SUMMARY
The human IgG 2G12 recognizes high-mannose carbohydrates on the HIV-1 envelope glycoprotein gp120. Its two antigen-binding fragments (Fabs) are intramolecularly domain exchanged, resulting in a rigid (Fab)2 unit including a third antigen-binding interface not found in antibodies with flexible Fab arms. We determined crystal structures of dimeric 2G12 IgG created by intermolecular domain exchange, which exhibits increased breadth and >50-fold increased neutralization potency compared with monomeric 2G12. The four Fab and two Fc regions of dimeric 2G12 were localized at low resolution in two independent structures, revealing IgG dimers with two (Fab)2 arms analogous to the Fabs of conventional monomeric IgGs. Structures revealed three conformationally-distinct dimers, demonstrating flexibility of the (Fab)2–Fc connections that was confirmed by electron microscopy, small-angle X-ray scattering, and binding studies. We conclude that intermolecular domain exchange, flexibility, and bivalent binding to allow avidity effects are responsible for the increased potency and breadth of dimeric 2G12.
INTRODUCTION
Difficulties in generating broadly neutralizing antibodies against human immunodeficiency virus type 1 (HIV-1) lie in structural features of the gp120-gp41 envelope spike trimer (Bartesaghi et al., 2013; Julien et al., 2013; Lyumkis et al., 2013). Briefly, the spike’s variable loops are highly susceptible to rapid mutation (Starcich et al., 1986), its few conserved regions are often sterically occluded via conformational masking (Kwong et al., 2002), and a host-derived glycan shield covers much of the spike surface, making gp120 one of the most heavily glycosylated proteins in nature (Poignard et al., 2001). As such, surface carbohydrates contribute to roughly 50% of gp120’s molecular weight (Botos and Wlodawer, 2005). Despite the fact that most antibodies elicited against HIV-1 are strain-specific, there exists a small set of broadly neutralizing antibodies that have demonstrated efficacy across strains (Kwong and Mascola, 2012; Mascola and Haynes, 2013). Isolated from the blood of infected individuals, these antibodies have been found to target conserved epitopes on either the gp120 or gp41 subunits of the envelope spike protein.
Human monoclonal antibody 2G12 recognizes clusters of N-linked high-mannose (Man8-9GlcNAc2) glycans on the surface of gp120 (Calarese et al., 2005; Sanders et al., 2002; Scanlan et al., 2002) and effectively neutralizes many Clade B and some Clade A strains of HIV-1 (Trkola et al., 1996). 2G12’s ability to target glycan clusters is facilitated by three-dimensional domain swapping (Liu and Eisenberg, 2002) of its heavy chains to form a rigid (Fab)2 antigen-binding unit (Calarese et al., 2003). By contrast, two independent antigen-binding units are found in conventional IgGs, which contain two heavy chains and two light chains arranged into two fragment antigen-binding (Fab) regions and a single fragment crystalline (Fc) region (Figure 1A). In a typical Fab, the two domains of the light chain (the variable light (VL) and constant light (CL) domains) pair with the variable heavy (VH) and constant heavy 1 (CH1) domains, respectively, of a single heavy chain. In the domain-swapped (Fab)2 unit of monomeric 2G12, each light chain is associated with both heavy chains, such that the CL domain of a light chain contacts CH1 from one heavy chain, and the VL domain from that light chain contacts the VH domain from the second heavy chain. The result is a single rigid (Fab)2 unit that contains a third antigen binding site at the VH/VH′ interface in addition to the two primary antigen binding sites formed by the VH/VL interfaces (Figure 1B). The unique VH/VH′ site allows for increased glycan contacts at the binding surface of 2G12, allowing it to target densely clustered carbohydrates that might be inaccessible to conventional IgG antibodies (Calarese et al., 2003).
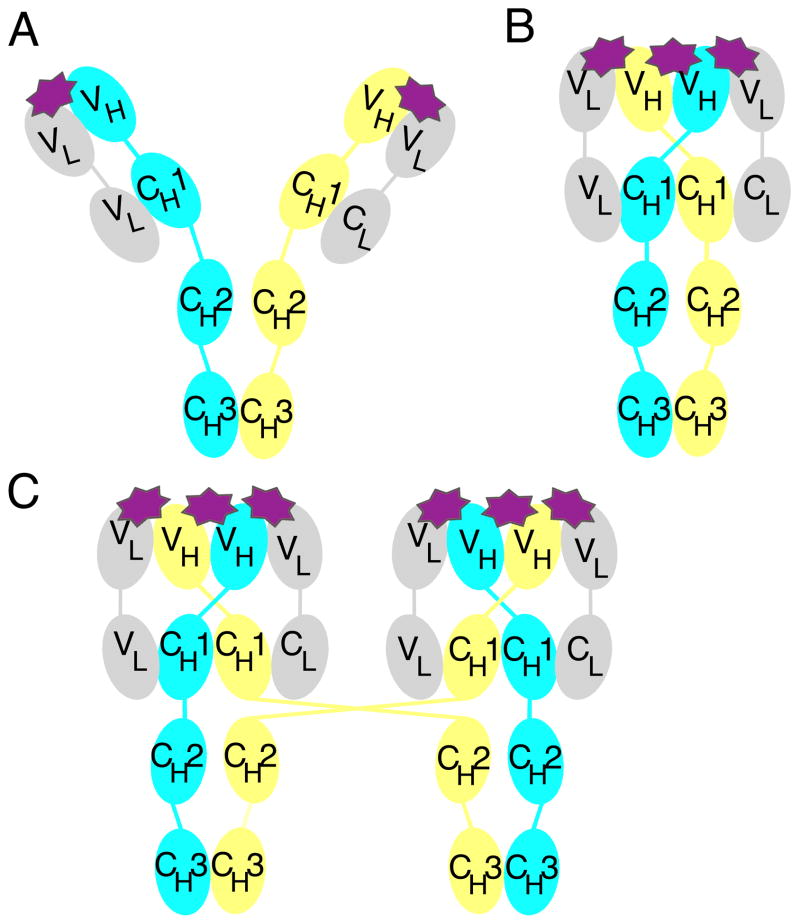
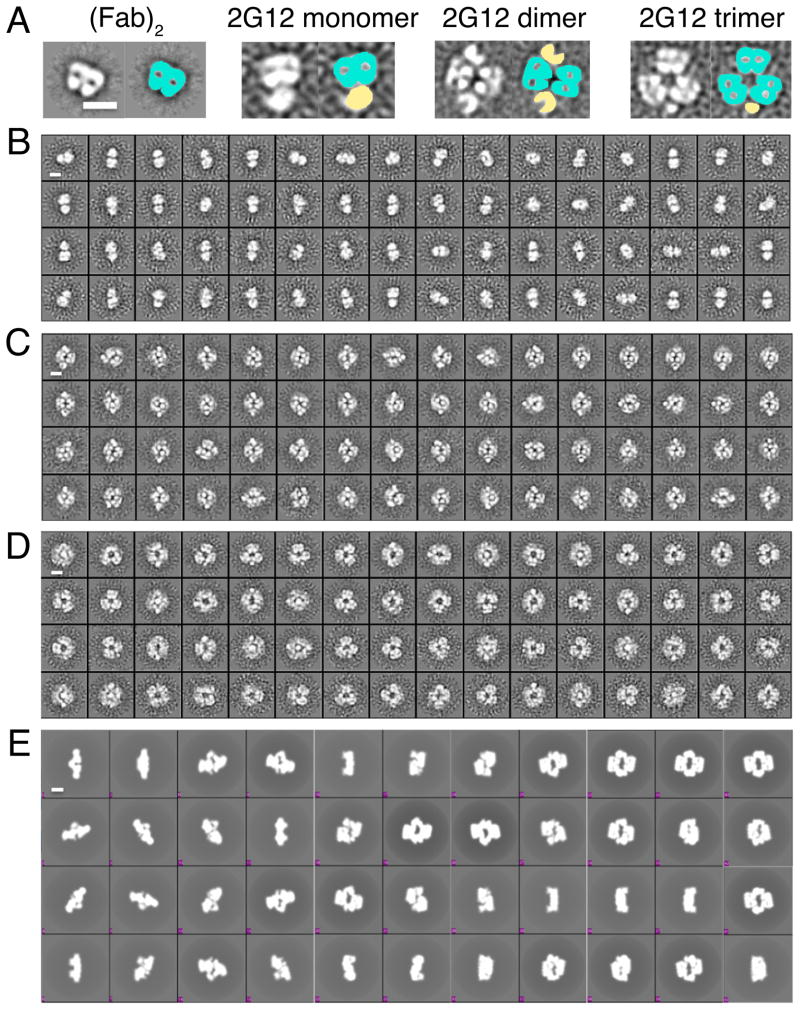
Figure 1. Schematic IgG structures.
Heavy chains (identical polypeptide chains, but colored differently for illustrating domain swapping) are cyan and yellow, light chains are gray, and antigen binding sites are shown as starbursts. (A) Conventional IgG with flexible Fab arms and two antigen binding sites at the VH/VL interfaces. (B) 2G12 monomer with domain-swapped (Fab)2 unit and an additional antigen binding site at the VH/VH′ interface. (C) 2G12 dimer with two domain-swapped (Fab)2 units and six antigen binding sites. See also Figure S1.
During purification of recombinant 2G12, we previously found that 2G12 formed monomers (two Fabs and one Fc) plus a small fraction of a higher molecular weight species, which was 50- to 80-fold more potent than 2G12 monomer in neutralization of clade A and B HIV-1 (West et al., 2009). This species, identified as 2G12 dimer (four Fabs, two Fcs), was proposed to form through intermolecular domain swapping rather than the intramolecular domain swapping that results in 2G12 monomer (Figure 1C; Figure S1A). Both monomer and dimer were produced during expression in mammalian cells and could be separated by size-exclusion chromatography into purified samples that did not interconvert upon storage or concentration.
Because 2G12 dimer is extremely potent and some 2G12 monomer-resistant HIV strains were neutralized by dimer, increasing the proportion of 2G12 dimer could lead to a more effective reagent for gene therapy or passive immunization. Indeed, introduction of a higher dimer/monomer ratio 2G12 cell line (West et al., 2009) into HIV-1-infected humanized mice increased protection against infection compared with the wild-type 2G12-expressing cell line, and injection of 2G12 dimer, but not monomer, controlled HIV-1 infection in mouse models (Luo et al., 2010). In addition to its increased capacity for antigen recognition, 2G12 dimer also has the potential for protection via effector functions because it can mediate antibody-dependent cellular cytotoxicity (ADCC) in vitro (Klein et al., 2010), indicating that it retains binding to the CD16 Fc receptor on natural killer cells despite its unusual structure.
To investigate the structural and mechanistic basis of the increased potency of 2G12 dimer compared with the monomer, we solved two independent, low-resolution structures of 2G12 dimer by X-ray crystallography. We performed several structural validations to confirm the 2G12 dimer structures. Collectively the structures revealed three conformationally-distinct forms of the dimer, suggesting that the two (Fab)2 units can adopt different positions relative to the Fcs, analogous to the flexibility of the two Fab arms of a conventional IgG. Consistent with the crystal structures, electron microscopy and small-angle X-ray scattering studies confirmed the flexible nature of 2G12 dimer. Additionally, we showed that 2G12 dimer, but not 2G12 monomer, could bind bivalently to immobilized gp120 in a biosensor assay and confirmed that both Fc regions in the 2G12 dimer were accessible to an Fc receptor using binding and stoichiometry measurements. Our results provide a structural explanation for the superior neutralization potency of 2G12 dimer compared with monomer (West et al., 2009) and rationalize the dimer’s ability to mediate Fc-mediated effector functions (Klein et al., 2010).
RESULTS
Crystallization and structure determination of 2G12 dimer
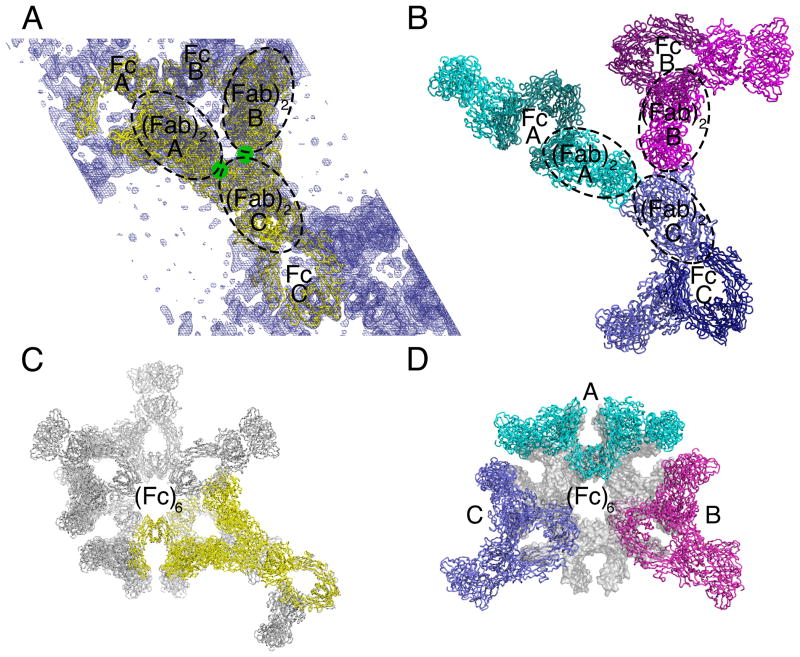
Structure determinations of intact antibodies are inherently limited by flexibility between domains, and in the case of 2G12, the existence of multiple oligomeric states. Despite these challenges, we were able to readily obtain crystals of intact purified 2G12 dimer. The best crystals (space group P6122) diffracted to only 7.4 Å (Table S1) despite optimizing crystallization conditions and screening >500 crystals. We obtained preliminary phases using molecular replacement with the 2G12 (Fab)2 (pdb entry 1OP3) and IgG Fc (pdb entry 1H3X) structures as search models and verified the solution using heavy atom derivative data (Figure 2; Figure S2, Table S1, Supplemental Experimental Procedures). Three 2G12 (Fab)2 units were initially located in the crystallographic asymmetric unit (Figure 2A). The Fc regions were found only in molecular replacement searches involving a fixed partial solution including the (Fab)2 units. Crystallographic R values after rigid body and B-factor domain refinement decreased from 0.50 to 0.37 after placing the Fc regions. The final model at 8.0 Å resolution (Rwork = 0.35; Rfree = 0.37) (Table S1) contained three (Fab)2 units and three Fc regions representing three separate half-dimers (Figure 2A). Applying crystallographic two-fold symmetry operations generated three physiological 2G12 dimers, each with two (Fab)2 units and two Fc regions (Figure 2B). The (Fab)2 units of the 2G12 dimers contacted each other at their antigen binding sites (Figure 2A). They were flanked by pairs of Fc regions that formed a hexamer via a six-fold non-crystallographic symmetry (NCS) axis coincident with a crystallographic 61 screw axis (Figure 2C). The Fc regions forming the hexamers contacted each other at the hinge between the CH2 and CH3 domains, the so-called “hot spot” on IgG Fc for interactions with receptors and other proteins (DeLano et al., 2000).
Figure 2. Packing in 2G12 dimer crystals.
(A) Asymmetric unit of a solvent flattened 8.0 Å resolution 2Fo−Fc electron density map contoured at 1.5 σ. The asymmetric unit contained three half-dimers; i.e., three Fc regions and three (Fab)2 units from three distinct 2G12 dimers (A, B, and C). Crystal contacts involved the antigen binding sites in the (Fab)2 units (green circles), see also Figure S2. (B) Application of symmetry operators to generate the second half of each 2G12 dimer. Dimer A (cyan) was structurally distinct from Dimers B (magenta) and C (indigo), which exhibited the same conformation. (C) Non-crystallographic six-fold symmetry axis showing a hexamer of Fc regions coincident with the 61 screw axis along the crystallographic c axis. The asymmetric unit is highlighted in yellow. (D) The three dimers in one layer of the crystal, represented by the cyan, magenta, and indigo dimers A, B, and C (panel B), as they fit into the Fc hexamer ring in panel C, see also Figure S2.
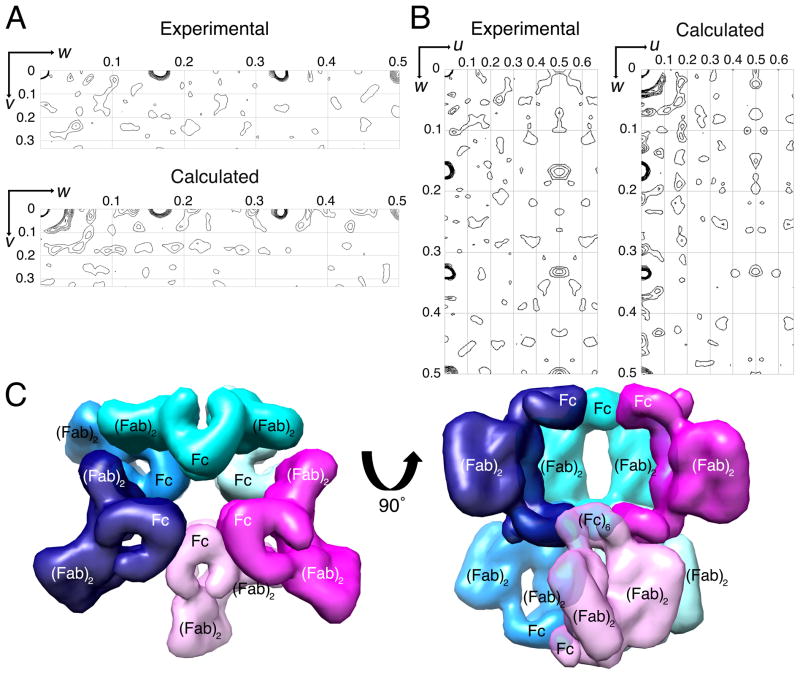
The placement of the (Fab)2 and Fc regions in the P6122 unit cell was dependent upon NCS relating the three half-dimers in the crystallographic asymmetric unit, which we sought to validate to confirm model accuracy. A self-rotation function yielded no peaks for NCS (data not shown). However, NCS axes that are parallel to crystallographic axes of the same or higher symmetry are not detected in self-rotation functions because their peaks coincide with peaks corresponding to crystallographic axes (Rupp, 2010). An NCS axis parallel to a crystallographic can be detected as a peak in a native Patterson map (Rupp, 2010). A native Patterson calculated using the 8 Å P6122 data showed peaks at positions 1/6, 1/3, and 1/2 along the w axis (Figure 3A,B). These peaks resulted from the six-fold rotational NCS axis formed by the hexamer of Fc regions along the crystallographic 61 screw axis (Figure 3C). Structure factors calculated from the model of 2G12 half-dimers in the P6 122 asymmetric unit reproduced these peaks in a native Patterson calculation (Figure 3A,B), validating the placement of the (Fab)2 and Fc regions in the low-resolution 2G12 dimer structure.
Figure 3. Native Patterson validation of packing in 2G12 dimer crystals.
(A,B) Native Patterson maps. Comparison of experimental and calculated Harker sections. (C) Two layers of 2G12 dimer molecules as they are packed into the P6122 crystals. Left: top-down view showing the cyan (dimer A), magenta (dimer B), and indigo (dimer C) 2G12 dimers (Figure 2) in the same layer. The magenta and indigo dimers are conformationally identical and the cyan dimer is distinct. The layer below contains three additional dimers (light green, light pink, and light blue), formed via a rotation and translation along the crystallographic 61 axis. Symmetry mates are indicated as a darker and lighter version of the same color. Right: a 90° rotation showing hexamer of Fc regions in the middle layer. See also Figure S3.
Overall structure of 2G12 dimer
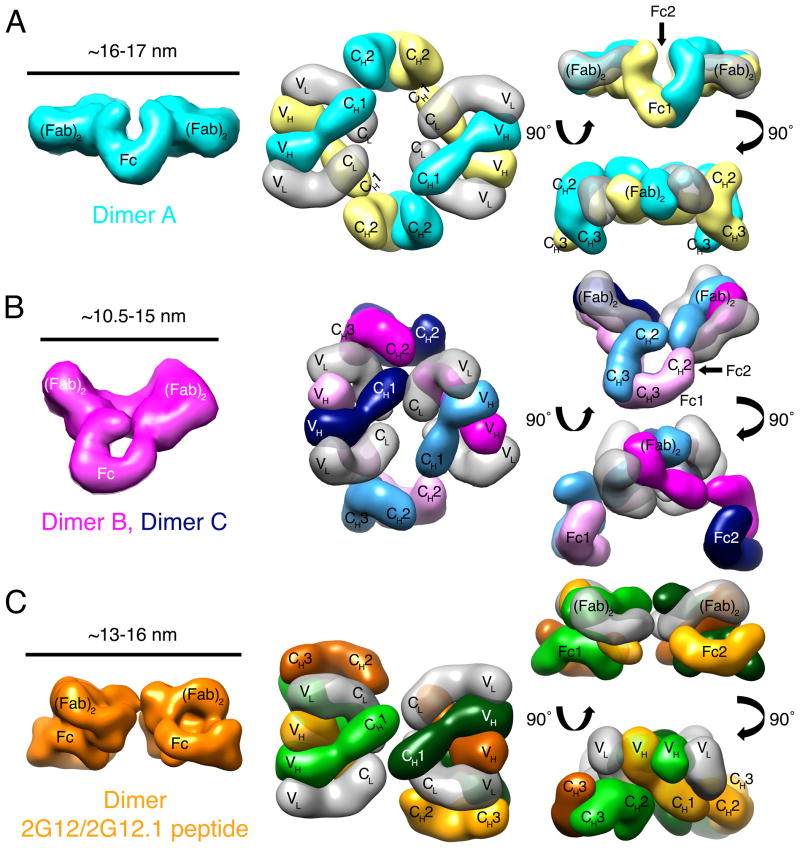
Two of the 2G12 dimers (dimers B and C) in the P6122 crystals were conformationally identical, whereas the third dimer (dimer A) was distinct (Figure 4A,B; Figure S4A,B). The (Fab)2 arms of dimers B and C were related by an angle of ~100°, similar to a typical angle between a pair of Fab arms in a conventional, non-domain-swapped IgG antibody (Roux, 1999), and the (Fab)2 arms of dimer A were related by ~180° (Figure 4A,B). The difference in relative positions of the (Fab)2 arms is consistent with flexibility between the two (Fab)2 portions of the 2G12 dimer structure, in contrast to the inflexibility of the single rigid (Fab)2 of 2G12 monomer. The span between the combining sites of the two (Fab)2 arms of 2G12 dimers, ranging from 105 to 170 Å, was similar to the combining sites of the two Fab arms of a conventional IgG, which are separated by ~150 Å (Klein and Bjorkman, 2010). These findings suggest that 2G12 dimer exhibits enhanced neutralization potency due to its ability to bind HIV spikes bivalently with flexible (Fab)2 arms, as compared with 2G12 monomer, which binds monovalently using a single (Fab)2 arm.
Figure 4. Structures of the three distinct 2G12 dimers from two independent crystal structures.
Dimer structures are shown as 25 Å electron density envelopes calculated from (Fab)2 and Fc coordinates located by molecular replacement. The approximate distance between the tips of the (Fab)2 units of each dimer is shown on the left. (A) Dimer A, one of two conformationally-distinct 2G12 dimers in the P6122 2G12 dimer structure. Middle and Right: Heavy chains are cyan and yellow, light chains are transparent gray. Middle: Top-down view of the 2G12 dimer with Fc regions pointing into the page. Right: Two rotated views showing the large distance between (Fab)2 units and Fc regions and an angle of 180° between (Fab)2 arms. (B) Dimers B and C, the second of the two distinct 2G12 dimers in the P6122 2G12 dimer structure. Middle and Right: Heavy chains are magenta, indigo, light pink, or light blue; light chains are transparent gray. Middle: Top-down view of the 2G12 dimer molecule with Fc regions pointing into the page. Right: Two rotated views showing an obtuse angle between (Fab)2 units. (C) 2G12 dimer in crystals of 2G12 bound to the 2G12.1 peptide. Middle and Right: Heavy chains are orange, green, light orange, or light green; light chains are light gray. Middle: Top-down view. The two (Fab)2 units are extended at 180° with respect to each other, as are the two Fc regions (not seen in this view). Right: Two rotated views showing the Fc regions under the (Fab)2 units. See also Figure S4.
Our data also support a novel mechanism for creating IgG dimers via 3-D domain swapping. In this model, each Fc is a hybrid, with each chain derived from a different monomer (Figure S1A). The heavy chain portions of each (Fab)2 unit in the 2G12 dimer each pair with the CH2-CH3 region (i.e., the Fc) extending from a neighboring (Fab)2. As in other intact IgG structures (Guddat et al., 1993; Harris et al., 1997; Harris et al., 1998; Kratzin et al., 1989; Saphire et al., 2001), the hinge regions of the 2G12 dimers were not resolved in the electron density maps. However, the orientation of the Fc regions in the three 2G12 dimers supported the connectivity model. The model was also consistent with the propensity of 2G12 to form higher order oligomers, including trimers (Figure S1B), which were evident in size-exclusion chromatography profiles of 2G12 (Figure S1C).
Crystallization and structure determination of 2G12 dimer/2G12.1 peptide complex
Packing in the P6122 2G12 dimer crystals involved contacts between the antigen binding sites of adjacent (Fab)2 units (Figure 2A). We reasoned that disrupting these contacts could allow 2G12 dimer to crystallize in a different packing arrangement that might diffract to higher resolution. To that end, we crystallized 2G12 dimer in the presence of 2G12.1, a 21-residue synthetic inhibitor peptide that binds to the combining site of 2G12 (Fab)2 (Menendez et al., 2008). We solved the 2G12 dimer/2G12.1 peptide complex structure in space group P6 to 6.5 Å using molecular replacement (Table S1). The angle between the (Fab)2 units and Fcs in the 2G12 dimer in this crystal form (Figure 4C) was different from either dimer A or dimer B in the P6122 2G12 dimer crystals (Figure 4A,B), thus molecular replacement worked only when we used separate (Fab)2 and Fc search models, resulting in an R-value decrease from 0.45 to 0.40 after the Fc region was correctly introduced into the model. Although low resolution precluded visualization of the 2G12.1 peptide in the antigen binding site, introduction of the peptide altered the packing such that there were no crystal contacts involving the (Fab)2 antigen binding site (Figure S3A). However, the Fc regions of 2G12 dimer were arranged as hexamers in the 2G12/2G12.1 crystals (Figure S3B), as also found in the packing of the 2G12 dimer crystals (Figure 2C; Figure 3C).
It was initially difficult to determine the connectivity between the (Fab)2 and Fc regions of the 2G12 dimer in the 2G12/2G12.1 structure because the Fc regions were arranged almost parallel to the (Fab)2 units (Figure 4C). The (Fab)2–Fc connections in the 2G12 dimer/2G12.1 complex structure were interpreted by assigning probable connectivity between the N-termini of the four heavy chains of the two Fc regions and the C-termini of the four heavy chains of the two (Fab)2 units. Using the shortest combinations of distances as the most plausible choice for connectivity between the chains (see Supplemental Results), we derived the same hybrid Fc model of intermolecular domain exchange as deduced for the formation of the 2G12 IgG dimer (Figure S1A). Assuming this connectivity, the 2G12 dimer in the 2G12/2G12.1 crystal structure adopted a conformation distinct from the two found in the 2G12 dimer alone structure: The Fc regions were oriented at 180° with respect to each other and were essentially parallel to the (Fab)2 units, which were also splayed ~180° apart (Figure 4C), similar to dimer A from the P6122 structure (Figure 4A). Comparison of the three 2G12 dimer conformations obtained from the 2G12 dimer and 2G12 dimer/2G12.1 complex structures demonstrated a large range of conformations accessible to the 2G12 dimer (Figure 4; Figure S4).
Electron microscopy (EM) studies of 2G12 dimer and higher-order oligomers
We studied the different fractions eluted from an SEC purification (Figure S1C) that corresponded in apparent molecular weights to 2G12 monomer, dimer, and trimer by negative stain EM and reference-free 2D classification. Reference-free 2D classes for each of the 2G12 species, including isolated 2G12 (Fab)2, were calculated (Figure 5). In many of the classes for 2G12 dimer and higher-order oligomers, the 2G12 (Fab)2 was discernable and the Fc less clear. The classes corresponding to the dimer and trimer were consistent with an intermolecular domain-swapping model as deduced from the crystal structures. The lack of well-defined class averages for 2G12 IgGs in comparison to a 2G12 (Fab)2 alone (Figure 5A) suggested that 2G12 IgG monomers, dimers, and trimers are flexible structures (Figure 5B–D). In the case of 2G12 monomer, the flexibility presumably resulted solely from the connection between the Fc and the rigid (Fab)2 unit, whereas the 2G12 multimers could adopt different positions of their (Fab)2 units with respect to their Fc regions and each other. Attempts to generate 3-D reconstructions of these particles produced maps that were difficult to interpret, also indicative of conformational heterogeneity. To assess the similarity of the 2G12 IgG dimer models analyzed via crystallography and negative stain EM, the crystal structure of dimer A (Figure 2B; Figure 3A) was filtered to 25 Å and back projected at a 20° Euler angle sampling (Figure 5E). The back projections of 2G12 dimer A showing different orientations of the two (Fab)2 pairs with respect to the Fcs consistently matched the independently generated EM class averages of the 2G12 dimer. Small-angle X-ray scattering (SAXS) data collected on the 2G12 IgG dimer were also consistent with the dimensions and flexibility observed by EM (Figure 5; Figure S5, Supplemental Experimental Procedures).
Figure 5. Negative stain EM of 2G12 oligomers.
Representative two-dimensional class averages (left) and false-colored images (right) of 2G12 (Fab)2, 2G12 monomer, 2G12 dimer, and 2G12 trimer showing the orientations of the domain-exchanged (Fab)2 units and the Fc regions. Fabs are colored in cyan and Fcs in yellow. Bar = 10 nm. (B–D) 2D class averages of 2G12 oligomers analyzed by negative stain EM. Bar = 10 nm. Panels of 64 class averages of the (B) 2G12 monomer, (C) 2G12 dimer, and (D) 2G12 trimer generated using reference-free Xmipp Clustering 2D alignment. (E) Back projections sampled at 20 Euler angles of the low-pass filtered 2G12 Dimer A crystal structure. Bar = 10 nm. Many of the back projections resembled the reference-free class averages of the 2G12 IgG dimer in panel C. See also Figure S5 for SAXS data supporting the EM and crystallographic results.
2G12 dimer binds bivalently to antigens and has fully accessible Fc regions
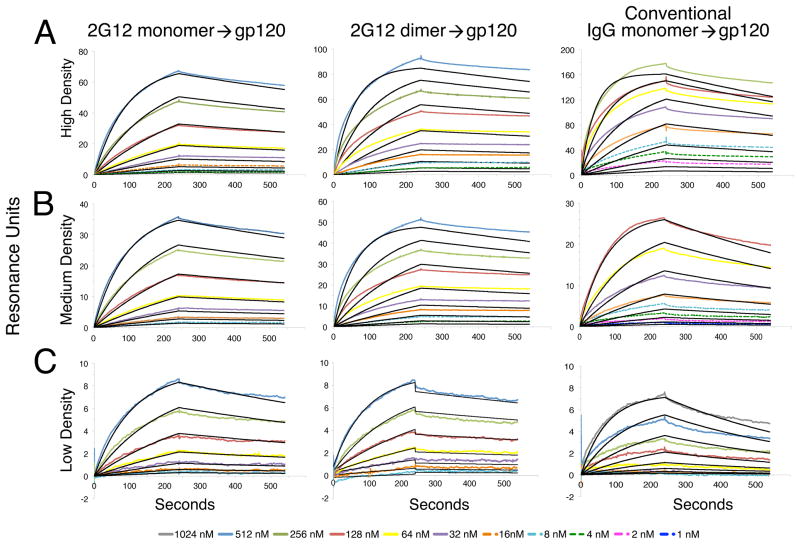
The crystal structures, single particle EM images and SAXS analyses of 2G12 dimer suggested that the (Fab)2 units of the IgG dimer are flexible with respect to the Fc region, analogous to the two Fab arms of a conventional IgG antibody. If so, then like a conventional IgG, 2G12 dimer should be capable of bivalent binding to a tethered antigen. By contrast, the intramolecular domain exchange in 2G12 monomer results in a rigid structure that does not have two independently movable Fab arms for bivalent binding (Figure 1B). Rather, the (Fab)2 unit behaves as a single unit using its conventional combining sites at the VH/VL interfaces plus the site created by the domain-swapped VH/VH′ region to bind to clusters of glycans (Calarese et al., 2003) and should therefore exhibit apparently monovalent binding. To verify these predictions, we compared the binding of 2G12 dimer, 2G12 monomer and conventional anti-gp120 IgGs to a clade B gp120 using a surface plasmon resonance (SPR) binding assay. 2G12 monomer, 2G12 dimer or monomeric anti-gp120 IgGs (NIH45-46G54W and 2909) (Diskin et al., 2011; Gorny et al., 2005) were injected over a gp120 protein from the clade B SF162 strain that was immobilized on the surface of a biosensor chip. The gp120 protein was coupled at high and medium coupling densities to permit bivalent binding and at a low density to reveal the potential loss of bivalent binding when the (Fab)2 arms of 2G12 dimer or the Fab arms of the conventional anti-HIV-1 IgGs could not cross-link between two gp120 molecules. To distinguish between monovalent and bivalent binding, we fit the sensorgram data to a 1:1 binding model and assessed the residuals for the goodness of fit (Figure 6; Figure S6). We found that 2G12 monomer exhibited apparently monovalent binding to gp120 at all coupling densities (Figure 6A–C). As predicted, 2G12 dimer exhibited apparently bivalent binding to gp120 at high and medium coupling densities (Figure 6A,B), but monovalent binding at low coupling density (Figure 6C). Conventional anti-gp120 IgGs bound with apparent bivalency at high coupling density but monovalently at medium and low densities, suggesting a smaller reach between combining sites than 2G12 dimer (Figure 6A–C).
Figure 6. Surface plasmon resonance-based binding studies.
Sensorgrams were derived from binding experiments in which 2G12 monomer, 2G12 dimer or a conventional monomeric anti-gp120 IgG was injected over immobilized gp120 from HIV-1 strain SF162. Experimental data (colored lines) were fit to a 1:1 binding model (black lines). Residual plots are shown in Figure S6. (A) 2G12 monomer, 2G12 dimer, and a conventional anti-gp120 IgG (NIH45-46G54W) injected over gp120 immobilized at high coupling density (~100 RU). 2G12 monomer, but not 2G12 dimer or the conventional IgG, could be fit to a 1:1 binding model. (B) 2G12 monomer, 2G12 dimer, and a conventional anti-gp120 IgG (NIH45-46G54W) injected over gp120 immobilized at medium coupling density (~50 RU). 2G12 monomer, but not 2G12 dimer or the conventional IgG, could be fit to a 1:1 binding model. (C) 2G12 monomer, 2G12 dimer, and a conventional anti-gp120 IgG (2909) injected over gp120 immobilized at low coupling density (<10 RU). All could be fit to a 1:1 binding model.
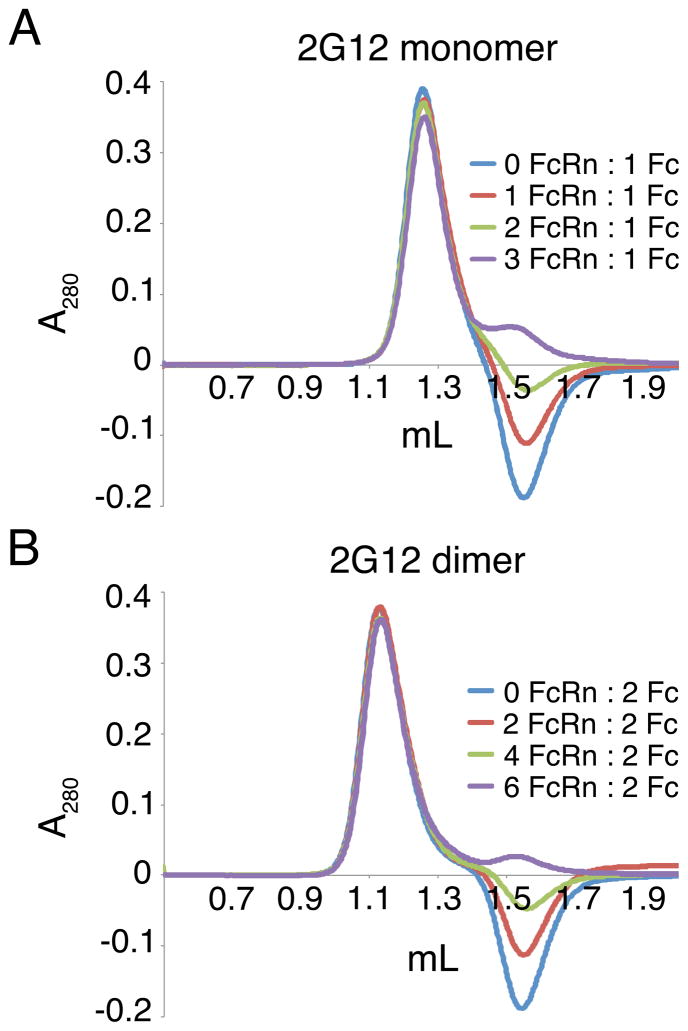
The 2G12 dimer structures suggested that the CH2-CH3 domain interface, the binding site for the neonatal Fc receptor (FcRn) (Burmeister et al., 1994) and the site of contacts that form the NCS 6-fold axis (Figure 2C; Figure 3C; Figure S3B), would be accessible for potential interactions. Two FcRn proteins bind to the Fc region of a conventional IgG monomer, one per polypeptide chain in the Fc (Huber et al., 1993; Sanchez et al., 1999). Since 2G12 dimer contains two Fc regions with four potential FcRn binding sites, up to four FcRn proteins could bind per 2G12 dimer. To characterize the binding interaction between FcRn and 2G12 dimer, we used equilibrium gel filtration, a technique that can be used to determine the stoichiometry of a complex that might dissociate during conventional gel filtration chromatography (Hummel and Dreyer, 1962; Sanchez et al., 1999; West and Bjorkman, 2000). For the 2G12 experiments, a gel filtration column was equilibrated with a buffer containing 5 μM FcRn, a concentration ~10- fold higher than the KD of the interaction between FcRn and IgG in solution (Huber et al., 1993). We then injected various ratios of FcRn to 2G12 monomer or dimer. In both chromatograms in Figure 7, the first peak corresponded to the 2G12 complex with FcRn, which migrated at an apparently higher molecular mass for the dimer complex than for the monomer complex. Both chromatograms also showed a peak or a trough at the position where unbound FcRn migrated: a peak in the case of the 3:1 ratio of FcRn per Fc region in 2G12 monomer and the 6:2 ratio of FcRn per Fc region in 2G12 dimer (indicating excess FcRn) and troughs in the cases of lower ratios of FcRn per Fc. Since excess FcRn was present when a 3:1 (or 6:2) ratio of FcRn per Fc was injected, the stoichiometry for both complexes is two FcRn proteins bound per Fc region. Thus two FcRn molecules were bound per 2G12 monomer, which contains one Fc region, and four FcRn molecules were bound per 2G12 dimer, which contains two Fc regions.
Figure 7. Equilibrium gel filtration analyses of FcRn-2G12 complexes.
(A) FcRn was incubated with 2.5 μM 2G12 monomer in the presence of 0, 1, 2, or 3 (additional) equivalents of FcRn in a buffer containing 5 μM FcRn (equilibration buffer). Samples were then injected onto a column equilibrated in the equilibration buffer. The peak that eluted first elution volume of free FcRn. (B) FcRn was incubated with 1.25 μM 2G12 dimer in the presence of 0, 2, 4, or 6 (additional) equivalents of FcRn in a buffer containing 5 μM FcRn (equilibration buffer). Samples were then injected onto a column equilibrated in the equilibration buffer. The peak that eluted first corresponds to an FcRn-2G12 dimer complex. The second peak or trough was at the elution volume of free FcRn.
DISCUSSION
Among known broadly neutralizing anti-HIV-1 antibodies, 2G12 is unusual in two respects: first, it recognizes a carbohydrate epitope on gp120 (Botos and Wlodawer, 2005), and second, its Fab arms are domain swapped to form a rigid single (Fab)2 unit (Calarese et al., 2003). We hypothesized that intermolecular domain swapping could create a dimeric species of 2G12 IgG that was observed by size-exclusion chromatography (West et al., 2009). Compared with 2G12 monomer, purified 2G12 dimer is 50- to 80-fold more potent in in vitro neutralization assays (West et al., 2009), more effective in protection from HIV infection in in vivo experiments (Luo et al., 2010), and mediates ADCC at lower concentrations (Klein et al., 2010). The present structural studies were initiated to investigate the structural mechanism for the increased activity of 2G12 dimer versus monomer.
Although crystal structures of intact IgGs are rare, with few examples in the literature (Guddat et al., 1993; Harris et al., 1997; Harris et al., 1998; Kratzin et al., 1989; Saphire et al., 2001), intact 2G12 IgG dimer formed crystals, both alone and complexed with the 2G12.1 inhibitor peptide. Both crystal forms diffracted weakly to low resolution, but we could solve crystal structures by molecular replacement for 2G12 dimer and the 2G12 dimer/2G12.1 complex at resolutions between 6.5 and 8 Å and verify the structures by heavy atom methods and native Patterson calculations.
The low-resolution structures revealed three distinct conformations of 2G12 dimer. In each conformation, the two (Fab)2 units in the dimer were arranged as arms at an obtuse angle, analogous to the Fab arms of a conventional IgG monomer that can bind bivalently to tethered antigens. The two Fc regions were separated from each other, suggesting they would not block each other’s interactions with Fc receptors. Indeed, 2G12 dimer interacted with FcRn with proportional stoichiometry as 2G12 monomer in equilibrium gel filtration studies, corroborating the idea that Fc regions in 2G12 dimer are as accessible for effector functions as the Fc regions in conventional IgGs. 2-D class averages from negative stain electron microscopy provided unbiased, reference-free profiles that confirmed the flexibility and orientations of the 2G12 dimers observed in crystal structures. The presence of three distinct conformations of 2G12 dimer indicated that different conformational intermediates were trapped in the two crystal forms. Consistent with this suggestion, 2G12 dimer could bind bivalently to immobilized gp120, allowing avidity effects to increase the apparent affinity of 2G12 dimer for its carbohydrate epitope. By contrast, the single rigid (Fab)2 unit of monomeric 2G12 bound monovalently to immobilized gp120, regardless of the density of the gp120 on the surface.
These results are relevant to a hypothesis that a general inability of many anti-HIV-1 antibodies to bind with avidity is one reason that the antibody response against HIV-1 is generally ineffective (Klein and Bjorkman, 2010). Briefly, we proposed that most anti-Env antibodies do not bind bivalently to HIV-1 spike trimers because (i) the small number and low density of HIV spike trimers does not normally permit inter-spike crosslinking, and (ii) the distance between the same epitope on individual monomers within a spike trimer usually exceeds the armspan of a conventional IgG, thereby preventing most instances of intra-spike crosslinking (Bartesaghi et al., 2013; Julien et al., 2013; Lyumkis et al., 2013). To assess potential bivalent binding by anti-viral IgGs, we defined the molar neutralization ratio (MNR) as the concentration at which a Fab achieves 50% inhibition of viral infectivity (IC50) divided by the IC50 for the parental IgG in an in vitro neutralization assay. An antibody that is unable to bind with avidity would exhibit an MNR of 2, because the IgG has twice the amount of antigen-binding capacity as the Fab. MNRs greater than 2 suggest avidity effects resulting from the IgG cross-linking epitopes on the virus. Results from published studies showed high MNRs for antibodies against respiratory syncytial virus and influenza (Edwards and Dimmock, 2001; Schofield et al., 1997; Wu et al., 2005), suggesting that antibodies can take advantage of avidity effects to bind to enveloped viruses other than HIV-1. However, a compilation of the highest reported MNRs for antibodies against HIV showed that neutralizing antibodies yielded relatively low MNRs (Klein and Bjorkman, 2010).
Although the (Fab)2 unit of monomeric 2G12 interacts with at least three high-mannose glycans, the binding surface is a single unit, thus the 2G12 monomer does not have the capacity to bind antigen with avidity as it is conventionally defined. However, 2G12 dimer can be up to 2500-fold more potent in neutralization than monomeric 2G12 on a molar basis (West et al., 2009), thus the high MNRs for 2G12 dimer compared to its monomeric form are similar to the large effects seen for conventional IgGs against non-HIV viruses (Klein and Bjorkman, 2010). The discovery that 2G12 dimer is a flexible molecule that behaves like an oversized version of a conventional IgG antibody now allows us to postulate that avidity effects, rather than simply doubling the surface area of a single combining site, account for the mechanism by which 2G12 dimer achieves its greater potency compared with 2G12 monomer.
The 2G12 dimer structure, taken together with what is known about the 2G12 epitope, permits speculation as to its mode of binding with avidity to HIV spike trimers. 2G12 has been reported to bind to a specific high-mannose patch on gp120 that has been called “intrinsic” due to its conserved and unusual avoidance of α-mannosidase processing while being trafficked through the ER and Golgi (Bonomelli et al., 2011). We propose that one of the (Fab)2 units of 2G12 dimer first tethers with high affinity to the intrinsic mannose patch before sampling the abundant high-mannose landscape in the surrounding glycan shield for a second binding opportunity. Because the intrinsic mannose patch on an adjacent gp120 monomer within a spike trimer would be too far away to be reached by the second (Fab)2 unit of 2G12 dimer, we believe that the second 2G12 “epitope” would be a suboptimal glycan cluster distinct from the intrinsic mannose patch. The finding that 2G12 dimer binds monovalently to immobilized gp120 coupled at low density suggests that two (Fab)2 units cannot bind simultaneously to a single gp120, thus the suboptimal second epitope would likely be on an adjacent gp120 within a spike trimer.
Our studies provide important information for therapeutic anti-HIV efforts in that they provide a direct demonstration of the importance of avidity in HIV-1 neutralization. Here we show, for the first time, a comparison of the structures of a monomeric and dimeric form of a carbohydrate-only anti-HIV-1 antibody. Unlike anti-HIV-1 antibodies against protein-only epitopes, the dimeric form of 2G12 can take advantage of avidity to increase its neutralization potency and breadth, thus is a promising candidate for therapeutic efforts to combat HIV-1. Despite initially discouraging results with less potent antibodies (Poignard et al., 1999), passive delivery of antibodies has been gaining new attention (reviewed in (Klein et al., 2013), with encouraging results from a cocktail of three or five broadly neutralizing antibodies in humanized mice infected with HIV-1 (Diskin et al., 2013; Klein et al., 2012). 2G12 dimer would be very effectively employed in such a cocktail, as it shares its primary epitope with no other known antibody and it can potentially exploit surrounding glycans as a secondary epitope. Of direct relevance to potential therapeutic use of antibodies for passive immunotherapy, bivalent binding can serve as a buffer against Env mutations (Klein and Bjorkman, 2010), thus 2G12 dimer is a good candidate for therapeutic efforts against clade A and clade B HIV strains.
EXPERIMENTAL PROCEDURES
Protein expression and purification
2G12 IgG was expressed in transiently transfected HEK293-6E suspension cells as described (West et al., 2009). A mixture of 2G12 monomer and dimer was isolated from harvested supernatants by FcRn affinity chromatography (Huber et al., 1993) using 5 mL of HiTrap NHS-activated resin (GE Healthcare) coupled to 12 mg of rat FcRn. Supernatants were loaded on the FcRn column in 250 mM MES, 150 mM NaCl, 0.02% Sodium azide, pH 5.7 and eluted in 250 mM HEPES, 150 mM NaCl, 0.02% NaN3, pH 8. The eluent was subjected to size-exclusion chromatography in 20 mM Tris pH 8.0, 150 mM NaCl using a Superdex 200 16/60 gel filtration column (GE Healthcare). Fractions corresponding to 2G12 dimer or monomer were pooled and subjected to SEC an additional two times. Further experimental details, including crystallization of 2G12 dimer and X-ray data collection, are in the Supplemental Experimental Procedures.
Data processing and structure determination
Diffraction data were processed using XDS (Kabsch, 2010) or HKL2000 (Otwinowski and Minor, 1997) and were indexed, integrated and scaled using POINTLESS and SCALA. Rather than Rmerge statistics, we used a combination of CC1/2 in the range of 90% (Karplus and Diederichs, 2012) and I/σ >1.5 in the highest resolution shell to determine the resolution cutoffs for our data sets. Data for 2G12 dimer were corrected for anisotropy using the UCLA-MBI Anisotropy Server (Strong et al., 2006). Molecular replacement was carried out using Phaser-MR (McCoy et al., 2007; Winn et al., 2011) and Molrep (Lebedev et al., 2008; Vagin and Teplyakov, 2010; Winn et al., 2011). All molecular replacement solutions in space group P6122 yielded higher translation function Z-scores (10 and above for initial placement of three (Fab)2 units) as well as lower initial R factors (0.54 versus over 0.60) than solutions in the enantiomeric space group P6522. Native Patterson calculations were done using POLARRFN (Winn et al., 2011) in the the UCLA-MBI Self Rotation Function Server, sfall (Winn et al., 2011) and Patterson (Winn et al., 2011). Rigid body refinement and domain B-factor refinement were carried out using CNS (Brunger, 2007; Brunger et al., 1998) or Phenix.refine (Adams et al., 2010). Model and electron density visualization were done using COOT (Emsley et al., 2010) and PyMol (Schroedinger, LLC). Map truncation was done using Phenix.map_box (Adams et al., 2010). Structure figures were prepared using PyMol. Globular structures in Figures 3, 4, and S4 were prepared by conversion of coordinate files into 25 Å mrc map volumes using pdb2mrc in the EMAN2.0 suite (Tang et al., 2007), which were then rendered in UCSF Chimera (Pettersen et al., 2004).
Electron Microscopy (EM)
2G12 species purified via SEC were analyzed by negative stain EM. A 3 μL aliquot of 1–2.5 μg/ml of 2G12 (Fab)2 or 2G12 monomer, dimer or trimer was applied to a freshly glow discharged carbon coated 400 Cu mesh grid and stained with 2% uranyl formate for 20 s. Grids were imaged using a FEI Tecnai T12 electron microscope operating at 120 kV at 52,000 x magnification and electron dose of 25 e−/Å2, which resulted in a pixel size of 2.05 Å at the specimen plane. Images were acquired with a Tietz 4k x 4k CCD camera using LEGINON (Suloway et al., 2005) at a defocus range of 700 to 1000 nm.
Particles were picked automatically using DoG Picker and put into a particle stack using the Appion software package (Lander et al., 2009; Voss et al., 2009). Initial reference-free 2D class averages were calculated using unbinned particles via the Xmipp Clustering 2D Alignment and sorted into classes (Sorzano et al., 2010). Particles corresponding to only 2G12 species were selected into a substack and another round of reference-free alignment was carried out using Xmipp Clustering 2D alignment and IMAGIC softwares to generate 64 classes (van Heel et al., 1996). A total of 6583, 10341, and 4230 particles went into the final 2D classes of the monomer, dimer and trimer, respectively. Back projections of 2G12 IgG dimer A were calculated using EMAN software (Tang et al., 2007) as follows: the X-ray coordinates were converted from PDB to MRC format using pdb2mrc, low pass filtered to 25 Å resolution to generate a density map, and the map was projected at 20° Euler angle increments using project3d and displayed in e2display.
Biosensor studies of 2G12 interactions with gp120
A Biacore T200 biosensor system (GE Healthcare) was used to evaluate the interactions of 2G12 monomer, 2G12 dimer, and control anti-gp120 antibodies NIH45-46G54W (Diskin et al., 2011) and 2909 (Gorny et al., 2005) with recombinant gp120 from the strain SF162 (gift of Leo Stamatatos, Seattle Biomed). Response units (RUs) of gp120 ranging from fewer than 10 to approximately 100 were covalently immobilized on flow cells of CM5 biosensor chips using standard primary amine coupling chemistry (Biacore T200 manual, GE Healthcare). A concentration series of either 2G12 monomer, dimer, or control anti-gp120 IgG was injected at 25°C in 10 mM HEPES with 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) surfactant P20 at pH 7.4. Sensor chips were regenerated using 10 nM glycine, pH 2.5. After subtracting the signal from the reference flow cell, we globally fit the kinetic data from each experiment to a 1:1 binding model (Biacore evaluation software). When an obvious refractive index change occurred when switching from the association to the dissociation buffer, the sensorgrams were also fit with a refractive index correction (third row, middle panel of Figures 6 and S6). We evaluated the fit to the 1:1 binding model of each binding interaction by calculating and plotting residuals between the experimental and modeled curves.
Equilibrium gel filtration
Equilibrium gel filtration chromatography (Hummel and Dreyer, 1962) was performed to determine the stoichiometry of the association of 2G12 monomer and 2G12 dimer with FcRn. Briefly, chromatography was performed at a flow rate of 100 μL/min using a SMART micropurification system (Pharmacia) monitoring the absorbance at 280 nm. The experiment was run on a Superdex 200 PC 3.2/30 gel filtration column, which was equilibrated with and run in equilibrium buffer (10 mM PIPES, 0.05% Sodium azide, pH 6.1) containing 5 μM soluble FcRn. All IgG samples and various concentrations of FcRn were buffer exchanged into the PIPES buffer via dialysis prior to injection over the column. 2.5 μM of 2G12 monomer was mixed with equilibration buffer containing 5 μM FcRn plus no additional FcRn, 2.5 uM additional FcRn, 5 μM additional FcRn, or 7.5 μM additional FcRn. 1.25 μM of 2G12 dimer was mixed with equilibration buffer containing 5 μM FcRn plus no additional FcRn, 2.5 uM additional FcRn, 5 μM additional FcRn, or 7.5 μM additional FcRn.
Supplementary Material
HIGHLIGHTS.
Structures of a dimeric form of 2G12 IgG, an anti-HIV-1 carbohydrate antibody
2G12 IgG dimer formed by intermolecular domain swapping to create two (Fab)2 arms
Three distinct 2G12 dimer conformations in crystal structures demonstrate flexibility
Increased potency of 2G12 dimer vs. monomer rationalized by greater flexibility
Acknowledgments
We thank Jost Vielmetter and the Caltech Protein Expression Center for assistance with Biacore studies and protein expression, Jens Kaiser, Pavle Nikolovski, the Caltech Molecular Observatory, the 2012 APS Data Collection Workshop and the CCP4 School at Argonne National Laboratory for help with crystallographic studies, the staff at SSRL Beamlines 4-2 and 12-2 and for assistance with SAXS studies, the staff at Beamlines 12-2 (SSRL) and APS GM/CAT Beamlines 231D-B and 231D-D for assistance with SAXS and crystallographic data collection, Justin Chartron for help with SAXS and crystallographic data analysis, the developers of PHENIX, CCP4, and XDS software for assistance and advice concerning X-ray crystallography, Leo Stamatatos, Zara Fulton, Reza Khayat, Gloria Tran, and members of the Bjorkman laboratory for protein reagents, William Lange, Marta Murphy and Maria Politzer for help making figures and models, and Andrew Davenport and Beth Stadtmueller for critical reading of the manuscript. This work was supported by a Collaboration for AIDS Vaccine Discovery grant from The Bill and Melinda Gates Foundation (grant ID 1040753 to P.J.B.), the National Institutes of Health (2 R37AI041239-06A1 to P.J.B.), and startup funds from the Scripps Research Institute (to A.B.W.).
Footnotes
Accession Numbers
Coordinates and X-ray crystallographic data for 2G12 IgG dimer and 2G12 IgG dimer/2G12.1 peptide complex have been deposited in the Protein Data Bank with accession numbers 4NHG and 4NHH, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nature structural & molecular biology. 2013 doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PloS one. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botos I, Wlodawer A. Proteins that bind high-mannose sugars of the HIV envelope. Progress in biophysics and molecular biology. 2005;88:233–282. doi: 10.1016/j.pbiomolbio.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica Section D, Biological crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- Calarese DA, Lee HK, Huang CY, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong CH, Wilson IA. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- Diskin R, Klein F, Horwitz JA, Halper-Stromberg A, Sather DN, Marcovecchio PM, Lee T, West AP, Jr, Gao H, Seaman MS, et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. The Journal of experimental medicine. 2013;210:1235–1249. doi: 10.1084/jem.20130221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MJ, Dimmock NJ. Hemagglutinin 1-specific immunoglobulin G and Fab molecules mediate postattachment neutralization of influenza A virus by inhibition of an early fusion event. Journal of virology. 2001;75:10208–10218. doi: 10.1128/JVI.75.21.10208-10218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. Journal of virology. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guddat LW, Herron JN, Edmundson AB. Three-dimensional structure of a human immunoglobulin with a hinge deletion. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4271–4275. doi: 10.1073/pnas.90.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ, Larson SB, Hasel KW, McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997;36:1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- Harris LJ, Skaletsky E, McPherson A. Crystallographic structure of an intact IgG1 monoclonal antibody. Journal of molecular biology. 1998;275:861–872. doi: 10.1006/jmbi.1997.1508. [DOI] [PubMed] [Google Scholar]
- Huber AH, Kelley RF, Gastinel LN, Bjorkman PJ. Crystallization and stoichiometry of binding of a complex between a rat intestinal Fc receptor and Fc. Journal of molecular biology. 1993;230:1077–1083. doi: 10.1006/jmbi.1993.1220. [DOI] [PubMed] [Google Scholar]
- Hummel JP, Dreyer WJ. Measurement of protein-binding phenomena by gel filtration. Biochimica et biophysica acta. 1962;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS pathogens. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Webster A, Gnanapragasam PN, Galimidi RP, Bjorkman PJ. A dimeric form of the HIV-1 antibody 2G12 elicits potent antibody-dependent cellular cytotoxicity. AIDS (London, England) 2010;24:1633–1640. doi: 10.1097/qad.0b013e32833ad8c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzin HD, Palm W, Stangel M, Schmidt WE, Friedrich J, Hilschmann N. The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I. Biological chemistry Hoppe-Seyler. 1989;370:263–272. [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. Journal of structural biology. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev AA, Vagin AA, Murshudov GN. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta crystallographica Section D, Biological crystallography. 2008;64:33–39. doi: 10.1107/S0907444907049839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein science : a publication of the Protein Society. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XM, Lei MY, Feidi RA, West AP, Jr, Balazs AB, Bjorkman PJ, Yang L, Baltimore D. Dimeric 2G12 as a potent protection against HIV-1. PLoS pathogens. 2010;6:e1001225. doi: 10.1371/journal.ppat.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunological reviews. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez A, Calarese DA, Stanfield RL, Chow KC, Scanlan CN, Kunert R, Katinger H, Burton DR, Wilson IA, Scott JK. A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:1380–1392. doi: 10.1096/fj.07-8983com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Poignard P, Sabbe R, Picchio GR, Wang M, Gulizia RJ, Katinger H, Parren PW, Mosier DE, Burton DR. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity. 1999;10:431–438. doi: 10.1016/s1074-7613(00)80043-6. [DOI] [PubMed] [Google Scholar]
- Poignard P, Saphire EO, Parren PW, Burton DR. gp120: Biologic aspects of structural features. Annual review of immunology. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- Roux KH. Immunoglobulin structure and function as revealed by electron microscopy. Int Arch Allergy Imm. 1999;120:85–99. doi: 10.1159/000024226. [DOI] [PubMed] [Google Scholar]
- Rupp B. Biomolecular crystallography : principles, practice, and application to structural biology. New York: Garland Science; 2010. [Google Scholar]
- Sanchez LM, Penny DM, Bjorkman PJ. Stoichiometry of the interaction between the major histocompatibility complex-related Fc receptor and its Fc ligand. Biochemistry. 1999;38:9471–9476. doi: 10.1021/bi9907330. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. Journal of virology. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield DJ, Stephenson JR, Dimmock NJ. Variations in the neutralizing and haemagglutination-inhibiting activities of five influenza A virus-specific IgGs and their antibody fragments. The Journal of general virology. 1997;78(Pt 10):2431–2439. doi: 10.1099/0022-1317-78-10-2431. [DOI] [PubMed] [Google Scholar]
- Sorzano CO, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernandez G, Li M, Xu G, Marabini R, Carazo JM. A clustering approach to multireference alignment of single-particle projections in electron microscopy. Journal of structural biology. 2010;171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. Journal of virology. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta crystallographica Section D, Biological crystallography. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. Journal of structural biology. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. Journal of structural biology. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Bjorkman PJ. Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor(,) Biochemistry. 2000;39:9698–9708. doi: 10.1021/bi000749m. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Galimidi RP, Foglesong CP, Gnanapragasam PN, Huey-Tubman KE, Klein JS, Suzuki MD, Tiangco NE, Vielmetter J, Bjorkman PJ. Design and expression of a dimeric form of human immunodeficiency virus type 1 antibody 2G12 with increased neutralization potency. Journal of virology. 2009;83:98–104. doi: 10.1128/JVI.01564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Pfarr DS, Tang Y, An LL, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. Journal of molecular biology. 2005;350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.