Abstract
Superoxide dismutases (SODs) are widely distributed enzymes that convert superoxides to hydrogen peroxide and molecular oxygen, using various metals as cofactors. Many actinobacteria contain genes for both Ni-containing (sodN) and Fe-containing (sodF) SODs. In Streptomyces coelicolor, expression of the sodF and sodN genes is inversely regulated by nickel-specific Nur, a Fur-family regulator. With sufficient nickel, Nur directly represses sodF transcription, while inducing sodN indirectly. Bioinformatic search revealed that a conserved 19-nt stretch upstream of sodN matches perfectly with the sodF downstream sequence. We found that the sodF gene produced a stable small-sized RNA species (s-SodF) of ∼90 nt that harbors the anti-sodN sequence complementary to sodN mRNA from the 5′-end up to the ribosome binding site. Absence of nearby promoters and sensitivity to 5′-phosphate-specific exonuclease indicated that the s-SodF RNA is a likely processed product of sodF mRNA. The s-SodF RNA caused a significant decrease in the half-life of the sodN mRNA. Therefore, Nur activates sodN expression through inhibiting the synthesis of sodF mRNA, from which inhibitory s-SodF RNA is generated. This reveals a novel mechanism by which antagonistic regulation of one gene is achieved by small RNA processed from the 3′UTR of another gene’s mRNA.
INTRODUCTION
Superoxide dismutases (SODs) are ubiquitous enzymes that catalyze conversion of superoxide to molecular oxygen and hydrogen peroxide, using catalytic metal ions. Depending on their structure and metal cofactors, three classes of SODs have been reported; Cu/Zn-SOD found in eukaryotes and some bacteria, Fe- or Mn-containing SOD present in bacteria, mitochondria and chloroplasts, and Ni-containing SOD found in some bacteria (1–4). They are present both in aerobes as well anaerobes, protecting and preparing cells against superoxide toxicity in the presence of O2 (5). Cells usually contain more than two types of SODs in diverse combinations, whose production is regulated tightly in response to metabolic and environmental cues.
In Escherichia coli, where sodA, sodB and sodC genes encode Mn-SOD, Fe-SOD and Cu,Zn-SOD, respectively, the presence of metal, oxygen and redox-active compounds, as well as growth phase regulate their gene expression via transcriptional regulators such as Fur, Fnr, ArcA, SoxR and RpoS (6,7). Regulation of sodA and sodB genes encoding cytoplasmic SODs is intricately inter-connected by a global regulator Fur in response to iron availability. In the presence of iron, Fur represses transcription of sodA and small regulatory RNA RyhB that inhibits translation and stability of sodB RNA, allowing the production of Fe-SOD. In the absence of iron, expression of sodA and RyhB is induced, resulting in the production of MnSOD (8–10). This mode of regulation is conserved in Pseudomonads, where the inverse regulation of sodA and sodB genes in response to iron is exerted by Fur, and sodB is activated by Fur through inhibiting the transcription of small regulatory RNA PrrF1 and PrrF2, functional homologs of RyhB (11–13). In Bacillus subtilis, production of Fe-containing proteins is activated by Fur through repressing transcription of yet another small RNA FsrA (14). These examples support the presence of an evolutionarily robust regulatory circuit mediated by an iron-specific regulator Fur and small RNAs in coordinated synthesis of iron-requiring proteins across distantly related bacteria.
NiSOD and its encoding gene (sodN) were first discovered in Streptomyces spp. (15). The sodN gene was subsequently found in the genome of nearly all streptomycetes, various actinomycetes (16,17), diverse marine cyanobacteria (18–21), and some distantly related proteobacteria and eukaryotic green algae (17). The SodN protein is processed at its N-terminal region by its cognate peptidase to produce active Ni-SOD (22), which consists of homohexameric polypeptides with bound nickel at the N-terminal hook of each monomer (23,24). Bioinformatic analyses predicted the presence of NiSOD either alone in some actinobacteria and marine cyanobacteria or in combination with FeSOD, MnSOD or CuZnSOD (16,25).
In Streptomyces coelicolor, where Fe-SOD and Ni-SOD are present, the two enzymes are produced in an antagonistic fashion in response to the presence of nickel in the media (26,27). Expression of the sodF gene encoding Fe-SOD is inhibited by nickel through a nickel-specific Fur-family regulator Nur that binds to and inhibits expression from the sodF promoter in the presence of nickel (26,28,29). Nur also negatively regulates nickel-uptake genes, justifying its name as nickel-uptake-regulator (28). On the other hand, expression of the sodN gene requires Nur as a positive regulator in the presence of nickel (28). However, Nur does not bind to the sodN gene, most likely acting via an indirect way that needs to be revealed (28,30).
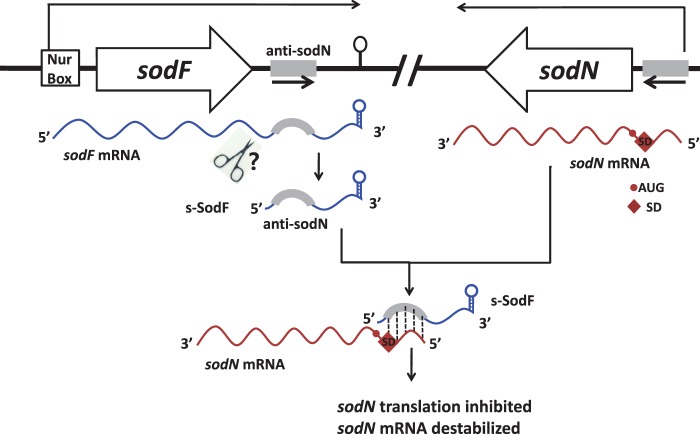
In this article, we investigated how the antagonistic regulation of sodF and sodN is achieved through Nur. We found that a small regulatory RNA is produced from sodF mRNA by endonucleolytic cleavage of about 90 nt from its 3′UTR. This provides a novel example of small regulatory RNA produced from the 3′UTR of functional mRNA by cleavage. Very recently, the presence of 3′UTR-generated processed small RNAs associated with Hfq in Salmonella has been demonstrated (31,32). However, their role as regulatory molecules, rather than intermediates in mRNA degradation pathway, awaits experimental validation. We now present the first example of a small regulatory RNA that is generated from the 3′UTR of a functional mRNA without affecting its coding region. These findings led us to propose a conclusive model where Nur activates sodN expression by inhibiting the production of small sodF RNA, which pairs with sodN mRNA, blocks its translation and facilitates sodN mRNA decay.
MATERIALS AND METHODS
Strains and growth conditions
Streptomyces coelicolor A3(2) M145 strain and its derivatives were grown and maintained according to standard procedures (33). For liquid culture, YEME medium (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 1% glucose, 10 or 34% sucrose, 5 mM MgCl2 separately autoclaved) was used. For nickel treatment, 50 μM NiSO4 was added to the culture when inoculating seed culture. The Δnur, ΔsodF and ΔsodF2 strains had been previously generated in our laboratory (28,34,35).
Construction of s-SodF overproducing strain
The sodF downstream fragment (from +10 to +142 nt from the end of the sodF stop codon) was generated from M145 chromosomal DNA by polymerase chain reaction (PCR) using primer pairs as described in Supplementary Table S1. The PCR product was fused by overlapping PCR to another 240 bp PCR-generated fragment containing the strong ermE* promoter (36). The 399-bp final PCR product was cut with EcoRI/XbaI restriction enzymes and was cloned into pSET162, an integration vector containing thiostrepton marker (37). The recombinant plasmid was confirmed by nucleotide sequencing and was then transformed into the ΔsodF strain. As a negative control, the parental vector pSET162 was introduced to ΔsodF in parallel.
S1 mapping and northern analyses
RNAs were isolated from wild type (M145) and various mutant cells grown in YEME to OD600 of 0.2 to 1.5. Harvested cells were disrupted in modified Kirby mixture (33) using an ultrasonicator with a microtip at 20% of the maximum amplitude (600 W, 20 kHz). Following extraction with phenol/chloroform, the supernatant was precipitated with isopropanol. The RNA pellet was dissolved in diethyl pyrocarbonate-treated distilled water and quantified by measuring its absorbance at 260 nm. To visualize rRNAs and check for contamination by genomic DNA, RNA samples (10 μg each) were electrophoresed in 1.3% agarose gel in 3-(N-morpholino) propanesulfonic acid buffer. For S1 mapping, DNA probes for sodN and sodF transcripts were generated by PCR using M145 chromosomal DNA as a template. The PCR-generated sodN probe spans from −175 to +127 nt relative to the start codon of the sodN coding region. To generate S1 probes for sodF RNA, the PCR-generated sodF fragments (−140 to +60 nt and −205 to +141 nt relative to the end of the sodF stop codon) were cloned into the pGEM-T easy vector (Promega), generating pGEM-sodF200 and pGEM-sodF346, respectively. From pGEM-sodF200, the probe DNA was generated by second PCR, using a T7 forward primer and sodF (+60) reverse primer (Supplementary Table S1), generating a 278-bp DNA fragment containing the sodF gene (−140 to +60) linked to 78 bp of vector sequence (Figure 1). From pGEM-sodF346, the probe DNA was generated by PCR, using a SP6 forward primer and sodF(+78) reverse primer (Supplementary Table S1), generating a 383-bp DNA fragment containing the sodF gene (−205 to +78) linked to 100 bp of vector sequence. The probe DNAs were radiolabeled at their 5′-ends with [γ-32P]-ATP by T4 polynucleotide kinase. For each RNA sample (25 μg), probe DNA was hybridized and digested with S1 nuclease according to the standard procedures. For 3′-end mapping of the sodF RNA, the probe was generated by PCR from pGEM-sodF346 as a template, using T7 and SP6 primer. BssSI-cut PCR product (199 bp) containing the sodF gene (+21 to +141) and 78 bp of vector sequence was labeled with [α-32P]-dATP. The protected fragments were analyzed on a 6% polyacrylamide gel containing 7 M urea. For high-resolution S1 mapping, a sequencing ladder was generated using sequenase version 2.0 as recommended by the manufacturer (USB corporation), using the sodF(+78) oligonucleotide primer and the template pGEM-sodF346 DNA in a labeling mix with [α-35S]-dATP. For northern analysis of sodF RNAs, a 22-nt single-stranded DNA probe (+18 to +39 nt relative to the end of the sodF stop codon; Supplementary Table S1) was synthesized, and labeled with [γ-32P]-ATP by T4 polynucleotide kinase. Each sample of 70 μg total RNA was resolved on 12.5% polyacrylamide gel containing 7 M urea, and transferred to a Zetaprobe-GT-membrane (Bio-Rad). Radioactive signals were detected and quantified by phosphor screen and image analyzer (FLA-2000; Fuji).
Figure 1.
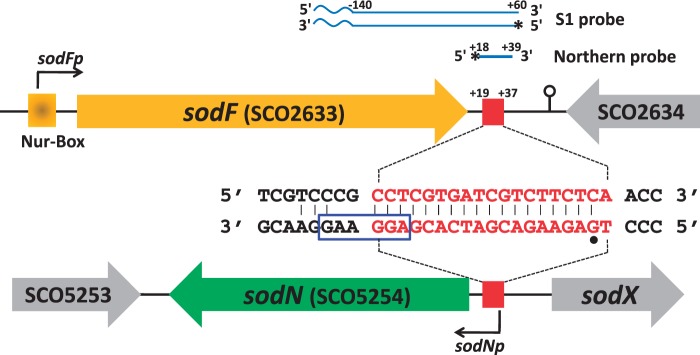
Complementarity between sense strands of sodF and sodN genes. Gene structures around sodF and sodN genes are schematically shown. The same 19 bp DNA sequence downstream and upstream of sodF and sodN genes, respectively, are shown as red square. The position of promoters (sodFp and sodNp) and Nur-binding site (Nur-box) was marked. The complementary sequence in the 3′ and 5′UTR of sodF and sodN transcripts, respectively, was shown in the middle. The position of 5′-end and ribosome binding site of sodN mRNA was indicated with a dot and square box, respectively. Probes for S1 mapping and northern analysis of sodF RNAs were presented with radiolabeled positions (asterisks) along with relative distances from the end of the stop codon of sodF ORF. The waved segment of S1 probe indicates non-related vector sequence.
5′ RACE
5′-rapid amplification of cDNA ends (RACE) was carried out using the FirstChoice® RLM-RACE kit (Ambion), following the manufacturer’s instructions with modifications for bacterial RNA (38). Briefly, 10 μg of total RNA, extracted from S. coelicolor M145 was treated with tobacco acid pyrophosphatase (TAP) before being ligated to a 5′-RACE adapter using T4 RNA ligase. This ligated product was then used as template for reverse transcription using primers complementary to sodF and sodN, together with SuperScript™ III reverse transcriptase (Invitrogen). The resulting cDNA then served as template for 5′-end PCR amplification, using an adaptor-specific outer primer (Supplementary Table S1) and the same oligonucleotide was used to prime the reverse transcription reaction. The second PCR was done with a pair of inner primer specific for adaptor and RNA, respectively. The final PCR products were separated on an agarose gel, excised and purified using the Qiagen® Gel extraction kit, and then sequenced.
Exonuclease digestion of RNA
RNA samples were prepared from the wild type and Δnur cells grown exponentially in YEME. Treatment of RNA samples with TAP and the terminator 5′-phosphate-dependent exonuclease (5′-exo) was done as described previously (39). Total RNAs (5 μg per each sample) were incubated in 10 μl reaction volume containing 1 μl of 10X TAP reaction buffer (0.5 M sodium acetate, pH 6.0, 10 mM ethylenediaminetetraacetic acid, 1% β-mercaptoethanol, 0.1% Triton X-100), 0.5 μl of TAP (5 μ; Epicentre) or water, and 1 μl of RNasin (40 μ; Promega) for 3 h at 37°C. The reaction mixture was extracted with phenol/chloroform, and the RNA was precipitated by ethanol. Further digestion with Terminator Exonuclease (Epicenter) was carried out in 20 μl reaction volume containing 2 μl of 10X reaction buffer (500 mM Tris Cl, pH 8.0, 20 mM MgCl2, 1 M NaCl), 1 μl of RNasin (40 U; Promega) and 1 μl of either Terminator Exonuclease (1 U; Epicentre) or water, for 3 h at 30°C. The reaction mixture was extracted with phenol/chloroform, ethanol precipitated in the presence of glycogen and analyzed by northern blotting as described above.
RESULTS
Presence of complementarity between the sense strands of sodF and sodN genes
The intergenic region between the sodN (SCO5254) gene and the sodX (SCO5255) gene, where SodX encodes a cognate peptidase for SodN, is highly conserved among streptomycetes (Supplementary Figure S1). A 19-bp sequence stretch within this conserved region also appears in the downstream of the sodF gene (17). This 19-nt stretch is complementary between the sense strands of sodF and sodN genes (Figure 1). Previously mapped 3′- and 5′-ends of sodF and sodN mRNAs, respectively, in S. coelicolor Müller (26,27), led us to hypothesize that a significant fraction of the complementary sequence can actually form base pairs between the 3′UTR of sodF and 5′UTR of sodN transcripts in the S. coelicolor A3(2) M145 strain that we used in this study. We determined the 5′-end of the sodN mRNA in S. coelicolor by 5′ RACE, and found that it starts at the same G nucleotide as determined for the Müller strain (27) (Figure 1). The complementary base-pairing region spanned the first 18 nt of the sodN mRNA, encompassing part of the ribosome binding site (Figure 1). We investigated whether the complementary base pairing between sodF and sodN transcripts was responsible for the inverse regulation of the two genes, and if so, how it was mediated.
Small-sized sodF RNAs encompassing the anti-sodN sequence are produced in a Nur-dependent manner
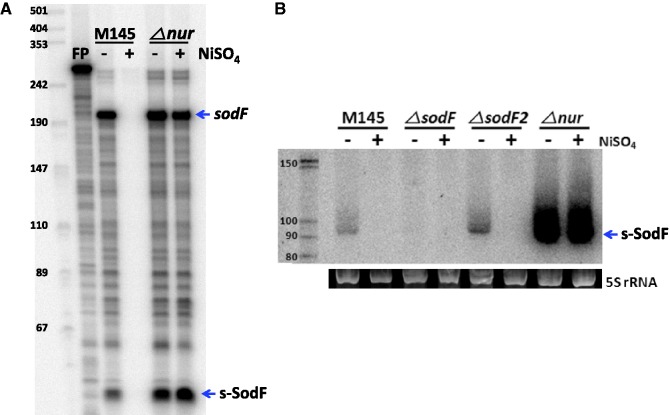
Previously determined positions of the 5′- and 3′-ends of the sodF mRNA in S. coelicolor Müller strain lay 38 nt upstream from the start codon and 86 nt downstream from the stop codon, respectively (26). The 19 nt sodN-complementary (anti-sodN) sequence in M145 started 19 nt downstream from the stop codon of the sodF ORF (+19 to +37 relative to the end of the stop codon; Figure 1). In order to monitor the presence and the boundary of transcripts generated from the sodF downstream region, we performed S1 mapping and northern analyses. For S1 mapping, we used a 5′-end-labeled DNA probe that contained part of the sodF coding sequence and its downstream region (−140 to +60 relative to the end of the stop codon, Figure 1). This probe detected the presence of two kinds of RNA; one producing a fully protected band of ∼200 nt (sodF) and the other (s-SodF) protecting ∼50 nt band (Figure 2A). The amount of both RNAs decreased significantly in the wild-type (M145) cell when nickel was added to the culture and were produced in a constitutive manner in a Δnur mutant. The longer protecting transcript most likely corresponded to the sodF mRNA encoding Fe-SOD, whereas the short-protecting RNA (s-SodF) lacked the sodF coding sequence but could contain the anti-sodN sequence.
Figure 2.
Small-sized sodF RNA that contains the 19 nt sodN-complementary (anti-sodN) sequence is produced in a Nur-dependent manner. (A) S1 mapping of sodF RNAs with DNA probe 5′-end labeled at +60 position relative to the stop codon of the sodF ORF. RNAs were prepared from the wild-type (M145) and △nur mutant grown in the presence and absence of 50 μM NiSO4. The arrows indicate the presence of sodF RNAs that fully protected the probe (sodF) and partly protected (s-SodF). FP denotes free probe with plasmid vector sequence attached. (B) Northern blot analysis of sodF RNAs. RNAs were prepared from the wild type (M145), △sodF, △sodF2 and △nur cells grown in the presence and absence of 50 μM NiSO4. RNAs were run on 12.5% PAG to resolve small-sized RNAs and hybridized with 22 nt single-stranded DNA probe (+18 to +39 nt relative to the sodF stop codon) labeled at 5′-end by 32P (Figure 1). The 5S rRNA band electrophoresed on agarose gel was shown in parallel to demonstrate the quantity and quality of RNA samples.
Since S1 mapping detected only the 5′-end point of the s-SodF RNA, we performed northern blot analysis to determine its size. Using a short DNA probe of 22 bp that encompassed the anti-sodN sequence, we detected a small RNA of about 90 nt in a 12.5% polyacrylamide gel specified for resolving small RNAs (Figure 2B). The amount of this RNA decreased in the presence of nickel and was constitutively enhanced in Δnur mutant, consistent with the observation by S1 mapping. The amount of s-SodF RNA was greatly reduced in a ΔsodF mutant. In contrast, deletion of sodF2, a paralog of sodF in S. coelicolor (35), did not affect the production of s-SodF, suggesting that the s-SodF RNA is primarily produced from the sodF gene. Northern blot analysis clearly revealed that there existed a small RNA species produced from the sodF downstream region, justifying the name s-SodF. The results also clearly indicated that the production of s-SodF RNA is inhibited by nickel and Nur.
The 5′ and 3′ boundaries of s-SodF RNA
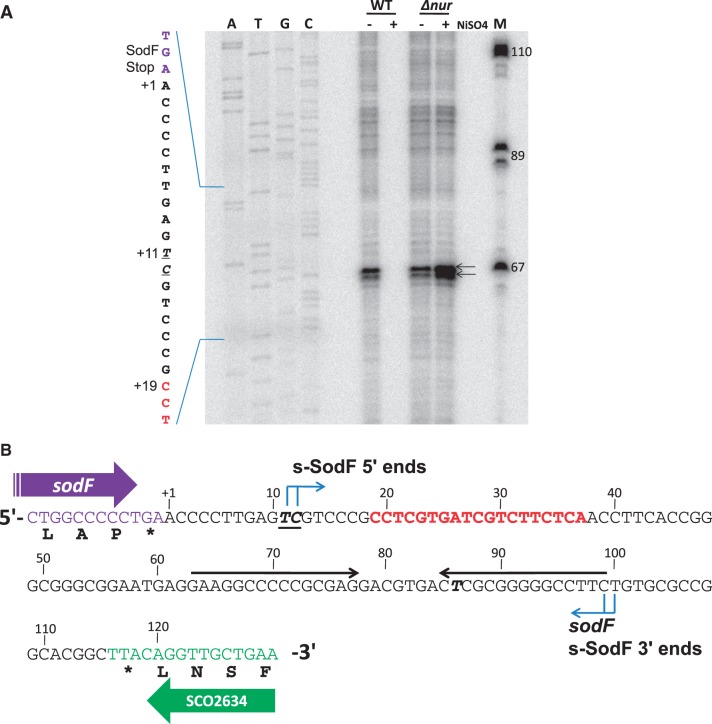
In order to determine the end points and the exact size of the s-SodF RNA, we performed high-resolution S1 mapping. The 5′-end mapping was done with the probe labeled at +78. The result showed that the 5′-end of s-SodF was localized at nucleotides T and C (+11 and +12 relative to the end of the stop codon), 7–8 nt upstream of anti-sodN sequence (Figure 3A). The 3′-end mapping was done with the probe labeled at +21. However, we were not able to localize the exact 3′-end point because the sequencing ladder from the template DNA was compressed severely around the protected band size. This indicated the presence of stable secondary structure near the 3′-end. End-mapping with 3′ RACE was not successful either. The presence of an inverted repeat of 15 nt stretch with 80% GC suggested a stable stem and loop structure (ΔG° = −34.5 kcal), which could hinder nucleotide sequencing ladder. This stem-loop structure likely served as an instrinsic transcription terminator. This coincided with the most prevalent type of intrinsic terminators in actinobacteria, with stem and loop structure of ΔG between −15 and −25 kcal, followed by less than two U residues (40). From the approximate size estimation of s-SodF by S1 mapping and northern analysis, combined with the presence of a potential intrinsic terminator sequence, we propose that the 3′ boundary of s-SodF RNA lies at the end of the inverted repeat near +100, and thus we estimate s-SodF to be 88–90 nt long (Figure 3B).
Figure 3.
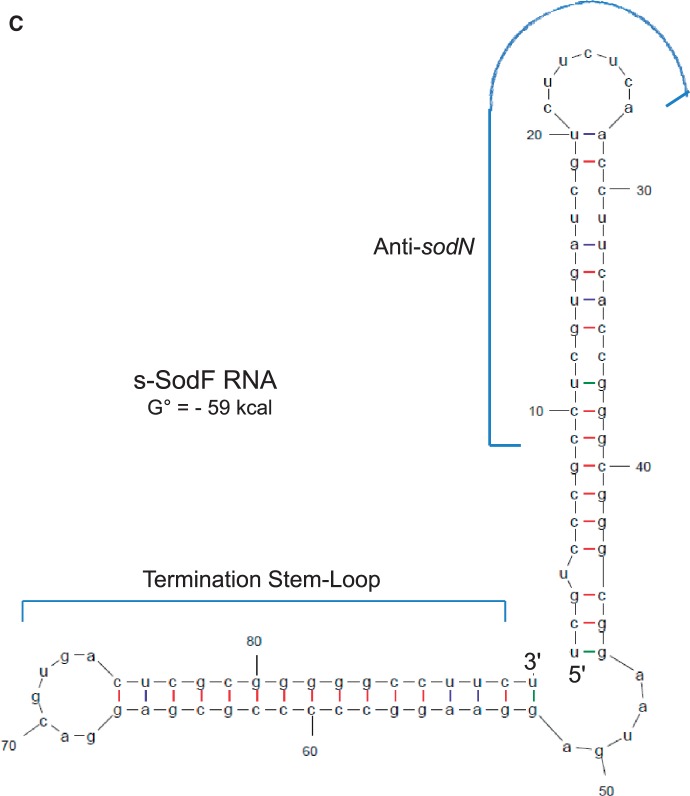
Determination of 5′-ends of s-SodF RNA. (A) Determination of 5′-ends by high-resolution S1 analysis. S1 mapping of sodF RNAs with a DNA probe 5′-end labeled at +78 nucleotide position relative to the stop codon of the sodF ORF. RNAs were prepared from the wild type (M145) and △nur mutant grown in the presence and absence of 50 μM NiSO4. The position of 5′-ends of the transcripts is underlined. The sequencing ladder was obtained with the sodF oligonucleotide primer (5′-CCT CGC GGG GGC CTT CCT CA-3′) and the template pGEM-sodF346. (B) Sequence information of intergenic region between sodF (SCO2633) and SCO2634. The nucleotide position was numbered relative to the end of the stop (TGA) codon of sodF ORF. The 5′-end position of s-SodF was marked (bold italic underlined) along with 19 nt anti-sodN sequence (red), 15 nt inverted repeats (horizontal arrows), and the possible 3′-end position of sodF and s-SodF RNAs. (C) Predicted secondary structure for s-SodF RNA. The secondary structure of 90 nt long s-SodF RNA (from +11 to +100; as in (B)), as well as stability, were predicted by mfold program (41). The 5′ proximal stem-loop contains the anti-sodN sequence.
We predicted the secondary structure of 90 nt s-SodF RNA (+11 to +100; Figure 3B) using mfold program (41). Figure 3C demonstrated that s-SodF can form a structure with two stem loops, with an overall ΔG° value of about −59 kcal. The 5′-proximal stem-loop has the anti-sodN sequence that spans from the middle of the stem to the entire loop. This stem-loop contains bubbles and a bulge in the stem with an estimated ΔG° of about −22 kcal. The predicted structure suggests that the 5′-end of sodN mRNA can easily start pairing with s-SodF through the anti-sodN sequence in the loop region. The termination stem-loop has a perfectly paired 16 bp stem (including the terminal G-U base pairing) with estimated ΔG° of about −36 kcal.
The s-SodF is produced from processing of the sodF mRNA
The observation that s-SodF RNA is produced in a Nur-dependent manner led us to search for the presence of Nur binding sequence in the genome near the starting position of the small RNA. No sequence matching the proposed Nur-box consensus (tTGCaa-N5-ttGCAA) was found (29). Electrophoretic mobility shift assay with the 200-bp DNA probe used for S1 mapping analysis (Figure 1) did not detect any binding protein present in cell extracts prepared from wild-type cells grown in Ni-supplemented YEME medium (data not shown). Therefore, the possibility of initiating transcription from its own Nur-dependent promoter for s-SodF synthesis appeared very low.
The 5′ phosphorylation status of s-SodF RNA was then examined using 5′-phosphate-dependent exonuclease. Total RNAs isolated from either wild type (M145) or Δnur mutant cells were treated with 5′-monophosphate-dependent exonuclease as described previously (39), and subjected to 12.5% PAGE and northern analysis. Results in Figure 4 (upper panel) demonstrated that 5′ exonuclease digested s-SodF RNA efficiently as it did the bulk of 23 S and 16 S rRNAs (bottom panel). Treatment with TAP that removes pyrophosphates from 5′ triphosphates of de novo transcribed bacterial mRNA did not make any difference. Therefore, it is highly likely that the 5′-end of s-SodF RNA is monophosphorylated, and hence was generated not by de novo transcription but from the cleavage of a longer RNA. Since the sodF mRNA spanned not only the coding region but also the downstream UTR that encompasses the anti-sodN sequence, it was most likely that s-SodF is generated from the full-length sodF mRNA by cleavage at the 5′ side of T (+11) or C (+12) residues (Figure 3).
Figure 4.
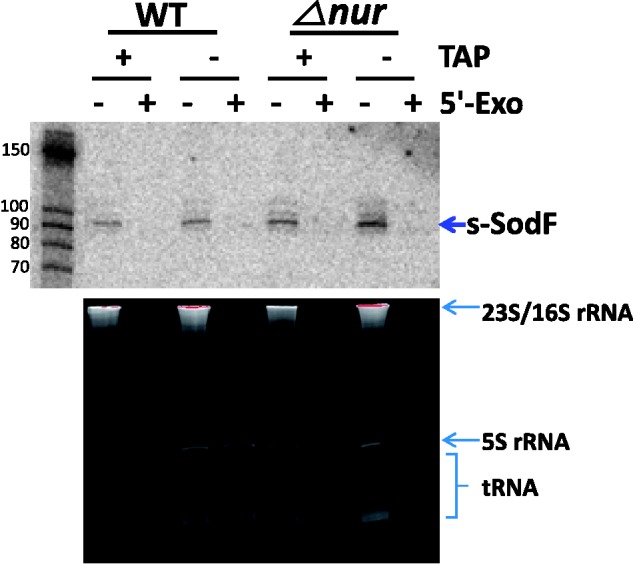
Sensitivity of s-SodF RNA toward 5′ monophosphate-dependent exonuclease. The phosphorylation state of the s-SodF RNA was monitored by treatment with a 5-monophosphate-dependent exonuclease (5′-Exo). Total RNA isolated from wild type (M145) and △nur cells were subjected to digestion with 5′-Exo and analyzed by gel electrophoresis (lower panel) and northern blotting with an s-SodF specific radiolabeled probe (upper panel). RNA samples were treated either with or without TAP that converts 5′-triphosphorylated RNA to 5′-monophosphorylated one prior to 5′-Exo treatment. The status of stable RNAs in each sample is shown in the lower panel.
s-SodF RNA decreases the stability of sodN mRNA
We then explored whether s-SodF RNA affected sodN gene expression. We estimated the half-life of sodN mRNA under various genetic background. Depending on growth conditions, the ratio of sodF to sodN mRNAs varied (see below). We measured the RNA decay rates, following the inhibition of transcription initiation by rifampicin. Cells grown in YEME to OD600 of 0.8 were treated with rifampicin (300 μg/ml), and were harvested at different time points (0–20 min) to prepare RNA. S1 mapping of sodF mRNA, s-SodF and sodN mRNA in each sample was done with RNA-specific probes in one reaction tube. As demonstrated in Figure 5, the sodN mRNA in the wild-type strain decayed at t1/2 of ∼3 min. The sodF mRNA decayed with half-life of about 10 min, whereas the s-SodF RNA was relatively stable with t1/2 of >>20 min. In ΔsodF mutant where no sodF transcripts are produced, the half-life of sodN mRNA increased to about 16 min, suggesting that sodF transcripts decrease the stability of sodN mRNA. A residual sodF-sized band observed in ΔsodF mutant is thought to be non-specific, and it is not detectable in the mutant introduced with pSET-derived plasmids (see below).
Figure 5.
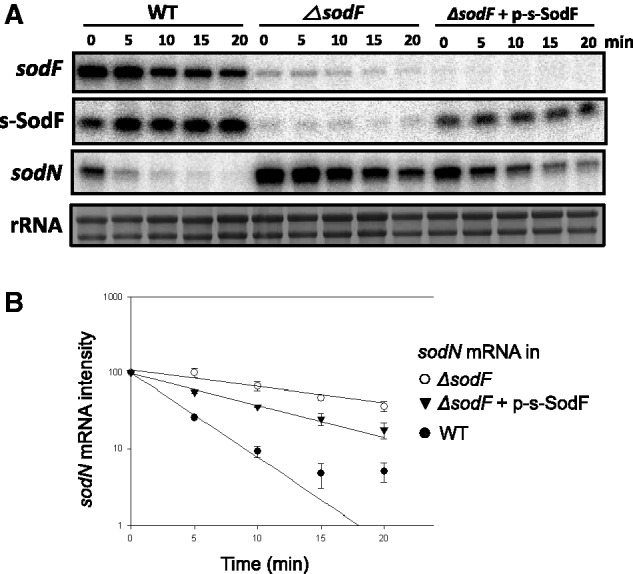
Effect of s-SodF RNA on the stability of sodN mRNA. (A) S1 mapping of sodF and sodN RNAs following rifampicin treatment. Wild-type (M145) and △sodF cells with or without pSET-based recombinant plasmid expressing s-SodF RNA from ermE* promoter (p-s-SodF) in the chromosome were grown in YEME medium to OD600 of 0.8 and treated with rifampicin (300 μg/ml). At 5, 10, 15 and 20 min after rifampicin treatment, cells were harvested and fixed with methanol. RNA samples were analyzed by S1 mapping with 5-end-labeled probes. A representative result was presented. (B) The relative amount of each RNA species was plotted to estimate half-life.
In order to verify the effect of s-SodF RNA alone, we introduced to ΔsodF mutant an overexpression plasmid for s-SodF RNA whose expression was driven by a strong ermE* promoter on the integrating vector pSET162. Figure 5 demonstrated that when s-SodF RNA was stably expressed in the absence of sodF mRNA, the half-life of sodN mRNA was about ∼7 min which was significantly shorter than ∼16 min observed in ΔsodF. Since the expression level of s-SodF RNA in ΔsodF was lower than the level in the wild type, it is thought that the sodN half-life was not brought down to the wild-type level (∼3 min). This experiment clearly demonstrated that the production of s-SodF RNA alone was sufficient to decrease the amount of sodN mRNA.
Mutations in the anti-sodN region of s-SodF inactivate its inhibitory function
In order to test whether the anti-sodN sequence in s-SodF was indeed critical to reduce the amount of sodN mRNA, we created mutants in this region. Variants with changes in the predicted 7-nucleotide loop region (V1), and the subsequent seven nucleotides (V2) were made by site-directed mutagenesis (Figure 6) using mutagenic primers. The secondary structure prediction by mFOLD suggested that V1 would assume almost identical structure to the wild type with ΔG° of −59 kcal, and V2 a similar structure with a larger loop with ΔG° of −55 kcal. We cloned the mutant s-SodF genes in the integration vector (pSET162) and introduced them into the chromosome of ΔsodF strain of S. coelicolor, in the same way as we made the expression construct for the wild type s-SodF (Figure 5). The results demonstrated that the introduction of wild-type s-SodF decreased the level of sodN mRNA to about 40% of that in the control strain without any s-SodF gene. On the other hand, V1 and V2 variants of s-SodF, even though they were expressed to higher levels than the wild type, did not affect the level of sodN mRNA (Figure 6B). The levels of V1 and V2 s-SodF RNAs were about 7-fold and 3-fold higher than the wild-type level, respectively. This differential expression could arise from the difference in the copy number of incorporated plasmids as well as RNA stability. We tried another stem-variant mutant (V3) that changed the last five nucleotides in the anti-sodN sequence from CCTCG to AACAT. This variant, however, was expressed too low to examine its effect, presumably due to a dramatic decrease in stability, as expected from the central bulge generated in the stem (data not shown). All together, these experiments verify that sequences in the anti-sodN region of s-SodF, at least the first 14 nt including the predicted loop, were critical for the inhibitory action of s-SodF.
Figure 6.
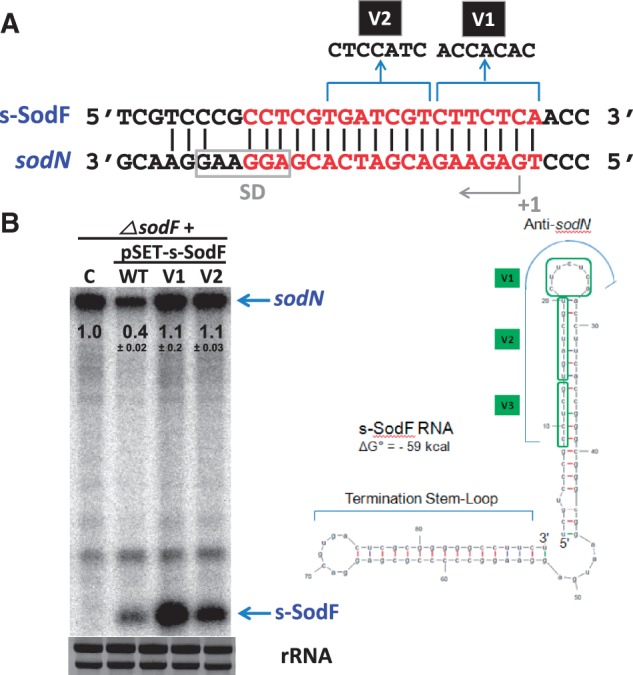
Sequence-specificity of s-SodF RNA to inhibit sodN expression. (A) Variants of s-SodF were created by changing sequences in the anti-sodN region. V1 harbor changes of seven nucleotides that correspond to the predicted loop region in anti-sodN sequence. V2 harbor changes in the subsequent seven nucleotides. (B) Effect of s-SodF mutations on the level of sodN mRNA. The mutated s-SodF genes were cloned in pSET152-based vector pSET162 with ermE* promoter, and incorporated into the chromosome of ΔsodF strain through phage attachment site, as done for the wild-type s-SodF construct used in Figure 5. The ΔsodF cells with parental vector control (C), wild type (WT) and variants (V1, V2) of s-SodF genes were grown in YEME to OD of 0.8. RNA samples were obtained from cells as in Figure 5, without rifampicin chase. A representative gel from three independent experiments was presented, and marked with the quantified values of the average ± SD for sodN mRNA.
Growth phase-dependent antagonistic expression of sodN and sodF
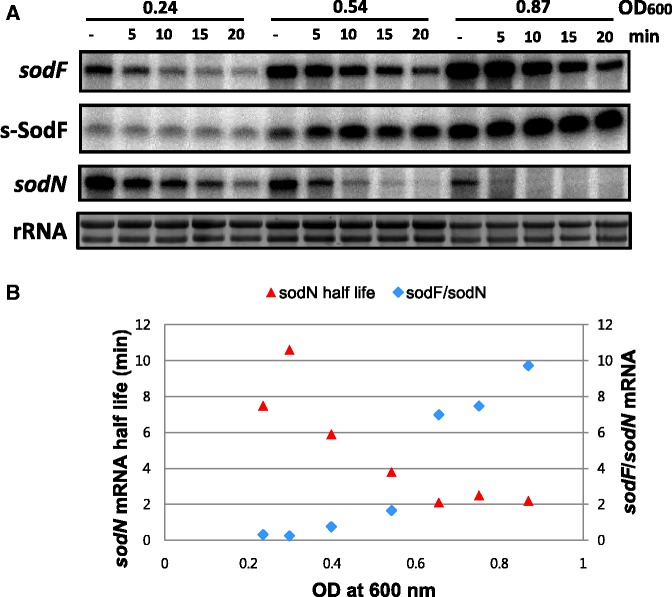
When we tried to measure the half-life of sodN mRNA in the wild-type strain, we experienced extensive variation. Soon we found that the amount of sodN mRNA decreased, whereas sodF RNA increased as cell growth proceeded from early to late stages of exponential growth. At early exponential phase (OD600 < 0.5), sodN RNA was more prevalent than sodF RNA, whereas the relative amount reversed at later growth phase (Supplementary Figure S2). We then measured the half-life of sodN mRNA at different OD by rifampicin chase experiment. A representative result presented in Figure 7 demonstrated clearly that at later phase of growth, where the amount of sodF RNA increased, the half-life of sodN mRNA became shorter. We plotted the change in sodN mRNA half-life as growth progresses in the exponential phase, along with the relative amount of sodF mRNA to sodN mRNA. Results in Figure 7B coincided well with the proposal that sodF transcripts lower the stability of sodN mRNA.
Figure 7.
Correlation between sodF/sodN ratio and the stability of sodN mRNA during growth. (A) Measurement of sodF and sodN mRNA stability at different growth phases in WT. S. coelicolor cells were grown in YEME medium to OD600 of 0.2 to 1.0 and treated with rifampicin (300 μg/ml) at specific OD600. At 5, 10, 15 and 20 min after rifampicin treatment, cells were harvested and fixed with methanol. Their RNAs were analyzed by S1 mapping with 5′-end-labeled probes for sodF and sodN RNAs. The radioactivity of the S1-protected bands was quantified by Phosphor Imager (Bio-Rad) and presented as relative values to the untreated sample. Representative results from cells grown to OD600 of 0.24, 0.54 and 0.87 were shown. (B) The change in the relative amount of sodF and sodN mRNA (sodF/sodN) during exponential growth was plotted, along with the changing half-lives of sodN mRNA, as batch culture proceeds. The sodF/sodN ratio was taken from the quantified amount of each mRNA-specific band in untreated samples at specific growth stage, as represented in (A).
DISCUSSION
Small regulatory RNA produced from a functional mRNA inhibits the expression of an antagonistically regulated gene
Accumulating lists of small non-coding RNAs have been shown to play a variety of regulatory roles (42,43). In many cases, especially in Gram-negative bacteria, these sRNAs function in association with the RNA-binding modulator Hfq to control expression of single to multiple genes, showing a broad range of specificity. Actinomycetes, Gram-positive bacteria with high GC content, along with cyanobacteria and deinococci, do not contain any apparent Hfq-homologues (38,44). Quite a number of small RNAs have been identified in actinomycetes, primarily in streptomycetes and mycobacteria, through bioinformatics combined with experimental verifications, cloning of isolated small RNAs and genome-scale deep sequencing of RNA (38,45–48). Even though some correlation with growth phases and differentiation have been demonstrated, direct targets and physiological functions of small RNAs from actinomycetes have been identified only in a very limited number of examples (46,49). This reflects difficulties not only in identifying functional small regulatory RNAs but also in predicting and/or validating their physiological targets in bacteria with genomes of high GC content. Based on sequence complementarity, we were able to identify a unique trans-acting small RNA, a cleaved 3′UTR product of a functional mRNA, which facilitates the decay of another mRNA and possibly inhibits translation, resulting in antagonistic regulation. The model depicted in Figure 8 summarized the antagonistic regulatory mechanism of sodF and sodN gene expression. According to this model, under nickel-limited conditions as experienced in later phases of growth in batch culture, Nur without corepressor nickel loses its binding affinity for the Nur-consensus (Nur-box) sequence that overlaps the sodF promoter (28). Induction of sodF gene transcription ensues, producing full-length sodF mRNA, from which a 90 nt-long 3′UTR segment that contains the anti-sodN sequence of 19 nt can be cleaved off to function as a stable small regulatory RNA (s-SodF). The s-SodF RNA is able to form perfect base pairing with the 5′ terminal portion of sodN mRNA by up to 18 bp. The base-pairing facilitates degradation of sodN mRNA and may possibly inhibit translation by occluding ribosome binding, resulting in a rapid decrease in the production of Ni-SOD. In this way, cells can ensure production of the non-nickel enzyme (Fe-SOD) and at the same time rapidly turn off the synthesis of nickel-requiring enzyme (Ni-SOD).
Figure 8.
A model for Nur-dependent inverse regulation of sodF and sodN. This model describes that the small processed sodF RNA negatively regulates the translation and stability of sodN mRNA, enabling the antagonistic regulation of sodN and sodF genes through nickel-specific Nur. Under nickel-limited conditions, Nur without nickel loses its binding activity to sodF promoter. Induction of sodF gene transcription ensues, producing full length sodF mRNA, from which a 90 nt-long 3′UTR segment that contains anti-sodN sequence of 19 nt can be cleaved off to function as a stable small regulatory RNA (s-SodF). The s-SodF RNA is able to form perfect base pairing with the 5′-end segment of sodN mRNA by up to 18 bp. The base-pairing can inhibit translation by occluding ribosome binding, and facilitates degradation of sodN mRNA, resulting in rapid decrease in the production of Ni-SOD.
Features of s-SodF RNA and possible mechanism of regulation
Most small non-coding RNAs identified so far are transcribed as independent units from their own promoter to the terminator (42,50). In some cases, the transcribed small RNA undergoes further processing to result in a more stable and/or functional form (51,52). Recently, deep sequencing of Hfq-bound small RNAs in Salmonella revealed that a significant proportion of them are produced from the 3′UTR (downstream of the coding region) of a transcribed gene (31). This raised the possibility of finding sRNAs which are transcribed in parallel with the mRNA downstream of the coding region, or are processed from existing mRNA. In one characterized example in Salmonella, DapZ was found to be transcribed from its own promoter located downstream of the coding region of dapB, which encodes a lysine biosynthetic enzyme (31). DapZ small RNA expression is independent of dapB expression, and it acts to translate at least two mRNAs encoding ABC transporters. A few sRNAs were suggested to be processed from existing mRNAs, based on the lack of promoters and the presence of 5′ monophosphate ends. However, all the cleavages sites occurred in the coding region, making the parental mRNAs inactive (31). Currently, s-SodF RNA is the only example that we know of to be produced as a processed 3′UTR product without affecting its corresponding coding sequence.
The predicted secondary structure of s-SodF RNA (Figure 3C) revealed two stem-loops, the 5′-proximal one with anti-sodN sequence and the termination stem-loop. The anti-sodN sequence was located in such a way that the final 6 nt that could pair with the 5′ terminal residues of sodN mRNA is present in the loop region. Therefore, it was plausible to speculate that the seed-pairing between s-SodF and sodN mRNA occurs through this single-stranded loop region. The stem region where the rest of the complementary sequence resides consists of relatively weak base-pairing with bubbles, easily breakable to form hydrogen bonds with sodN mRNA. Loss of inhibitory activity by changing sequences in the predicted loop region (V1) and the subsequent stem region (V2) predicts that base-pairing of at least 14 bp may be required for the inhibitory function of s-SodF (Figure 6). Whether the base pairing between the two molecules requires any RNA-binding protein modulators or not awaits further investigation. In S. coelicolor, an example has been reported for the interaction of a trans-acting small RNA (scr5239) and its target (dagA mRNA encoding an agarase), where a base-pairing was suggested to involve a 17 nt-long near-perfect match with a bulge of 1 nt (49). For RNAs with GC content of >75%, strict base pairing between sRNA and mRNA may be required to compete with prevalent intramolecular secondary structures with low ΔG values. The search for relatively long complementary sequence stretches in the genome may increase the possibility of identifying possible interacting partners of sRNAs. The RNases that are responsible for processing s-SodF RNA and degrading sodN mRNA await further investigation. Our preliminary experiments showed that mutation of genes for RNase III (53) and RNase E (54) did not affect the production of s-SodF and stability of sodN mRNA, suggesting the involvement of other RNases (data not shown).
Predicted occurrence of similar regulation
From public genome databases, it is predicted that several groups of bacteria possess both sodN and sodF genes. They include most Streptomyces spp., some actinomycetes, cyanobacteria, chlamydiae, planctomycetes, gamma-, delta- and alpha-proteobacteria (Supplementary Table S2). Except S. cattleya that contains only the sodN gene, all 10 Streptomyces species whose genomes were examined exhibited the complementary sequence between the sense strands of sodF and sodN genes. They range from 16 to 19 nt of complementarity between sodF 3′UTR and sodN 5′UTR (Supplementary Table S3). Whether these other streptomycetes produce a similar s-SodF RNA and regulate sodN in a similar fashion as in S. coelicolor is an interesting question to be investigated. A recent genome-wide analysis of non-coding RNAs in three Streptomyces spp. revealed the presence of s-SodF RNA in S. coelicolor and its homologs in S. avermitilis and S. venezuelae (48). This supports the possibility that similar regulation will be in action in these organisms. In S. bingchenggensis BCW-1, there exist two sodF paralog genes as in S. coelicolor (Supplementary Table S3). However, unlike the two sodF genes (sodF and sodF2) which encode functional SodF proteins in S. coelicolor, only one gene (sodF; SBI_02504) in S. bingchenggensis encodes a functional SodF, followed by a degenerate anti-sodN sequence with 8 nt mismatch, which is unlikely to serve an inhibitory function. On the other hand, the sodF2 gene (SBI_01257) has a big in-frame deletion of the coding region but maintains a relatively good anti-sodN sequence (Supplementary Table S3). If there are no sequencing errors, this suggests that the regulatory pathway with s-SodF may have been maintained throughout the evolution of Streptomyces spp. even when the adjacent coding sequence no longer produces a functional SodF protein. Whether there are other gene pairs or regulons that utilize this type of 3′UTR-generated small regulatory RNA requires further bioinformatic and experimental investigations. Search for complementarity between distant regions in the genome, especially between 3′UTR regions and anywhere around distant coding region, is expected to give fruitful clues. This will facilitate excavating small regulatory RNAs with putative targets.
Inverse regulation of isoenzymes and antagonistic proteins
Antagonistic regulation between Fe-containing and Ni-containing isoforms of enzymes is not unprecedented. In Helicobacter mustelae, a carnivore-colonizing species, Fe-containing urease (UreA2B2) is produced under nickel-depleted condition, whereas Ni-containing urease (UreAB) prevails under nickel-sufficient condition (55). In this case, the nickel-specific regulator NikR regulates both operons, directly repressing and activating ureA2B2 and ureAB transcription, respectively (56). It is interesting to note the existence of inverse regulation between FeSOD and its iso-enzymes with different metals, MnSOD and NiSOD. Since SODs are abundant proteins and are important for air-exposed life, antagonistic regulation in response to the availability of specific preferred metal and oxidative conditions should be beneficial for the economy of the cell. For soil-dwelling streptomycetes, abundant nickel in the aerobic soil environment would fit the evolution of a gene regulatory system where sodF is repressed such that nickel is utilized before iron. For facultative E. coli, life under anaerobic iron-rich condition could have evolved a gene system where SodB is a preferred enzyme under anaerobic condition, and SodA is induced in the presence of oxygen and oxidative stress.
The observation that the inverse regulation exerted by Fur family regulators involves small regulatory RNAs as mediators is intriguing. RyhB-mediated regulation by Fur in E. coli, PrrF1/F2-mediated regulation by Fur in Pseudomonas and FsrA-mediated regulation by Fur in B. subtilis all enable inverse regulation of Fe- versus non-Fe proteins. The discovery of s-SodF in S. coelicolor adds to this list of small RNA-coupled regulation by Fur family members. These examples support the presence of an evolutionarily robust regulatory circuit mediated by metal-specific Fur family regulators and small RNAs in coordinated synthesis of iso-proteins with specific metal cofactors across distantly related bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [57–58].
ACKNOWLEDGEMENTS
We are grateful for Drs Narry Kim and Young-Kook Kim for their helpful advices on detecting small RNAs. We also thank Dr Dmitri Rodionov who informed about the presence of repeating sequence in sod genes. Hae Mi Kim, J.H. Shin and Y.B. Cho were recipients of BK21 Fellowship for Life Sciences at SNU.
FUNDING
Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology [2011-0031960 to J.-H.R.]. Funding for open access charge: Intelligent Synthetic Biology Center of Global Frontier Project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fridovich I. Superoxide anion radical (O2-.), superoxide dismutases, and related matters. J. Biol. Chem. 1997;272:18515–18517. doi: 10.1074/jbc.272.30.18515. [DOI] [PubMed] [Google Scholar]
- 2.Perry JJ, Shin DS, Getzoff ED, Tainer JA. The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta. 2010;1804:245–262. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller AF. Superoxide dismutases: active sites that save, but a protein that kills. Curr. Opin. Chem. Biol. 2004;8:162–168. doi: 10.1016/j.cbpa.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Miller AF. Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 2012;586:585–595. doi: 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storz G, Imlay JA. Oxidative stress. Curr. Opin. Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 6.Gort AS, Ferber DM, Imlay JA. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 1999;32:179–191. doi: 10.1046/j.1365-2958.1999.01343.x. [DOI] [PubMed] [Google Scholar]
- 7.Hassan HM, Schrum LW. Roles of manganese and iron in the regulation of the biosynthesis of manganese-superoxide dismutase in Escherichia coli. FEMS Microbiol. Rev. 1994;14:315–323. doi: 10.1111/j.1574-6976.1994.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 8.Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 2000;182:3802–3808. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tardat B, Touati D. Two global regulators repress the anaerobic expression of MnSOD in Escherichia coli::Fur (ferric uptake regulation) and Arc (aerobic respiration control) Mol. Microbiol. 1991;5:455–465. doi: 10.1111/j.1365-2958.1991.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 11.Hassett DJ, Howell ML, Ochsner UA, Vasil ML, Johnson Z, Dean GE. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 1997;179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polack B, Dacheux D, Delic-Attree I, Toussaint B, Vignais PM. The Pseudomonas aeruginosa fumc and soda genes belong to an iron-responsive operon. Biochem. Biophys. Res. Commun. 1996;226:555–560. doi: 10.1006/bbrc.1996.1393. [DOI] [PubMed] [Google Scholar]
- 13.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl Acad. Sci. USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl Acad. Sci. USA. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Youn HD, Kim EJ, Roe JH, Hah YC, Kang SO. A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem. J. 1996;318(Pt 3):889–896. doi: 10.1042/bj3180889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupont CL, Neupane K, Shearer J, Palenik B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 2008;10:1831–1843. doi: 10.1111/j.1462-2920.2008.01604.x. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt A, Gube M, Kothe E. In silico analysis of nickel containing superoxide dismutase evolution and regulation. J. Basic Microbiol. 2009;49:109–118. doi: 10.1002/jobm.200800293. [DOI] [PubMed] [Google Scholar]
- 18.Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Duprat S, Galperin MY, Koonin EV, Le Gall F, et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl Acad. Sci. USA. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palenik B, Brahamsha B, Larimer FW, Land M, Hauser L, Chain P, Lamerdin J, Regala W, Allen EE, McCarren J, et al. The genome of a motile marine Synechococcus. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 20.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 21.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 22.Eitinger T. In vivo production of active nickel superoxide dismutase from Prochlorococcus marinus MIT9313 is dependent on its cognate peptidase. J. Bacteriol. 2004;186:7821–7825. doi: 10.1128/JB.186.22.7821-7825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Nickel superoxide dismutase structure and mechanism. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 24.Wuerges J, Lee JW, Yim YI, Yim HS, Kang SO, Djinovic Carugo K. Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proc. Natl Acad. Sci. USA. 2004;101:8569–8574. doi: 10.1073/pnas.0308514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priya B, Premanandh J, Dhanalakshmi RT, Seethalakshmi T, Uma L, Prabaharan D, Subramanian G. Comparative analysis of cyanobacterial superoxide dismutases to discriminate canonical forms. BMC Genomics. 2007;8:435. doi: 10.1186/1471-2164-8-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim EJ, Chung HJ, Suh B, Hah YC, Roe JH. Expression and regulation of the sodF gene encoding iron- and zinc-containing superoxide dismutase in Streptomyces coelicolor Muller. J. Bacteriol. 1998;180:2014–2020. doi: 10.1128/jb.180.8.2014-2020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim EJ, Chung HJ, Suh B, Hah YC, Roe JH. Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Muller. Mol. Microbiol. 1998;27:187–195. doi: 10.1046/j.1365-2958.1998.00674.x. [DOI] [PubMed] [Google Scholar]
- 28.Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- 29.An YJ, Ahn BE, Han AR, Kim HM, Chung KM, Shin JH, Cho YB, Roe JH, Cha SS. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res. 2009;37:3442–3451. doi: 10.1093/nar/gkp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung HJ, Choi JH, Kim EJ, Cho YH, Roe JH. Negative regulation of the gene for Fe-containing superoxide dismutase by an Ni-responsive factor in Streptomyces coelicolor. J. Bacteriol. 1999;181:7381–7384. doi: 10.1128/jb.181.23.7381-7384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl Acad. Sci. USA. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieser T. Practical Streptomyces Genetics. Norwich: John Innes Foundation; 2000. [Google Scholar]
- 34.Han A-R. 2007 MS thesis. Graduate School, Seoul National University, Seoul. [Google Scholar]
- 35.Chung HJ, Kim EJ, Suh B, Choi JH, Roe JH. Duplicate genes for Fe-containing superoxide dismutase in Streptomyces coelicolor A3(2) Gene. 1999;231:87–93. doi: 10.1016/s0378-1119(99)00088-8. [DOI] [PubMed] [Google Scholar]
- 36.Bibb MJ, Janssen GR, Ward JM. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 37.Kim MS, Hahn MY, Cho Y, Cho SN, Roe JH. Positive and negative feedback regulatory loops of thiol-oxidative stress response mediated by an unstable isoform of sigmaR in actinomycetes. Mol. Microbiol. 2009;73:815–825. doi: 10.1111/j.1365-2958.2009.06824.x. [DOI] [PubMed] [Google Scholar]
- 38.Swiercz JP, Hindra, Bobek J, Haiser HJ, Di Berardo C, Tjaden B, Elliot MA. Small non-coding RNAs in Streptomyces coelicolor. Nucleic Acids Res. 2008;36:7240–7251. doi: 10.1093/nar/gkn898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Celesnik H, Deana A, Belasco JG. PABLO analysis of RNA: 5′-phosphorylation state and 5′-end mapping. Methods Enzymol. 2008;447:83–98. doi: 10.1016/S0076-6879(08)02205-2. [DOI] [PubMed] [Google Scholar]
- 40.Mitra A, Angamuthu K, Jayashree HV, Nagaraja V. Occurrence, divergence and evolution of intrinsic terminators across eubacteria. Genomics. 2009;94:110–116. doi: 10.1016/j.ygeno.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panek J, Bobek J, Mikulik K, Basler M, Vohradsky J. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics. 2008;9:217. doi: 10.1186/1471-2164-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vockenhuber MP, Sharma CM, Statt MG, Schmidt D, Xu Z, Dietrich S, Liesegang H, Mathews DH, Suess B. Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor. RNA Biol. 2011;8:468–477. doi: 10.4161/rna.8.3.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tezuka T, Hara H, Ohnishi Y, Horinouchi S. Identification and gene disruption of small noncoding RNAs in Streptomyces griseus. J. Bacteriol. 2009;191:4896–4904. doi: 10.1128/JB.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moody MJ, Young RA, Jones SE, Elliot MA. Comparative analysis of non-coding RNAs in the antibiotic-producing Streptomyces bacteria. BMC Genomics. 2013;14:558. doi: 10.1186/1471-2164-14-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vockenhuber MP, Suess B. Streptomyces coelicolor sRNA scr5239 inhibits agarase expression by direct base pairing to the dagA coding region. Microbiology. 2012;158:424–435. doi: 10.1099/mic.0.054205-0. [DOI] [PubMed] [Google Scholar]
- 50.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 2007;65:373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 53.Price B, Adamidis T, Kong R, Champness W. A Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J. Bacteriol. 1999;181:6142–6151. doi: 10.1128/jb.181.19.6142-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee K, Cohen SN. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 2003;48:349–360. doi: 10.1046/j.1365-2958.2003.03435.x. [DOI] [PubMed] [Google Scholar]
- 55.Stoof J, Breijer S, Pot RGJ, van der Neut D, Kuipers EJ, Kusters JG, van Vliet AHM. Inverse nickel-responsive regulation of two urease enzymes in the gastric pathogenHelicobacter mustelae. Environ. Microbiol. 2008;10:2586–2597. doi: 10.1111/j.1462-2920.2008.01681.x. [DOI] [PubMed] [Google Scholar]
- 56.Stoof J, Kuipers EJ, van Vliet AH. Characterization of NikR-responsive promoters of urease and metal transport genes of Helicobacter mustelae. Biometals. 2010;23:145–159. doi: 10.1007/s10534-009-9275-7. [DOI] [PubMed] [Google Scholar]
- 57.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]