Abstract
Circular chromosomes can form dimers during replication and failure to resolve those into monomers prevents chromosome segregation, which leads to cell death. Dimer resolution is catalysed by a highly conserved site-specific recombination system, called XerCD-dif in Escherichia coli. Recombination is activated by the DNA translocase FtsK, which is associated with the division septum, and is thought to contribute to the assembly of the XerCD-dif synapse. In our study, direct observation of the assembly of the XerCD-dif synapse, which had previously eluded other methods, was made possible by the use of Tethered Particle Motion, a single molecule approach. We show that XerC, XerD and two dif sites suffice for the assembly of XerCD-dif synapses in absence of FtsK, but lead to inactive XerCD-dif synapses. We also show that the presence of the γ domain of FtsK increases the rate of synapse formation and convert them into active synapses where recombination occurs. Our results represent the first direct observation of the formation of the XerCD-dif recombination synapse and its activation by FtsK.
INTRODUCTION
In most Bacteria and Archaea, genetic information is found on circular chromosomes that, after replication, can form dimers by homologous recombination. In Escherichia coli, conversion of chromosomes into dimers occurs on average once every six generations (1). As a consequence, the two sister chromosomes that constitute this dimer cannot be segregated to daughter cells unless the dimer is turned back into monomers. Failure to resolve chromosome dimers leads to defects in chromosome segregation and subsequent cell death (2). Resolution of dimeric chromosomes (CDR) is performed by the XerCD-dif site-specific recombination system. The importance of this function for faithful chromosome segregation explains its high conservation and the Xer system is now considered as one of the most conserved structural feature of circular chromosomes in Bacteria and Archaea (3,4).
In E. coli, XerC and XerD act jointly to recombine two copies of the dif site located in the replication termination region (ter) of the chromosome (Figure 1A and B). Although limited structural data are available for these proteins, several related tyrosine recombinase proteins (YR) such as Cre, λint or Flp have been crystalized alone or bound to DNA, allowing to derive a general model for this enzyme family (5). The XerCD-dif recombination process is sketched in Figure 1A and B as it is hypothesized to occur today. The dif site is composed of two protein-binding arms, dif XerC and dif XerD, separated by a central region, dif cent. XerC and XerD, respectively, bind specifically to dif XerC and dif XerD (6). It is proposed that two XerCD-dif complexes then interact to form the XerCD-dif synapse. Within this synapse, only one type of recombinase, either XerC or XerD, is expected to be active and each of the two units of that recombinase cuts the DNA strand at the dif site to which it is bound (7). This nucleophilic attack of DNA, mediated by a conserved tyrosine residue, forms a covalent link between the recombinase and the dif site. The second step of the reaction is a strand exchange between the two dif copies in the central region followed by ligation, which creates a Holliday junction (HJ1). This intermediate isomerizes into a second one (HJ2), thereby activates the second pair of recombinases, which cut and exchange the second pair of strands, finishing the recombination reaction (8). In this process, the two pairs of recombinases are sequentially activated to catalyse the exchange of the two DNA strands. Therefore, the selection of the first active pair of recombinases controls the reaction (8). It has been proposed that, within the XerCD-dif synapse, XerC is the one initially active while XerD is initially inactive. As a consequence, the reaction is blocked at the HJ1 and thus tends to be reversible, without recombination (9). In order to catalyse a complete recombination process, XerD must be activated. This activation is part of a cell cycle checkpoint that is achieved by FtsK, a division septum-associated DNA translocase, which is essential for cell division (10–12). The amino-terminal part of FtsK is composed of transmembrane helices that anchor the protein in the membrane and of a linker that interacts with other proteins of the division septum (13,14). The translocase activity of FtsK is contained in its carboxy-terminal part, which is composed of three sub-domains: α, β and γ (15). The ‘motor’ part of this translocase, constituted by the α and β sub-domains, is related to the large AAA+ ATPase family, known to hydrolyse ATP for multiple purposes including DNA translocation but also substrate remodelling (For reviews: 15–18). The γ domain is the ‘driver’ of the translocase activity. By recognizing KOPS sequences, which are oriented towards dif on each chromosome replichores, the γ domain imposes the direction of DNA translocation towards dif (19–23). Upon reaching the dif site, the γ domain activates XerCD-dif recombination through a specific contact with the carboxy-terminal part of XerD (10,24–27). A current hypothesis proposes that FtsK could be involved in the formation of the XerCD-dif synapse and its remodelling into a ‘XerD-active’ conformation in which XerC is made inactive and XerD is ready to be activated by contact with the γ domain (9,27).
Figure 1.
XerCD-dif recombination analysed with TPM. (A) Sequence of dif site with the DNA binding site for XerC (dif XerC) for XerD (dif XerD) and the central region (dif cent) represented. (B) Model for XerCD-dif recombination (9). Recombinases (XerC or XerD) are represented as grey circles, with Y indicating the active tyrosine. DNA molecules are oriented with uppercase and lowercase A and B letters. The reaction is sketched in five steps: synapse formation; first strand cleavage, exchange and ligation to form the first holiday junction (HJ1); isomerization of HJ1 into HJ2; resolution of HJ2; and dissociation. If XerC cuts first, the process is blocked at HJ1 step and goes backward. If XerD is activated to cut first, the recombination can be complete. (C) Scheme of TPM setup to measure the length of a DNA molecule. A glass coverslip (g) is coated with PEG (p) and neutravidin (n). A DNA molecule is attached to that surface by biotin bound to one of its 5′ end. A latex bead coated with antidigoxigenin (ad) is bound to the other extremity of the DNA molecule thanks to the presence of digoxigenin on this 5′ end. As explained in the text, the amplitude of the Brownian motion of the bead (Aeq) depends on the size of the DNA molecule that tethers the observed bead to the glass surface (for details see text and M&M). (D) Scheme explaining how XerCD-dif synapse formation may reduce the apparent size of a DNA molecule that contains two dif sites (black box: dif XerD; white box: dif XerC). (E) DNA molecules used in this work. The representation of dif sites is the same as in (D) and the sizes (bp) of the different segments of these molecules and the 5′ modifications are indicated (b = biotin, d = digoxigenin).
Although this model highlights the role of the assembly of the XerCD-dif synapse for the control of the reaction, it remains speculative since synaptic complexes have not been observed to date. The well-established Electrophoretic Mobility Shift Assay (EMSA) has allowed the description of synapses formed with some YR other than XerCD (28,29), and of complexes related to XerCD-dif but not of XerCD-dif synapses suggesting that those are not stable enough to be detected in EMSA gels (6). For the Cre-loxP system, EMSA and structural analysis have shown that synapses are preferentially assembled in an antiparallel structure including a bending of the loxP site as a key control of the recombinase activation (30,31). Atomic Force Microscopy enabled the visualization of synapses of the λint-att, Cre-loxP and Flp-FRT systems and confirmed the bending of DNA within these complexes (32,33). More recently, synapse formation and recombination catalysed by these same three recombinases were all examined by single molecule techniques based on tethered DNA molecules (34–37). These studies showed that productive and unproductive synapses are formed and that the rate-limiting steps of the reaction are synapse assembly together with complex dissociation after recombination. Overall these data strengthen the idea of synapse assembly being a crucial step in the control of YR’s catalysed site-specific DNA recombination.
Here we report the characterization of the XerCD-dif synapse by using a single molecule approach, Tethered Particle Motion (TPM). This method has previously been fruitful in studying the synaptic assembly of λint-att and Cre-loxP (34,35,37). This force-free experimental approach allows monitoring of individual events occurring simultaneously on several DNA molecules. We show that the synapse can be assembled without FtsK. The minimal requirement for the synapse assembly is XerC, XerD and two dif sites. This XerCD-dif synapse, however, appears to be inactive for strand exchange unless the γ domain of FtsK is also present. Kinetic analyses showed that the presence of the γ domain increases the rate of XerCD-dif synapse assembly and increases its stability. These results allow us to discuss the current XerCD-dif recombination model.
MATERIALS AND METHODS
Protein and DNA
xerC, xerCYF, xerD and xerDYF genes (Genescript) are cloned into pet32b plasmid (pROUT06, pROUT16, pROUT07 and pROUT15, respectively) under pT7. Proteins production was performed in BL21-DE3 strain (LB, 37°C to OD600 = 0.6). After 42°C heat chock for 10 min, cultures were transferred to 16°C and induced with 0.1 mM IPTG for at least 2 h. Lysis was performed in buffer-A (50 mM Tris pH = 8; 300 mM NaCl; 10% glycerol) complemented with 0.05 mg lysozyme and protease inhibitor cocktail (ROCHE®) for 30 min on ice. After centrifugation (1 h at 27.000 g), the soluble fraction was submitted to chromatography on nickel-sepharose (1 ml His-trap column; GE®) using a 10–500 mM imidazole gradient in buffer-A. Eluted tagged-Xer proteins were then further purified by a chromatography on heparin (1 ml Hep-Trap, column; GE®) using a 300–1500 mM NaCl gradient in buffer-B (50 mM Tris pH = 8; 300 mM NaCl; 10% glycerol, 1 mM DTT; 1 mM EDTA). Eluted proteins were then submitted to size exclusion chromatography (HiLoad 16/60 Superdex 200, GE®). After concentration (Vivaspin®), proteins were dialysed against buffer-S (50 mM Tris pH = 8; 500 mM NaCl; 20% glycerol, 1 mM DTT; 1 mM EDTA), tested for aggregation by a Dynamic Light Scattering device (Dynapro) and stored as individual aliquots at −80°C. XerCγ and XerDγ were purified using the same procedure but produced from pBAD plasmids kindly provided by Sherratt’s lab. The purified Cre protein was purchased from NEB®.
DNA molecules used in TPM were produced by PCR on pFX346 and pFX347 plasmids that Barre’s lab kindly provided to us. Modified oligonucleotides, carrying the tags at the 5′ end, were Biot-OCD1 (CATTGCTACAGGCATCGTGG); Dig-OCD2 (TATTCAGGCGTAGCACCAGG); Biot-OCD3-Rev (CGGCGTCAATACGGGAT); and Dig-OCD4 (CAGTGAGCGCAACGCAATTAA) (Sigma-Aldrich). PCR products were separated by electrophoresis on 1% low-melting agarose gels and purified as described elsewhere (38).
Microscope slides and coverslips passivation
Slides (RS-France) and coverslips (24 × 36 mm2 thickness n°1) (Menzel-Gläser) were immerged into a sulfochromic acid solution for 20 min, extensively rinsed with deionized water and dried under a nitrogen flow. The cleaned glasses were thiolated by an overnight incubation at room temperature in a methanol mixture [91% methanol (VWR-Prolabo), 4% MilliQ H2O, 4% (3-mercaptopropyl) trimethoxysilane (Sigma-Aldrich) and 1% acetic acid (VWR-Prolabo)]. After treatment, they were rinsed with methanol and isopropanol and dried under a nitrogen flow. Silanized glasses were incubated for 3 h at room temperature with a polyethylene glycol (PEG) mixture. Slides were treated with PEG-maleimide (Fluka-Biochemika) (1:1) whereas coverslips were treated with PEG-maleimide and PEG-maleimide-biotin (Nanocs) at a ratio of 10:1. After passivation, glasses were washed extensively with deonized water, dried under a nitrogen flow and stored within a silica gel-containing desiccator for further use.
Formation of the DNA–bead complexes
The beads were functionalized with anti-DIG as described in Plénat et al. (39). DNA molecules and beads at an equimolar concentration of 40 pM were mixed for 1 h at room temperature in the PBB buffer composed of a phosphate buffer (1 mM KH2PO4, 3 mM Na2HPO4, 150 mM NaCl, pH 7.4, Euromedex, France), BSA (1 mg/ml, Sigma-Aldrich) and Pluronic F-127 (1 mg/ml, Sigma-Aldrich).
Assembly of the fluidic observation chamber for TPM experiments
A 0.5 mm-thick adhesive spacer (Grace Bio Labs) was cut to dimensions fitting to the coverslip, to obtain a 20 mm-long and 3 mm-wide channel in the middle and used to stick the coverslip to a microscope slide into which two holes spaced by 20 mm were previously drilled. The flow cell was first rinsed with PBB buffer, then with PBB supplemented with 20 µg/ml neutravidin.
After 20 min of incubation, the flow cell was rinsed with the PBB buffer. DNA–bead complexes were then incubated for 1 h in the passivated flow cell. The flow cell was rinsed extensively with the protein buffer PBP (20 mM Tris pH8, 50 mM NaCl, 1 mM DTT, 1 mM MgCl2, BSA (0.1 mg/ml, Sigma-Aldrich), Pluronic F-127 (1 mg/ml, Sigma-Aldrich) before starting the observations.
Single particle tracking and calculation of the amplitude of motion of the particles
The tethered beads of 300 nm diameter were visualized at 21 ± 1°C by using a dark-field microscope (Axiovert 200, Zeiss) equipped with a x32 objective and an additional x1.6 magnification lens, on a CMOS camera Falcon 1.4M100 (pixel size: 7.4 µm, Dalsa) at a recording frequency of 25 Hz.
The acquisitions were all performed continuously on the same field of view for 25 min or more. For all samples, the initial step of 1 min corresponds to the tracking of the DNA–bead complexes in the presence of the buffer PBP only. It is followed by a single injection of the desired protein mix whose effect on the DNA–bead complexes is analysed between the 3rd and the 20th minute. It is then followed by the injection of PBB complemented with SDS 0.1%. The concentration of proteins in the mixes ranged between 40 nM and 80 nM for each recombinase. For each protein preparation, we performed preliminary experiments and used the maximal concentration that did not result unspecific binding events leading to extremely low value of Aeq such as described by Laurens et al. (40).
The software developed by Magellium (France) tracks all the visible beads in the dark field and computes their amplitudes of motion including corrections for experimental drift. The detailed calculations of the amplitude of motion of the beads can be found in (41).
Three criteria of validity of the DNA–bead complexes are defined. The first one concerns the symmetry of the trajectories (41). The second one excludes any trace containing a single Aeq inferior to 1 nm that usually indicates a positioning problem. The third one discards trajectories with Aeq inferior to 100 nm on more than 10% of the experiment duration, which may be attributed to unspecific binding. The validity of the DNA–particles complexes was established during the incubation with the proteins. After the injection of SDS, some of the complexes detached from the coverslips or were immobilized, which led to a decrease in the percentage of DNA-particle still under examination after the injection of SDS.
The probability distributions of Aeq were fitted by one or two Gaussians using the Prism software (Graphpad, La Jolla, USA). Standard errors on the centres of these Gaussians were given by the fit procedure.
The kinetics analysis of the TPM traces were carried out using the simple threshold method, with averaging over 2 s frames (periods shorter than 3 s) were discarded (34). The formation and dissociation rates were obtained by fitting the histograms of the duration of the unsynapsed and synapsed states with a monoexponential function, A exp(-kformation OR dissociationt) +B, in Prism software (Graphpad, La Jolla, USA).
RESULTS
TPM experimental setup to study XerCD-dif synapsis
For the TPM technique, we relied on tracking 300 nm diameter-sized particles attached at one end of the DNA molecules while the other extremity of these molecules is immobilized on a passivated coverslip (Figure 1C and M&M). The 2D projection of the bead displacement relative to the anchoring point of the DNA molecule can be characterized over time by its mean-square radius, noted Aeq for amplitude of motion at equilibrium. Aeq directly depends on the apparent length of the monitored DNA (Supplementary Figure S1), such that changes as small as 100 bp can be discriminated (39,41,42). In order to track the formation of a synapse between two dif sites, we constructed two DNA molecules, containing either one dif site (DNA-dif1, 2255 bp) or two dif sites (DNA-dif2, 2271 bp, Figure 1E and M&M). Those two dif sites were separated by 1074 bp, a distance large enough to unambiguously detect apparent shortening of the DNA due to synapse formation (Figure 1D). We constructed and purified tagged version of the Xer proteins. We checked that their DNA-binding and recombination activities were not altered by the presence of the tag (Supplementary Figure S2).
XerCD-dif synapse formation
The formation of a synapse between the dif sites on the DNA-dif2 molecule should decrease the apparent length of the DNA molecule by roughly the length separating the two dif sites and lead to a reduction of ∼90 nm according to our calibration curve (Supplementary Figure S1). We measured the Aeq of 38 DNA-dif2 molecules before and after the addition of proteins. In all experiments, beads were tracked for 19 min. We then constructed histograms of their Aeq displayed as a probability density graph (Figure 2A). In the absence of protein, based on the calibration curve, we expected a theoretical Aeq (Aeqtheo) of 259.2 ± 18.5 nm, (Supplementary Figure S1) and we observed a single peak centred on 233.8 ± 0.1 nm (mean ± se as described in M&M, Figure 2A; black histograms, N = 38). This difference of 25 nm between experimental Aeq (Aeqexp) and Aeqtheo was observed in all experiments. We hypothesize that this could be due to sequence-dependent structural properties of this DNA molecule such as an intrinsic DNA curvature.
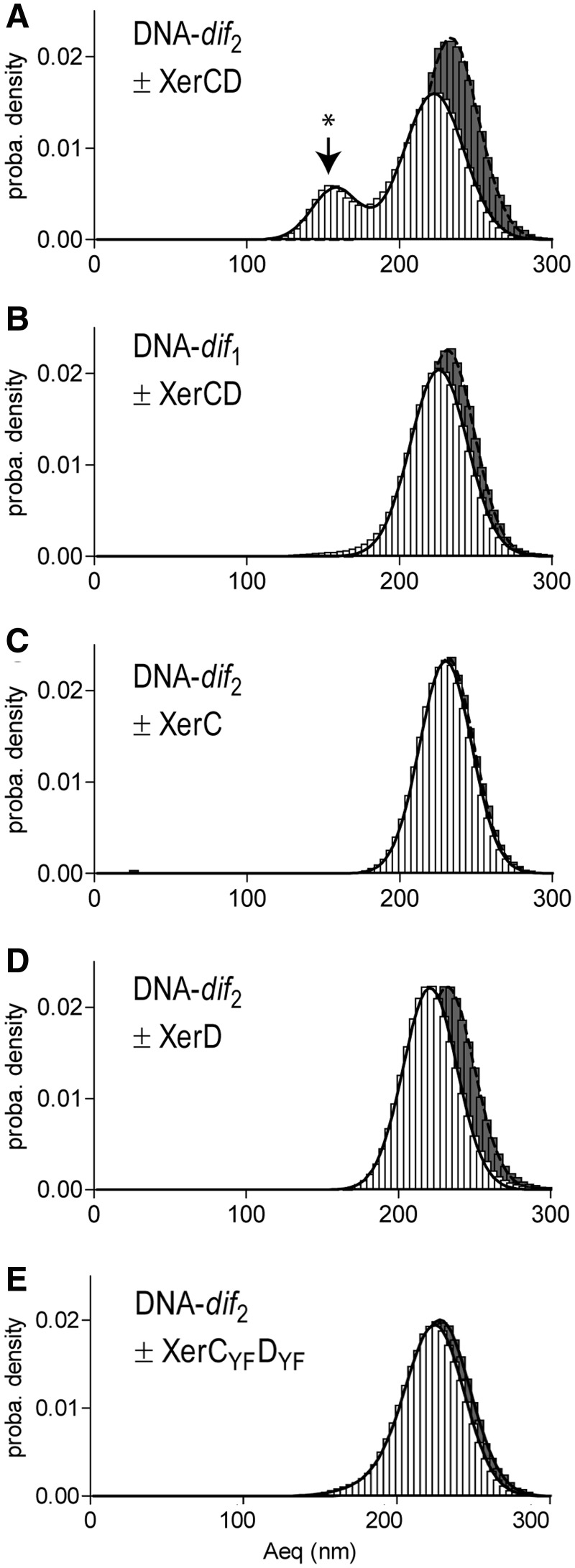
Figure 2.
XerCD-dif synapse formation. Probability distributions of Aeq, before protein injection (black histograms), or during the 19 min following protein injection (white histograms). (A) DNA-dif2 molecules incubated XerCD mix (N = 38). Peak attributed to the formation of XerCD-dif synapses in indicated by *. (B) DNA-dif1 molecules incubated with XerCD mix (N = 179). (C) DNA-dif2 molecules incubated with XerC (N = 118). (D) DNA-dif2 molecules incubated with XerD (N = 117). (E) DNA-dif2 molecules incubated with XerCYFDYF mix (N = 42).
In the presence of XerC and XerD, a nucleoprotein complex was expected to form on the DNA molecule and assemble the two distant dif sites into the XerCD-dif synapse. The XerCD-dif synapse-containing DNA molecule would then resemble a DNA molecule deleted of 1102 bp (28 bp corresponding to one dif site + 1074 pb corresponding to the intervening fragment between the two dif sites; Figure 1E) and should thus behave as a DNA molecule measuring only 1169 bp. The Aeqtheo for this length of DNA is 165.1 ± 12.5 nm (Supplementary Figure S1), but if we consider that the synapsed DNA molecule may be shortened by sequence-dependent properties similarly to the protein-free molecule, the DNA molecules containing XerCD-dif synapses should exhibit an Aeq lying between 140 nm and 165 nm. Recombined or HJ-containing molecules should also exhibit an Aeq lying between 140 nm and 165 nm. After injection of a mix of XerC and XerD on the previously observed DNA molecules, we observed two peaks on the Aeq distribution graph (Figure 2A; white bars). The first peak was centred on 158.0 ± 0.4 nm (22% of the density probability; Figure 2A, noted by *), compatible with XerCD-dif synapse formation. The second peak was centred on 222.7 ± 0.1 nm (79% of the density probability), which represents a sizeable reduction of ∼10 nm compared to the Aeq measured for protein-free DNA molecules. While this difference is clearly too small to reflect synapse formation, it may rather correspond to other protein-induced structural changes of DNA-dif2.
As a control for the detection of a XerCD-dif synapse, we performed the same experiment with DNA-dif1 molecules, which contain only one dif site. In the absence of proteins, we observed a single peak centred on 231.7 ± 0.1 nm (N = 179), which is a good match to that measured for DNA-dif2. After addition of a mix of XerC and XerD proteins, we still observed a single peak centred on 226.0 ± 0.1 nm (N = 179) (Figure 2B). This 5 nm reduction observed after injection of XerCD on DNA-dif1 corresponds to half the value of the reduction observed after addition of XerCD on DNA-dif2 on the second peak. These small Aeq reductions are very likely due to the sole binding of XerC and/or XerD to one or two dif sites. In contrast, the large Aeq reductions, which are seen for DNA-dif2 but not DNA-dif1, can be attributed to synapse formation.
We next asked if XerC and XerD are both required for synapse formation. We repeated TPM experiments using DNA-dif2 incubated with either XerC or XerD (Figures 2C and D). In the absence of protein, the measured Aeqs were in good agreement with the previous experiment (XerC experiment: 231.5 ± 0.1 nm; N = 118 and XerD experiment: 232.1 ± 0.1 nm; N = 117). After XerC addition, we did not observe a significant change of the Aeq (230.0 ± 0.1 nm; N = 118; Figure 2C). When XerD was added, we also observed a single peak, but showing a 12 nm reduction of the Aeq compared to the protein-free DNA (220.8 ± 0.1 nm; N = 117; Figure 2D). Again, this reduction was too small to reflect synapse formation and may rather correspond to protein-induced structural changes of the DNA. We concluded that neither XerC nor XerD alone are able to provoke the important Aeq reduction, which probably corresponds to XerCD-dif synapses.
The important reduction of Aeq observed following the incubation of DNA-dif2 molecules with a mix of XerC and XerD might be due either to XerCD-dif synapses or to XerCD-catalysed strand exchange between the two dif sites (Figure 1B). To decipher between these two possibilities, we constructed some catalysis-defective variants of XerC and XerD. To do so, we mutated the xerC and xerD genes in order to change catalytic tyrosines (Y275 for XerC and Y279 for XerD) to phenylalanines (F275 for XerC and F279 for XerD). The modified proteins, XerCYF and XerDYF, were purified following the same procedure as for the wild-type proteins (M&M). We then repeated the TPM experiments with DNA-dif2 incubated with a mix of XerCYF and XerDYF (Figure 2E). In the absence of proteins, we observed a single peak centred on 230.6 ± 0.1 nm (N = 42). After injection of the XerCYFDYF mix, two peaks were measured: one centred on 225.6 ± 1.8 (87% of the probability density) and the other centred on 198.2 ± 58.9 (12% of the probability density). This Aeq reduction (∼30 nm) was too large to only correspond to protein-induced structural changes of the DNA and may rather reflect synapse formation. This suggests that XerCYF and XerDYF are capable of forming the XerCYFDYF-dif synapses, albeit much less efficiently compared to unmodified proteins. Catalysis does not seem to be required for synapse formation.
From the above data, we conclude that the important reduction of Aeq observed is due to the formation of the XerCD-dif synapse. It follows that synaptic complex formation requires XerC, XerD and two dif sites but can form in the absence of other factors such as FtsK.
XerCD-dif synapses are inactive
To explore if strand exchange occurred in the XerCD-dif synapses, we tested if the complexes formed between XerC, XerD and two dif sites could be disassembled using detergent. Indeed, because it breaks non-covalent interactions in DNA–protein complexes, SDS addition (0.1% final concentration) should allow us to discriminate synapses based only on non-covalent interactions from those containing at least one strand exchange (Figure 2B).
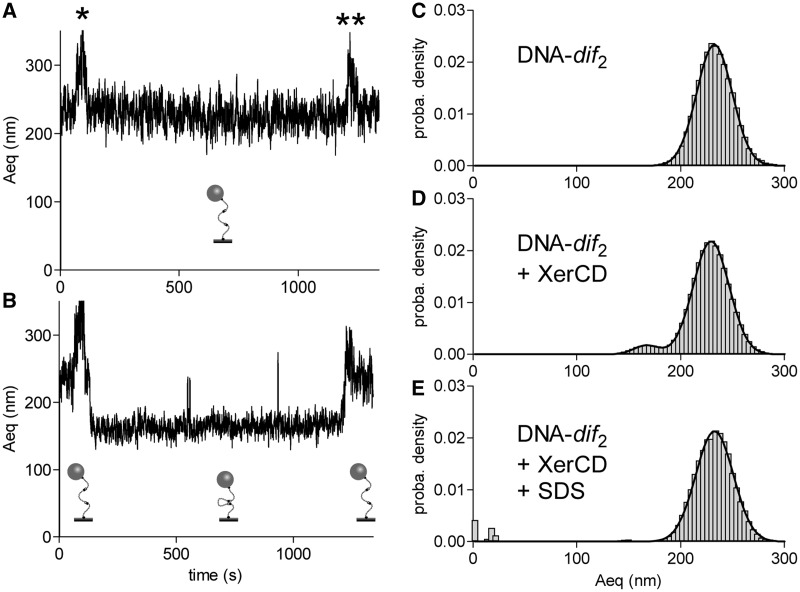
To this end, we analysed individual particle traces by plotting Aeq as a function of time for all the particles in an experiment in which DNA-dif2 was incubated with a XerCD mix. On Figure 3A, a typical trace observed for a DNA molecule remaining unsynapsed is presented: the Aeq stayed unchanged after protein addition (noted by *). Figure 3B shows a typical trace observed for a DNA molecule showing XerCD-dif synapse formation. Aeq was reduced by ∼65 nm after addition of the XerCD mix (noted by *). On the 62 molecules recorded, 12 (19%) exhibited this strong Aeq reduction. This Aeq reduction was, however, not caused by strand exchange in the XerCD-dif synapse since addition of SDS in the reaction (noted by **) restored an Aeq comparable to that of an unsynapsed-DNA. Histograms of the Aeq measured for 62 DNA molecules before and after SDS addition show that all complexes attributed to synapses were disassembled by the SDS treatment (Figure 3C–E). In this experiment, the peak corresponding to unsynapsed-DNA was centred on 229.4 ± 0.1 nm (94% of the probability density) and the peak corresponding to synapsed-DNA was centred on 166.8 ± 0.3 nm (5% of the probability density corresponding to 12 DNA molecules). After the injection of SDS, only one peak was observed centred on 232.7 ± 0.2 nm. This shows that strand exchange either does not occur within XerCD-dif synpases formed in our TPM setup or rapidly reverse upon SDS challenging.
Figure 3.
XerCD-dif synapses are non-productive. (A, B) Typical traces (Aeq = f(time)) observed for non-synapsed (A) and reversibly synapsed (B) DNA molecules. Stars indicate the times of injection of XerCD mix (*) and SDS (**). Schemes of the proposed structures of the DNA are shown underneath each trace. (C–E) Probability distributions of Aeq before protein addition (C), during the 19 min following XerCD mix addition (D) and after SDS injection (E).
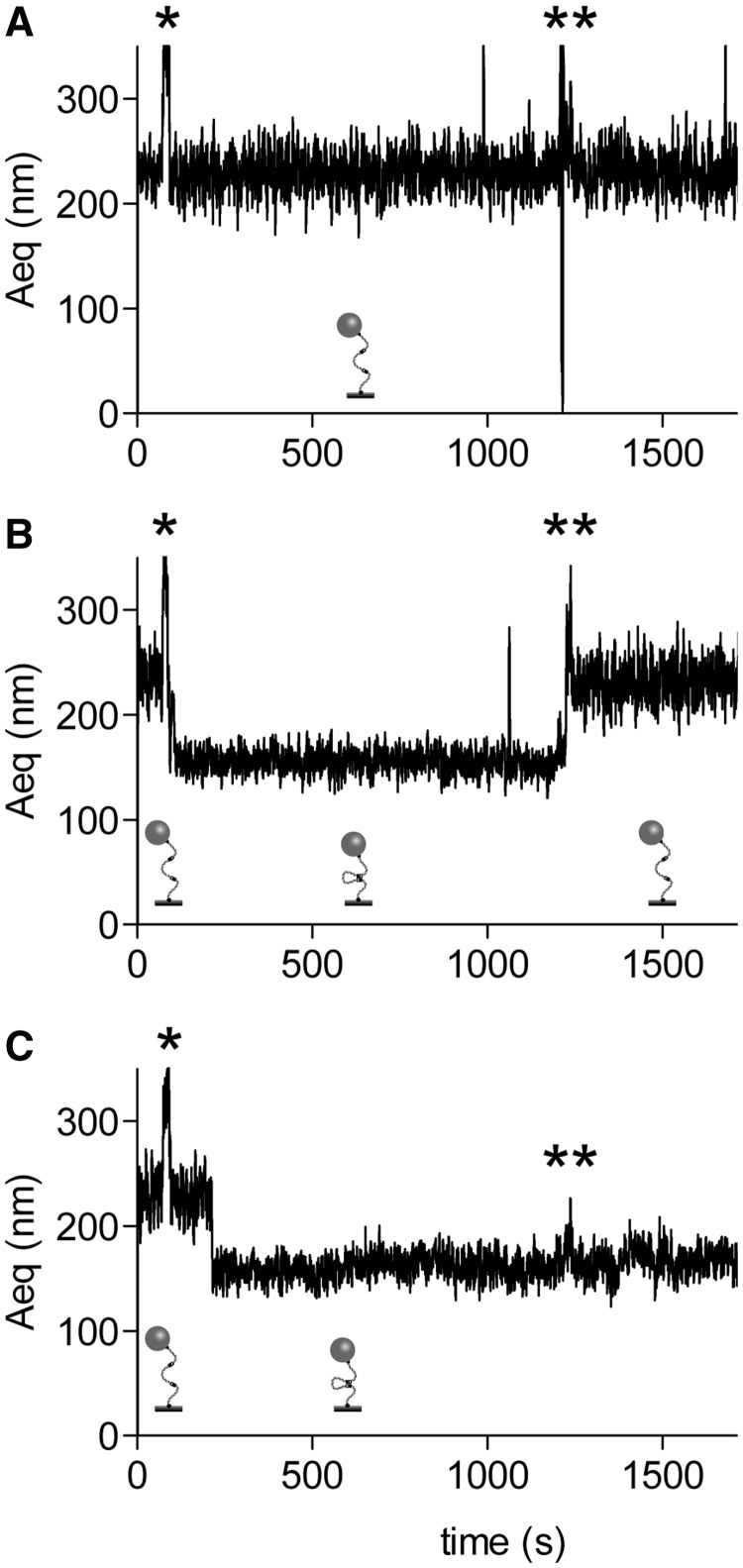
To control that recombination would effectively lead to a SDS-resistant Aeq reduction, we performed TPM experiments using Cre and a DNA fragment containing loxP sites. Unlike XerCD-dif, Cre-loxP recombination only needs Cre and two loxP sites and has been shown to occur in similar assays (35–37). The DNA-dif2 molecule we used also contains two directly repeated loxP sites separated by 1068 bp (loxP sites are 34 bp long and are present 11 bp before each dif sites on DNA-dif2). Thus, similarly to XerCD-dif, a Cre-recombined DNA-dif2 molecule should show an 1102 bp deletion and behave as an 1169 bp DNA molecule. We expected that such a recombined molecules show Aeq between 140 nm and 165 nm. On Figure 4A, a typical trace observed for an unaffected DNA molecule is presented. The Aeq stayed unchanged even after protein addition (noted by *), and after addition of SDS in the reaction (noted by **). Figure 4B shows a clear Aeq reduction following Cre addition. On this example, the Aeq reduction is reversible since addition of SDS in the reaction restored an Aeq similar to unaffected DNA. This suggests that, in this case, the two loxP sites were synapsed but recombination did not occur. On Figure 4C, however, the Aeq reduction that followed protein addition was resistant to SDS challenging, confirming that DNA strand exchange indeed occurred within this Cre-loxP synapse and that we can detect such an event with our experimental procedure. Among the 115 DNA traces recorded in that experiment, 47 showed synapsis and 2 of those exhibited resistance to SDS suggesting strand exchange.
Figure 4.
Cre-loxP synapse formation and recombination. Typical traces (Aeq = f(time)) observed for non-synapsed (A), reversibly synapsed (B) and irreversibly synapsed (C) DNA molecules. Stars indicate the times of injection of Cre (*) and SDS (**). Schemes of the proposed structures of the DNA are shown underneath each trace.
Taken together, these results show that no strand exchange could be observed in a XerCD-dif synapse. This is consistent with XerD being inactive in the absence of FtsK (9). However in the absence of FtsK, reversible HJ1 formation should be possible by XerC catalysis (Figure 1B). The fact that we did not observe such HJ1 suggests that they are not formed or that the SDS challenging occurs in a timescale that allows the HJ1 to reverse before synapse denaturation.
XerCD-dif synapses are productive in the presence of the γ-domain of FtsK
The results obtained with Cre support the idea that the inactivity of the XerCD-dif synapses we observed may be due to a lack of activation by FtsK. To test this, we used chimeric XerCγ and XerDγ proteins, consisting of the fusion of the γ domain of FtsK on either XerC or XerD. XerCγ and XerDγ have been recently reported as being competent for the activation of XerCD-dif recombination (27).
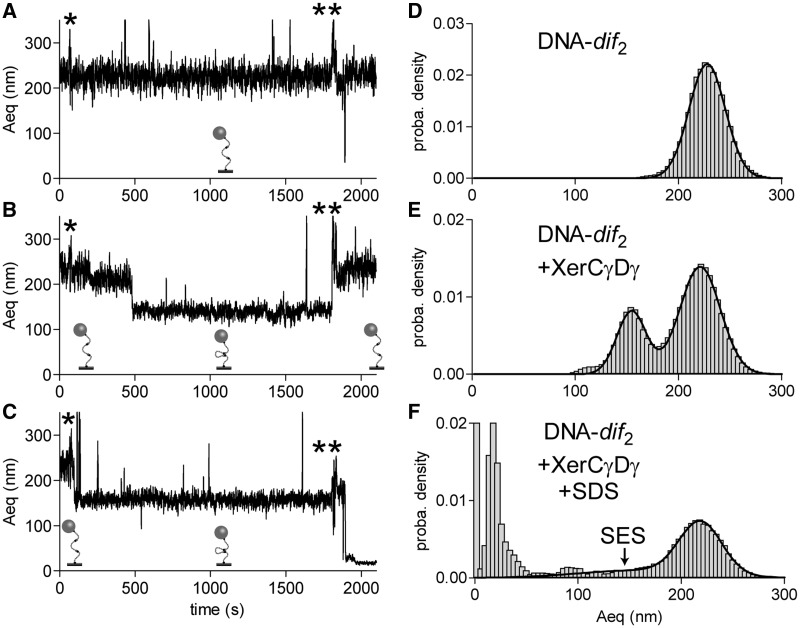
We purified XerCγ and XerDγ and used them in our TPM setup with DNA-dif2. For 10 of the 123 DNA molecules observed, the Aeq reduction, which followed injection of the XerCγDγ mix was found to be resistant to SDS challenge, strongly suggesting that strand exchange had occurred (Figure 5A–C). Histograms of the Aeq recorded show that the peak corresponding to unsynapsed-DNA was centred on 221.0 ± 0.2 nm (69% of the probability density) and that the peak corresponding to synapsed-DNA molecule was centred on 154.6 ± 0.2 nm (29% of the probability density corresponding to 70 DNA molecules, Figure 5D and E). We concluded that XerCγ and XerDγ form XerCγDγ-dif synapses comparable to the XerCD-dif synapses. After the injection of SDS, signal became noisier because some DNA–bead complexes disappeared, and others transiently and unspecifically bound to the surface (Figure 5C). Despite this noise, however, one peak was still clearly detectable, centred on 218.0 ± 0.3 nm (35% of the probability density) whereas a broad shoulder was spread out ∼160.3 ± 8.1 nm (Figure 5F, noted by SES, 15% of the probability density). This population corresponds to the 10 molecules that showed SDS-resistant Aeq reduction (Figure 5C) and, since a similar population was not recorded with the mix of unmodified XerC + XerD (Figure 3E), we conclude that these molecules have most probably undergone at least one XerCD-mediated strand exchange.
Figure 5.
XerCγDγ-dif synapse formation and recombination. (A–C) Typical traces (Aeq = f(time)) observed for non-synapsed (A), reversibly synapsed (B) and irreversibly synapsed (C). Stars indicate the times of injection of XerCγDγ mix (*) and SDS (**). Schemes of the proposed structures of the DNA are shown underneath each trace. (D–F) Probability distributions of Aeq before protein addition (D) during the 30 min following XerCγDγ mix addition (E) and after SDS injection (F). SES, for ‘strand exchange-synapse’, indicates the part of the curve corresponding to such events.
The γ-domain of FtsK increases the rate of XerCD-dif synapse assembly
Interaction of XerCD, XerCγDγ or XerCYFDYF with DNA-dif2 molecules fluctuated between synapsed and unsynapsed states (Supplementary Figure S3). The kinetics of these transitions were analysed by measuring the dwell time in these two states on recorded traces. In the three conditions, the histograms of the dwell time could be fitted by single exponential functions characteristics of a first-order mechanism (Figure 6A–C).
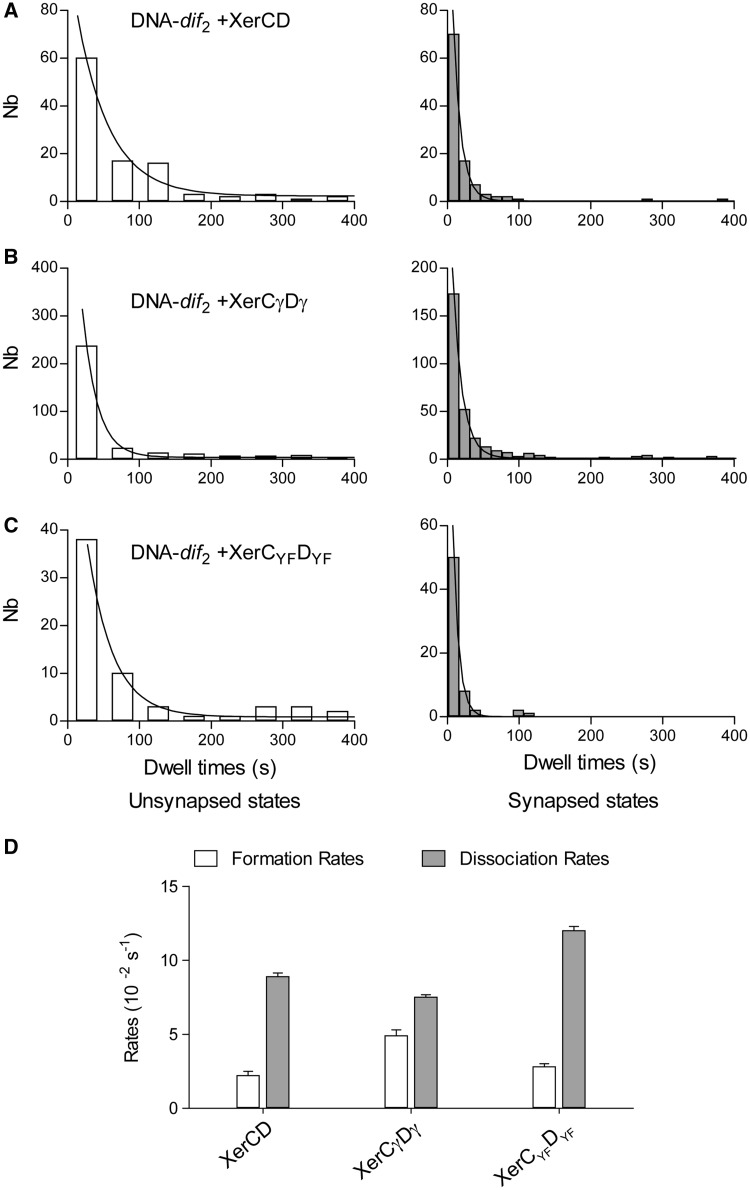
Figure 6.
Kinetics analysis. Histogram of the dwell times of unsynapsed states (left panel, white histograms) and of the synapsed states (right panel, grey histograms) measured for: (A) XerCD (N = 100); (B) for XerCγDγ (N = 123); and (C) for XerCYFDYF (N = 42). (D) The formation and dissociation rates were obtained by fitting the histograms of the duration of the unsynapsed and synapsed states with a monoexponential function (M&M).
The rates of formation of the looped states (34) were found equal to (2.2 ± 0.3) × 10−2 s−1 (R2 = 0.937, N = 100) in the presence of XerCD, (4.9 ± 0.4) x 10−2 s−1 (R2 = 0.934, N = 123) in the presence of XerCγDγ and (2.8 ± 0.2) x 10−2 s−1 (R2 = 0.989, N = 42) in the presence of XerCYFDYF (Figure 6D). The measured rates for XerCD and its catalytic mutants are altogether quite similar to one another, suggesting that modification of the catalytic state of XerCD has little impact on synapse formation. In contrast, the rate measured in presence of the γ domain (XerCγ and XerDγ) is two times higher than the two other ones. This could explain that 21% of the DNA molecules were synapsed immediately after the injection of the XerCγDγ mix (Supplementary Figure S3), while this only happened for 5–8% of the molecules in the case of XerCD and XerCYFDYF.
The dissociation rates of the looped states were calculated to be (8.9 ± 0.2) × 10−2 s−1 (R2 = 0.996, N = 100) in the presence of XerCD, (7.5 ± 0.2) × 10−2 s−1 (R2 = 0.934, N = 123) in the presence of XerCγDγ and (12.0 ± 0.3) × 10−2 s−1 (R2 = 0.998, N = 42) in the presence of XerCYFDYF. The presence of γ domain thus clearly decreased the dissociation rate, whereas mutation of the catalytic tyrosines increased it, explaining the low amount of synaptic states observed on Figure 2E. A likely explanation for the stabilization of the synaptic state is the occurrence of DNA strands exchange.
DISCUSSION
Here we report the first experimental setup allowing the detection of the assembly of a synapse between dif sites mediated by XerC and XerD recombinases. This approach relies on shortening of the apparent length of a DNA molecule, which contains two dif sites, in a TPM setup. In the presence of a XerCD mix, we observed shortening of the DNA length, corresponding to the predicted length of DNA molecule containing XerCD-dif synapses. This was not observed when the DNA molecules were only incubated with XerC or XerD, or when they contained only one dif site. In addition, this apparent reduction of the DNA length did not need catalytically active recombinases to occur. We conclude that the formation of XerCD-dif synapses is responsible for this DNA shortening.
Our results show that XerC, XerD and two dif sites are the minimal requirements to form a XerCD-dif synapse. FtsK is thus not required for XerCD-dif synapse formation. This contrasts with models positing that the translocation activity of FtsK is involved in XerCD-dif synapsis (9). It follows that in vivo, dif sites may synapse before being reached by FtsK. This view is consistent with the fact that the dif-containing ter region of the chromosome stays localized at mid-cell during the last third of the cell cycle (43). In addition, we have recently shown that the dif site is the last point of contact between sister chromosomes, independently of their monomeric or dimeric state, suggesting that XerCD-dif synapse may take advantage of the long period of time which separates termination of replication from cell division to form (12).
No recombination was detected within the XerCD-dif synapses in our TPM setup. This conclusion can be drawn based on the fact that the Aeq reduction we observed is not resistant to SDS challenging, implying that no covalent link had been formed to maintain the nucleoproteic complex responsible for Aeq reduction. Inactivity of the synapse seems to be a specific property of the XerCD-dif synapse since other recombination synapses like Cre-loxP and λint-att are productive once formed in equivalent setups (29,31,34–37). Compared to Cre-loxP, the efficiency of XerCD-dif synapse formation may appear low since it only concerns a minority of the DNA molecules (35). Such a comparison is, however, complicated by the fact that XerCD-dif synapses are inactive so that recombination products do not accumulate during the experiment. Based on previous reports, XerCD-dif synapses are expected to exhibit a reversible XerC-dependent HJ-intermediate formation activity, even in the absence of FtsK (9). In vitro, however, this is only observable under particular conditions that trap HJ intermediates such as the presence of ethydium bromide in the reaction (10). The fact that we have not observed HJ1 in our TPM experiments may either be due to HJ1 reversion on timescales shorter than SDS challenging or to XerC inactivity within XerCD-dif synapses. This last hypothesis would imply that the XerCD-dif synapses have a conformation that does not allow XerC activity under the conditions we used. However, we cannot rule out the eventuality that HJ1 formation is too rare to be observed in our setup.
The current model proposes that two types of XerCD-dif synapses could be assembled: one XerC-active into which XerC is active but XerD is not and one XerD-active into which neither XerC nor XerD are active. The function of FtsK would consist in two distinct actions: remodelling the XerC-active synapses into XerD-active ones and activating XerD in XerD-active synapses (9,27). Such a model implies that FtsK differentiates the two synapses isoforms, and eventually induces the dissociation of XerC-active synapses to promote the assembly of XerD-active ones. Since current models are proposing that XerC-active and XerD-active synapses may be structurally different (Figure 1B), these may provoke different Aeq reduction in TPM setup. However, no experimental information is yet available to predict the importance of such structural differences and we did not observe evidence for two synapses in our TPM experiments. Indeed, none of the histograms of the Aeqs corresponding to synapses-containing DNA molecules appeared bimodal. In addition, the mean Aeq observed for inactive XerCD-mediated synapses differs only slightly from active ones observed with XerCγDγ (158.0 ± 0.4 nm versus 154.6 ± 0.2 nm). Thus, either the XerD-active synapse most preferentially forms in the TPM setup or the two types of synapse cannot be differentiated in this setup. Control experiments performed with XerD incubated with DNA-dif2 or with XerC and XerD incubated with DNA-dif1 suggested the presence of a structural-change of the DNA molecule, which could not be attributed to synapse formation. This structural change was dependent upon the presence of XerD (Aeq reduced by 12 nm; Figure 2D) and seemed to increase with the number of dif sites present on the DNA molecule used (XerCD induced an Aeq reduction of 10 nm for DNA-dif2 versus of 5 nm for DNA-dif1; Figure 2A and B). This structural effect could probably be attributed to a DNA bending event similar to the one occurring with Cre-loxP (44). These results suggest that our TPM measurements are able to detect slight structural differences such as DNA bending. Since the only proposed difference between XerC-active and XerD-active synapses comes from the bending of DNA, TPM should presumably be able to discriminate between them. We therefore postulate that the absence of detection of two different types of synapses is likely to be due to the fact that only the XerD-active synapse forms. However, in absence of precise prediction for these synapses structures, we cannot rule out the fact that both XerC-active and XerD-active synapses are formed and that our setup cannot discriminate them.
Using chimeric XerCγ and XerDγ, consisting of the fusion of the γ domain of FtsK either to XerC or to XerD, we have managed to obtain active XerCγDγ-dif synapses. This conclusion is based on the fact that SDS-resistant Aeq reductions corresponding to synapses are observed when DNA-dif2 is incubated with a XerCγDγ mix. This strand exchange activity (∼8%) is efficient when compared to previously reported efficiency for in vitro XerCD-dif strand exchange or recombination (8,27). Because HJ-containing or fully recombined DNA molecules accumulate in the presence of XerCγ and XerDγ but not in presence of XerC and xerD, we observed more XerCγDγ-dif synapses than XerCD-dif synapses. The different efficiencies of synapse formation observed between XerCD and XerCγDγ are thus certainly due to the accumulation of HJ-containing and recombined DNA in the case of XerCγDγ (Compare Figure 3 with Figure 5). In the kinetic analysis of the process, it appears that XerCγDγ-dif synapses are more stable than XerCD-dif ones, which also reflects that recombination occurred in the first instance but not in the second. As explained earlier, the actual rate measured for XerCγDγ-dif assembly is higher than the XerCD-dif one, suggesting that the γ domain of FtsK helps in the synapsis process. Thus, γ domain of FtsK appears to increase the rate of synapse assembly and to activate XerD within the synapse.
The current model proposes that the XerC-active synapse is preferentially formed in absence of FtsK. This conclusion is based on the observation that XerC catalyses the formation of HJ1 intermediate between dif sites in vivo and in vitro in absence of the carboxy-terminal part of FtsK (9,10). However, these data rely on the detection of recombination products, which accumulate because XerC is the only active subunit in the absence of FtsK, despite the fact that the recombination reaction could be extremely rare. Our data show that XerCD-dif synapse readily forms in absence of FtsK but we were unable to detect any DNA strand exchange within this complex. When the γ domain of FtsK was fused to XerC and XerD, we observed strand exchange activity but we were unable to detect any conformational differences between XerCD-dif and XerCγDγ-dif synapses. Taken together, although based on negative results, these data suggest that only one synapse conformation forms most preferentially and that this complex is ready for XerD catalysis, not for XerC. Assembling only XerD-active synapses may represent an advantage for the cell since XerC-active synapses would induce uncontrolled single-strand breaks and HJs formation on the chromosome, which could be harmful for chromosome stability. As already proposed, the function of FtsK as a checkpoint for the synchronization between septum closure and chromosome dimer resolution would then lie within its capacity to activate XerD for the first DNA strand cleavage (11,27). When septum-tethered FtsK translocates towards the dif site, it stops on a XerD-active synapse and activates it. Alternatively it can favour the assembly of a XerCD-dif synapse. The γ domain mediates both of these actions and the spatio-temporal regulation of the CDR may thus be due to the fact that this γ domain is on FtsK, which is localized at the septum, and not on XerD. If the γ domain was fused to XerD, this would indeed lead to a constitutively active recombination system. In order to confirm this model, it will be important to determine if XerC-active synapses do or do not exist. Coupling FRET to TPM, as it has been done in the case of Cre-loxP studies, would be the best way to answer that question (36).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [27].
FUNDING
CNRS, University of Toulouse 3, ANR-BLAN-1327-01 ‘DYNAMO’ and ANR-11-NANO-010 ‘TPM On a Chip’; ‘Ministère de l’Enseignement Supérieur et de la Recherche’ on a special program from the university of Toulouse at the ITAV [USR3505 to C.T.D.]. Funding for open access charge: ANR ANR-BLAN-1327-01 ‘DYNAMO’ and ANR-11-NANO-010 ‘TPM On a Chip’.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the members of ‘Genetic Recombination and the cell cycle’ and ‘Functional dynamics of biological membranes and DNA molecules’ groups at LMGM and IPBS laboratories in Toulouse for helpful discussions. We especially thank E. Joly for precious advice on the manuscript. We are grateful to the laboratories of D. Sherratt and F.-X. Barrefor providing us with plasmids. We thank the TRI platform (IPBS, Mourey’s Laboratory) for biophysical analysis of our proteins. A special thought to Guy Duval-Valentin.
REFERENCES
- 1.Steiner WW, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J. Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lesterlin C, Barre FX, Cornet F. Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol. Microbiol. 2004;54:1151–1160. doi: 10.1111/j.1365-2958.2004.04356.x. [DOI] [PubMed] [Google Scholar]
- 3.Carnoy C, Roten C-A. The dif/Xer recombination systems in proteobacteria. PLoS ONE. 2009;4:e6531. doi: 10.1371/journal.pone.0006531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kono N, Arakawa K, Tomita M. Comprehensive prediction of chromosome dimer resolution sites in bacterial genomes. BMC Genomics. 2011;12:19. doi: 10.1186/1471-2164-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 6.Blakely GW, Sherratt DJ. Interactions of the site-specific recombinases XerC and XerD with the recombination site dif. Nucleic Acids Res. 1994;22:5613–5620. doi: 10.1093/nar/22.25.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blakely GW, Davidson AO, Sherratt DJ. Binding and cleavage of nicked substrates by site-specific recombinases XerC and XerD. J. Mol. Biol. 1997;265:30–39. doi: 10.1006/jmbi.1996.0709. [DOI] [PubMed] [Google Scholar]
- 8.Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol. Cell. 1999;4:949–959. doi: 10.1016/s1097-2765(00)80224-5. [DOI] [PubMed] [Google Scholar]
- 9.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 10.Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grainge I. FtsK–a bacterial cell division checkpoint? Mol. Microbiol. 2010;78:1055–1057. doi: 10.1111/j.1365-2958.2010.07411.x. [DOI] [PubMed] [Google Scholar]
- 12.Stouf M, Meile J-C, Cornet F. FtsK actively segregates sister chromosomes in Escherichia coli. Proc. Natl Acad. Sci. USA. 2013;110:11157–11162. doi: 10.1073/pnas.1304080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubarry N, Possoz C, Barre F-X. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol. Microbiol. 2010;78:1088–1100. doi: 10.1111/j.1365-2958.2010.07412.x. [DOI] [PubMed] [Google Scholar]
- 14.Dubarry N, Barre F-X. Fully efficient chromosome dimer resolution in Escherichia coli cells lacking the integral membrane domain of FtsK. EMBO J. 2010;29:597–605. doi: 10.1038/emboj.2009.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Crozat E, Grainge I. FtsK DNA translocase: the fast motor that knows where it’s going. ChemBioChem. 2010;11:2232–2243. doi: 10.1002/cbic.201000347. [DOI] [PubMed] [Google Scholar]
- 17.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Ann. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 18.Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 19.Bigot S, Saleh O, Cornet F, Allemand JF, Barre FX. Oriented loading of FtsK on KOPS. Nat. Struct. Mol. Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- 20.Sivanathan V, Allen MD, de Bekker C, Baker R, Arciszewska LK, Freund SM, Bycroft M, Löwe J, Sherratt DJ. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat. Struct. Mol. Biol. 2006;13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Löwe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham JE, Sherratt DJ, Szczelkun MD. Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA. Proc. Natl Acad. Sci. USA. 2010;107:20263–20268. doi: 10.1073/pnas.1007518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JY, Finkelstein IJ, Crozat E, Sherratt DJ, Greene EC. Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK. Proc. Natl Acad. Sci. USA. 2012;109:6531–6536. doi: 10.1073/pnas.1201613109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates J, Zhekov I, Baker R, Eklund B, Sherratt DJ, Arciszewska LK. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol. Microbiol. 2006;59:1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonné L, Bigot S, Chevalier F, Allemand J-F, Barre F-X. Asymmetric DNA requirements in Xer recombination activation by FtsK. Nucleic Acids Res. 2009;37:2371–2380. doi: 10.1093/nar/gkp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham JE, Sivanathan V, Sherratt DJ, Arciszewska LK. FtsK translocation on DNA stops at XerCD-dif. Nucleic Acids Res. 2010;38:72–81. doi: 10.1093/nar/gkp843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grainge I, Lesterlin C, Sherratt DJ. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 2011;39:5140–5148. doi: 10.1093/nar/gkr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segall AM, Nash HA. Synaptic intermediates in bacteriophage lambda site-specific recombination: integrase can align pairs of attachment sites. EMBO J. 1993;12:4567–4576. doi: 10.1002/j.1460-2075.1993.tb06145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh K, Lau CK, Gupta K, Van Duyne GD. Preferential synapsis of loxP sites drives ordered strand exchange in Cre-loxP site-specific recombination. Nat. Chem. Biol. 2005;1:275–282. doi: 10.1038/nchembio733. [DOI] [PubMed] [Google Scholar]
- 30.Lee L, Chu LC, Sadowski PD. Cre induces an asymmetric DNA bend in its target loxP site. J. Biol. Chem. 2003;278:23118–23129. doi: 10.1074/jbc.M302272200. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh K, Guo F, Van Duyne GD. Synapsis of loxP sites by Cre recombinase. J. Biol. Chem. 2007;282:24004–24016. doi: 10.1074/jbc.M703283200. [DOI] [PubMed] [Google Scholar]
- 32.Cassell G, Moision R, Rabani E, Segall A. The geometry of a synaptic intermediate in a pathway of bacteriophage lambda site-specific recombination. Nucleic Acids Res. 1999;27:1145–1151. doi: 10.1093/nar/27.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vetcher AA, Lushnikov AY, Navarra-Madsen J, Scharein RG, Lyubchenko YL, Darcy IK, Levene SD. DNA topology and geometry in Flp and Cre recombination. J. Mol. Biol. 2006;357:1089–1104. doi: 10.1016/j.jmb.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 34.Mumm JP, Landy A, Gelles J. Viewing single lambda site-specific recombination events from start to finish. EMBO J. 2006;25:4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan H-F. Real-time single-molecule tethered particle motion experiments reveal the kinetics and mechanisms of Cre-mediated site-specific recombination. Nucleic Acids Res. 2012;40:6208–6222. doi: 10.1093/nar/gks274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinkney JNM, Zawadzki P, Mazuryk J, Arciszewska LK, Sherratt DJ, Kapanidis AN. Capturing reaction paths and intermediates in Cre-loxP recombination using single-molecule fluorescence. Proc. Natl Acad. Sci. USA. 2012;109:20871–20876. doi: 10.1073/pnas.1211922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan H-F, Ma C-H, Jayaram M. Real-time single-molecule tethered particle motion analysis reveals mechanistic similarities and contrasts of Flp site-specific recombinase with Cre and λ Int. Nucleic Acids Res. 2013;41:7031–7047. doi: 10.1093/nar/gkt424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausubel FM, Brent B, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Hoboken, NJ, États-Unis: John Wiley & Sons, Inc; 1987. [Google Scholar]
- 39.Plénat T, Tardin C, Rousseau P, Salomé L. High-throughput single-molecule analysis of DNA-protein interactions by tethered particle motion. Nucleic Acids Res. 2012;40:e89. doi: 10.1093/nar/gks250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurens N, Bellamy SRW, Harms AF, Kovacheva YS, Halford SE, Wuite GJL. Dissecting protein-induced DNA looping dynamics in real time. Nucleic Acids Res. 2009;37:5454–5464. doi: 10.1093/nar/gkp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manghi M, Tardin C, Baglio J, Rousseau P, Salomé L, Destainville N. Probing DNA conformational changes with high temporal resolution by tethered particle motion. Phys. Biol. 2010;7:046003. doi: 10.1088/1478-3975/7/4/046003. [DOI] [PubMed] [Google Scholar]
- 42.Pouget N, Turlan C, Destainville N, Salomé L, Chandler M. IS911 transpososome assembly as analysed by tethered particle motion. Nucleic Acids Res. 2006;34:4313–4323. doi: 10.1093/nar/gkl420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bates D, Epstein J, Boye E, Fahrner K, Berg H, Kleckner N. The Escherichia coli baby cell column: a novel cell synchronization method provides new insight into the bacterial cell cycle. Mol. Microbiol. 2005;57:380–391. doi: 10.1111/j.1365-2958.2005.04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gopaul DN, Guo F, Van Duyne GD. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.