed a dramatic increase in peak height on agonist activation, suggesting that the ligand-bound receptor may be more efficiently recruited to its genomic target loci. It will therefore be of interest to test the ligand-dependent binding profile also in adipocytes. LXRs have been shown to play critical roles in the regulation of overall cholesterol catabolism, absorption and transport in the intestine, macrophages and liver (42). Furthermore, the LXR target gene MYLIP that inhibits the LDLR pathway by targeting LDLR to proteasomal degradation (64) is a likely target gene of miR-222 according to our microarray and seed analysis, exposing a CVD-relevant novel regulatory factor in the cholesterol intercellular trafficking pathway.
The miRNA with most interaction with TFs (including regulation of PPARG) is miR-27a. The regulation of GPAM by miR-27b was recently described in mouse liver (56) and is supported by our microarray and heptamer analysis in human adipocytes. Moreover, we observe multiple other genes that are posttranscriptionally regulated along the pathway. These target associations include the LDLR, LPL and LRP5 that function in lipid transport. It is worth pointing out that also LPIN1 is among genes that have a modest downregulation on miR-27a transfection, in agreement with existence of a well-conserved binding site in its 3′-UTR. However, further validation is required to ascertain its regulation. Participation of carbohydrate metabolism in fueling the triacylglycerol synthesis is supported by the switch in regulation of GPD genes. The upstream enzyme hexokinase-2 that phosphorylates and thereby activates glucose, similar to GPAM, LPIN1 or LPL, was identified among the list of target genes associated with all TFs and miR-27a. Based on the earlier reports and our combined microarray and 3′-UTR heptamer analysis, miR-27 family is establishing itself as a key miRNA regulator of the triacylglycerol metabolism.
Interestingly, both miR-222 and miR-29a regulate the BCKD complex in the BCAA catabolism. Recently, increased levels of the BCAAs were shown to play an important role in diabetes (65). In a model proposed by Newgard (66), and supported by experiments applying in vivo mouse models (67), an obesity-related decline in BCAA catabolism in adipose tissue drives the rise of circulating levels of these amino acids. The model suggests that readily usable glucose and lipid substrates may obviate the need for amino acid catabolism in adipose tissue. However, the mechanism by which increased supply of these substrates causes downregulation of the BCAA catabolic enzymes is unknown. Drugs that activate PPARγ (thiazolidindiones or TZDs) can restore expression of the catabolic genes to normal (68), already suggesting a role of suppressed PPAR signaling in this metabolic adaptation. The miR-29 family is implicated in diabetes based on studies of hepatic gluconeogenesis in diabetic rat models (69). Based on our data, it will be relevant not only to study PPARγ, but to include CEBPα, miR-27a, miR-29a and miR-222 as other key regulators of this pathway, and by that potentially further elucidate the novel link of the BCAA pathway to diabetes.
Both miRNAs targeting the DBT subunit of the BCKD complex according to our data (miR-29a and miR-222) are upregulated in the adipose tissue of diabetic rats and are induced by increased glucose levels in mouse adipocytes (70,71). Moreover, the targeting of DBT by miR-29 family has already been validated in other cell types (72), making the combinatorial repression of DBT by the miRNAs one likely explanation for lowered catabolism of BCAAs in diabetic adipose tissue. In addition to the initial steps of BCAA catabolism, also the BCAA transport step mediated by SLC7A5 appears to be a highly regulated node possibly contributing to the diabetic phenotype. On top of being associated as a PPARγ target in our analysis, SLC7A5 appears to be targeted by miR-27a and miR-29a, both glucose responsive and induced in diabetic condition (71). The complexity of SLC7A5 regulation is further increased when looking at HUVEC data that reveal it as a potential target of as many as 9 TFs and an additional miRNA (miR-663) in the endothelial cells and in context of recent literature implicating it in key metabolic changes required for T-cell differentiation (73).
As an initial means to discover key pathways, we used the gene expression levels as soft constraints to obtain predictions for metabolic activity in Recon1, a generic model of human metabolism (2,6). Several other methods for the integration of expression data on genome scale metabolic networks have been and are currently being developed (74) and will be important to benchmark and consolidate the prediction results in future studies. Transcript-level measurements address the space of available network states that translational control and posttranslational modifications further fine tune [for HMGCR this is well established (53)]. The metanodes enable mapping and visualization of further data onto metabolic pathways, facilitating data exchange and hypothesis-driven research in context of the metabolic network. Here, two trends in transcriptional regulation were observed: (i) shared and high-occupancy binding nearby gene loci of the initial and terminal steps of a pathway (the cholesterol synthesis and the BCAA pathways), a type of transcriptional regulation that has been reported advantageous for fast responses to environmental conditions in pathways with low protein synthesis cost (75), and (ii) tight regulation spread along the entire pathway (triacylglycerol synthesis), which might link to the tight transcriptional regulation on pathways spanning high cost enzymes (75).
In conclusion, the analysis of genomic and transcriptomic data linked with a metabolic network model is useful as a means to explore high-throughput data in a global manner, revealing genes implicated in disease as convergence points of regulation. To focus on metabolic pathways that differ in activity comparing two phenotypes, constraint-based modeling to predict active metabolic pathways can be included. The putative shared TF and miRNA target genes from pathways activated during human adipocyte differentiation that our new data sets revealed were ACADM, DBT, GPAM, HK2, LPL and SLC7A5. Genes associated with all adipocyte TFs studied further include ABCA1, ACSL1, the acetyl-CoA acetyltransferase 2 (ACAT2), the BCAT1; two more genes from the branched-chain alpha-keto acid dehydrogenase complex, namely BCKDHB and the DLD; four genes from the cholesterol synthesis pathway DHCR7, HMGCS2, HMGCLL1, HMGCR; and three other lipase genes from triglyceride metabolism, namely lipase C, lipase G and LPIN1. These data can now be compared with published data sets such as those from HUVECs using the IDARE tool. Our workflow extends from current routines in which these disparate but complementary types of cellular information are kept apart and further motivates study of biological processes from an integrative point-of-view.
ACCESSION NUMBERS
The microarray and deep sequencing data from this publication have been submitted to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) and assigned the identifier GSE41578 and can be explored in context of the pathways described using the web resource at http://systemsbiology.uni.lu/idare.html, including a track hub for UCSC Genome Browser that allows fast visualization of ChIP-seq signal tracks at any gene locus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds National de la Recherche Luxembourg [AFR, AM2c and C08/BM/01]; funding from the University of Eastern Finland and the Fondation du Pélican de Mie et Pierre Hippert-Faber under the aegis of Fondation de Luxembourg. Funding for open access charge: University of Luxembourg.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Dr Anna-Liisa Levonen, Prof. Ilya Shmulevich and Prof. Carsten Carlberg for comments and suggestions to this manuscript, Prof. Martin Wabitsch for kindly providing the SGBS cell line, Dr Echardt Treuter for kindly providing the LXR antibody, Martine Schmitz for testing the antibodies used for ChIP-seq, Dr Patrick May, Fotis Georgatos, Christian Hutter and Dr Reinhard Schneider for providing access to the LCSB-Portal server and the high-throughput data service providers at Microarray and sequencing facility of Turku Centre for Biotechnology, at DNA Vision, Charleroi, Belgium and at EMBL Genomics Core at Heidelberg. M.H. and L.S. designed and performed the microarray time series and ChIP-seq experiments. L.S. designed and performed the miRNA array and miRNA mimic transfection experiments and ChIP-qPCR experiments. M.G. performed the constraint-based modeling jointly implemented together with T.S. M.H. performed the statistical analysis of the microarray data and P.B. designed and performed the miRNA motif enrichment and target association analysis. M.G. and L.S. designed and performed the RT-qPCR validations. M.H. and M.G. analyzed the ChIP-seq data. J.L., M.G., M.H. and T.S. designed the online web tool that J.L. and M.G. implemented. All authors participated in designing the integrative analysis and writing of the manuscript.
REFERENCES
- 1.World Health Organization. Global status report on noncommunicable diseases 2010. 2000. World Health Organization, Geneva, Switzerland. http://www.who.int/nmh/publications/ncd_report2010/en/
- 2.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiele I, Swainston N, Fleming RMT, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolfsson O, Palsson BØ, Thiele I. The human metabolic reconstruction Recon 1 directs hypotheses of novel human metabolic functions. BMC Syst. Biol. 2011;5:155. doi: 10.1186/1752-0509-5-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suarez RK, Moyes CD. Metabolism in the age of “omes”. J. Exp. Biol. 2012;215:2351–2357. doi: 10.1242/jeb.059725. [DOI] [PubMed] [Google Scholar]
- 6.Shlomi T, Cabili MN, Herrgård MJ, Palsson BØ, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 7.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Børgesen M, Francoijs K-J, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol. Cell. Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho N, Momose Y. Peroxisome proliferator-activated receptor gamma agonists as insulin sensitizers: from the discovery to recent progress. Curr Medic Chem. 2008;8:1483–1507. doi: 10.2174/156802608786413474. [DOI] [PubMed] [Google Scholar]
- 12.Ono K. Current concept of reverse cholesterol transport and novel strategy for atheroprotection. J Cardiol. 2012;60:339–343. doi: 10.1016/j.jjcc.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Mudhasani R, Imbalzano AN, Jones SN. An essential role for Dicer in adipocyte differentiation. J. Cell. Biochem. 2010;110:812–816. doi: 10.1002/jcb.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl Acad. Sci. USA. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, Dani C, Amri E-Z, Scheideler M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 2009;390:247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 16.Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee J-W, Lee YS, Kim JB. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 2010;392:323–328. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009;276:2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol. Endocrinol. 2009;23:925–931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John E, Wienecke-Baldacchino A, Liivrand M, Heinäniemi M, Carlberg C, Sinkkonen L. Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucleic Acids Res. 2012;40:1–15. doi: 10.1093/nar/gks025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wabitsch M, Brenner RE, Melzner I, Braun M, Möller P, Heinze E, Debatin KM, Hauner H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 22.Francke U, Brown MS, Goldstein JL. Assignment of the human gene for the low density lipoprotein receptor to chromosome 19: synteny of a receptor, a ligand, and a genetic disease. Proc. Natl Acad. Sci. USA. 1984;81:2826–2830. doi: 10.1073/pnas.81.9.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Péterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 24.Harlan WR, Winesett PS, Wasserman AJ. Tissue lipoprotein lipase in normal individuals and in individuals with exogenous hypertriglyceridemia and the relationship of this enzyme to assimilation of fat. J. Clin. Invest. 1967;46:239–247. doi: 10.1172/JCI105526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eichler GS, Huang S, Ingber DE. Gene expression dynamics inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 26.Newman AM, Cooper JB. AutoSOME: a clustering method for identifying gene expression modules without prior knowledge of cluster number. BMC Bioinf. 2010;11:117. doi: 10.1186/1471-2105-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castoldi M, Schmidt S, Benes V, Hentze MW, Muckenthaler MU. miChip: an array-based method for microRNA expression profiling using locked nucleic acid capture probes. Nat. Protoc. 2008;3:321–329. doi: 10.1038/nprot.2008.4. [DOI] [PubMed] [Google Scholar]
- 28.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berninger P, Gaidatzis D, Van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr. Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 32.Bauer-Mehren A, Rautschka M, Sanz F, Furlong LI. DisGeNET: a Cytoscape plugin to visualize, integrate, search and analyze gene-disease networks. Bioinformatics. 2010;26:2924–2926. doi: 10.1093/bioinformatics/btq538. [DOI] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hebenstreit D, Gu M, Haider S, Turner DJ, Liò P, Teichmann SA. EpiChIP: gene-by-gene quantification of epigenetic modification levels. Nucleic acids Res. 2011;39:e27. doi: 10.1093/nar/gkq1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ignarro LJ. Endothelium-derived nitric oxide: pharmacology and relationship to the actions of organic nitrate esters. Pharm. Res. 1989;6:651–659. doi: 10.1023/a:1015926119947. [DOI] [PubMed] [Google Scholar]
- 38.Geraghty MT, Vaughn D, Nicholson AJ, Lin WW, Jimenez-Sanchez G, Obie C, Flynn MP, Valle D, Hu CA. Mutations in the Delta1-pyrroline 5-carboxylate dehydrogenase gene cause type II hyperprolinemia. Hum. Mol. Genet. 1998;7:1411–1415. doi: 10.1093/hmg/7.9.1411. [DOI] [PubMed] [Google Scholar]
- 39.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt SF, Jørgensen M, Chen Y, Nielsen R, Sandelin A, Mandrup S. Cross species comparison of C/EBPα and PPARγ profiles in mouse and human adipocytes reveals interdependent retention of binding sites. BMC Genomics. 2011;12:152. doi: 10.1186/1471-2164-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soccio RE, Tuteja G, Everett LJ, Li Z, Lazar MA, Kaestner KH. Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol. Endocrinol. 2011;25:694–706. doi: 10.1210/me.2010-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siersbæk MS, Loft A, Aagaard MM, Nielsen R, Schmidt SF, Petrovic N, Nedergaard J, Mandrup S. Genome-wide profiling of peroxisome proliferator-activated receptor γ in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol. Cell. Biol. 2012;32:3452–3463. doi: 10.1128/MCB.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanter JE, Tang C, Oram JF, Bornfeldt KE. Acyl-CoA synthetase 1 is required for oleate and linoleate mediated inhibition of cholesterol efflux through ATP-binding cassette transporter A1 in macrophages. Biochim Biophys Acta. 2012;1821:358–364. doi: 10.1016/j.bbalip.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregersen LH, Jacobsen A, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC cancer. 2012;12:232. doi: 10.1186/1471-2407-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwee SA, Hernandez B, Chan O, Wong L. Choline kinase alpha and hexokinase-2 protein expression in hepatocellular carcinoma: association with survival. PloS One. 2012;7:e46591. doi: 10.1371/journal.pone.0046591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rome S, Lecomte V, Meugnier E, Rieusset J, Debard C, Euthine V, Vidal H, Lefai E. Microarray analyses of SREBP-1a and SREBP-1c target genes identify new regulatory pathways in muscle. Physiol. Genomics. 2008;34:327–337. doi: 10.1152/physiolgenomics.90211.2008. [DOI] [PubMed] [Google Scholar]
- 49.Boergesen M, Pedersen TÅ, Gross B, Van Heeringen SJ, Hagenbeek D, Bindesbøll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin Y, Dalen KT, Gustafsson J-A, Nebb HI. Regulation of hepatic fatty acid elongase 5 by LXRalpha-SREBP-1c. Biochim. Biophys. Acta. 2009;1791:140–147. doi: 10.1016/j.bbalip.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Goto T, Nagai H, Egawa K, Kim Y-I, Kato S, Taimatsu A, Sakamoto T, Ebisu S, Hohsaka T, Miyagawa H, et al. Farnesyl pyrophosphate regulates adipocyte functions as an endogenous PPARγ agonist. Biochem. J. 2011;438:111–119. doi: 10.1042/BJ20101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Rogers PM, Stayrook KR, Su C, Varga G, Shen Q, Nagpal S, Burris TP. The selective Alzheimer’s disease indicator-1 gene (Seladin-1/DHCR24) is a liver X receptor target gene. Mol. Pharmacol. 2008;74:1716–1721. doi: 10.1124/mol.108.048538. [DOI] [PubMed] [Google Scholar]
- 53.Gibson DM, Parker RA, Stewart CS, Evenson KJ. Short-term regulation of hydroxymethylglutaryl coenzyme A reductase by reversible phosphorylation: modulation of reductase phosphatase in rat hepatocytes. Adv. Enzyme Regul. 1982;20:263–283. doi: 10.1016/0065-2571(82)90020-6. [DOI] [PubMed] [Google Scholar]
- 54.Ni C-W, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am. J. Physiol.: Heart Circ. Physiol. 2011;300:H1762–H1769. doi: 10.1152/ajpheart.00829.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 56.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2012;01:1–10. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dircks LK, Sul HS. Mammalian mitochondrial glycerol-3-phosphate acyltransferase. Biochim. Biophys. Acta. 1997;1348:17–26. doi: 10.1016/s0005-2760(97)00106-9. [DOI] [PubMed] [Google Scholar]
- 58.McNamara JP, Azain M, Kasser TR, Martin RJ. Lipoprotein lipase and lipid metabolism in muscle and adipose tissues of Zucker rats. Am. J. Physiol. 1982;243:R258–R264. doi: 10.1152/ajpregu.1982.243.3.R258. [DOI] [PubMed] [Google Scholar]
- 59.Brockmöller SF, Bucher E, Müller BM, Budczies J, Hilvo M, Griffin JL, Orešič M, Kallioniemi O, Iljin K, Loibl S, et al. Integration of metabolomics and expression of glycerol-3-phosphate acyltransferase (GPAM) in breast cancer–link to patient survival, hormone receptor status and metabolic profiling. J. Proteome Res. 2011;11:850–860. doi: 10.1021/pr200685r. [DOI] [PubMed] [Google Scholar]
- 60.Tang W, Zhu J, Su S, Wu W, Liu Q, Su F, Yu F. MiR-27 as a prognostic marker for breast cancer progression and patient survival. PLoS One. 2012;7:e51702. doi: 10.1371/journal.pone.0051702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basel-Vanagaite L, Zevit N, Zahav AH, Guo L, Parathath S, Pasmanik-Chor M, McIntyre AD, Wang J, Albin-Kaplanski A, Hartman C, et al. Transient infantile hypertriglyceridemia, fatty liver, and hepatic fibrosis caused by mutated GPD1, encoding glycerol-3-phosphate dehydrogenase 1. Am. J. Hum. Genet. 2012;90:49–60. doi: 10.1016/j.ajhg.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feldmann R, Fischer C, Kodelja V, Behrens S, Haas S, Vingron M, Timmermann B, Geikowski A, Sauer S. Genome-wide analysis of LXRα activation reveals new transcriptional networks in human atherosclerotic foam cells. Nucleic Acids Res. 2013;41:3518–3531. doi: 10.1093/nar/gkt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiraki T, Kamiya N, Shiki S, Kodama TS, Kakizuka A, Jingami H. Alpha,beta-unsaturated ketone is a core moiety of natural ligands for covalent binding to peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 2005;280:14145–14153. doi: 10.1074/jbc.M500901200. [DOI] [PubMed] [Google Scholar]
- 64.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCormack SE, Shaham O, McCarthy MA, Deik AA, Wang TJ, Gerszten RE, Clish CB, Mootha VK, Grinspoon SK, Fleischman A. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr. Obes. 2012;8:52–61. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herman MA, She P, Peroni OD, Lynch CJ, Kahn BB. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA levels. J. Biol. Chem. 2010;285:11348–11356. doi: 10.1074/jbc.M109.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsiao G, Chapman J, Ofrecio JM, Wilkes J, Resnik JL, Thapar D, Subramaniam S, Sears DD. Multi-tissue, selective PPARγ modulation of insulin sensitivity and metabolic pathways in obese rats. Am. J. Physiol.: Endocrinol. Metab. 2011;300:E164–E174. doi: 10.1152/ajpendo.00219.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang J, Liu C, Qiao A, Cui Y, Zhang H, Cui A, Zhang S, Yang Y, Xiao X, Chen Y, et al. MicroRNA-29a-c decrease fasting blood glucose levels by negatively regulating hepatic gluconeogenesis. J. Hepatol. 2012;58:535–542. doi: 10.1016/j.jhep.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 70.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 71.Herrera BM, Lockstone HE, Taylor JM, Ria M, Barrett A, Collins S, Kaisaki P, Argoud K, Fernandez C, Travers ME, et al. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53:1099–1109. doi: 10.1007/s00125-010-1667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mersey BD, Jin P, Danner DJ. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum. Mol. Genet. 2005;14:3371–3377. doi: 10.1093/hmg/ddi368. [DOI] [PubMed] [Google Scholar]
- 73.Sinclair LV, Rolf J, Emslie E, Shi Y-B, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blazier AS, Papin JA. Integration of expression data in genome-scale metabolic network reconstructions. Front. Physiol. 2012;3:299. doi: 10.3389/fphys.2012.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wessely F, Bartl M, Guthke R, Li P, Schuster S, Kaleta C. Optimal regulatory strategies for metabolic pathways in Escherichia coli depending on protein costs. Mol. Syst. Biol. 2011;7:515. doi: 10.1038/msb.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.