Abstract
The HDM2-p53 loop is crucial for monitoring p53 level and human pathologies. Therefore, identification of novel molecules involved in this regulatory loop is necessary for understanding the dynamic regulation of p53 and treatment of human diseases. Here, we characterized that the ribosomal protein L6 binds to and suppresses the E3 ubiquitin ligase activity of HDM2, and subsequently attenuates HDM2-mediated p53 polyubiquitination and degradation. The enhanced p53 activity further slows down cell cycle progression and leads to cell growth inhibition. Conversely, the level of p53 is dramatically decreased upon the depletion of RPL6, indicating that RPL6 is essential for p53 stabilization. We also found that RPL6 translocalizes from the nucleolus to nucleoplasm under ribosomal stress, which facilitates its binding with HDM2. The interaction of RPL6 and HDM2 drives HDM2-mediated RPL6 polyubiquitination and proteasomal degradation. Longer treatment of actinomycin D increases RPL6 ubiquitination and destabilizes RPL6, and thereby putatively attenuates p53 response until the level of L6 subsides. Therefore, RPL6 and HDM2 form an autoregulatory feedback loop to monitor the level of p53 in response to ribosomal stress. Together, our study identifies the crucial function of RPL6 in regulating HDM2-p53 pathway, which highlights the importance of RPL6 in human genetic diseases and cancers.
INTRODUCTION
The tumor suppressor p53 plays a pivotal role in monitoring genomic stability and preventing malignant transformation, and the frequent loss of function of p53 is associated with a majority of human cancers (1). p53 induces cell cycle arrest, apoptosis or senescence depending on the stimuli; thus, the cellular p53 expression must be maintained at a low level under normal condition. HDM2, an E3 ubiquitin ligase, has been identified as a crucial negative regulator of p53 via inhibiting p53 transcriptional activity and promoting ubiquitination and degradation of p53 (2,3). In turn, HDM2 is transcriptionally activated by p53 (4–6). Therefore, HDM2 and p53 form a tight autoregulatory feedback loop. So far, numerous mechanisms have been implicated in understanding the regulation of HDM2-p53 axis in DNA damage, cell cycle progression and multiple cellular conditions (7). Notably, increasing evidence has connected p53 with ribosome biogenesis, an essential process that maintains cells growth and individual development (8–10).
Ribosome biogenesis is a complex and orchestrated process. Changes in ribosome biogenesis have been implicated in human pathologies (11–14). Evidence supporting this concept results from the finding that patients with Diamond–Blackfan anemia, a human genetic disease characterized by congenital erythroblastopenia, are harboring heterozygous loss-of-function mutations in ribosomal protein (RP) genes (12,14,15). In addition, depletion of RPS14 results in the 5q− syndrome with a characteristic defect in erythroid differentiation (12,16,17). Patients suffering from these diseases have propensities to develop cancers (11). Given the importance of RPs in human pathologies, it is critical to identify the underlying mechanism that is responsible for the role of RPs in these diseases. Mounting evidence has suggested that the RP-HDM2-p53 loop is a crucial linker between ribosome and human pathologies (8,18–20). The importance of this finding has been proved in human Diamond–Blackfan anemia and 5q− syndrome, for which the aberrant activation of the p53-dependent cell cycle checkpoint in erythroid precursors is responsible for the associated macrocytic anemia (11). For the underlying mechanism, the activation of p53 is due to the binding of RPs to HDM2 upon ribosomal stress.
Ribosomal stress or nucleolar stress, a situation indicating disruption of ribosome biogenesis, can be induced by low doses of actinomycin D (ActD), a chemical reagent that specially inhibits RNA polymerase I activity and consequently reduces ribosomal RNA synthesis (21). Upon ribosomal stress, the nucleolus unassembled RPs, such as RPL5 (22), RPL11 (2,7,23–25), RPL26 (2), RPS7 (3,26) and RPS14 (19,27), are released into the nucleoplasm where they are captured by HDM2 and attenuate HDM2-mediated p53 ubiquitination and degradation (7,8,28,29). Notably, the physiological significance of the RP–HDM2 interaction has been demonstrated in mice that carry a C305F knockin in MDM2 (30). This mutation disrupted the binding of MDM2 to RPL5 and RPL11. Mice harboring this mutation retained their normal p53 response to DNA damage, while impaired the p53 response to ribosomal stress (30). Furthermore, disruption of the RP–MDM2 interaction significantly accelerated Eμ-Myc-induced lymphomagenesis (30). This finding strongly illustrates that the RP-HDM2-p53 axis plays an important role in response to ribosomal stress and tumorigenesis. Therefore, unveiling the extraribosomal function of other RPs will help us to further understand the essential role of RPs in human pathologies.
RPL6 is found to associate with the Noonan syndrome (31,32), an autosomal dominant developmental disorder characterized by short stature, typical faces with hypertelorism and webbing of the neck (32). Besides, unusual expression of RPL6 has been identified in human gastric cancer (33,34). The close connection of p53 with tumorigenesis encouraged us to explore whether disease-associated RPL6 has a regulatory role in the HDM2-p53 axis. In this study, we found that RPL6 is essential for p53 stabilization. In response to ribosomal stress, RPL6 relocates into the nucleoplasm, where it interacts with HDM2 and subsequently attenuates HDM2-mediated p53 ubiquitination and degradation. Furthermore, we also demonstrated that RPL6 is a substrate for HDM2-mediated ubiquitination and proteasomal degradation. RPL6 inhibits HDM2 E3 ligase activity and finally is degraded by HDM2. Therefore, RPL6 and HDM2 form a feedback regulatory loop. Collectively, our findings identify that RPL6 is a novel and essential regulator in the HDM2-p53 axis and characterize a mechanism whereby RPL6 stabilizes p53.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney (HEK) 293T, A549, U2OS, HCT116 (gifted by Dr Jun Gu, Peking University) and mouse embryonic fibroblast cells (MEF, p53−/−, MDM2−/−) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 humidified atmosphere.
Plasmids and reagents
HA-p53, His-ubiquitin and N-terminal-tagged Myc-HDM2 were described previously (27). N-terminal tagged HA-MDMX was provided by Dr Lingqiang Zhang (State Key Laboratory of Proteomics, Beijing Proteome Research Center, Beijing Institute of Radiation Medicine). The complementary DNA of full-length RPL6 was amplified by polymerase chain reaction and inserted into the pcDNA-3Flag vector at EcoRI and XhoI restriction sites. Anti-HA, anti-Flag, anti-Myc, anti-human HDM2 (SMP14), anti-HA-HRP, anti-β-actin and anti-p53 antibodies were previously described (27). Rabbit polyclonal antibody of RPL6 was generated by immunizing a rabbit with the purified protein of full-length RPL6. Drugs used in this study are as follows. ActD was from Sigma; MG132 and cycloheximide (CHX) were from Calbiochem.
Transfection and Co-immunoprecipitation analysis
Cells were transfected with plasmids using MegaTran 1.0 transfection reagent (Origene) according to the manufacturer's instructions. At 48 h after transfection, cells were harvested and lysed in the Nonidet P-40 lysis buffer (50 mM Tris–HCl, pH 8.0, 1.0% NP-40, 1 mM EDTA, 150 mM NaCl). The supernatant was incubated with indicated antibody at 4°C overnight and then protein G was added to the samples and incubated for another 4 h at 4°C. Beads were washed three times with the lysis buffer. The binding proteins were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and detected by immunoblotting with indicated antibodies.
Immunofluorescence analysis
HeLa cells grown on coverslips were transfected with Flag-RPL6 expressing plasmid. Twenty-four hours after transfection, cells were treated with ActD (5 nM) for the indicated time and washed three times with pre-cold phosphate buffered saline. Cells were then fixed and permeabilized with absolute methanol at −20°C for 10 min and blocked with 1% bovine serum albumin for 1 h at room temperature. Cells were incubated with primary antibodies for 2 h at 37°C, and then washed three times with pre-cold phosphate-buffered saline followed by incubation with specific fluorescently labeled secondary antibodies for 1 h at 37°C. Images were visualized with a confocal laser scanning microscope (Leica TCS SP5).
In vivo ubiquitination assay
HCT116 cells were transfected with indicated combinations of plasmids as shown in the figure legends. In vivo ubiquitination assay was performed as previously described (27). Eluted proteins were analyzed by immunoblotting with indicated antibodies.
RNA interference
The individual designed short hairpin RNA (shRNA) sequence targeting RPL6 is 1#5′- CGGGTGGTTAAACTTCGCAA and 2#5′-CAGAAAGCTGTGGACTCACA. Transfection was performed with the InCella transfection reagent according to the manufacturer’s instructions (Genaxxon bioscience GmbH).
Cell cycle analysis
To evaluate the effect of RPL6 on cell cycle progression, cells were transfected with plasmid expressing Flag-RPL6 and harvested with trypsinization at 24 h after transfection. Cells were fixed in 70% ethanol at 4°C overnight, treated with 1 mg/ml RNase at 37°C for 30 min, stained with propidium iodide (50 μg/ml) at 4°C for 30 min and analyzed by Fluorescence Activating Cell Sorter (FACS).
Cell proliferation assay
To monitor the effect of RPL6 on cell growth, cells transfected with Flag-RPL6 were seeded in 96-well plates. Cell proliferation was measured by the CellTiter 96® AQueous nonradioactive cell proliferation assay according to the manufactory instruction (Promega). The absorbance of samples was determined at 492 nm.
RESULTS
RPL6 interacts with HDM2
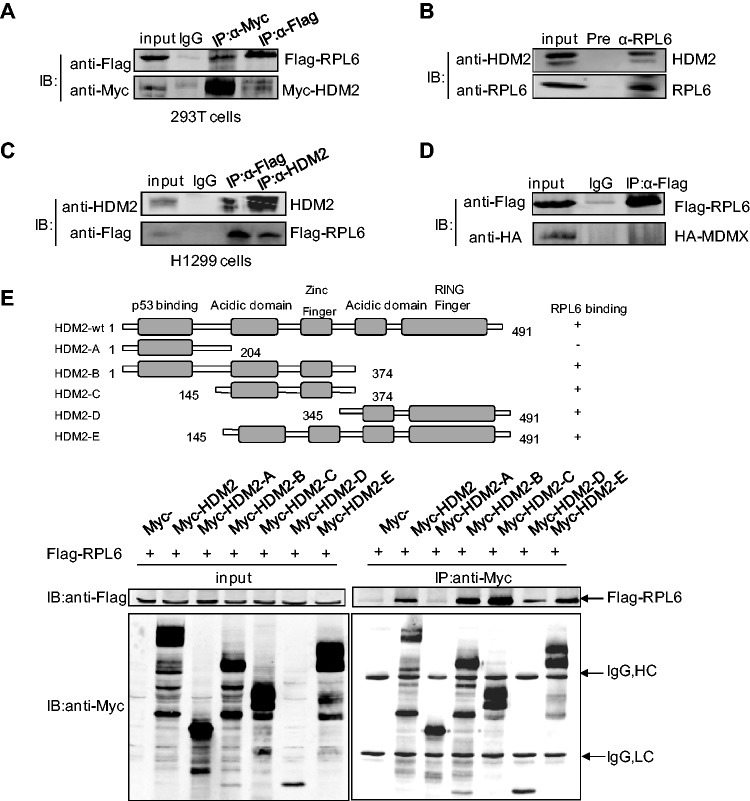
To detect whether RPL6 is involved in the HDM2-p53 pathway, 293T cells were transfected with Flag-RPL6 and Myc-HDM2 and the interaction between RPL6 and HDM2 was examined by co-immunoprecipitation (Co-IP) assay. Flag-RPL6 was specifically immunoprecipitated with HDM2 by anti-Myc antibody (Figure 1A). Conversely, Myc-HDM2 was also co-immunoprecipitated with RPL6 by anti-Flag antibody comparing with the control IgG (Figure 1A). Similar results were obtained in HCT116 cells (Supplementary Figure S1A). To avoid the artificial effect caused by overexpression, the level of ectopically expressed RPL6 was compared with endogenous RPL6 (Supplementary Figure S1B), which indicates that the ectopic RPL6 is moderate. To further confirm the association of HDM2 and RPL6, binding of endogenous RPL6 and HDM2 was examined in HCT116 cells with anti-RPL6 antibody. As shown in Figure 1B, the RPL6 immunoprecipitate contained HDM2. Notably, because of the tight relationship between HDM2 and p53, the interaction of RPL6 and p53 was also examined and observed in 293T cells (Supplementary Figure S1C). To test whether the interaction between RPL6 and HDM2 is mediated by p53, Co-IP assay was performed in p53-null H1299 cells. The result showed that RPL6 still interacted with HDM2 in the absence of p53 (Figure 1C), which suggests that p53 is not required for the interaction of RPL6 and HDM2.
Figure 1.
RPL6 interacts with HDM2. (A) Interaction between exogenous RPL6 and HDM2. HEK293T cells were transfected with Flag-RPL6 and Myc-HDM2. Thirty-six hours after transfection, cells were harvested and lysed with 1 ml NP-40 cell lysis buffer. The cell lysate was divided equally into the three IPs, and 3% input was loaded as the input and the remaining was immunoprecipitated with control IgG, anti-Flag or anti-Myc antibody followed by immunoblotting (IB) with indicated antibodies. (B) Interaction of endogenous HDM2 and RPL6 in HCT116 cells. The lysates of HCT116 were immunoprecipitated with anti-RPL6 or preimmune serum followed by IB with anti-HDM2 and anti-RPL6 antibodies. (C) RPL6 interacts with HDM2 in p53-null H1299 cells. Cell lysates were prepared from H1299 cells and Co-IP assay was performed with anti-Flag, anti-HDM2 or control IgG followed by IB with anti-HDM2 and anti-Flag antibodies. (D) Flag-RPL6 does not interact with HA-MDMX in HEK293T cells. Cells were transfected with Flag-RPL6 and HA-MDMX and harvested for Co-IP assay with the indicated antibodies. This experiment has been performed three times. (E) Mapping of the HDM2 domains for RPL6 binding. HCT116 cells were transfected with plasmids expressing full-length Flag-RPL6 and different truncation constructs of Myc-HDM2. Immunoprecipitation (IP) was performed with anti-Myc antibody followed by IB with anti-Flag antibody. A diagram for the truncations of HDM2 used in this assay was shown on top.
MDMX (also named MDM4), the homolog of HDM2, has been reported to play a critical role in facilitating the regulation of p53 by HDM2. Previous studies also showed that RPs do not bind directly to MDMX but may interact indirectly through HDM2 (35,36). To probe whether RPL6 could bind to MDMX, we examined the interaction between these two proteins by Co-IP, however, no interaction between RPL6 and MDMX could be detected (Figure 1D). These data demonstrate that RPL6 specifically interacts with HDM2.
To further map the key domains of HDM2 applied to its interaction with RPL6, Co-IP assay was performed with full-length RPL6 and several HDM2 truncation mutants (Figure 1E). HDM2 mutant A that lacks of the central acidic domain failed to co-immunoprecipitate with RPL6, whereas other HDM2 mutants that contain the central acidic domain retained their ability to bind to RPL6. These data demonstrate that RPL6 associates with HDM2, and this is primarily mediated through the central acid domain of HDM2.
RPL6 enhances the abundance and activity of p53
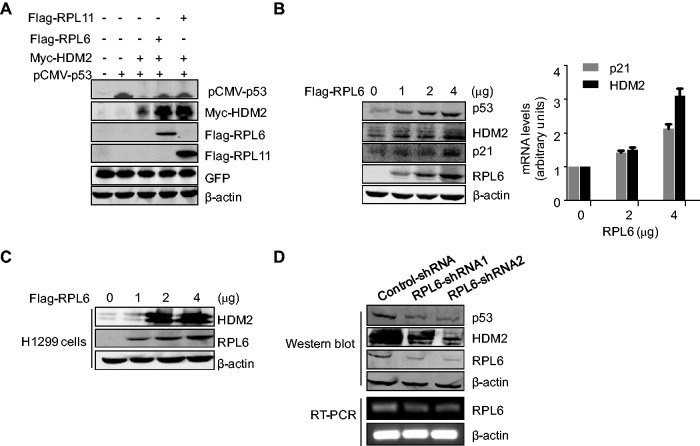
As the major negative regulator of p53, HDM2 mediates p53 ubiquitination and degradation due to its E3 ligase activity. To validate whether the interaction of RPL6 and HDM2 affects p53 level, U2OS cells were transfected with combinations of plasmids expressing Flag-RPL6, Myc-HDM2 and pCMV-p53, and the level of p53 was examined. As shown in Figure 2A, overexpression of RPL6 dramatically increased ectopic p53 level, which is as efficient as that of RPL11, a well-studied important regulator of the HDM2-p53 pathway (25). Strikingly, the endogenous levels of p53, as well as its downstream target genes p21 and HDM2, were also elevated by RPL6 in a dose-dependent manner (Figure 2B, left panel). Consistent with this finding, the messenger RNA (mRNA) levels of p21 and HDM2 were also promoted following the increase of RPL6 expression (Figure 2B, right panel), indicating that p53 is transcriptionally activated by RPL6.
Figure 2.
RPL6 enhances the abundance and activity of p53. (A) Ectopic expression of RPL6 increases the abundance of exogenous p53 and HDM2. U2OS cells were transfected with pCMV-p53, Myc-HDM2, Flag-RPL6 and an irrelevant plasmid Green Fluorescent Protein (GFP) as indicated. Cell lysate was subjected to IB with indicated antibodies. (B) RPL6 enhances the level of endogenous p53 and its downstream target genes HDM2 and p21. U2OS cells were transfected with increasing amount of Flag-RPL6. The amount of plasmid DNA kept constant (4 μg) in all lanes. Cells were harvested at 36 h after transfection and subjected to IB with indicated antibodies (left panel). Reverse Transcript-qPCR (RT-qPCR) was performed to detect the mRNA levels of p21 and HDM2 (right panel). (C) RPL6 enhances the abundance of HDM2 in the absence of p53. p53-null H1299 cells transfected with pCMV-HDM2 and RPL6 were subjected to IB with anti-HDM2 antibody. (D) Knockdown of endogenous RPL6 decreases p53 abundance. A549 cells were transfected with RPL6-shRNA1, RPL6-shRNA2 or control-shRNA. After 96 h, cell lysates were immunoblotted with anti-p53, anit-HDM2, anti-RPL6 or anti-β-actin antibodies (upper). The knockdown efficiency of RPL6 was also validated at mRNA level (lower).
Notably, transfection with vectors encoding RPL6 and RPL11 dramatically increased HDM2 expression (Figure 2A). As the expression of Myc-HDM2 was under the control of Cytomegalovirus (CMV) promoter that does not contain the binding site of p53, the increased level of Myc-HDM2 is not caused by the enhanced p53 transcriptional activity. To further validate this hypothesis, RPL6 and HDM2 were transfected into the p53-deficient H1299 cells, and as expected, an enhanced abundance of HDM2 was observed with the addition of RPL6 (Figure 2C). This result demonstrates that the functional p53 is not required for RPL6-mediated upregulation of HDM2.
To determine whether RPL6 is essential for maintaining p53 abundance, we examined the level of endogenous p53 in A549 cells with depleted RPL6. As shown in Figure 2D, knockdown of RPL6 led to a marked decrease of p53, as well as HDM2, which indicates that RPL6 is required for p53 in sustaining its abundance. Collectively, these results demonstrate that RPL6 is a novel and crucial regulator of the tumor suppressor p53.
RPL6 stabilizes p53 by attenuating HDM2-mediated p53 polyubiquitination and degradation
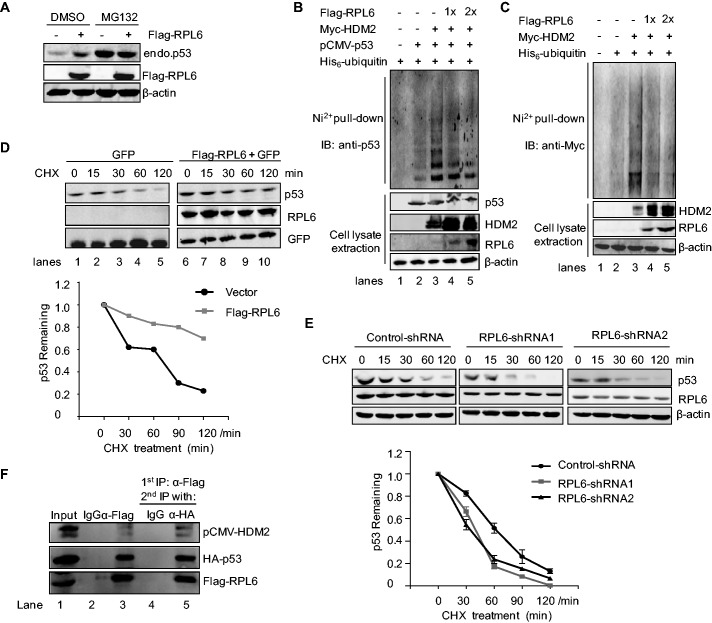
HDM2-mediated p53 polyubiquitination is essential for the regulation of p53 stability. The observation that RPL6 interacts with HDM2 and enhances the abundance of p53 encouraged us to further elucidate the underlying mechanism. To assess whether RPL6 increases p53 abundance via inhibiting HDM2-mediated polyubiquitination and degradation, HCT116 cells transfected with Flag-RPL6 were treated with MG132. As shown in Figure 3A, ectopically expressed RPL6 dramatically increased endogenous p53 level. In contrast, treatment of MG132 abolished this effect, as the level of p53 remained unchanged even with high level of RPL6. Furthermore, in vivo ubiquitination assay was carried out to confirm this observation. Overexpression of HDM2 increased p53 polyubiquitination (Figure 3B, lane 3); however, this effect was dramatically attenuated by RPL6 in a concentration-dependent pattern (Figure 3B, lane 4 and 5). Consistently, we also observed that overexpression of RPL6 attenuated HDM2 autoubiquitination (Figure 3C), which is consistent with the observation that the level of HDM2 was dramatically increased with the ectopic RPL6 (Figure 2A).
Figure 3.
RPL6 stabilizes p53 through inhibiting HDM2-mediated polyubiquitination and degradation. (A) RPL6 enhances the abundance of p53 at the post-translational level. HCT116 cells were transfected with Flag-vector or Flag-RPL6. At 24 h after transfection, cells were subjected to MG132 treatment for 8 h and dimethyl sulfoxide (DMSO) was used as the control. Cells were then harvested and subjected to IB with anti-p53, anti-Flag and anti-β-actin antibody, respectively. (B and C) RPL6 inhibits HDM2-mediated p53 polyubiquitination and HDM2 autoubiquitination. HCT116 cells were transfected with indicated plasmids and treated with 30 μM MG132 for 8 h, and ubiquitination of p53 or Myc-HDM2 was detected by in vivo ubiquitination assay with anti-p53 and anti-Myc antibody, respectively. (D) RPL6 extends the half-life of endogenous p53. U2OS cells were transfected with pcDNA-3Flag or Flag-RPL6. GFP was used as the transfection control. Twenty-four hours after transfection, cells were treated with CHX at different time points, and the level of p53 was detected by IB with anti-p53 antibody. (E) Downregulation of RPL6 shortens p53 half-life. A549 cells were transfected with control-shRNA or RPL6-shRNA for 80 h and then treated with CHX for indicated time, and the level of p53 was detected with anti-p53 antibody (upper panel). The relative levels of p53 were quantified and plotted as the mean ± s.e. from three independent experiments (lower panel). (F) RPL6, HDM2 and p53 form a complex in vivo. HEK293T cells transfected with Flag-RPL6, HA-p53 and pCMV-HDM2 were subjected to IPs with control IgG or anti-Flag antibody, followed by elution with Flag peptide. A total of 10% elution was loaded (lane 2 and 3), and the remaining 90% elution was immunoprecipitated with anti-HA antibody or control IgG, followed by IB with indicated antibodies.
To further confirm that the effect of RPL6 on p53 occurred at the post-translational level, HCT116 cells were transfected with Flag-RPL6 followed by exposing to the protein synthesis inhibitor CHX for indicated times, and the p53 level was examined by western blot analysis. As shown in Figure 3D, cells transfected with RPL6 significantly extended the half-life of endogenous p53 comparing with that of cells transfected with the control vector. Similar experiment was also performed in A549 cells with depleted RPL6. As shown in Figure 3E, depletion of RPL6 dramatically shortened the half-life of p53. Consistently, these results demonstrate that RPL6 stabilizes p53 at the post-translational level, which is mainly through inhibiting HDM2-mediated p53 polyubiquitination.
The inhibition of HDM2 by RPL6 may attribute to the formation of a complex among RPL6, HDM2 and p53. To illustrate this possibility, sequential Co-IP analysis was performed. 293T cells transfected with plasmids encoding Flag-RPL6, pCMV-HDM2 and HA-p53 were first subjected to IP with anti-Flag antibody, followed by elution with Flag peptide (Figure 3F, lane 3). The elution containing RPL6-associated proteins was then subjected to IP with anti-HA antibody (Figure 3F, lane 5). As shown in Figure 3F, both HDM2 and p53 were detected in the immunoprecipitates with anti-Flag and anti-HA antibodies, suggesting the formation of a complex among RPL6, HDM2 and p53. Collectively, these results indicate that RPL6 stabilizes p53 through inhibiting p53 ubiquitination and degradation by binding to HDM2.
RPL6 induces cell growth inhibition
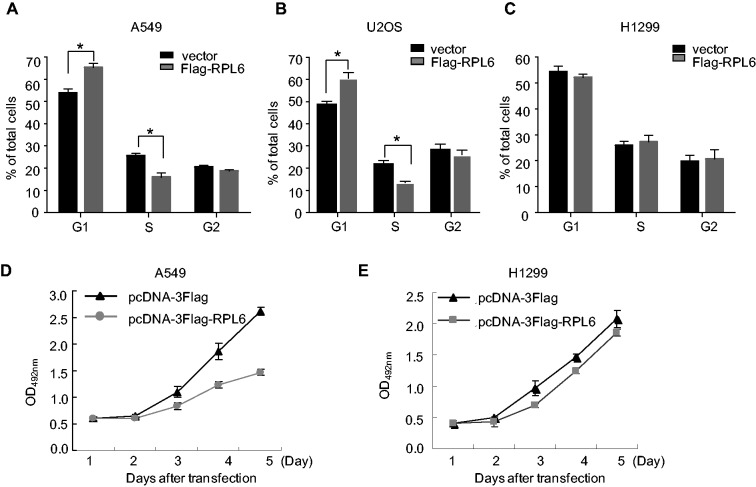
As p53 is tightly linked to cell cycle progression and cell growth, we sought to detect the consequence of RPL6 in these two physiological progresses. To this end, A549 cells were transfected with Flag-RPL6 or control vector and then FACS analysis was performed. As shown in Figure 4A, ectopic RPL6 moderately induced G1 arrest. Similar results were also obtained in U2OS cells (Figure 4B). Consistently, the moderate increased p21 was also observed in A549 and U2Os cells with ectopic RPL6 (Supplementary Figure S2B). However, no obvious effect of RPL6 on cell cycle progression was observed in p53-deficient H1299 cells (Figure 4C). The same experiment was also performed in HCT116 cells with wild-type or null p53, and the result showed that ectopic RPL6 caused G2 arrest in a p53-dependent manner (Supplementary Figure S2A and C). To further confirm the effect of RPL6 on cell cycle progression is p53-dependent, we also performed the same assay in A549 cells with depleted p53. As shown in Supplementary Figure S2D, no obvious changes in cell cycle distribution were observed when RPL6 was overexpressed in p53-depleted A549 cells, indicating that the inhibition of cell cycle progression by ectopic RPL6 is in a p53-dependent manner. Moreover, we also evaluated the effect of RPL6 on cell growth by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. As shown in Figure 4D and E, ectopic RPL6 induced cell growth inhibition in the p53-positive A549 cells but not in p53-deficient H1299 cells. While in p53-null HCT116 cells, a moderate inhibition of RPL6 on cell growth was also observed (Supplementary Figure S2E). These results indicate that RPL6 seems to act differently in different cell lines/types: RPL6 can induce p53-mediated growth arrest under certain conditions in a cell-type dependent manner, and the p53-independent mechanisms may also be involved in some other cell lines.
Figure 4.
RPL6 induces p53-dependent cell growth inhibition. (A, B and C) A549, U2OS and H1299 cells were transfected with Flag-vector or Flag-RPL6, and 24 h after transfection, cells were subjected to FACS analysis. The percentage of cell in each phase was obtained from three independent experiments. Bars represent standard deviations. *P < 0.05. (D and E) Overexpression of RPL6 inhibits cell growth. A549 (D) and H1299 (E) transfected with pcDNA-3Flag or Flag-RPL6 were cultured in 96-well plates and the MTT assay was performed.
RPL6 regulates p53 in response to ribosomal stress
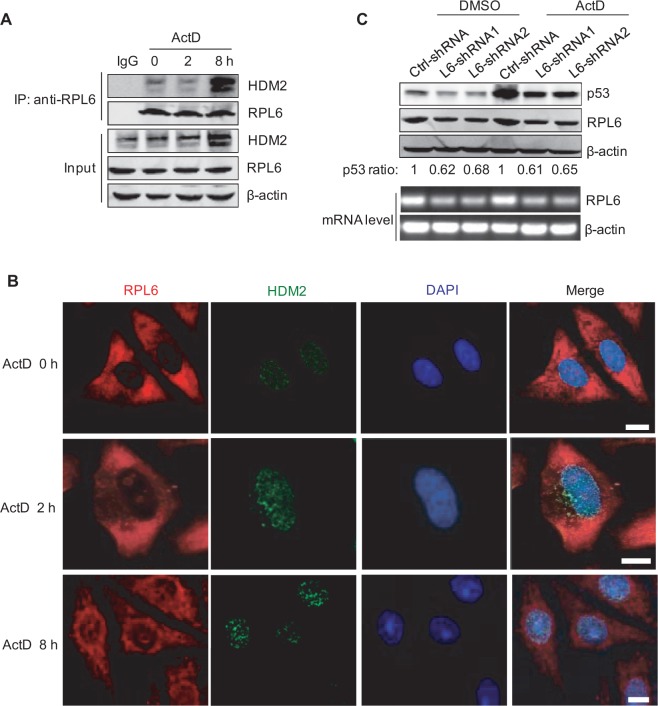
Increasing evidence has shown that the HDM2-p53 circuit could respond to low dose ActD-induced ribosomal stress. As a novel regulator of HDM2-p53 pathway, we wondered whether RPL6 also plays crucial roles in regulating HDM2-p53 loop under stressed condition. To test this idea, HCT116 cells were treated with a low dose of ActD, and the interaction of endogenous RPL6 with HDM2 was investigated by Co-IP. As shown in Figure 5A, the interaction between RPL6 and HDM2 monitored at 2 h after ActD treatment did not increase significantly. However, when the ActD treatment was extended to 8 h, the interaction between these two proteins was dramatically enhanced (Figure 5A). To investigate whether the increased interaction between HDM2 and RPL6 is because of the increased HDM2, HCT116 cells were treated with the proteasome inhibitor MG132 to block the degradation of HDM2, and then the effect of ActD treatment on HDM2-RPL6 interaction was examined. As shown in Supplementary Figure S3A, ActD treatment increased the binding of RPL6 with HDM2, although the amount of HDM2 was maintained at similar level. This result indicates that the enhanced association of RPL6 and HDM2 is not because of the increased level of HDM2 but may result from the relocalization of RP upon ribosomal stress. To validate this point, immunofluorescence assay was performed to detect the localization of RPL6 in response to ribosomal stress. As shown in Figure 5B, RPL6 was absent in the nucleoplasm at 2 h after ActD treatment, however, when the treated duration was extended to 8 h, RPL6 translocated from the nucleolus to the nucleoplasm (Figure 5B). To further illustrate that the relocation of RPL6 contributed to its interaction with HDM2, the location of RPL6 at 4 h after ActD treatment was also detected. As shown in Supplementary Figure S3B, RPL6 has already translocated into the nucleoplasm at 4 h after ActD treatment. Moreover, no significantly increased interaction between RPL6 and HDM2 was observed at this time point (Supplementary Figure S3C), which suggested that the relocation of RPL6 precedes its interaction with HDM2.
Figure 5.
RPL6 is necessary for maintaining the abundance of p53 in response to ribosomal stress. (A) The interaction between RPL6 and HDM2 under ribosomal stress. HCT116 cells were treated with ActD for indicated time. Co-IP was performed to detect the interaction between RPL6 and HDM2. (B) RPL6 translocates from the nucleolus to nucleoplasm in response to ActD treatment. HeLa cells were treated with ActD (5 nM) for indicated time followed by immunofluorescent staining. Scale bar, 10 μm. (C) Depletion of RPL6 decreases ActD-induced p53 abundance. A549 cells were transfected with control-shRNA or RPL6-shRNA. Ninety hours after transfection, cells were incubated with or without ActD (5 nM) for 8 h; the cell lysates were subjected to IB with indicated antibodies. Decreases of p53 level by RPL6 knockdown under homeostatic condition and stress condition were quantified (upper panel). The knockdown efficiency of RPL6 was validated with reverse transcriptase-polymerase chain reaction as well (lower panel).
As a novel regulator of p53, we hypothesized that RPL6 is necessary for maintaining the abundance of p53 in response to ribosomal stress. To test this point, we monitored the level of p53 in cells with depleted RPL6. Downregulation of RPL6 decreased the abundance of p53 similarly in both ActD-untreated and ActD-treated cells, although the fold increase in p53 abundance upon ActD treatment is unaffected by RPL6-shRNA (Figure 5C). These results suggest that RPL6 is required for maintaining p53 abundance under both normal and stressed conditions.
RPL6 is a substrate of HDM2
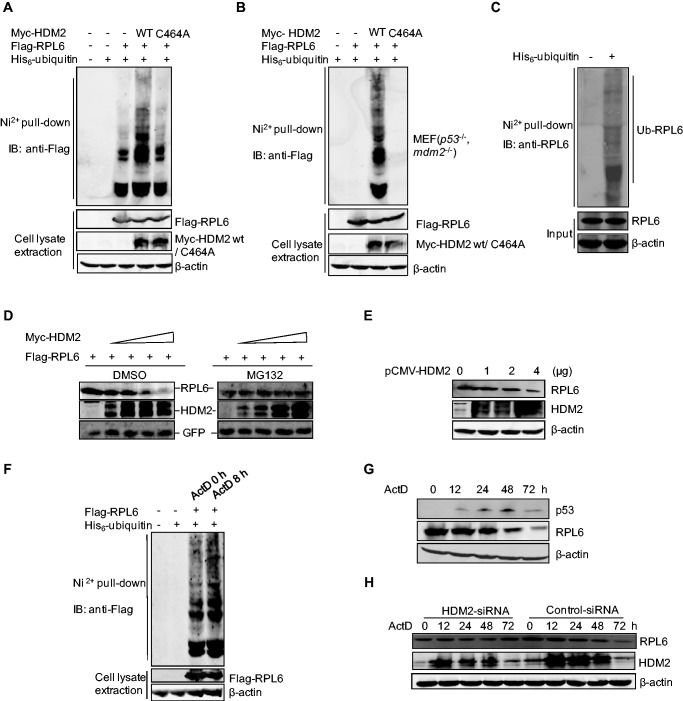
HDM2 is a well-known Really Interesting New Gene (RING)-finger E3 ubiquitin ligase for p53. Based on the strong interaction between RPL6 and HDM2, we accessed whether RPL6 served as an ubquitination substrate of HDM2. HCT116 cells were transfected with plasmids expressing Flag-RPL6, His-ubiquitin together with wild-type HDM2 or mutant HDM2 C464A, and the ubiquitination of RPL6 was investigated by in vivo ubiquitination assay. The result showed that wild-type HDM2 markedly increased RPL6 ubiquitination; in contrast, HDM2 C464A did not show the same effect (Figure 6A). To get further insight into the essential role of HDM2 in RPL6 ubiquitination, in vivo ubiquitination assay was also performed with MEFs cells in the absence of p53 and mdm2 (p53−/−, mdm2−/−). Consistently, no band of RPL6 mono- or polyubiquitination was observed in the absence of MDM2 (Figure 6B). However, overexpression of wild-type HDM2 but not HDM2 C464A greatly rescued RPL6 ubiquitination (Figure 6B). Furthermore, endogenous RPL6 was also ubiquitinated (Figure 6C) and HDM2 played an important role in the ubiquitination of endogenous RPL6 (Supplementary Figure S5A). Collectively, these results demonstrate that RPL6 is a substrate of the E3 ubiquitin ligase HDM2.
Figure 6.
RPL6 is a substrate of the HDM2 E3 ubiquitin ligase. (A and B) Ubiquitination of RPL6 increases in the presence of HDM2. HCT116 cells (A) and p53−/−, mdm2−/− MEFs (B) were transfected with plasmids as indicated for 20 h. Cells were incubated with 30 µM MG132 for 8 h before harvesting. Cell lysates were then subjected to in vivo ubiquitination assay. (C) Endogenous RPL6 is ubiquitinated. HCT116 cells were transfected with His-ubiquitin and in vivo ubiquitination assay was performed to detect the ubiquitination of endogenous RPL6 with anti-RPL6 antibody. (D) HDM2 decreases the abundance of ectopically expressed RPL6 in a dose-dependent manner. HCT116 cells were transfected with increasing amount of Myc-HDM2 (0, 1, 2, 3, 4 µg) and Flag-RPL6 (1 µg) as indicated. Twenty-four hours later, cells were treated with MG132 (30 μM) for 9 h. (E) HDM2 decreases the level of endogenous RPL6. A549 cells were transfected with increasing amount of pCMV-HDM2. Forty-two hours after transfection, cells were harvested and the level of RPL6 was detected with anti-RPL6 antibody. (F) The ubiquitination of RPL6 increases in response to ActD treatment. HCT116 cells were transfected with plasmids as indicated. 24 h after transfection, cells were incubated with ActD (5 nM) for indicated time before harvesting. Cell lysates were subjected to in vivo ubiquitination assay. (G) The abundance of p53 reduces following the decrease of RPL6 after prolonged ActD treatment. U2OS cells were treated with 5 nM ActD for indicated times. Cells were harvested and subjected to IB with anti-p53, anti-RPL6 and anti-β-actin antibodies. (H) Decrease of RPL6 in response to ActD treatment is HDM2-dependent. A549 cells were transfected with Small Interfering RNA (siRNA)-control or siRNA-HDM2 and cells were subjected to ActD treatment for indicated times. The levels of RPL6 and HDM2 were examined by IB with anti-RPL6 and anti-HDM2 antibodies, respectively.
To investigate whether HDM2-mediated RPL6 ubiquitination would lead to its proteasomal dependent degradation, HCT116 cells were transfected with Flag-RPL6 and increasing amounts of Myc-HDM2, and the accumulation of RPL6 with and without the treatment of proteasome inhibitor MG132 was compared. As shown in Figure 6D, the level of RPL6 was gradually reduced following elevated expression of Myc-HDM2 in the absence of MG132, suggesting a rapid degradation of RPL6. On the contrary, MG132 treatment blocked the degradation of RPL6 (Figure 6D, right). To further confirm this conclusion, we detected the endogenous level of RPL6 with the increasing amount of HDM2. As shown in Figure 6E, RPL6 abundance was decreased obviously in the presence of high dose of HDM2, and similar results were obtained in U2OS cells (Supplementary Figure S5B). These data indicate that HDM2-mediated RPL6 polyubiquitination leads to proteasomal dependent degradation of RPL6.
The translocation of RPL6 from the nucleolus to the nucleoplasm under ribosomal stress (Figure 5B) drove us to further explore whether the subcellular localization change of RPL6 favored its HDM2-mediated ubiquitination. An in vivo ubiquitination assay was carried out to detect the ubiquitination of RPL6 under ActD-induced ribosomal stress. The ubiquitination of both exogenous and endogenous RPL6 was increased in cells treated with ActD (Figure 6F and Supplementary Figure S5C). To gain further insight into the dynamic regulation of RPL6 in response to ribosomal stress, U2OS cells were treated with ActD for a longer time, and then the level of RPL6 was investigated. As shown in Figure 6G, the level of RPL6 remained unchanged within 24 h, whereas the level of p53 was dramatically enhanced. However, at 48 h after ActD treatment, RPL6 was decreased. This phenomenon may be because of the ActD-induced translocation of RPL6 to the nucleoplasm, where RPL6 is exposed to HDM2-mediated ubiquitination and proteasomal degradation. To validate this point, immunofluorescent assay was performed to detect the localization of RPL6 at different time points in response to ActD-induced ribosomal stress. We found that the nucleoplasmic relocalization of endogenous RPL6 almost reached its maximum extent at about 8 h after ActD treatment, no more obvious translocation of RPL6 was observed when we extend the ActD treatment time up to 16 h and 24 h (Supplementary Figure S4). The relocalization of RPL6 induced by ActD treatment facilitates its interaction with HDM2. Given that RPL6 is relatively stable and maintained at a high level under normal conditions, a dramatic decrease of RPL6 may not be observed at the early stage of ribosomal stress, instead, when the treatment is long enough, the ubiquitination of RPL6 is accumulated and more RPL6 are subjected to degradation (Supplementary Figure S5C), and thus a significant decrease of RPL6 is detected at 48 h after ActD treatment (Figure 6G). Moreover, the observed decrease in RPL6 protein levels upon ActD treatment is HDM2-dependent, as HDM2 depletion blocked the decrease of RPL6 even after longer treatment of ActD (Figure 6H). As RPL6 is a novel regulator of p53 and downregulation of RPL6 induces the decrease of the p53 abundance, consistently, a significant decrease of p53 was observed at 72 h following ActD treatment, which occurred after the decrease of RPL6 (Figure 6G). Thus, the decrease of RPL6 attenuates its role in regulating the HDM2-p53 axis and finally makes p53 return to the basal level. Although low doses of ActD can specially inhibit RNA polymerase I activity and disrupt ribosomal RNA synthesis, which leads to the perturbation of ribosome biogenesis and protein synthesis; however, we did not observe a significant reduced protein synthesis (Supplementary Figure S5D).
DISCUSSION
As structural components, the functions of RPs in ribosome assembly are well studied. However, more extraribosomal functions of RPs are gradually unveiled during the past decade (8). Among them, the critical role of RPs in activating p53 in the lesion of ribosome biogenesis has drawn wide attention (2,3,19,20,23,25,26,29,37–40). The importance of this finding has been underscored by the evidence that many genetic diseases, such as Treacher–Collins syndrome (41), Dyskeratosis congenital (42) and 5q− syndrome (17,43), are tightly associated with mutations in RPs and elevated p53 level. Given the important involvement of p53 in these diseases caused by the dysfunction of RPs, it is crucial to identify the molecular mechanism that is responsible for the role of RPs in regulating p53.
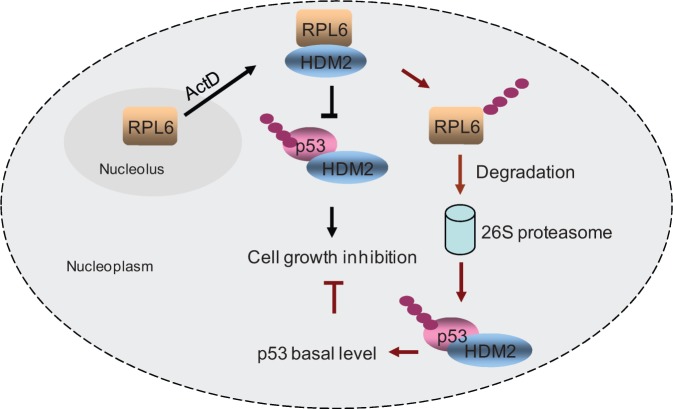
In this study, we established RPL6 as a novel and essential regulator of p53. As summarized in Figure 7, when cells are subjected to ribosomal stress, RPL6 relocates to the nucleoplasm, where it interacts with HDM2 and attenuates HDM2-mediated p53 polyubiquitination and degradation. The enhanced p53 leads to cell growth inhibition in a p53-dependent manner. As a feedback loop, the binding of RPL6 and HDM2 triggers the ubiquitination and instability of RPL6. The reduced level of RPL6 reflects a putative consequence that the inhibition of HDM2 by RPL6 is relieved and makes p53 return to the basal level. Strikingly, RPL26, another large subunit of RP, is capable of regulating p53 via interacting with HDM2 (2,37), but it can also augment p53 through binding to the 5′ Untranslating Region (UTR) of p53 mRNA (44). Whether the regulation of p53 by RPL6 also occurs at the post-transcriptional level deserves further study. Although RPs bind to HDM2 and inhibit p53 ubiquitination, more research about the exact underlying molecular mechanism for this observation is needed. As most of the recognized HDM2-binding RPs bind to the central acid domain of HDM2 but not the C-terminal RING domain that is responsible for the E3 ligase activity of HDM2, RP is unlikely to function through directly inhibiting the E3 ligase activity HDM2. One possibility is that binding of RPs with HDM2 may induce conformational change of HDM2, which makes the interaction between p53 and the RING domain of HDM2 more difficult, thus impairs the normal polyubiquitinaton of p53 by HDM2. To prove this hypothesis, a 3D structure of the RP-HDM2-p53 ternary complex is urgently needed.
Figure 7.
A schematic model for the function of RPL6 in regulating the HDM2-p53 circuit. In response to ribosomal stress, RPL6 relocates into the nucleoplasm, where it interacts with HDM2 and inhibits HDM2-mediated p53 ubiquitination and degradation. Consequently, overexpression of RPL6 leads to p53-dependent cell growth inhibition. In turn, the binding of RPL6 with HDM2 triggers RPL6 ubiquitination and degradation, thus forming an autoregulatory loop. The decreased level of RPL6 makes p53 come back to the basal level.
As a novel and crucial regulator of p53, we find that RPL6 translocates from the nucleolus into nucleoplasm in response to ribosomal stress, where it binds to HDM2 and blocks HDM2-mediated p53 polyubiquitination. These findings are similar to that of RPS7 (3), RPS14 (19,27) and RPL26 (2,37), which help us to realize the essential role of RPs in response to cellular stresses. However, this also raises an interesting question, why mammalian cells need so many RPs to antagonize the negative regulation of HDM2 to p53. One possible explanation is that it may be related with the complex and multiple functions of p53 in maintaining normal physiological dynamic balance under different stimuli. Certain RPs may be required in some cases occurred at different time points, although the underlying mechanism whereby how these RPs are selected and regulated under certain circumstances needs more in-depth research.
Another striking finding here is that RPL6 is a substrate for HDM2 E3 ubiquitin ligase. Previous studies have shown that RPL11 (45,46) and RPS14 (27) can be modified with NEDD8. NEDDylation of RPS14 and RPL11 influences their subcellular localization, stability and activation of p53 in response to ribosomal stress (45,46). In the case of RPL6, we observed a strong ubiquitination of RPL6 by HDM2, which ultimately leads to its proteasome-dependent degradation. The HDM2-mediated decreasing of RPL6 provides a new mechanism by which the decreased RPL6 allows p53 to return to the basal state and terminates the response of p53 to ribosomal stress. Given that not all the RPs involved in HDM2-p53 loop are ubiquitinated, it may reflect a regulated rather than a passive event. Wan et al., (47) reported that phosphorylation of RPS3 mediates its nuclear import when cells are subjected to TNFα stimuli. Nevertheless, it remains to be seen whether these post-translational modifications, such as ubiquitination, phosphorylation or NEDDylation, can also occur in other RPs. If so, what are the physiological functions of these modifications, whether the modification can influence the interaction between RP and HDM2. These are amusing questions for future research.
In this study, we found that overexpression of RPL6 induces cell growth inhibition in p53-positive cells. Similarly, other RPs including RPS7, RPS14, RPL26 and RPL11 all activate p53 and induce cell growth inhibition in the presence of p53 (2,3,7,19). A recent study by Wu et al., (48) reported that knockdown of RPL6 inhibits cell growth in human gastric cancer cell SGC7901. This observation seemingly contradict with our putative observation that downregulation of RPL6 may promote cell growth. One possible explanation is that the cell line SGC7901 harbors mutation in p53, thus the growth effect caused by RPL6 in this cell line is not through p53 but may be via other mechanism, i.e. partly though regulating Cyclin E, instead of p53 (33). And in the current study, the inhibitory effect of RPL6 on cell growth in p53-null HCT116 cells indicates that p53-independent mechanisms might also be involved. Thus, RPL6 regulates cell growth through different mechanisms in different cells, either in a p53-mediated manner or via p53-independent regulating mechanisms.
HDM2 is amplified in many types of tumors including breast cancer, sarcoma, glioma and blood cancers (8,49). HDM2 has become a novel target for cancer therapy (50–52). In this study, we identify that RPL6 binds to HDM2 and stabilizes HDM2. High expression of RPL6 has been identified in human cancers (33,34), yet it remains to be seen whether the interaction between RPL6 and HDM2 has changed in tumorigenesis. Studies designed to explore these problems will provide more evidence for the application of RPL6 in cancer therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors sincerely thank Dr Jian Wang from the State Key Laboratory of Proteomics, Beijing Proteome Research Center for providing plasmids containing HDM2 and its truncation constructs, and Dr Liangqiang Zhang for the gift of HA-MDMX plasmid.
FUNDING
National Science Foundation of China [31170709, 30930020]; National High Technology and Development Program of China [2010CB911804] and the International Centre for Genetic Engineering and Biotechnology [CRP/CHN09-01]. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding for open access charge: National Science Foundation of China [31170709].
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, Jin C, Chen H, Zhang L, Yang X, et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 2010;38:6544–6554. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 6.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 7.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell. Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montanaro L, Trere D, Derenzini M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shenoy N, Kessel R, Bhagat T, Bhattacharya S, Yu Y, McMahon C, Verma A. Alterations in the ribosomal machinery in cancer and hematologic disorders. J. Hematol. Oncol. 2012;5:32. doi: 10.1186/1756-8722-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito E, Konno Y, Toki T, Terui K. Molecular pathogenesis in Diamond–Blackfan anemia. Int. J. Hematol. 2010;92:413–418. doi: 10.1007/s12185-010-0693-7. [DOI] [PubMed] [Google Scholar]
- 15.Boria I, Garelli E, Gazda HT, Aspesi A, Quarello P, Pavesi E, Ferrante D, Meerpohl JJ, Kartal M, Da Costa L, et al. The ribosomal basis of diamond-blackfan anemia: mutation and database update. Hum. Mutat. 2010;31:1269–1279. doi: 10.1002/humu.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barlow JL, Drynan LF, Trim NL, Erber WN, Warren AJ, McKenzie AN. New insights into 5q- syndrome as a ribosomopathy. Cell Cycle. 2010;9:4286–4293. doi: 10.4161/cc.9.21.13742. [DOI] [PubMed] [Google Scholar]
- 17.Boultwood J, Pellagatti A, McKenzie AN, Wainscoat JS. Advances in the 5q- syndrome. Blood. 2010;116:5803–5811. doi: 10.1182/blood-2010-04-273771. [DOI] [PubMed] [Google Scholar]
- 18.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2012;32:388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget. 2011;2:234–238. doi: 10.18632/oncotarget.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 23.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, Itoh B, Wang J, Komatsu Y, Yang YR, et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat. Med. 2011;17:944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Bai D, Ma X, Guan J, Zheng X. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene. 2012;33:246–254. doi: 10.1038/onc.2012.560. [DOI] [PubMed] [Google Scholar]
- 28.Lindstrom MS, Deisenroth C, Zhang Y. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle. 2007;6:434–437. doi: 10.4161/cc.6.4.3861. [DOI] [PubMed] [Google Scholar]
- 29.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenmochi N, Yoshihama M, Higa S, Tanaka T. The human ribosomal protein L6 gene in a critical region for Noonan syndrome. J. Hum. Genet. 2000;45:290–293. doi: 10.1007/s100380070018. [DOI] [PubMed] [Google Scholar]
- 32.Ion A, Crosby AH, Kremer H, Kenmochi N, Van Reen M, Fenske C, Van Der Burgt I, Brunner HG, Montgomery K, Kucherlapati RS, et al. Detailed mapping, mutation analysis, and intragenic polymorphism identification in candidate Noonan syndrome genes MYL2, DCN, EPS8, and RPL6. J. Med. Genet. 2000;37:884–886. doi: 10.1136/jmg.37.11.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gou Y, Shi Y, Zhang Y, Nie Y, Wang J, Song J, Jin H, He L, Gao L, Qiao L, et al. Ribosomal protein L6 promotes growth and cell cycle progression through upregulating cyclin E in gastric cancer cells. Biochem. Biophys. Res. Commun. 2010;393:788–793. doi: 10.1016/j.bbrc.2010.02.083. [DOI] [PubMed] [Google Scholar]
- 34.Du JP, Jin XH, Shi YQ, Cao YX, Zhao YQ, Liu CJ, Yin F, Hu WH, Chen BJ, Qiao TD, et al. [Differential expression of RPL6/Taxreb107 in drug resistant gastric cancer cell line SGC7901/ADR and its correlation with multiple-drug resistance] Zhonghua Zhong Liu Za Zhi. 2003;25:21–25. [PubMed] [Google Scholar]
- 35.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilkes DM, Chen J. Distinct roles of MDMX in the regulation of p53 response to ribosomal stress. Cell Cycle. 2007;6:151–155. doi: 10.4161/cc.6.2.3719. [DOI] [PubMed] [Google Scholar]
- 37.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- 39.Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, Lu H. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J. Biol. Chem. 2006;281:24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 43.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 45.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahata B, Sundqvist A, Xirodimas DP. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2011;31:3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- 47.Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 2011;12:335–343. doi: 10.1038/ni.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Q, Gou Y, Wang Q, Jin H, Cui L, Zhang Y, He L, Wang J, Nie Y, Shi Y, et al. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS One. 2011;6:e26401. doi: 10.1371/journal.pone.0026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol. Cancer Res. 2004;2:1–8. [PubMed] [Google Scholar]
- 50.Ooi MG, Hayden PJ, Kotoula V, McMillin DW, Charalambous E, Daskalaki E, Raje NS, Munshi NC, Chauhan D, Hideshima T, et al. Interactions of the Hdm2/p53 and proteasome pathways may enhance the antitumor activity of bortezomib. Clin. Cancer Res. 2009;15:7153–7160. doi: 10.1158/1078-0432.CCR-09-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azmi AS, Beck FW, Sarkar FH, Mohammad RM. Network perspectives on HDM2 inhibitor chemotherapy combinations. Curr. Pharm. Des. 2011;17:640–652. doi: 10.2174/138161211795222612. [DOI] [PubMed] [Google Scholar]
- 52.Patel S, Player MR. Small-molecule inhibitors of the p53-HDM2 interaction for the treatment of cancer. Expert Opin. Investig. Drugs. 2008;17:1865–1882. doi: 10.1517/13543780802493366. [DOI] [PubMed] [Google Scholar]