Abstract
Type I toxin–antitoxin loci consist of two genes: a small, hydrophobic, potentially toxic protein, and a small RNA (sRNA) antitoxin. The sRNA represses toxin gene expression by base pairing to the toxin mRNA. A previous bioinformatics search predicted a duplicated type I locus within Escherichia coli O157:H7 (EHEC), which we have named the gene pairs zorO-orzO and zorP-orzP. We show that overproduction of the zorO gene is toxic to E. coli; co-expression of the sRNA OrzO can neutralize this toxicity, confirming that the zorO-orzO pair is a true type I toxin–antitoxin locus. However, OrzO is unable to repress zorO in a strain deleted for RNase III, indicating that repression requires cleavage of the target mRNA. Sequence analysis and mutagenesis studies have elucidated a nucleotide sequence region (V1) that allows differential recognition of the zorO mRNA by OrzO and not OrzP, and a specific single nucleotide within the V1 of OrzO that is critical for repression of zorO. Although there are 18 nt of complementarity between the OrzO sRNA and the zorO mRNA, not all base pairing interactions are needed for repression; however, the amount needed is dependent on whether there is continuous or discontinuous complementarity to the target mRNA.
INTRODUCTION
As a newly recognized class of gene expression regulators, small noncoding RNAs have been found in all kingdoms of life. In bacteria, these RNAs are referred to as small RNAs (sRNAs) and are often 50–300 nt in length. Some sRNAs bind to proteins and modulate their functions, as exemplified by the CsrB RNA of Escherichia coli, which directly binds and sequesters the CsrA protein, thus affecting carbon metabolism in E. coli (1). However, the majority of characterized sRNAs function by base pairing to target mRNAs, either regulating the translation or altering the stabilities of the targets.
These base pairing sRNAs fall into two categories: trans-encoded sRNAs and antisense sRNAs. Trans-encoded sRNAs are located at chromosomal positions distal from the genes they regulate. These sRNAs typically interact with their targets by limited base pairing and often require the RNA chaperone Hfq to function [reviewed in (2)]. True antisense sRNAs are encoded on the opposite strand of DNA to their targets. They share extensive base pairing potential (usually ≥60 nt) to their target mRNAs [reviewed in (3,4)]. Although antisense sRNAs were initially discovered on mobile genetic elements, several pairs have been described on bacterial chromosomes [reviewed in (5)].
A subset of antisense sRNAs in bacteria can repress the expression of small proteins <60 amino acids in length. These proteins are usually hydrophobic and toxic when overproduced. The sRNA (antitoxin) base pairs with the toxin mRNA and affects the translation and/or the stability of the mRNA. Gene pairs consisting of a toxic protein-encoding gene and a corresponding antitoxin sRNA-encoding gene are referred to as type I toxin–antitoxin systems. The first type I pair to be identified was the hok/sok locus of the R1 plasmid in E. coli, whereby Hok is the toxin, and Sok the antitoxin (6). The hok mRNA has a long half-life, whereas the Sok sRNA has a short half-life. When cell division occurs, a daughter cell that has not inherited the R1 plasmid will die owing to the quick degradation of the Sok sRNA and the translation of the more stable hok mRNA. Besides the hok/sok locus, several type I toxin–antitoxin gene pairs have been described on other plasmids in gram negative bacteria and were also shown to play a role in plasmid maintenance [reviewed in (7)]. Additionally, the RNAI-RNAII locus of the pAD1 plasmid of Enterococcus faecalis clearly showed that similar loci were present in gram positive bacteria (8–10). Homologs of these plasmid loci were later found within bacterial chromosomes (11–14).
In recent years, several type I toxin–antitoxin systems that do not have homology to plasmid sequences have been identified and characterized in a number of bacterial chromosomes. Although both the toxin and antitoxin-encoding genes are located at the same locus, their relative positioning varies among the chromosomally encoded type I pairs. Some antitoxins overlap with the 5′ untranslated region (UTR) of their targets, such as the Rdl sRNAs of the Ldr-Rdl family in E. coli (15), or the 3′ UTR, like RatA and SR4 of the TxpA-RatA and BsrG/SR4 loci of Bacillus subtilis (16,17). Others directly overlap with the coding sequences of the target mRNAs, as exemplified by the Ibs-Sib family of E. coli in which Sib sRNA completely overlaps with the ibs mRNA coding region (18).
Along with these, there are a few unconventional type I loci in which the antitoxins are encoded divergent from the toxin-encoding genes [reviewed in (19)]. Unlike the traditional type I loci that can have ≥60 nt base paring potential, the unconventional type I loci have limited base pairing, often only 18–21 nt. These loci include the TisAB-IstR and the ShoB-OhsC pairs in E. coli (18,20).
A previous bioinformatics search identified two highly homologous loci encoded in tandem in the chromosome of E. coli O157:H7 EDL933 (EHEC) (14). Each locus includes two genes: one encodes a small hydrophobic protein, and the other encodes an sRNA that is located divergently from the protein-encoding gene. This genetic organization is similar to that of the type I toxin–antitoxin pairs ShoB-OhsC and TisAB-IstR. In the two newly identified loci, the small protein-encoding genes (29 amino acids) were previously annotated as z3289 and z3290. We have since renamed z3289 and z3290 as zorO (Z protein often repeated) and zorP, respectively. Additionally, we have denoted the sRNA gene divergent from zorO as orzO (overexpression represses zor toxicity) and the sRNA gene divergent from zorP as orzP (Figure 1A). The sRNAs contain regions of complementarity (18–19 nt) to the mRNAs, suggesting that they could base pair. In addition, the zor-orz loci are highly conserved in most commensal and pathogenic E. coli and Shigella strains but not in the common lab strains, like E. coli MG1655 (14).
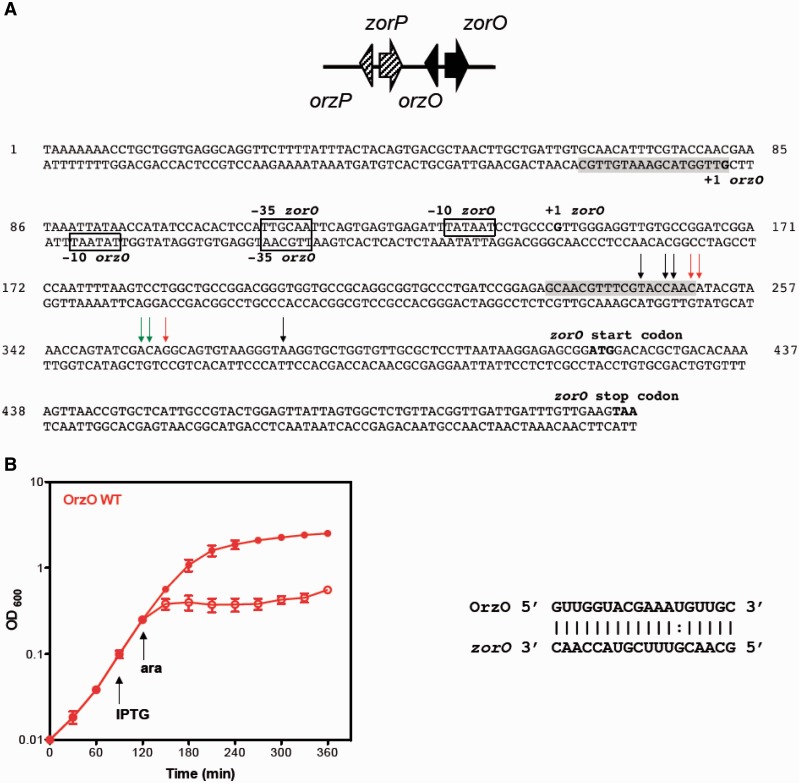
Figure 1.
The zorO-orzO gene locus is a true type I toxin–antitoxin system. (A) Genetic organization of the duplicated zor-orz locus in E. coli EDL933. Sequence details of the zorO-orzO genes; region of base pairing is shaded, and previously mapped transcription starts are indicated in bold (14). Predicted −35 and −10 promoter elements are boxed. Black arrows indicate RNA processing products found in both the wild type and Δrnc strain; green arrows, wild type strain only; red arrows, Δrnc strain only. (B) Overexpression of OrzO can repress zorO-mediated toxicity. E. coli strain UTK007 (see Supplementary Table S1 for details) was transformed with pBR-plac-orzO (induced by IPTG) and pBAD-zorO (induced by arabinose). The strain was grown to OD600 ≈ 0.1, and the culture was split into two. IPTG (1 mM final concentration) was added to one half of the culture. Arabinose (0.2% final concentration) was added 30 min later to each culture and OD600 was measured over time. Solid symbols indicate the addition of IPTG; open symbols, no IPTG. Shown are the mean values ± standard deviations for three independent cultures. Predicted region of base pairing is shown.
Overexpression of ZorO was shown to be toxic; however, it was never demonstrated that the sRNA OrzO could repress zorO-induced toxicity (14). Thus, is zorO-orzO, a true type I toxin–antitoxin locus? Given the high homology between the zorO-orzO and zorP-orzP loci, is there ‘cross-talk’ in the regulation? In this study, we confirmed that the zorO-orzO locus is indeed a true type I toxin–antitoxin system. Using the inherent toxicity associated with zorO overproduction, we performed mutational analyses to further investigate the requirements for successful repression by the OrzO antitoxin. Through these analyses, we revealed that the 5′ end of the OrzO sRNA can solely repress zorO and the V1 region within 5′ end of the Orz sRNA determines the specific recognition of the zor target.
MATERIALS AND METHODS
Bacteria strains and plasmids
All bacterial strains and plasmids used in this work are listed in Supplementary Table S1. The sequences of all oligonucleotides are listed in Supplementary Table S2.
Growth conditions
Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium with shaking. Antibiotics were added when necessary at the following concentration: 100 μg/ml ampicillin, 25 μg/ml chloramphenicol, 25 μg/ml kanamycin. Arabinose was added to a final concentration of 0.2 or 0.002% as indicated. Isopropyl β-D-1-thiogalactopyranoside (IPTG), when used, was at a final concentration of either 0.1 or 1 mM, as indicated.
Generation of strains for rescue experiments
All strains used in the described rescue experiments are derived from DJ624, which contains ΔlacX74 and does not possess the zor-orz locus (21). To better control expression from the PBAD promoter across the population of cells, the araE gene was placed under control of the PCP18 constitutive promoter through P1 transduction of the kan-PCP18-araE allele (22). The kanamycin cassette was then removed using pCP20, resulting in strain UTK007 (23). Generation of the hfq and rnc deletions was performed using the mini-λ-Red recombination system (24–26). In each case, pKD4 served as a polymerase chain reaction (PCR) template for amplifying the kanamycin cassette using primers containing 20 nt of homology to the kanamycin cassette, flanked by ∼40 nt of homology to the chromosomal region (25).
Plasmid construction
For overexpression of the sRNAs OrzO, OrzP, OhsC, IstR, the genes were amplified from genomic DNA (E. coli O157:H7 EDL933) and ligated into the AatII and EcoRI sites of pBR-plac generating pBR-plac-orzO, pBR-plac-orzP pBR-plac-ohsC and pBR-plac-istR (21). The zorO gene was ligated into the PstI and HindIII sites of pEF21 from the mapped +1 of its transcription, generating pEF21-zorO (pBAD-zorO in the text) (14,18). Mutations of individual nucleotide residues for all constructs were performed by site-directed mutagenesis of pBR-plac-orzO, pBR-plac-orzP or pEF21-zorO. For generation of pBR-pLac-orzO 6(1)10(1) and pBR-plac-orzO (1)5(1)11, pBR-plac-orzO 6(1)11 served as the template. The two chimeric constructs, pBR-plac-orzO-ohsC and pBR-plac-orzO-istR, were generated using SOE PCR (27,28). PCR product A amplified the first 18 nt of orzO from pBR-plac-orzO along with 100 nt upstream of vector sequence (including the AatII cut site); the 3′ end of the product contained 15 nt of PCR product B. PCR product B amplified either ohsC or istR from E. coli O157:H7 EDL933 genomic DNA; this product began immediately following the region of base pairing that these RNAs would have for their respective targets (18,20). The 5′ end of PCR B contained 15 nt of homology to the 5′ end of orzO. The products were then spliced together via PCR, digested with AatII and EcoRI and ligated into pBR-plac. Plasmid DNA was isolated using the Qiagen Mini Plasmid Kit; PCR products were isolated using the Qiagen PCR Purification Kit.
Rescue experiments
Rescue experiments were as described previously (15,18). The pBR-plac plasmid carrying the orz wild type/mutant gene under an IPTG-induced PLlacO-1 promoter was initially transformed into UTK007 or one of the derivatives described above, through electroporation (21). Afterward, the pEF21 plasmid containing the zor wild type/mutant gene under an arabinose-induced PBAD promoter was transformed into the same cells (29). The resulting transformants harboring two plasmids were grown overnight and then diluted to OD600 of 0.01. When the OD600 reached ≈ 0.1, the cultures were split and IPTG was added to half the culture to a final concentration of either 0.1 or 1 mM for the overexpression of the orz gene. Thirty minutes after IPTG induction, arabinose was added to a final concentration of either 0.002 or 0.2% for the overproduction of the zor gene. OD600 was taken and recorded every 30 min. Shown are averages ± standard deviations for a minimum of three independent experiments.
RNA extraction
To examine RNA levels of zorO and OrzO, cells harboring the appropriate plasmids were grown to an OD600 ≈ 0.2, and arabinose was added as indicated to induce zorO (time 0). Five minutes after induction with arabinose, the culture was split and IPTG (0.1 or 1 mM) was added to half. RNA was extracted via direct lysis as previously described with some modifications (30). Cells (750 μl aliquots) from time 0, 5, 15, 30 and 60 min after arabinose were harvested and incubated at 65°C with 500 μl acid phenol: chloroform (preheated to 65°C) and 102 μl of direct lysis solution (320 mM sodium acetate, 8% SDS, 16 mM EDTA) for 10 min. After incubation, the mixtures were centrifuged at 13 000 rpm at room temperature for 10 min. Supernatants were transferred to tubes containing 500 μl of 65°C phenol: chloroform, mixed thoroughly and spun again for 10 min at room temperature. The supernatants were transferred and extracted twice with 400 μl phenol: chloroform. The RNA was then ethanol precipitated and resuspended in RNase-free water. When needed, RNA was treated with Turbo DNaseI (Life Technologies) following manufacturer’s instructions.
Northern analysis
Total RNA (10 μg) isolated at indicated time points following arabinose induction was separated on a denatured 6 (for the separation of zor mRNA) or 8% (for the separation of orz sRNA) polyacrylamide-urea gel and transferred to a Zeta-Probe Genomic GT membrane (Bio-Rad). Specific probes were generated by end-labeling oligonucleotides with γ-32P by T4 polynucleotide kinase. Incubation of the membrane and washes were performed as outlined by Opdyke et al. (31). Northern analysis was performed from a minimum of three independent experiments for every construct examined.
Quantification of RNA levels
The relative intensities of the resulting bands from northern analysis detected via autoradiography were quantified using ImageJ (32). The intensity of the band for WT at T15 (induced with the level of IPTG indicated) was set to a value of ‘1’ as a standard for comparison. Relative intensities to WT at T15 for the other bands on the same northern blot were thus calculated. Averages and standard deviations were determined, and the statistical significance was calculated using Student’s t-test.
Primer extension
Total RNA (5 μg) isolated 30 min after arabinose induction from rescue experiments was separated on a denatured 8% polyacrylamide-urea gel as described previously (14,31).
RESULTS
zorO-orzO is a bona fide type I toxin–antitoxin locus
The zorO gene of E. coli O157:H7 EDL933, initially annotated as z3289, was predicted to be type I toxin (14). Overexpression of the small protein from a high-copy plasmid in E. coli MG1655 led to cell growth stasis as well as a decrease in colony forming units, indicating that ZorO is toxic (14). In the same study, the sRNA OrzO (denoted at that time as sRNA-1) was detected via northern analysis. This sRNA, though encoded divergent from the toxin gene, could potentially base pair with the 5′ UTR of the zorO mRNA (Figure 1A and B).
Despite these data, the crucial question remained whether the zorO-orzO pair is a true type I toxin–antitoxin locus. If so, then the OrzO sRNA should be able to repress the toxicity of ZorO overproduction. Rescue experiments were used to test if OrzO can repress ZorO. We expressed zorO from an arabinose-inducible promoter (PBAD) on one plasmid and orzO from an IPTG-inducible promoter (PLlacO-1) on a second compatible plasmid (15,18). These experiments were performed in a strain derived from E. coli MG1655 that does not possess the zor-orz loci so as to eliminate any possible repression from a chromosomally encoded OrzO. While the induction of zorO led to cell growth stasis, indicating that ZorO is toxic, the co-expression of OrzO prevented this cell stasis, suggesting that OrzO serves as an antitoxin for ZorO (Figure 1B). Thus, we conclude that the zorO-orzO pair is a true type I toxin–antitoxin locus.
RNase III but not Hfq is required for repression of zorO-induced toxicity
The majority of trans-encoded sRNAs characterized to date require the RNA chaperone Hfq to function efficiently, whereas the antisense sRNAs typically do not (2,7,33). However, given that there is limited complementarity between the zorO mRNA and the corresponding OrzO sRNA (18 nt perfect complementarity), we wanted to examine whether Hfq is required for the observed repression of the ZorO toxin. A similar rescue experiment was performed as outlined above in a strain deleted for hfq. The results showed that the OrzO sRNA can fully repress zorO in both the wild type and the Δhfq strain (Supplemental Figure S1), indicating that Hfq is not needed for the OrzO sRNA to regulate zorO.
RNase III, a double-stranded ribonuclease, has been shown to be critical for cleaving paired toxin–antitoxin complexes including hok/sok, tisB-istR, bsrG/sr4, txpA-ratA, as well as for the sRNA GadY and its mRNA target gadX-gadW (16,20,34–36). To determine whether repression by OrzO was dependent on expression of RNase III, we performed our rescue experiment in a strain deleted for rnc, the gene encoding RNase III. The results (Figure 2A) show that OrzO is incapable of rescuing zorO-induced toxicity in this mutant, even at low levels of inducing agent (0.002% arabinose, see below; data not shown). When zorO is expressed with OrzO in a wild-type strain, we detect major transcripts of ∼280 and 310 nt in length via northern analysis (Figure 2B). When examining zorO in the Δrnc strain, we noted an accumulation of these full-length transcripts in comparison with expression in the wild-type strain, suggesting that the full-length mRNAs are more stable in the deletion strain (Figure 2B). This implies that cleavage of the zorO-OrzO RNA duplex by RNase III is important for repression of zorO-induced toxicity. Surprisingly, OrzO levels (induced by 1 mM IPTG) appear reduced in the Δrnc strain (Supplementary Figure S2A).
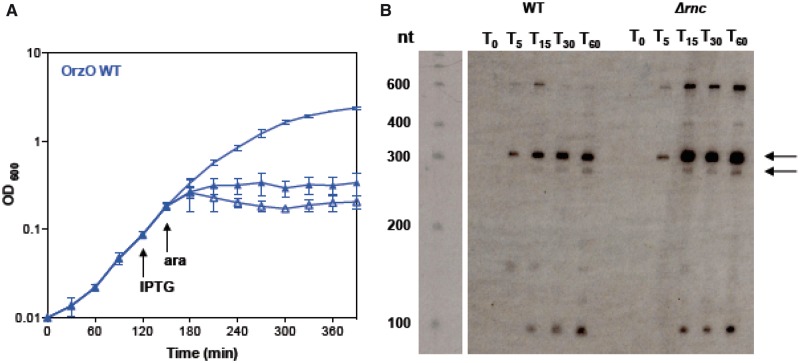
Figure 2.
RNase III is critical for OrzO repression of zorO. (A) Overexpression of OrzO cannot repress zorO-mediated toxicity in a strain deleted for rnc. E. coli strain UTK011, which is deleted for rnc, was transformed with pBR-plac-orzO and pBAD-zorO. The strain was grown to OD600 ≈ 0.1, and split into three. One of the three cultures received no supplementation. IPTG (1 mM) was added as indicated to one culture. After 30 min, arabinose (0.2%) was added to two of the cultures and cell growth was monitored by measuring the OD600 over time. Solid symbols indicate the addition of arabinose and IPTG; open symbols, arabinose and no IPTG; no symbol indicates unsupplemented culture. Shown are the averages ± standard deviations from three independent experiments. (B) Accumulation of full-length zorO mRNA in the rnc deletion strain. Total RNA was isolated from a wild-type strain (UTK007) and an rnc deletion strain (UTK011) harboring pBAD-zorO 0, 5, 15, 30, 60 min after the addition of 0.2% arabinose. Arrows indicate the predicted full-length zorO transcripts.
We attempted to map the cleavage sites of zorO when OrzO is expressed in a wild type and a Δrnc strain using primer extension. Similar to our northern analyses, we noted an accumulation of full-length zorO mRNA in the Δrnc strain even when OrzO is expressed (Supplementary Figure S2B). However, we are able to map OrzO-specific cleavage sites within the zorO mRNA in the Δrnc strain (Figure 1A, Supplementary Figure S2B), similar to what has been reported for cleavage of gadX-gadW by GadY and cleavage of cII-O by the OPP RNA (35,37,38). This finding suggests that while RNase III may be critical for regulation of zorO repression, there may be additional factors involved (see ‘Discussion’ section). As summarized in Figure 1A and shown in Supplementary Figure S2B, the major processing sites lie within the region of base pairing for both the wild type and Δrnc strain. We did note that in the Δrnc strain, there were sites that appear to be shifted from the wild-type cleavage sites, as well as decreased cleavage at some sites (Figure 1A, Supplementary Figure S2B). We also detected processing outside the region of base pairing for both strains (Figure 1A, Supplementary Figure S2B), but it is not clear at this point the origin of these products.
5′ end of the OrzO sRNA can solely repress zorO
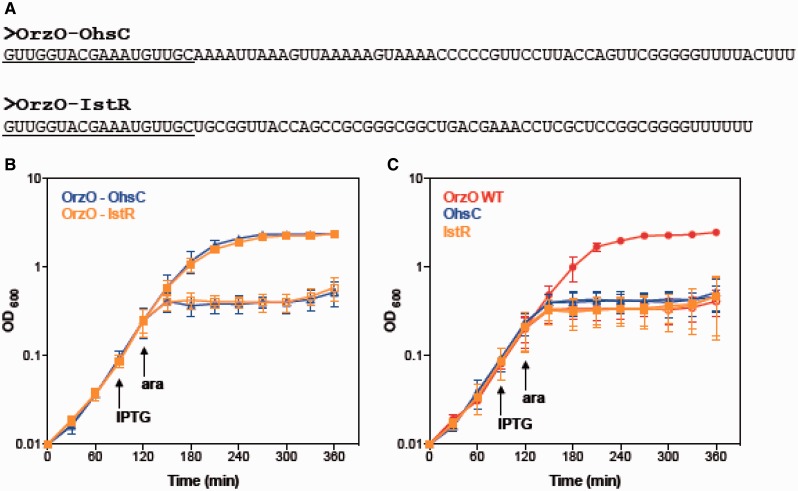
A previous study has shown that the antitoxin IstR-1 of E. coli regulates its toxin target TisB by 5′ end pairing (20). Similarly, the OhsC antitoxin is predicted to base pair to its target ShoB through its 5′ end (18). In zorO-orzO pair, the longest stretch of potential base pairing between OrzO and the zorO mRNA also occurs in the immediate 5′ end of OrzO. To test if just the 5′ end of OrzO can repress zorO, we first constructed two OrzO mutants truncated at the 3′ end. However, we were unable to detect the expression of these truncated sRNAs (data not shown). We hypothesized that the inability to detect these truncated RNAs was owing to their instability; therefore, we generated chimeric sRNAs to the OhsC and IstR-1 sRNAs. The base pairing regions that OhsC and IstR-1 had to their own toxins were replaced with the first 18 nt of the 5′ end of the OrzO sRNA (Figure 3A). Consequently, only the 5′ end of the chimeras can base pair with the zorO mRNA. Rescue experiment results showed that although the wild-type OhsC and IstR sRNAs failed to repress zorO, both chimeras successfully repressed the expression of zorO (Figure 3B and C). This demonstrates that the first 18 nt of OrzO are sufficient for full repression.
Figure 3.
The 5′ end of OrzO is sufficient for inhibition of zorO-mediated toxicity. (A) Generation of an OrzO-OhsC and OrzO-IstR chimeras. The 5′ ends of the type I antitoxins OhsC and IstR were replaced with the 5′ end of OrzO, the longest region of complementarity to zorO. Underlined sequences indicate the OrzO nucleotides. (B) Overexpression of both OrzO-OhsC and OrzO-IstR chimeras repress zorO. Rescue experiment was performed as outlined in Figure 1B with either OrzO-OhsC (blue triangles) or OrzO-IstR (orange squares) expressed from the PLlacO-1 promoter of pBR-plac. Solid symbols indicate the addition of IPTG; open symbols, no IPTG. The averages of three independent cultures ± standard deviations are shown. (C) Neither wild type OhsC nor IstR can repress zorO-induced toxicity. Rescue experiment was performed with pBR-plac-orzO (red circles), pBR-plac-ohsC (blue triangles) or pBR-plac-istR (orange squares) as outlined in Figure 1B. Solid symbols indicate addition of IPTG; open symbols, no IPTG. Shown are the averages ± standard deviations for three independent cultures.
Orz regulation is target specific
The 5′ end of OrzO contains a continuous stretch of 18 nt that can base pair to the zorO mRNA (Figure 1B). The OrzP sRNA, which is a homolog of OrzO, also shares 15 nt complementarity to zorO (Figure 4B). Because of the sequence similarities between OrzO and OrzP, it is possible that OrzP is capable of regulating ZorO (Figure 4A).
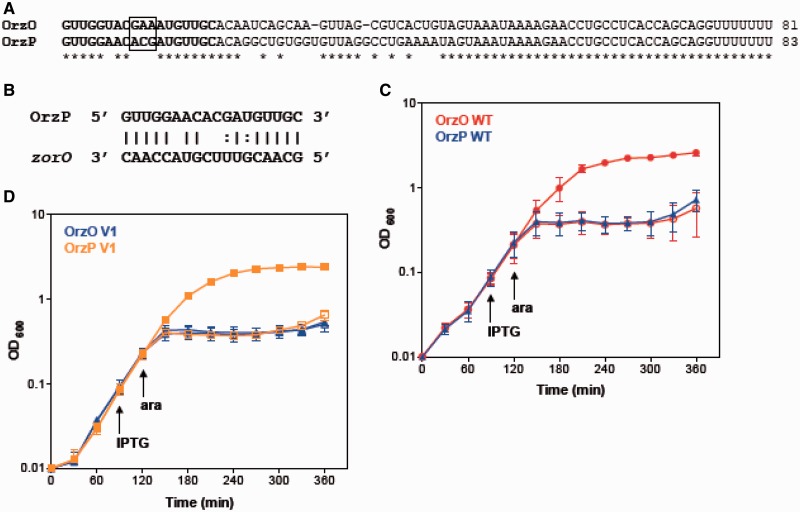
Figure 4.
The V1 region is responsible for target discrimination by OrzO. (A) Alignment of OrzO and OrzP sRNAs. The V1 region is indicated in the black box; the entire base pairing region is in bold. (B) Potential base pairing interactions between OrzP and zorO in the predicted binding region. (C) OrzO, not OrzP, can repress zorO. Rescue experiment was performed as outlined in Figure 1B with either wild-type OrzO (red circles) or OrzP (blue triangles) expressed from the PLlacO-1 promoter of pBR-plac. Solid symbols indicate the addition of IPTG; open symbols, no IPTG. Averages ± standard deviations of three independent cultures are shown. (D) An OrzP V1 mutant can rescue cells from zorO overexpression. Rescue experiment was performed as outlined in Figure 1B using pBR-plac-orzP V1 (orange squares), in which its V1 region was mutated to match OrzO, and pBR-plac-orzO V1 (blue triangles), in which its V1 was mutated to match OrzP. Solid symbols indicate addition of IPTG; open symbols, no IPTG. Averages ± standard deviations for three independent cultures are shown.
Rescue experiments were used to determine whether the OrzP sRNA can repress the expression of zorO. When OrzO was co-expressed with zorO, the strain was able to grow. However, when OrzP was co-expressed with zorO, cell stasis occurred (Figure 4C), suggesting that only OrzO, but not OrzP, can repress zorO. We also confirmed that OrzP was being expressed at levels equivalent to OrzO by northern analysis (Supplementary Figure S3), which indicates that the failure of OrzP to rescue is not due to expression differences. Thus, repression by the Orz sRNAs is target-specific.
V1 region determines OrzO specificity for zorO
Although OrzO and OrzP share the same predicted base pairing region with zorO, only OrzO is able to repress zorO. To further investigate what dictates this specificity, we compared the sequences of the two sRNAs (Figure 4A). Despite the great sequence similarity between OrzO and OrzP, there are several differences. In particular, the nucleotide sequences from +9 to +11 compose a variable region of 3 nt between OrzO (GAA) and OrzP (ACG). This variable region (referred to as V1) is the most prominent difference observed within the 5′ regulatory domain of the two sRNAs.
To determine if this V1 region is responsible for specificity in the OrzO regulation of zorO, the three nucleotides in the V1 region of OrzO were changed from ‘GAA’ to ‘ACG’, the sequence found in OrzP (Table 1). We then tested the ability of the resulting mutant strain (15 nt of base pairing potential) to rescue cells from zorO-induced toxicity. Our results showed that the OrzO V1 mutant completely failed to rescue cells (Figure 4D), suggesting that the V1 region is critical for repression by the OrzO sRNA.
Table 1.
Summary of the mutations generated and their repressive abilities
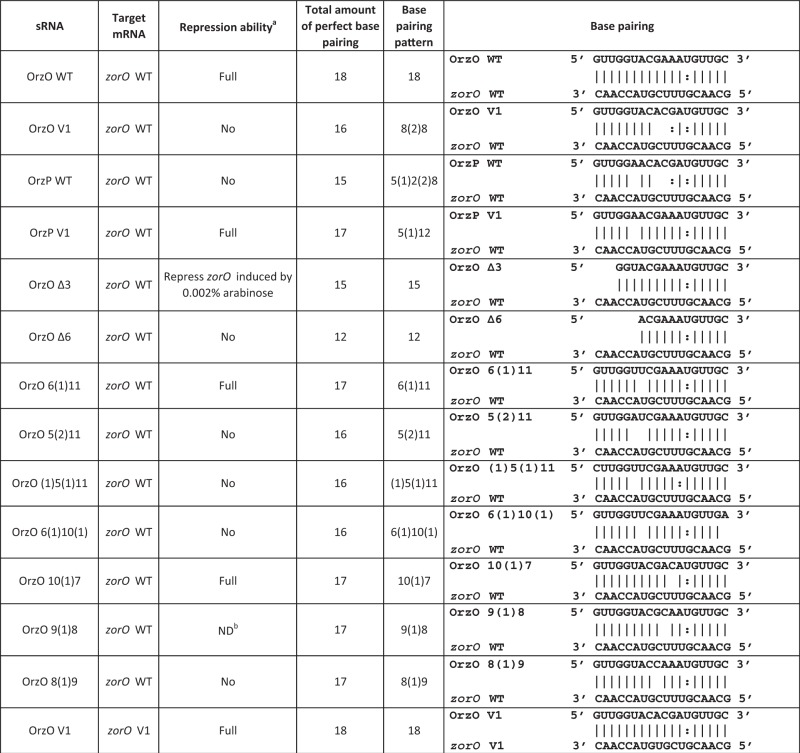 |
aRepression ability when zorO induced by 0.2% arabinose unless otherwise indicated.
bDespite generation of this mutant independently multiple times, we never obtained consistent results when analyzing its ability to repress zorO induced either with 0.2 or 0.002% arabinose.
To confirm these results, we generated compensatory mutations in zorO (referred to as zorO V1) so as to restore complementarity to the OrzO V1 mutant. Restoring the base pairing allowed the OrzO V1 to fully repress ZorO V1-induced toxicity (Supplementary Figure S4). These data implied that the V1 region is important for OrzO to recognize its target.
Furthermore, we engineered an OrzP V1 mutant, in which its V1 region was changed from ‘ACG’ to ‘GAA’, the sequence found in OrzO. Consequently, the OrzP V1 mutant has increased ability to base pair to zorO, as it could perfectly match zorO in the V1 region (Table 1). Unlike the OrzP WT, which cannot inhibit zorO mRNA expression, the OrzP V1 mutant fully represses zorO (Figure 4D). These data, in addition to the observations from the zorO WT-OrzO V1 pair and the zorO V1-OrzO V1 pair, further support the hypothesis that the V1 region is responsible for the Orz sRNA specificity to its zor target.
The first base of the V1 region is critical for OrzO regulation of zorO
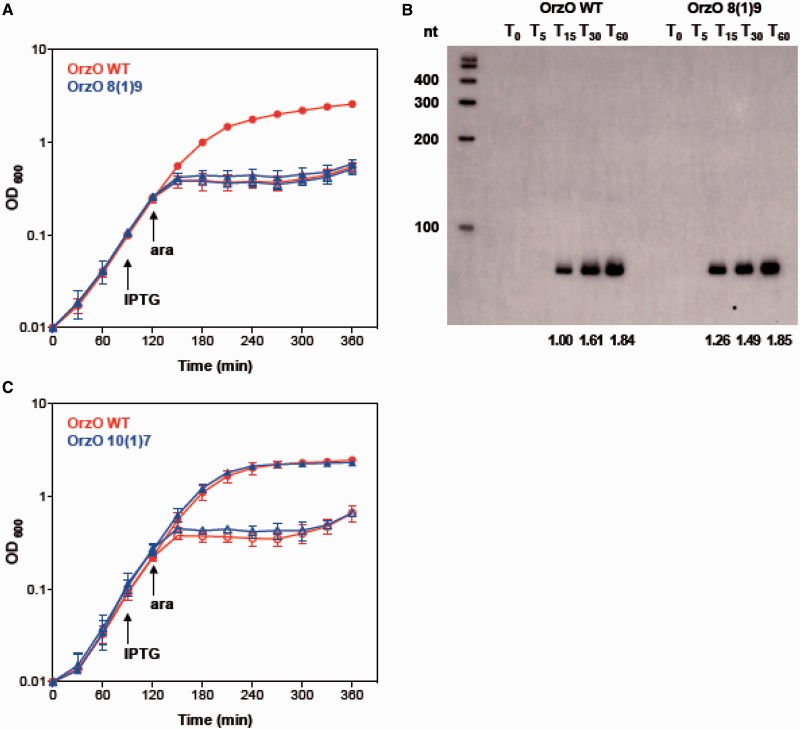
Since the V1 region of the 5′ end of OrzO is composed of three nucleotides (GAA), we then asked whether all three nucleotides contribute equally to the ability of OrzO to regulate zorO. Each nucleotide of the V1 region was mutated individually and tested for the ability to rescue cells from zorO toxicity. The resulting mutants are named OrzO 8(1)9, OrzO 9(1)8 and OrzO 10(1)7. These are named such that the number in the parentheses indicates the number of unpaired nucleotides, while the numbers on the two sides of the parentheses represent the number of the nucleotides within the 5′ end of OrzO that are capable of base pairing with zorO (Table 1). For example, in OrzO 8(1)9, the first 8 nt of OrzO can pair to zorO, followed by a single unpaired nt, and then 9 nt of pairing.
Rescue experiments were used to assess the repression abilities of the three mutants. Interestingly, the repression ability of the single mutants was not equivalent. The point mutation in the first nucleotide of the V1 region of OrzO completely abolished the repression ability of OrzO sRNA (Figure 5A), as the OrzO 8(1)9 mutant failed to rescue cells even at a low induction level of zorO (0.002% arabinose, see below). Northern analysis further indicated that the OrzO 8(1)9 mutant showed levels equivalent to that of the OrzO wild type (WT) at T15 and T60, suggesting that the failure of the OrzO 8(1)9 mutant to rescue is due to mutating the first nucleotide of the V1 region (Figure 5B, Supplementary Figure S7A). Mutation of the third nucleotide of the V1 region did not affect the repression ability of OrzO, as the OrzO 10(1)7 mutant still fully repressed zorO (Figure 5C).
Figure 5.
A specific residue within the V1 region of OrzO is necessary for zorO repression. (A) Mutation of the first nucleotide of the V1 region fails to rescue E. coli from ZorO toxicity. Rescue experiment was performed as outlined in Figure 1B except that arabinose was added to a final concentration of 0.002%. Shown are the averages ± standard deviations of pBR-plac-orzO (red circles) or pBR-plac-orzO 8(1)9 (blue triangles), which has the first nucleotide of the V1 region mutated. Solid symbols indicate addition of IPTG; open symbols, no IPTG. (B) Expression of OrzO 8(1)9 is similar to wild type OrzO. Total RNA was isolated from E. coli UTK007 overexpressing zorO (0.002% arabinose) and either OrzO (1 mM IPTG) or OrzO 8(1)9 (1 mM IPTG) as described in ‘Materials and Methods’ section. Quantification of RNA levels is described in the ‘Materials and Methods’ section, with OrzO WT at T15 set to a value of 1. Numbers below the panel indicate the average relative intensities. (C) Mutation of the third nucleotide of the V1 region has no impact on the ability of OrzO to repress zorO expression. The third nucleotide of the V1 region of OrzO was mutated to generate OrzO 10(1)7. A rescue experiment was performed as outlined in Figure 1B with wild-type OrzO (red circles) or OrzO 10(1)7 (blue triangles) expressed from the PLlacO-1 promoter of pBR-plac. Solid symbols indicate addition of IPTG; open symbols, no IPTG. Averages ± standard deviations for three independent cultures are shown.
The OrzO 9(1)8 mutant (second nucleotide of the V1 mutated) was highly variable in its ability to repress zorO (Supplementary Figure S6A), despite generation of the mutant independently multiple times. To clarify whether this may be due to expression problems, we performed a more in-depth expression comparison to wild type OrzO. We first examined the basal level of OrzO induction required to repress zorO, and determined that this occurs at 0.1 mM IPTG induction (Supplementary Figure S5A–C). We then compared expression of OrzO 9(1)8 when induced at 1 mM IPTG to wild type OrzO expressed at 0.1 mM IPTG (Supplementary Figure S6B and C). Indeed, expression of OrzO 9(1)8 is higher than the basal levels of wild-type OrzO that is needed for zorO repression. This suggests that the attenuated repression ability observed in OrzO 9(1)8 is likely due to the nucleotide change rather than the expression difference. Regardless, our results imply that the first nucleotide of the V1 region is most critical for the OrzO sRNA to regulate zorO.
15 nt of continuous base pairing interactions are sufficient for OrzO regulation
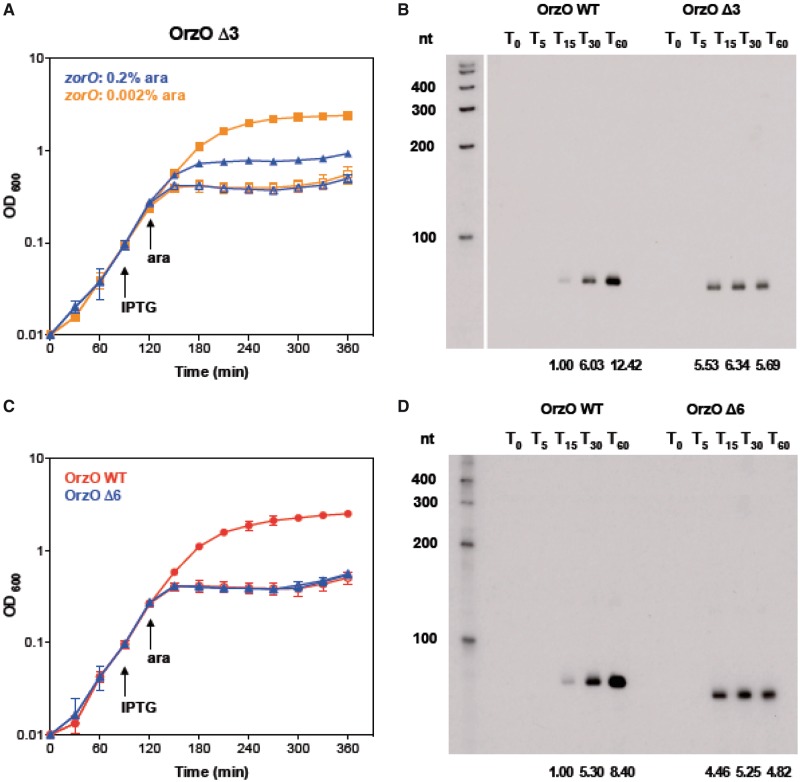
We have shown that the 5′ base pairing domain of OrzO is critical for regulating zorO, and the V1 region dictates the correct recognition of the cognate zor targets. However, it is not clear whether all nucleotides within the 5′ end of OrzO besides the V1 region are needed for repression of zorO. To better refine the requirements for successful base pairing, we generated two deletion constructs in which the first 3 and 6 nt were deleted from the 5′ end of OrzO while leaving the V1 intact. The OrzO Δ3 and Δ6 share a continuous stretch of 15 and 12 nt complementarity with zorO mRNA, respectively (Table 1).
Rescue experiments were used to assess the abilities of these two mutants to repress zorO. Neither the OrzO Δ3 nor Δ6 completely repressed zorO when we used 0.2% arabinose to induce expression of the toxin (Figure 6A, data not shown). We also inspected the levels of OrzO Δ3 and Δ6 and observed that their levels were decreased as compared with the level of full-length OrzO when all were induced with 1 mM IPTG (data not shown).
Figure 6.
Only 15 nt of continuous base pairing are required for OrzO inhibition of zorO. (A) OrzO Δ3 prevents zorO-induced toxicity at low levels of arabinose. Rescue experiment was performed as outlined in Figure 1B with OrzO Δ3 expressed from the PLlacO-1 promoter of pBR-plac. Note that zorO was induced by either 0.2 (blue triangles) or 0.002% (orange squares). Solid symbols indicate addition of IPTG; open symbols, no IPTG. Averages ± standard deviations for three independent cultures are shown. (B) Comparison of RNA levels between wild-type OrzO induced by 0.1 mM IPTG and OrzO Δ3 induced by 1 mM IPTG. Total RNA was isolated from E. coli UTK007 overexpressing OrzO or OrzO Δ3 from pBR-plac as described in ‘Materials and Methods’ section. Average intensities are shown as described in Figure 5B and in the ‘Materials and Methods’ section. (C) OrzO Δ6 fails to repress zorO-induced toxicity even at low levels of arabinose induction. Rescue experiment was performed as outlined in Figure 5A using either pBR-plac-orzO (red circles) or pBR-plac-orzO Δ6 (blue triangles) as indicated. Note that the arabinose concentration used was 0.002%. Solid symbols indicate addition of IPTG; open symbols, no IPTG. Averages ± standard deviations for three independent cultures are shown. (D) Comparison of RNA levels between the wild-type OrzO induced by 0.1 mM IPTG and the OrzO Δ6 mutant induced by 1 mM IPTG. Total RNA was isolated from E. coli UTK007 overexpressing OrzO or OrzO Δ6 from pBR-plac as described in ‘Materials and Methods’ section. Average intensities are shown as described in Figure 5B and in the ‘Materials and Methods’ section.
On closer examination of the rescue experiment using OrzO Δ3, we noticed a ‘transient’ rescue or a delay in growth stasis compared with expression of zorO alone when induced with 0.2% arabinose. When we reduced the arabinose concentration to 0.002%, we noted that overproduction of ZorO still caused cell growth stasis, in a pattern identical to the 0.2% arabinose induction group (Supplementary Figure S5D and E). In addition, co-expression of OrzO Δ3 relieved the toxicity caused by induction of zorO at 0.002%, suggesting that the failure of this mutant to rescue at higher concentration of arabinose was likely due to the reduced levels of this sRNA mutant (Figure 6A). However, OrzO Δ6 still could not repress toxicity when ZorO was induced by 0.002% arabinose (Figure 6C).
We observed that the RNA levels of OrzO Δ3 and Δ6 (induced by 1 mM IPTG) were higher than the basal levels of wild-type OrzO (induced by 0.1 mM IPTG) at T15 (Figures 6B, D, Supplementary Figure S7B and C). The levels of the wild type sRNA continued to increase over time (T30 and T60), whereas the levels of the truncated sRNAs remained relatively steady. Even though OrzO Δ3 did not accumulate to a level as high as the wild-type sRNA, it was capable of repressing zorO. OrzO Δ6, however, could not even transiently rescue cells, despite being induced initially to levels higher than wild type OrzO (Figure 6D, Supplementary Figure S7C). Combined, these data imply that 15 nt, but not 12 nt, of continuous base pairing is sufficient for the OrzO sRNA to repress zorO.
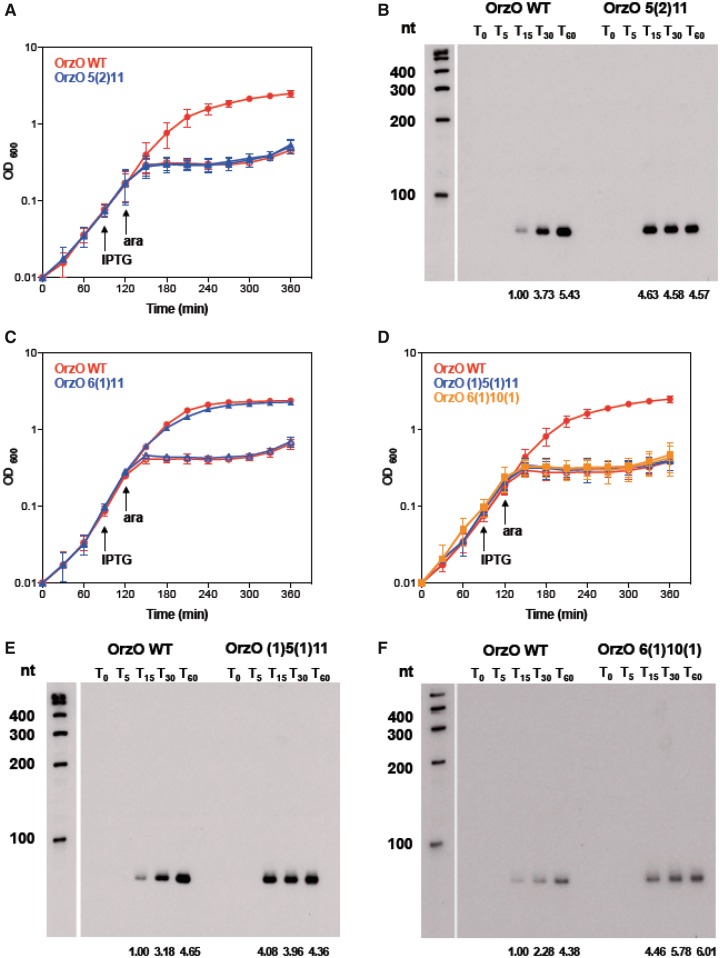
A minimum of 17 nt of discontinuous base pairing interactions is required for OrzO repression of zorO
As shown above, 15 nt of continuous complementarity to zorO is sufficient for repression. Our OrzP V1 mutant can also repress zorO, but it does not have 15 nt of continuous base pairing (Table 1). Instead, this mutant has 17 nt of discontinuous pairing. This suggests that the amount of complementary base pairs required for discontinuous base pairing interactions may be different from that for continuous base pairing interactions.
Given that the OrzP V1 [pairing: 5(1)12] can rescue cells from zorO-induced toxicity, we designed series of mutants with decreased base pairing potential as compared with OrzP V1 (Table 1). We constructed OrzO 5(2)11 with 16 nt of base pairing, which is composed of a 5-nt stretch of complementarity, followed by a 2-nt gap and then another 11-nt stretch of complementarity. The OrzO 5(2)11 mutant could not repress ZorO toxicity when the mRNA was induced with either 0.2 or 0.002% arabinose (Figure 7A and data not shown). We also confirmed that the OrzO 5(2)11 mutant (induced by 1 mM IPTG) was being expressed at levels higher than the basal levels of OrzO WT (induced by 0.1 mM IPTG) at T15 and T30 (Figure 7B, Supplementary Figure S7D), suggesting that the failure of OrzO 5(2)11 to rescue is likely not due to insufficient RNA levels, but rather base pairing differences.
Figure 7.
Repression of zorO requires 17 nt of discontinuous base pairing. (A) OrzO 5(2)11 fails to inhibit ZorO toxicity. Rescue experiment was performed as outlined in Figure 5A with either wild-type OrzO (red circles) or OrzO 5(2)11 (blue triangles) expressed from the PLlacO-1 promoter of pBR-plac. Solid symbols indicate addition of IPTG; open symbols, no IPTG. Averages ± standard deviations for three independent cultures are shown. (B) Comparison of RNA levels between the wild type OrzO induced by 0.1 mM IPTG and the OrzO 5(2)11 mutant induced by 1 mM IPTG. Average intensities are shown as described in Figure 5B and in the ‘Materials and Methods’ section. (C) OrzO 6(1)11 can repress zorO. Rescue experiment was performed as outlined in Figure 1B with either wild-type OrzO (red circles) or OrzO 6(1)11 (blue triangles) expressed from the PLlacO-1 promoter of pBR-plac. Solid symbols indicate addition of IPTG; open symbols, no IPTG. Averages ± standard deviations for three independent cultures are shown. (D) Neither OrzO 6(1)10(1) nor OrzO (1)5(1)11 can prevent zorO-induced toxicity. Rescue experiment was performed as outlined in Figure 5A with pBR-plac-orzO (red circles), pBR-plac-orzO (1)5(1)11 (blue triangles) or pBR-plac-orzO 6(1)10(1) (orange squares). Note that IPTG was added to 1 mM final concentration, and arabinose was added to 0.002% final concentration. Solid symbols indicate addition of IPTG; open symbols, no IPTG. Averages ± standard deviations for three independent cultures are shown. (E) Comparison of RNA levels between the wild-type OrzO induced by 0.1 mM IPTG and the OrzO (1)5(1)11 mutant induced by 1 mM IPTG. Average intensities are shown as described in Figure 5B and in the ‘Materials and Methods’ section. (F) Comparison of RNA levels between the wild-type OrzO induced by 0.1 mM IPTG and the OrzO 6(1)10(1) mutant induced by 1 mM IPTG. Average intensities are shown as described in Figure 5B and in the ‘Materials and Methods’ section.
Our OrzO 5(2)11 possesses 16 nt of pairing to zorO, whereas our OrzP V1 possesses 17 nt of pairing [5(1)12]. It maybe that a minimum of 17 nt of discontinuous base pairing is required for repression. To explore this further, we generated several more constructs. The OrzO 6(1)11 mutant has 17 nt of pairing, and indeed it can fully repress zorO (Figure 7C). We then mutated the last nucleotide of pairing of this construct, generating the OrzO 6(1)10(1) mutant, which possess 16 nt of pairing. We also generated the OrzO (1)5(1)11 mutant, in which the first nucleotide of pairing was mutated, and in total, possesses only 16 nt of pairing. When either mutant was used in a rescue experiment, they were unable to repress toxicity, even if zorO was induced with 0.002% arabinose (Figure 7D). Furthermore, northern analysis indicated that these mutants (induced by 1 mM IPTG) were expressed at levels equivalent to or even higher than OrzO WT induced by 0.1 mM IPTG (Figure 7E, F, and Supplementary Figure S7E, F). Thus, the failure to rescue is likely due to insufficient pairing and not due to insufficient production of the antitoxin.
DISCUSSION
In this study, we show that the zorO-orzO gene pair of E. coli O157:H7 is a true type I toxin–antitoxin locus. More specifically, our data are consistent with the properties of an unconventional type I system, as the OrzO antitoxin is encoded divergently from zorO and has limited base pairing potential with it. Overproduction of ZorO is toxic; however, co-expression of OrzO represses this toxicity and rescues the cells. We discovered that the 5′ end of OrzO can solely repress zorO-induced toxicity, and the V1 region dictates the correct recognition of zorO. Furthermore, the first base of the V1 region is critical for the OrzO sRNA to regulate the expression of zorO mRNA. Although there is 18-nt complementarity with zorO at the 5′ end of OrzO, not all base pairing interactions are needed for repression. For continuous complementarity, 15 nt of perfect base pairing is sufficient to function, while for discontinuous complementarity, a minimum of 17 nt of perfect base pairing is required to maintain the repression ability if a single non-pairing nucleotide is present within the base pairing region.
Role of RNase III in repression of zorO expression
Ribonuclease III (RNase III), encoded by the rnc gene, cleaves double-stranded RNA, and has been linked to processing of several type I toxin–antitoxin pairs found in E. coli and B. subtilis (16,20,34). While RNase III is not essential for growth in E. coli, deletion of the gene in B. subtilis is lethal. Work by Durand et al. has shown that the essentiality of RNase III in B. subtilis is because it cleaves the type I toxins txpA and yonT (36). Interestingly, while the RNA duplex of another type I toxin–antitoxin locus in B. subtilis, BsrG/SR4, is cleaved by RNAse III, repression of the toxin does not require this ribonuclease (16). Thus, while RNase III plays a role in the processing of several described type I pairs in both E. coli and B. subtilis, its essentiality for repression can be variable between the different systems.
RNase III is also involved in the processing of other RNA duplexes that are not type I toxin–antitoxin systems. These include the interaction of the mRNA gadX-gadW with the sRNA GadY and the interaction of the mRNA cII-O with the OOP RNA of λ (35,38,39). For the zorO-orzO pair, deletion of RNase III led to accumulation of the full-length zorO mRNA, and in this strain background, zorO-induced toxicity could not be repressed by co-expression of OrzO. Yet, we still could detect OrzO-dependent processing of zorO in a Δrnc strain. Similar observations were seen with gadX-gadW and cII-O; cleavage still occurred without RNase III, although the processing sites were slightly shifted (35,38,39). We observed a similar shift in the processing of zorO in a Δrnc strain (Figure 1A, Supplementary Figures S2B). There are other ribonucleases within E. coli that may be contributing to this processing [reviewed in (40)]. For example, RNaseE is critical for the function of many Hfq-dependent base pairing RNAs [reviewed in (41)]. Further analysis is needed to determine what other enzymes may be contributing to processing of zorO-OrzO; however, based on the in-depth analysis of the gadX-gadW processing by GadY, it is possible that an uncharacterized double-stranded ribonculease may exist (35).
We were surprised to note that the levels of OrzO were reduced in a Δrnc strain; we have observed that the chromosomal expression levels of OrzO within an E. coli O157:H7 deleted for rnc are also decreased in comparison with a wild-type strain (Won and Fozo, unpublished observations). We had anticipated that the levels of OrzO would either be unaffected by the loss of RNase III or perhaps higher, as there would be decreased cleavage of the zorO-OrzO RNA duplex. It maybe that RNase III cleaves OrzO directly, and that this cleavage impacts its stability and/or activity. However, we have been unable to detect alternative forms of OrzO, and cannot conclude that this is occurring.
Regulation of ZorO by the 5′ end of OrzO
To determine whether the 5′ end of OrzO was sufficient for regulating zorO, we generated two constructs expressing either the first 42 or 34 nt of OrzO. These truncated sRNAs did not rescue, nor were we ever able to detect their overexpression (data not shown). However, our chimeric sRNAs, OrzO-OhsC (75 nt) and OrzO-IstR (72 nt) were stable and able to rescue E. coli from ZorO toxicity. Thus, although the 5′ end of OrzO is sufficient for repression, additional RNA sequence at the 3′ end may be needed for stability. Many type I antitoxins are predicted to have extensive secondary structure (7). The reason for such extensive structure among the type I antitoxins has not been thoroughly examined, but our data suggest that it may be required to prevent quick degradation of the sRNA.
Our data show that the OrzO sRNA represses zorO expression through base pairing interactions between the 5′ end of OrzO and the 5′ UTR of zorO. The binding region in the 5′ end appears to be conserved across many sRNAs. For example, a previous study that examined 18 well-characterized E. coli/Salmonella Hfq-dependent sRNAs found that more than one-third used their 5′ ends to repress their targets (42). In addition, the Hfq-independent sRNA IstR-1 also acts by 5′ end pairing [reviewed in (19)]. Similar interactions have been predicted to occur with the shoB-ohsC pair, in which the 5′ end of OhsC shares 19 nt of complementarity to the 5′ UTR of shoB [(18) and reviewed in (19)]. Although the reason why so many sRNAs function through 5′ pairing remains unclear, it has been proposed that similar to eukaryotic microRNAs, some bacterial sRNAs may have a 5′ conserved ‘seed’ region that facilitates the selection for correct mRNA targets (42). Thus, the OrzO sRNA likely uses the first 18 nt of its 5′ end as the ‘seed’ region. Further sequence analysis studies of a larger group of sRNAs and their targets will be helpful to evaluate whether the existence of the ‘seed’ region can be applied as a general principle in the prediction of bacterial 5′ pairing sRNAs and their target mRNAs.
Recognition of zorO by the V1 region of OrzO
We have shown specifically that the V1 region of OrzO plays a crucial role in recognizing its target. Replacing the V1 region of OrzO with that of OrzP abrogates the ability of the sRNA to repress the zorO mRNA, whereas replacing the V1 region of OrzP with that of OrzO allows the mutated OrzP to repress zorO.
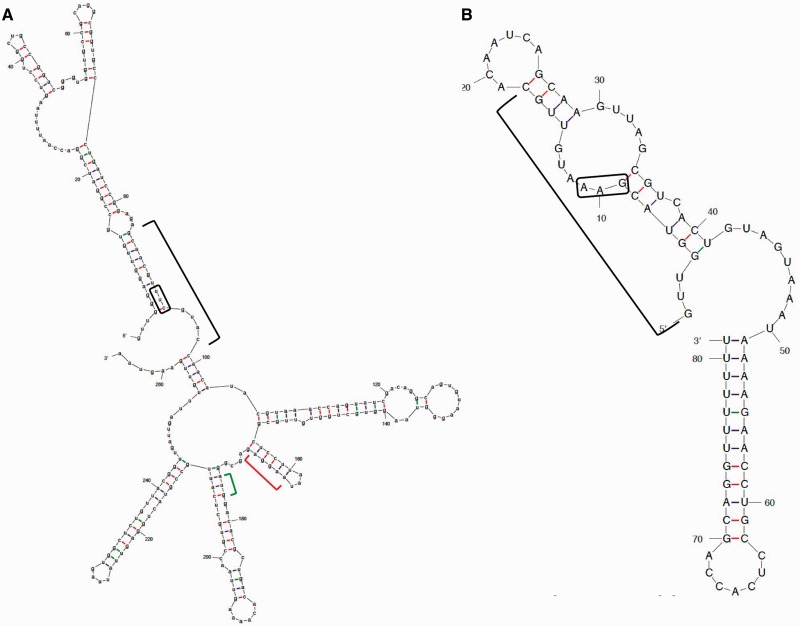
When inspecting the predicted structures of OrzO and zorO (Figure 8A and B), we find that both the V1 region of OrzO and the V1 target site in zorO are located in single-stranded structures. The relatively relaxed property of these single-stranded structures may allow for enhanced access of the single-stranded nucleotides to pair with their targets compared with those in a stem structure. Thus, it is likely that the pairing between the V1 region of OrzO sRNA and the corresponding site of zorO mRNA occurs before that of other complementary nucleotide regions.
Figure 8.
Predicted secondary structures of zorO mRNA and OrzO indicate the V1 regions are mostly single-stranded. (A) Putative RNA structure of zorO mRNA generated by M-fold (http://http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (43). The black bracket indicates the region of complementarity to OrzO; red brackets, the putative ribosome binding site; green bracket, the start codon. The black box is the V1 region binding site. (B) Putative RNA structure of OrzO sRNA generated by M-fold (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (43). The sequence indicated by the black bracket is the region of complementarity to the zorO mRNA; the black box is the V1 region.
In addition, when we mutated the bases of the V1 region, we noted that mutation of the first nucleotide of the V1 region caused a complete loss of the repression of zorO without affecting the levels of the sRNA. However, other single base mutants constructed in this study all successfully repress zorO (Table 1). These results suggested that as compared with other nucleotides, pairing via the first nucleotide of the V1 region is most critical for the OrzO sRNA to regulate zorO mRNA expression.
These observations suggest that OrzO and zorO may interact through a ‘kissing complex’. Generally in a ‘kissing complex’, the binding regions of the sRNA and the mRNA are located in loop structures (single-stranded). The initial base pairing occurs at the short exposed regions; thus, mutations of nucleotides involved the primary ‘kissing’ region impact the repression outcome, as exemplified by the RNAII-RNAI pair in ColE1-derived plasmid and the CopA-CopT pair in R1 plasmid (44,45). Once the initial pairing is established, the kissing complex can extend and propagate the formation of double-stranded RNA in the rest of the sequence. Similarly, this has been reported for the interaction of the ibs-Sib type I toxin–antitoxin pair in E. coli; however, in this system, there are two recognition sites, in zorO-orzO, there appears to be only one recognition domain (46).
While these previously described systems seem to corroborate the results of this study, structure-probing and kinetic experiments need to be performed to validate the predictions and to define the order of occurrence of base pairing in different regions.
Multiple factors contribute to the repression ability of Orz
Antitoxin sRNAs function by base pairing to their target mRNAs. To successfully repress the toxin mRNA, an adequate amount of functional antitoxin sRNA is required. Otherwise, even if the sRNA is capable of repressing its mRNA target, insufficient RNA levels will result in partial or complete loss of repression.
We observed this directly with our OrzO Δ3 mutant, the expression levels of which were lowered when compared with full-length OrzO. Because all of our Orz constructs are under the control of the same promoter (IPTG-inducible PLlac-O1), the decreased levels of the OrzO Δ3 are likely due to accelerated degradation. When performing the rescue experiment using the OrzO Δ3 mutant, an initial repression of zorO was observed when we induced the toxin with 0.2% arabinose, but this repression was not long lasting (Figure 6A). Because the antitoxin sRNA is induced 30 min before that of the toxin mRNA, it is likely that enough OrzO Δ3 sRNA had accumulated to initially repress zorO; however, as the ‘pre-made’ OrzO Δ3 was consumed through base pairing with zorO, the newly produced OrzO Δ3 was not sustained at a high enough level to maintain zorO repression. When the inducer concentration was decreased from 0.2 to 0.002% to reduce zorO levels, the OrzO Δ3 mutant was able to repress zorO, implying that the levels of the sRNA were now sufficient for repression.
It is also likely that the structure, and thus, specific nucleotide sequence, of OrzO contributes as well to its functional ability. For example, several of our mutants showed reduced expression levels at 1 mM IPTG when compared with the wild type at the same IPTG induction. As stated, all of our OrzO constructs were controlled by the same inducible promoter, so transcription should not be affected. Thus, it is more likely that these constructs are more unstable than wild-type OrzO. As shown in Figure 8, much of the 5′ end of OrzO (region of base pairing) is predicted to be in a stem structure. Disruption of this long stem, even by a single nucleotide, may impact overall stability of the sRNA. Further analysis of the effect of these mutations on the structure and degradation of OrzO is needed to resolve this.
Aside from RNA levels, the base pairing potential of the Orz sRNA also contributes to its ability to repress zorO. Our data show that 15 nt of continuous complementarity at 5′ end of OrzO are sufficient for the repression of zorO mRNA, as represented by the OrzO Δ3 mutant. However, unlike continuous base pairing, in which the pairing region of the sRNA perfectly matches the target sequence, unpaired nucleotides would interrupt the formation of the sRNA-mRNA duplex, affecting repression of the mRNA. Thus, it is likely that more base pairing interactions are needed to compensate for the instability of the RNA duplex caused by the unpaired base(s). The OrzO 6(1)10(1) and 6(1)11 mutants support this theory. Both RNAs were produced at levels similar to wild-type OrzO, but only Orzo 6(1)11 was able to successfully repress ZorO toxicity. The main difference between OrzO 6(1)10(1) and OrzO 6(1)11 comes from the total number of base pairing interactions, 16 versus 17, respectively. Additionally, the OrzP V1 mutant [pairing: 5(1)12] also successfully represses zorO. Combined, these data suggest that the total amount of pairing impacts the repression ability, rather than specific positioning of nucleotides (outside the V1 region).
How does binding to the 5′ UTR repress zorO toxicity?
The region of complementarity in the zorO mRNA is located in its long 5′ UTR, ∼73–91 nt upstream of the start codon. How could binding to this region prevent zorO expression? It is important to note that regulation of the type I toxins Hok and TisB by their respective antitoxins occurs through pairing far upstream of the start codons (47,48). Translation of hok requires the translation of a small peptide (encoded by mok) found within the 5′ UTR of the hok mRNA. The antitoxin, Sok, is complementary to mok, and hence binding of the sRNA prevents translation of mok, and consequently, hok (48). For tisB, its ribosome binding site is inaccessible owing to its secondary structure. However, within its 5′ UTR is a standby ribosome binding site; here the ribosome can bind, and as the structure breathes, move to the true ribosome binding site for translating tisB. The antitoxin, IstR-1, has sequence complementarity that overlaps the stand-by site; thus, formation of a tisB-IstR-1 RNA duplex would prevent ribosome binding (47).
The zorO mRNA has 5′ UTR that is ∼180 nt in length (Figure 1A); within this UTR are several potential small open reading frames and ribosome binding sites. It is possible that zorO translation is mediated in a manner similar to either hok or tisB, and that binding by OrzO may block either translation of an internal peptide or binding of the ribosome to a standby site. Ongoing experiments will help elucidate how binding of the antitoxin so far upstream from the ATG start site successfully represses zorO expression.
Role of the zor-orz locus?
A major question regarding many type I toxin-antitoxin loci is what is their true biological function? This is particularly true for the chromosomally encoded loci. Work has implicated the role of the type I toxin TisB in halting cell division during the SOS response, allowing the cell time to repair damaged DNA, as well as playing a role in persister cell formation (49,50). Several of the type I loci described in B. subtilis exist on prophages [reviewed in (51)]. In particular, txpA-ratA is located on the skin element, which is excised during sporulation (17). It has been suggested that TxpA helps maintain the skin element within B. subtilis. For the BsrG/SR4 locus, data have shown that degradation of the bsrG toxin mRNA is accelerated under high temperatures (16). How this may influence the function of the toxin has not yet been elucidated.
In the case of the zor-orz locus, it is not found beyond E. coli and Shigella species, and is absent from laboratory strains of E. coli like MG1655 (14). This implies that the locus may have been lost on domestication of E. coli and perhaps the zor-orz locus is needed for growth in the normal habitat of E. coli, the intestine. Furthermore, previous work showed that there are differences in the chromosomal expression of zorO and orzO during growth with glucose as the sole carbon source (14). We have confirmed that the endogenous expression of the sRNA and the mRNA is sensitive to carbon source, implying a possible role for the locus in cellular metabolism (Wen, Fozo, unpublished observations). Thus, the zor-orz locus may play a role in controlling cellular growth in response to changing nutrient conditions in the native environment. Current work examining the effects of deleting the locus on total cellular metabolism and gene expression are ongoing to decipher how these genes may impact overall cellular growth.
Overall, our data indicate that the 5′ end of OrzO is sufficient for repressing zorO; that the V1 region dictates Orz sRNA specificity for its target; and that there appears to be a minimum length required for base pairing. Further investigations are aimed at understanding the precise regulatory mechanisms of this untraditional type I toxin–antitoxin system to better our understanding of the intricacies of RNA regulation.
SUPLLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for this work was provided by start-up funds from the University of Tennessee. Funding for open access charge: University of Tennessee start-up funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank B. Miracle, T. Brandenburg, Z. Li and P. Dogra for technical assistance. We also are grateful to J. Wells and G. Storz for comments on the manuscript.
REFERENCES
- 1.Liu MY, Gui G, Wei B, Preston JF, III, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 2.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Georg J, Hess WR. cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Ann. Rev. Gen. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes K, Larsen JEL, Molin S. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 1985;161:292–298. doi: 10.1128/jb.161.1.292-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fozo EM, Hemm MR, Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver KE, Jensen KD, Colwell A, Sriram SI. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 1996;20:53–63. doi: 10.1111/j.1365-2958.1996.tb02488.x. [DOI] [PubMed] [Google Scholar]
- 9.Weaver KE, Tritle DJ. Identification and characterization of an Enterococcus faecalis plasmid pAD1-encoded stability determinant which produces two small RNA molecules necessary for its function. Plasmid. 1994;32:168–181. doi: 10.1006/plas.1994.1053. [DOI] [PubMed] [Google Scholar]
- 10.Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol. Microbiol. 2000;37:652–660. doi: 10.1046/j.1365-2958.2000.02035.x. [DOI] [PubMed] [Google Scholar]
- 11.Faridani OR, Nikravesh A, Pandey DP, Gerdes K, Good L. Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Res. 2006;34:5915–5922. doi: 10.1093/nar/gkl750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 13.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology. 2009;155:2930–2940. doi: 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 16.Jahn N, Preis H, Wiedemann C, Brantl S. BsrG/SR4 from Bacillus subtilis–the first temperature-dependent type I toxin-antitoxin system. Mol. Microbiol. 2012;83:579–598. doi: 10.1111/j.1365-2958.2011.07952.x. [DOI] [PubMed] [Google Scholar]
- 17.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fozo EM. New type I toxin-antitoxin families from “wild” and laboratory strains of E. coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 2012;9:1504–1512. doi: 10.4161/rna.22568. [DOI] [PubMed] [Google Scholar]
- 20.Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 22.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 2001;147:3241–3247. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- 23.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 24.Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 28.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 29.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 34.Gerdes K, Nielsen A, Thorsted P, Wagner EG. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J. Mol. Biol. 1992;226:637–649. doi: 10.1016/0022-2836(92)90621-p. [DOI] [PubMed] [Google Scholar]
- 35.Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA.J. Mol. Biol. 2011;406:29–43. doi: 10.1016/j.jmb.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durand S, Gilet L, Condon C. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012;8:e1003181. doi: 10.1371/journal.pgen.1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krinke L, Mahoney M, Wulff DL. The role of the OOP antisense RNA in coliphage lambda development. Mol. Microbiol. 1991;5:1265–1272. doi: 10.1111/j.1365-2958.1991.tb01900.x. [DOI] [PubMed] [Google Scholar]
- 38.Krinke L, Wulff DL. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987;1:1005–1013. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- 39.Krinke L, Wulff DL. RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA. Genes Dev. 1990;4:2223–2233. doi: 10.1101/gad.4.12a.2223. [DOI] [PubMed] [Google Scholar]
- 40.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 41.Lalaouna D, Simoneau-Roy M, Lafontaine D, Masse E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta. 2013;1829:742–747. doi: 10.1016/j.bbagrm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc. Natl Acad. Sci. USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hjalt T, Wagner EG. The effect of loop size in antisense and target RNAs on the efficiency of antisense RNA control. Nucleic Acids Res. 1992;20:6723–6732. doi: 10.1093/nar/20.24.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin-Chao S, Cohen SN. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell. 1991;65:1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- 46.Han K, Kim KS, Bak G, Park H, Lee Y. Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acids Res. 2010;38:5851–5866. doi: 10.1093/nar/gkq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Thisted T, Gerdes K. Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J. Mol. Biol. 1992;223:41–54. doi: 10.1016/0022-2836(92)90714-u. [DOI] [PubMed] [Google Scholar]
- 49.Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unoson C, Wagner EG. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008;70:258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]
- 51.Durand S, Jahn N, Condon C, Brantl S. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 2012;9:1491–1497. doi: 10.4161/rna.22358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.