SUMMARY
Resting mitochondrial matrix Ca2+ is maintained through a MICU1-established threshold inhibition of MCU activity. It is not known how MICU1 interacts with MCU to establish this Ca2+ threshold for mitochondrial Ca2+ uptake and MCU activity. Here, we show that MICU1 localizes to the mitochondrial matrix side of the inner mitochondrial membrane and MICU1/MCU binding is determined by a MICU1 N-terminal polybasic domain and two interacting coiled-coil domains of MCU. Further investigation reveals MICU1 forms homo-oligomers, and this oligomerization is independent of the polybasic region. However the polybasic region confers MICU1 oligomeric binding to MCU and controls mitochondrial Ca2+ current (IMCU). Moreover, MICU1 EF-hands regulate MCU channel activity but do not determine MCU binding. Loss of MICU1 promotes MCU activation leading to oxidative burden and a halt to cell migration. These studies establish a molecular mechanism for MICU1 control of MCU-mediated mitochondrial Ca2+ accumulation, and dysregulation of this mechanism likely enhances vascular dysfunction.
INTRODUCTION
Receptor-mediated cytosolic Ca2+ flooding is rapidly cleared by endoplasmic reticulum re-filling, plasma membrane efflux and mitochondrial Ca2+ uptake (Berridge et al., 2003; Soboloff et al., 2012). In most eukaryotic non-excitable and excitable cells, carriers, exchangers and channels regulate mitochondrial Ca2+ uptake during the resting and active states (Drago et al., 2011; Duchen et al., 2008; Nicholls, 2005; Rizzuto et al., 2012; Santo-Domingo and Demaurex, 2010; Williams et al., 2013). Following G-protein coupled receptor or receptor tyrosine kinase agonist stimulation, the Mitochondrial Ca2+ Uniporter (MCU) channel rapidly transports elevated cytosolic Ca2+ into the mitochondrial matrix utilizing the highly negative mitochondrial membrane potential (ΔΨm) (Bernardi, 1999; Gunter et al., 1994; Kirichok et al., 2004). Mitochondrial Ca2+ uptake stimulates bioenergetics through activation of Ca2+ sensitive dehydrogenases and thus modulates ATP synthesis (Babcock et al., 1997; Balaban, 2009; Denton and McCormack, 1980; Duchen et al., 2008; Hajnoczky et al., 1995; Hansford, 1994; McCormack et al., 1990). Reciprocally, mitochondrial Ca2+ overload promotes physical damage to mitochondria leading to bioenergetic crisis and cell death (Orrenius et al., 2003; Szalai et al., 1999). Pathophysiological dysregulation of mitochondrial Ca2+ uptake has been linked to organ damage via chronic oxidative burden, autophagy, and cellular dysfunction (Baines et al., 2005; Cardenas et al., 2010; Clapham, 2007; Joiner et al., 2012; Lemasters et al., 2009; Mallilankaraman et al., 2012b).
Mitochondrial Ca2+ uptake has been functionally described for decades, but only recently using integrative bioinformatics and RNAi screening approaches, three components of the MCU complex (MICU1, MCU and MCUR1) were identified that together constitute the activity of MCU (Baughman et al., 2011; De Stefani et al., 2011; Mallilankaraman et al., 2012a; Mallilankaraman et al., 2012b; Perocchi et al., 2010). Although the mitochondrial uniporter complex tightly regulates Ca2+ influx, the molecular mechanisms of uniporter regulation are unclear. This unknown regulatory mechanism stems from the observation that mitochondrial Ca2+ influx exhibits a finite trigger proposed as a “set point” in 1978 (Nicholls, 1978, 2005). The set point is overcome and the mitochondria begins Ca2+ uptake at cytosolic Ca2+ levels exceeding ~3 μM. The trigger was characterized as the function of MCU-associated protein MICU1 which is able to suppress mitochondrial Ca2+ uptake up to the cytosolic levels ~3 μM Ca2+ (Mallilankaraman et al., 2012b). Thus far, MICU1 is the only known protein that interacts with MCU to prevent mitochondrial Ca2+ uptake under resting conditions (Mallilankaraman et al., 2012b), but the MICU1 localization and the binding domains of the MICU1/MCU complex that establish the set point are unknown.
Here, we show that MICU1 interacts with MCU in the mitochondrial matrix and MICU1/MCU binding is determined by a MICU1 N-terminal polybasic domain and two coiled-coil MCU domains. MICU1 EF-hands regulate MCU channel activity but not MICU1/MCU binding. Additionally, loss of MICU1 induces MCU activity leading to vascular endothelial oxidative stress and diminished endothelial cell migration.
RESULTS
Protein Flux Analysis Establishes MICU1 is Compartmentalized in the Mitochondrial Matrix
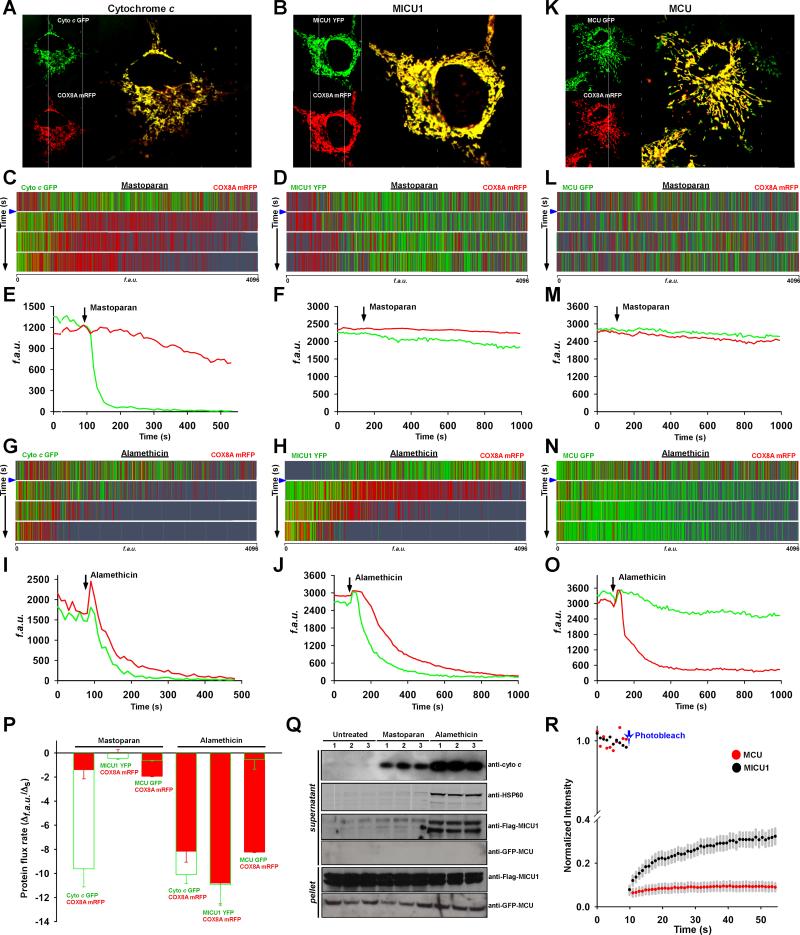
The MCU pore subunit is located in the inner mitochondrial membrane (IMM), and thus its regulator, MICU1, must be present either in the intermembrane space or the matrix side of the mitochondrial inner membrane. To determine the sub-organellar localization of MICU1, we developed a dynamic protein flux assay using confocal microscopy in which the outer mitochondrial membrane (OMM) or both the OMM and IMM of the mitochondria were selectively permeabilized and release of MICU1 was subsequently visualized from the mitochondrial compartments. HeLa cells were co-transfected with expression vector pairs encoding YFP-tagged MICU1 and mRFP-tagged mitochondrial targeting sequence of matrix-localized cytochrome c oxidase subunit 8 (COX8) (Figure 1B) or mitochondrial intermembrane space protein, GFP-tagged cytochrome c (cyto c) cotransfected with mRFP- COX8 (Figure 1A). Combinations of both cyto c/COX8 and MICU1/COX8 showed general mitochondrial localization (Figure 1A and 1B). To visualize the MICU1 compartmentalization, the plasma membrane was permeabilized using 0.002% digitonin (20 μg/ml) and permeabilized cells were bathed in digitonin-free intracellular medium (ICM) before exposure to mitochondrial permeabilizing agents.
Figure 1. Protein Flux Dynamics Illustrate MICU1 is Localized in the Mitochondrial Matrix Compartment.
(A) HeLa cells were co-transfected with cytochrome c-GFP and COX8A-mRFP (MTS) plasmid constructs.
(B) Similarly, HeLa cells were co-transfected with MICU1-YFP and COX8-mRFP (MTS).
(C and D) Plasma membranes of HeLa cells were briefly permeabilized with digitonin (0.002% wt/vol) containing intracellular medium for 7 min. Cells were treated with mastoparan (20 μg/ml) to trigger outer mitochondrial membrane permeabilization. Protein flux dynamics were visualized using confocal microscopy. The depletion of either cyto c-GFP or MICU1-YFP was depicted as representative green histogram lines from an individual experiment. The depletion of COX8A-mRFP was depicted as representative red histogram lines from the same individual experiment. Each row corresponds to different time intervals before (top row) and after (bottom three rows) mastoparan challenge (indicated by blue arrow). n = 6-9.
(E) Representative protein release traces from panel C of cytochrome c (green) and COX8A (red) following mastoparan. n = 6-9.
(F) Representative protein release traces from panel D of MICU1 (green) and COX8A (red) following mastoparan. n = 6-9.
(G and H) Release illustration of cytochrome c and COX8A or MICU1 and COX8A flux dynamics before (top row) and after (bottom three rows) alamethicin (20 μg/ml) addition (indicated by blue arrow). n = 6-9.
(I and J) Representative traces from panels G and H of cytochrome c and COX8A or MICU1 and COX8A depletion before and after alamethicin addition. n = 6-9.
(K-O) Similar to cytochrome c and MICU1, MCU-GFP flux assessment was performed. n = 6-9.
(P) Dissipation of fluorescence intensity was calculated as flux rate after mastoparan or alamethicin addition. Mean ± SEM; n = 6-9.
(Q) To assess the localization of MICU1, HeLa cells were permeabilized and exposed to mastoparan or alamethicin (20 μg/ml) for 5 min. Cytosolic (supernatant) and mitochondrial (pellet) fractions were subjected to immunoblotting to examine the release of cytochrome c (intermembrane space marker), HSP60 (matrix marker), MICU1-Flag and MCU-GFP from mitochondria. Lanes1, 2, 3 refer to triplicate samples. n = 3.
(R) Time course of FRAP from cells expressing MICU1-YFP or MCU-GFP. The FRAP recovery curve is a plot of the normalized fluorescence recovery versus time. Mean ± SEM; n = 6.
The cyto c/COX8 co-transfected permeabilized cells were subjected to mastoparan (20 μg/ml), a wasp venom peptide toxin that induces OMM permeabilization (Kluck et al., 1999; Pfeiffer et al., 1995). The release of intermembrane space resident proteins due to mastoparan was visualized by confocal microscopy in real-time. OMM permeabilization by mastoparan rapidly released cyto c but not matrix-localized COX8 (Figure 1C and 1E). Similarly, MICU1-YFP/COX8-mRFP permeabilized cells were exposed to mastoparan. We found that mitochondrial localized MICU1 and COX8 were not released from the mitochondrial intermembrane space (Figure 1D and 1F). We next asked whether permeabilization of both the OMM and the IMM promotes MICU1 release from the mitochondrial matrix. Permeabilized cells were exposed to a fungal peptide, alamethicin (20 μg/ml) that induces large pores in both mitochondrial membranes (Kluck et al., 1999). Permeabilization of both membranes induced the release of COX8 from the matrix in addition to cyto c from the intermembrane space (Figure 1G, 1I and 1P). It has been recently reported that MICU1 exists in the intermembrane space (Csordas et al., 2013), but similar to COX8, MICU1 was released only upon permeabilization of both the OMM and IMM by alamethicin (Figure 1H, 1J and 1P). The integral inner membrane channel pore subunit MCU was not released by mastoparan or alamethicin permeabilization (Figure 1K-1P). The dynamic protein flux assay was complemented by Western blot analysis. As expected, cyto c was released by both mastoparan and alamethicin and appeared in the cytosolic supernatant (Figure 1Q). MICU1 was only released by alamethicin while integral membrane MCU was not released by either mastoparan or alamethicin (Figure 1Q). A soluble mitochondrial matrix protein HSP60 was used as a substitute for COX8 (Figure 1Q). Further, we examined whether MCU and MICU1 diffuse rapidly after photobleaching. Photobleaching of MICU1 but not MCU showed rapid fluorescence recovery after photobleaching (FRAP). This result suggested that MICU1 is less associated with the IMM than MCU (Figure 1R). These results reveal that MICU1 is compartmentalized in the mitochondrial matrix side of the IMM.
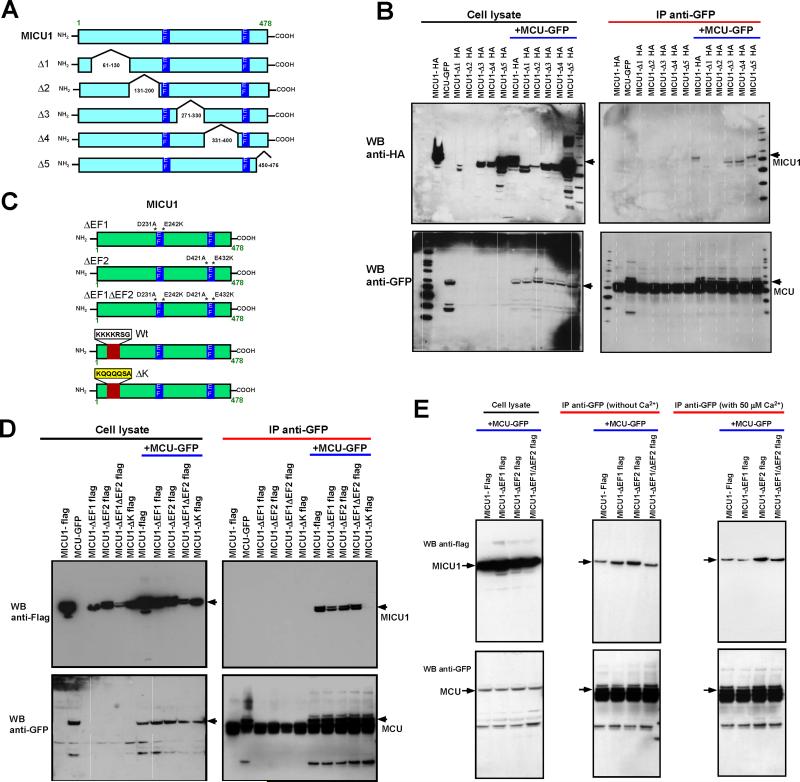
Mapping of MICU1 and MCU Binding Regions Reveals Conserved MICU1 Polybasic Motif but not EF-hand Domains are Essential for MCU Binding
Although MICU1 and MCU form a complex (Mallilankaraman et al., 2012b; Perocchi et al., 2010), it is not known which regions of MICU1 and MCU determine binding. To map the binding regions of MICU1 and MCU, we generated five HA-tagged MICU1 truncation mutants (Figure 2A) and transfected these into COS-7 cells stably expressing GFP-tagged full-length MCU. The transfected cell lysates were subjected to immunoprecipitation and Western blot analysis. Immunoprecipitation of GFP-tagged MCU pulled-down all MICU1 truncation mutants except deletion of amino acids 131-200 (MICU1-Δ2) (Figure 2B). Intriguingly, the MICU1-Δ1 (deletion of amino acids 61-130) partially interacts with wild-type MCU (Figure 2B). Although the effect of the MICU1-Δ2 deletion was profound, lack of any interaction suggests a conformational deformity of MICU1-Δ2 as a result of the truncation. In contrast, the partial interaction of MICU1-Δ1 suggests that the protein is maintaining an interacting tertiary structure that is recognized by MCU. Therefore, we examined the MICU1-Δ1 amino acid sequence for evolutionarily conserved interaction motifs and found a series of positively charged lysine residues (aa 99-110) (Figure S1A and S1B) which corresponds to the consensus sequence of a polybasic region motif which is known as not only an interaction motif but also as a membrane anchoring motif (Hancock et al., 1990; Heo et al., 2006; Papayannopoulos et al., 2005; Williams, 2003). The polybasic N-terminal region domain was mutated to poly-glutamine (MICU1-ΔK) (Figure 2C) and MICU1-ΔK reduced MCU interactions (Figure 2D), however MICU1-ΔK still localized to the mitochondria (Figure S1D and S1E). The marked but not complete loss of interaction with MCU suggests the MICU1 polybasic region motif is a determinant of MICU1/MCU interaction. The profound loss of binding in MICU1-Δ2 may be a consequence of the deleted region's proximity to an EF hand of MICU1. To determine if the EF hands are necessary for MCU binding, both EF hands were mutated (Figure 2C). Although functionally important (Mallilankaraman et al., 2012b; Perocchi et al., 2010), the EF hands of MICU1 did not determine MCU binding (Figure 2D). This result also demonstrates that although MICU1-ΔK has functional EF hands, it failed to interact with MCU (Figure 2D). We next asked whether EF-hands’ Ca2+ binding domains may participate in MCU/MICU1 binding. We therefore tested whether Ca2+ determines MICU1/MCU binding using MICU1 full-length and single and double EF1 and EF2 mutants. Our co-immunoprecipitation data demonstrated that MICU1/MCU binding is independent of [Ca2+] (Figure 2E). Together, these results indicate that the polybasic region, but not the EF-hands of MICU1, are determinant of MCU binding.
Figure 2. Mapping the MICU1 Interaction Domains with MCU.
(A) Scheme depicts full-length MICU1 and truncation constructs.
(B) Stably MCU-GFP expressing COS-7 cells were transfected with HA-tagged full-length MICU1 or truncations. Following immunoprecipitation with GFP antibody, total cell lysates and immunoprecipitated materials were subjected to Western blot analysis. Cell lysates were probed with anti-HA (top left) or anti-GFP antibodies (bottom left) to serve as inputs. Immunoprecipitated samples were probed with anti-HA (top right) or anti-GFP antibodies (bottom right). Anti-GFP antibodies co-immunoprecipitate full-length MICU1, Δ3, Δ4 and Δ5 MICU1-HA truncations but not MICU1-Δ1 and MICU1-Δ2. n = 3-5.
(C) Scheme depicts full-length MICU1 and MICU1-ΔEF1, MICU1-ΔEF2, MICU1-ΔEF1ΔEF2 and MICU1-ΔK mutant constructs.
(D) Stably MCU-GFP expressing COS-7 cells were transfected with Flag-tagged full-length MICU1 and mutants. Following immunoprecipitation with GFP antibody, total cell lysates and immunoprecipitated materials were subjected to Western blot analysis. Cell lysates were probed with anti-Flag (top left) or anti-GFP antibodies (bottom left) to serve as inputs. Immunoprecipitated samples were probed with anti-Flag (top right) or anti-GFP antibodies (bottom right). Anti-GFP antibodies co-immunoprecipitate full-length MICU1, MICU1-ΔEF1, MICU1-ΔEF2 and MICU1-ΔEF1ΔEF2 but not MICU1-ΔK mutant. n = 3.
(E) As described in D, stably MCU-GFP expressing COS-7 cells were transfected with Flag-tagged MICU1-ΔEF1, MICU1-ΔEF2 and MICU1-ΔEF1ΔEF2 mutants. Cell lysates were incubated with or without 50 μM Ca2+, immunoprecipitation was performed with GFP antibody, and were subjected to Western blot analysis. Cell lysates were probed with anti-Flag (top left) or anti-GFP antibodies (bottom left) to serve as inputs. Immunoprecipitated samples were probed with anti-Flag (top middle and right) or anti-GFP antibodies (bottom middle and right).
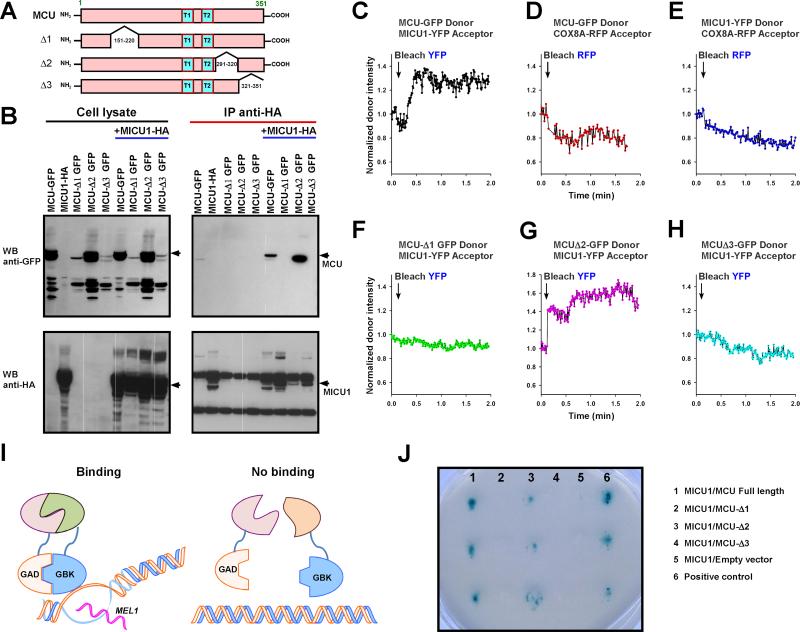
MCU Coiled-Coil Domains are Determinants of MICU1 Binding
Next to map which regions of MCU bind MICU1, we created three MCU truncation mutants stably expressing in COS-7 cells that were lacking aa 150-220 (MCU-Δ1), aa 291-320 (MCU-Δ2) or aa 321-351 (MCU-Δ3) (Figure 3A). Then, these stable cells were transfected with full-length MICU1-HA plasmid constructs. The pull-down of full-length MICU1-HA was unable to immunoprecipitate MCU-Δ1 and MCU-Δ3 suggesting that these regions corresponding to MCU coiled-coil domains are essential for MICU1 binding (Figure 3B and Figure S1C). To validate the co-immunoprecipitation data, we performed FRET acceptor photobleaching to determine whether MCU-Δ1 and - Δ3 interact with MICU1. Co-expression of mitochondrial marker COX8A-mRFP and MCU-GFP constructs revealed that MCU Δ1, Δ2 and Δ3 mutants did not alter the mitochondrial targeting (Figure S2). Comparable protein expressions of wild-type, Δ1, Δ2 and Δ3 MCU-GFP were selected for FRET analysis. Expectedly, MCU-GFP and MICU1-YFP interact, however neither MCU-GFP nor MICU1-YFP interact with matrix localized COX8A-RFP (Figure 3C-3E). Similar to co-immunoprecipitation analysis, MCU wild-type and MCU-Δ2, but not MCU-Δ1 and MCU-Δ3, interact with MICU1 (Figure 3C and 3F-3H). Further, we have also conducted interaction studies by Yeast Two-Hybrid assay (Figure 3I). In support of co-immunoprecipitation and FRET, MCU wild-type and MCU-Δ2 showed positive blue colonies, but MCU-Δ1 and MCU-Δ3 did not, indicating that MCU wild-type and MCU-Δ2 interact with MICU1 (Figure 3J). Collectively, these results demonstrate that MCU-Δ1 and MCU-Δ3, regions corresponding to the coiled-coil domains, are necessary for interaction with MICU1.
Figure 3. MCU Coiled-Coil Domains are Determinant of MICU1 Binding.
(A) Scheme depicts full-length MCU and truncation constructs.
(B) Stably MICU-HA expressing COS-7 cells were transfected with GFP-tagged full-length MCU or truncations. Following immunoprecipitation with HA antibody, total cell lysates and immunoprecipitated materials were subjected to Western blot analysis. 10% of cell lysates were probed with anti-GFP (top left) or anti-HA antibodies (bottom left) to serve as inputs. Immunoprecipitated samples were probed with anti-GFP (top right) or anti-HA antibodies (bottom right). Anti-HA antibodies co-immunoprecipitate full-length MICU1 and MCU-Δ2, but not MCU-Δ1 and MCU-Δ3. n = 3.
(C) HeLa cells were co-transfected with MCU-GFP donor and MICU1-YFP acceptor. MICU1-YFP acceptor was bleached and a representative trace of the MCU-GFP donor fluorescence is shown. n = 4
(D) Similarly, MCU-GFP donor and COX8A-RFP acceptor. n = 4
(E) MICU1-YFP donor and COX8A-RFP acceptor. n = 4
(F) MCUΔ1-GFP donor and MICU1-YFP acceptor. n = 3
(G) MCUΔ2-GFP donor and MICU1-YFP acceptor. n = 3
(H) MCUΔ3-GFP donor and MICU1-YFP acceptor. n = 3
(I) Scheme depicts Yeast Two-Hybrid system for analysis of MCU and MICU1 binding.
(J) Full-length MCU, MCU-Δ1, MCU-Δ2 or MCU-Δ3 and MICU1 were subcloned into pGBKT7 and pGADT7 vectors as bait and prey, respectively. After transformation, screening was performed by stringent selection (growth on quadruple dropout medium and resistance to the antibiotic Aureobasidin A) and blue colonies confirmed the MICU1/MCU interaction. Each column represents triplicates of MICU1/MCU, MICU1/MCU-Δ1, MICU1/MCU-Δ2, MICU1/MCU-Δ3, MICU1/empty vector and a positive control from the manufacturer (from left to right). n = 3.
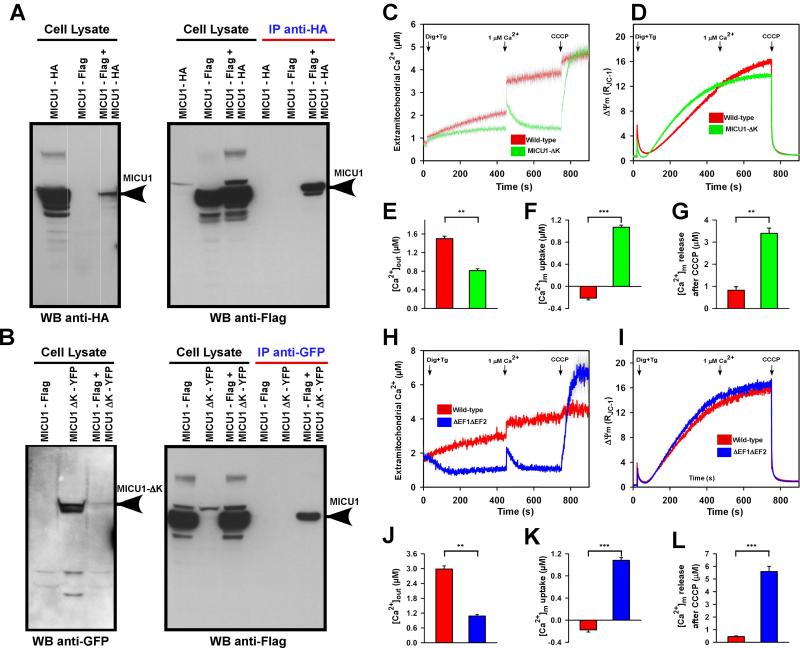
MICU1 Polybasic Region and EF-hands Regulate MCU-Mediated Mitochondrial Ca2+ Uptake
We next sought to characterize whether MICU1 undergoes oligomerization as a mechanistic step in regulation of MCU-mediated Ca2+ uptake. To evaluate the possibility of MICU1 oligomerization, we developed a co-immunoprecipitation strategy using co-transfection of two MICU1 constructs (MICU1-HA and MICU1-Flag) in COS-7 cells. Co-immunoprecipitation of MICU1-Flag with anti-HA antibody indicated that MICU1-HA and MICU1-Flag are able to form oligomeric complexes (Figure 4A right panel). Similarly, COS-7 cells were co-transfected with MICU1-Flag and MICU1-ΔK-YFP plasmid constructs. Co-immunoprecipitation of MICU1-Flag with anti-GFP antibody indicated that MICU1-Flag is still able to oligomerize with MICU1-ΔK (Figure 4B right panel). These results demonstrate that the polybasic region of MICU1 is not a MICU1/MICU1 interaction domain, but instead supports the polybasic region's role in MCU interaction.
Figure 4. MICU1 Binding and Functional Domains Regulate MCU-Mediated Ca2+ Uptake.
(A) COS-7 wild-type cells were co-transfected with HA-tagged full-length MICU1 and Flag-tagged full-length MICU1 plasmid constructs. Following immunoprecipitation with HA antibody, total cell lysates and immunoprecipitated materials were subjected to Western blot analysis. Cell lysates were probed with anti-HA (left) or anti-Flag antibodies (right panel first three lanes) to serve as inputs. Immunoprecipitated samples were probed with anti-Flag (right panel last three lanes). n = 3.
(B) COS-7 wild-type cells were co-transfected with Flag-tagged full-length MICU1 and YFP-tagged MICU1-ΔK plasmid constructs. Following immunoprecipitation with GFP antibody, total cell lysates and immunoprecipitated materials were subjected to Western blot analysis. Cell lysates were probed with anti-GFP (left) or anti-Flag (right panel first three lanes) antibodies to serve as inputs. Immunoprecipitated samples were probed with anti-Flag (right panel last three lanes). n = 3.
(C and D) HeLa cells were transfected with MICU1-ΔK plasmid and stable cells were generated after G418 selection. MICU1-ΔK and wild-type cells were permeabilized with 0.004% digitonin in ICM buffer and loaded with extramitochondrial ratiometric Ca2+ indicator Fura-2FF. These cells were also loaded with ratiometric ΔΨm indicator JC-1. Cells were then challenged with 1μM Ca2+ bolus. Finally, total mitochondrial Ca2+ uptake and ΔΨm dissipation were determined by uncoupler CCCP. Mean ± SEM; n = 3-6.
(E) Quantitation of wild-type and MICU1-ΔK extra-mitochondrial [Ca2+]out (to 450s). Mean ± SEM; **p < 0.01; n = 3-6.
(F) Quantitation of 1μM Ca2+ bolus uptake, [Ca2+]m. Mean ± SEM; ***p < 0.001, n = 3-6.
(G) Quantitation of mitochondrial Ca2+ release following CCCP. Mean ± SEM; **p < 0.01, n = 3-6.
(H-L) Similar to MICU1-ΔK, EF hand double mutant ΔEF1ΔEF2 was assessed for MCU-mediated Ca2+ uptake and ΔΨm dynamics. Mean ± SEM; **p < 0.01, ***p < 0.001, n = 3-6.
Having determined the binding regions of MCU and MICU1 through a series of deletion and mutation analysis, we next asked whether MICU1-ΔK and MICU1-ΔEF1ΔEF2 affect MCU-mediated [Ca2+]m uptake. Permeabilized control, MICU1-ΔK, and MICU1-ΔEF1ΔEF2 expressing HeLa cells were loaded with Fura-FF to determine [Ca2+]m uptake rate as a decay of extra-mitochondrial Ca2+ and JC-1 to simultaneously monitor mitochondrial membrane potential (ΔΨm) dynamics. MICU1 actively inhibits MCU at cytosolic Ca2+ levels < 3 μM (Mallilankaraman et al., 2012b). Therefore, to determine the phenotype of the MICU1-ΔK, HeLa cells were challenged with a 1 μM Ca2+ bolus, well within the functional inhibitory range of wild-type MICU1. We found that MICU1-ΔK cells rapidly cleared the 1 μM Ca2+ bolus under normal ΔΨm indicating the loss of MICU1 inhibition of MCU activity at < 3 μM. Conversely, control cells did not exhibit MCU-dependent Ca2+ uptake (Figure 4C-4F), because the MICU1 inhibition was intact. Next, we examined the total amount of Ca2+ buffered by control and MICU1-ΔK mitochondria using the mitochondrial uncoupler, CCCP. MICU1-ΔK overexpressing cells accumulated large amounts of Ca2+ (Figure 4G) suggesting that MICU1-ΔK is unable to properly interact with MCU to establish the set-point for MCU-mediated Ca2+ uptake. Functionally, MICU1 EF hands are the Ca2+ sensor of the MCU complex, and although EF hand mutants bind MCU (Figure 2D), the mutations of EF1, EF2, or EF1/EF2 (Figure 2C) led to dysfunction with a functional phenotype similar to MICU1 knockdown (KD) (Mallilankaraman et al., 2012b) in which the mitochondria constitutively accumulate Ca2+ that would otherwise remain as cytosolic Ca2+ (Figure 4H-4L). These data indicate that both polybasic and EF-hands are essential for MICU1 regulation of MCU.
MICU1 Polybasic and EF-hand Motifs Regulate IMCU
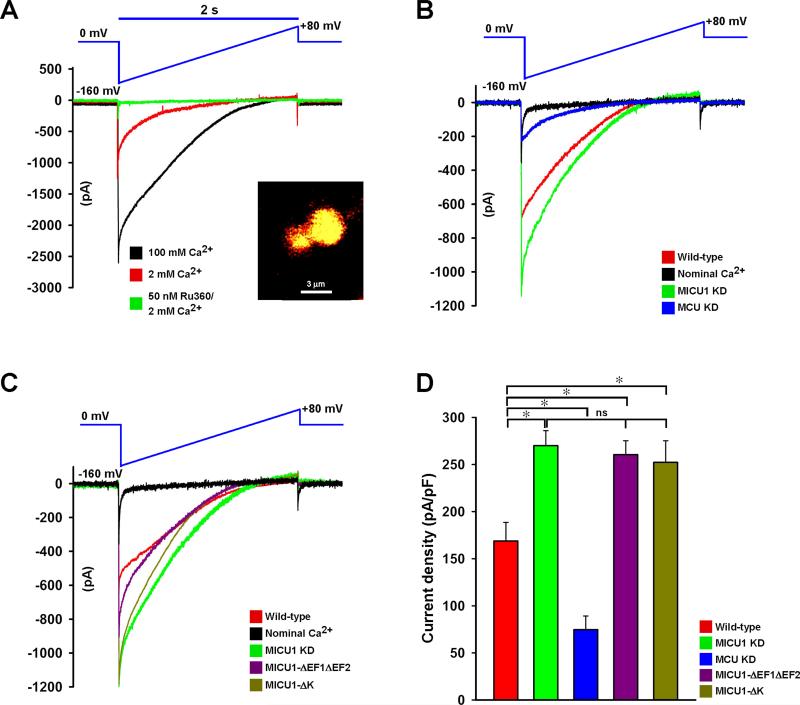
Because MCU-mediated mitochondrial Ca2+ uptake is regulated by MICU1, we examined whether MICU1-ΔK or MICU1-ΔEF1ΔEF2 mutants were able to regulate MCU current (IMCU) in HeLa cells. To measure IMCU, we utilized mitoplast patch-clamp (Chaudhuri et al., 2013; Fieni et al., 2012; Joiner et al., 2012; Kirichok et al., 2004). As a first step, freshly isolated mitochondria were loaded with ΔΨm indicator, rhodamine 123 and the mitoplast preparation was confirmed by confocal imaging (Figure 5A inset). In the patch-clamp of whole-mitoplast configuration, the addition of 2 mM Ca2+ to the bath elicited an inwardly rectifying Ca2+ current that is almost completely blocked by 50 nM Ru360 (Kirichok et al., 2004) (Figure 5A). Next, we examined if knockdown of human MICU1 or MCU proteins altered IMCU. Compared to control conditions, IMCU in mitoplasts from MCU knockdown (MCU KD) cells was significantly lower (Figure 5B and 5D) which is correlated with MCU-mediated Ca2+ uptake (Figure S3A-S3C). In sharp contrast, stable knockdown of MICU1 exhibited larger IMCU (Figure 5B and 5D). Finally, we conducted mitoplast patch-clamp recordings in MICU1-ΔK and MICU1-ΔEF1ΔEF2 mutants over-expressing HeLa cells. The electrophysiology revealed that MICU1-ΔK and ΔEF1ΔEF2 mitoplasts exhibited larger MCU current densities when compared to MCU currents from wild-type (Figure 5C and 5D). Having demonstrated the MCU Δ1 and Δ3 role in MICU1 binding, we next tested whether deletion of coiled-coil domains alter IMCU. MCU #861 shRNA insensitive MCU full-length and mutant constructs were stably expressed in MCU KD HeLa cells and IMCU was measured (Figure S3D-S3F). Reconstitution of full-length MCU in MCU KD mitoplasts reestablished IMCU (Figure S3E and S3F). Interestingly, MCU-Δ1 and MCU-Δ3 generated larger IMCU than MCU-Δ2 indicating that MCU-Δ1 and -Δ3 regions are necessary for MICU1-mediated regulation of MCU activity. Taken together, these data strongly support that loss of MICU1 polybasic or EF-hands increase the IMCU, exhibiting dominant negative phenotypes.
Figure 5. MICU1 Polybasic and EF-Hand Domains Regulate IMCU.
(A) Mitoplasts generated from HeLa mitochondria were loaded with ΔΨm indicator rhodamine 123 and visualized using confocal microscopy (inset). Mitoplast current (IMCU) was recorded before and after application of 2 (red) or 100 mM (black) Ca2+ to the bath medium. Currents were measured during a voltage-ramp as indicated. IMCU recorded in the presence of 2 mM Ca2+ was nearly completely inhibited by 50 nM of Ru360 (green trace). Traces are a representative single recording of IMCU. n = 6.
(B) Mitoplasts generated from HeLa wild-type cells (red), stable MICU1 KD (green), or stable MCU KD (blue) were used for IMCU measurements. IMCU was measured using 5 mM bath Ca2+ unless otherwise indicated. Nominal Ca2+-free buffer (no EGTA; contaminant Ca2+ ~6 μM) was used as a control and trace shown in black. Traces are a representative single recording of IMCU. n = 6-12.
(C) Similarly, IMCU from MICU1 mutants MICU1-ΔEF1ΔEF2 (purple) and MICU1-ΔK (brown) mitoplasts were measured and compared to wild-type (red) and MICU1 KD (green). IMCU was measured with 5 mM bath Ca2+ except for nominal Ca2+-free (black) medium. Traces are a representative single recording of IMCU. n = 6-12.
(D) IMCU densities (pA/pF) for wild-type (red), MICU1KD (green), MCU KD (blue), MICU1-ΔEF1ΔEF2 (purple), and MICU1-ΔK (brown). Mean ± SEM; *p < 0.05; ns = not significant, n = 6-12.
Loss of MICU1 Promotes Mitochondrial Ca2+ Accumulation and Oxidative Burden, Halting Endothelial Cell Migration
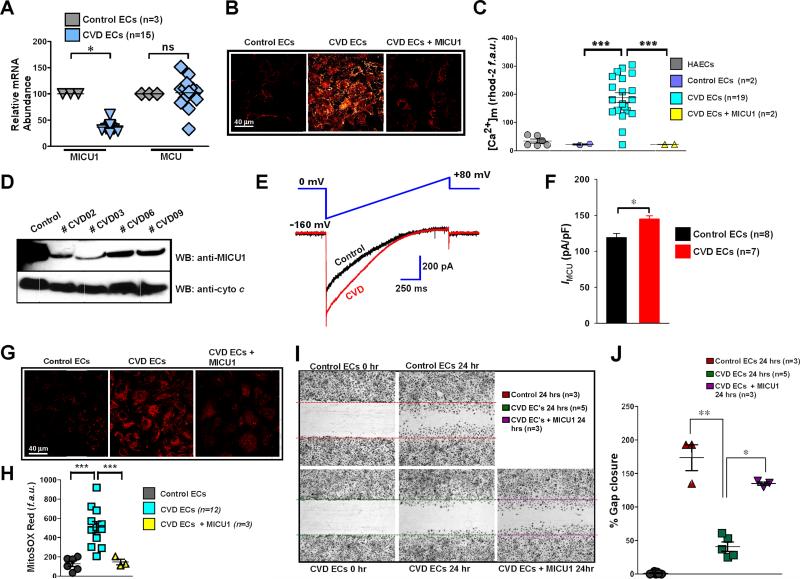
The ability of MICU1 to suppress MCU-mediated basal mitochondrial Ca2+ accumulation suggests that MICU1, rather than the MCU pore, is sensing Ca2+ uptake from the cytosol (Mallilankaraman et al., 2012b). Ablation of MICU1 causes mitochondrial Ca2+-dependent chronic oxidant elevation and cellular stress. The MICU1 and MCU expression profiles in cell lines and primary cells indicated that human aortic endothelial cells (HAECs) have relatively high levels of MICU1 mRNA when compared to other cell types (Figure S4A and S4B) (Aichberger et al., 2005). Nevertheless, MCU mRNA levels were relatively unchanged (Figure S4A and S4B). Therefore, we tested whether MICU1 downregulation impacts pathophysiological EC function. Because EC function is compromised in various cardiovascular disease states (Libby et al., 2006), we performed qRT-PCR to assess MICU1 expression in human control and cardiovascular vascular disease (CVD)-derived primary ECs (Table S1). MICU1 but not MCU mRNA levels were markedly downregulated in CVD ECs (Figure 6A). Basal [Ca2+]m was considerably elevated in ECs derived from CVD patients when compared to control ECs. Further, reconstitution of MICU1 in CVD ECs reduced the [Ca2+]m accumulation (Figure 6B and 6C). These results were further confirmed by the reduction of MICU1 protein in patients (Figure 6D). To examine if loss of MICU1 led to more MCU activity, mitochondria were isolated and mitoplast patch-clamp was performed. CVD ECs exhibited higher current density (Figure 6E and 6F) suggesting that the increased Ca2+ uptake is a result of increased open probability of MCU due to lower expression of MICU1 (Figure 6A and 6D). Aberrant basal mitochondrial Ca2+ uptake may lead to mitochondrial oxidative stress in the form of superoxide, which is produced when electrons leak from the electron transport chain to react with molecular oxygen. Ablation of MICU1 in the EC cell line, EA.hy926, increased mitochondrial superoxide levels (Figure S4C-S4E). In CVD ECs, loss of MICU1 resulted in higher [Ca2+]m, through altered MCU-mediated mitochondrial Ca2+ uptake which subsequently increased basal superoxide levels (Figure 6G and 6H). We next tested whether elevated ROS from MICU1 knockdown lowers antioxidant levels and alters EC proliferation (Balaban et al., 2005). Total reduced glutathione (GSH) content was assessed in EA.hy926 ECs expressing MICU1 shRNA or MICU1 shRNA + shRNA insensitive MICU1. Reconstitution of MICU1 in MICU1 KD cells prevented the chronic ROS-induced reduction of GSH levels (Figure S4F and S4G). Further, reduced cell proliferation in MICU1 silenced ECs was restored by reconstitution with shRNA insensitive MICU1cDNA (Figure S4H and S4I). Another key functional question is whether loss of MICU1 leads to altered angiogenesis in CVD. CVD-derived ECs exhibited diminished migration, which was rescued by stable expression of MICU1 (Figure 6I and 6J). We next tested whether CVD-derived ECs have an altered monocyte adhesion phenotype. We observed that CVD-derived ECs have an inherent propensity for monocyte adhesion (Figure S4J and S4K).
Figure 6. Loss of MICU1 Exacerbates MCU-mediated Ca2+ Uptake and Limits Endothelial Cell Migration.
(A) Human peripheral veins were obtained from control and coronary/peripheral artery disease subjects (CAD/PAD). Freshly isolated ECs were cultured and total RNA was used for detection of MICU1 and MCU mRNA expression levels by qRT-PCR. Mean ± SEM; *p < 0.05; ns = not significant, n = 3-15.
(B and C) Confocal imaging and quantitation of basal [Ca2+]m accumulation as indicated by mitochondrial rhod-2 fluorescence in control, CVD and CVD + MICU1 rescue ECs.Mean ± SEM; ***p < 0.001; n = 2-19 subjects.
(D) MICU1 protein expression in control and CVD ECs.
(E) IMCU is larger in human CVD endothelial cells derived mitoplasts. Traces are a representative single recording of IMCU. n = 7-8.
(F) Average IMCU densities at 5 mM Ca2+ in control and CVD ECs. Mean ± SEM; *p < 0.05; n = 7-8.
(G and H) Representative images and quantitation of MitoSOX Red fluorescence (mROS indicator) in control, CVD and CVD + MICU1 rescue ECs. Mean ± SEM; ***p < 0.001; n = 3-12.
(I and J) Representative images and quantitation of migratory scratch assay in CVD and CVD + MICU1 rescue ECs. Mean ± SEM; *p < 0.05, **p < 0.01; n = 3-5.
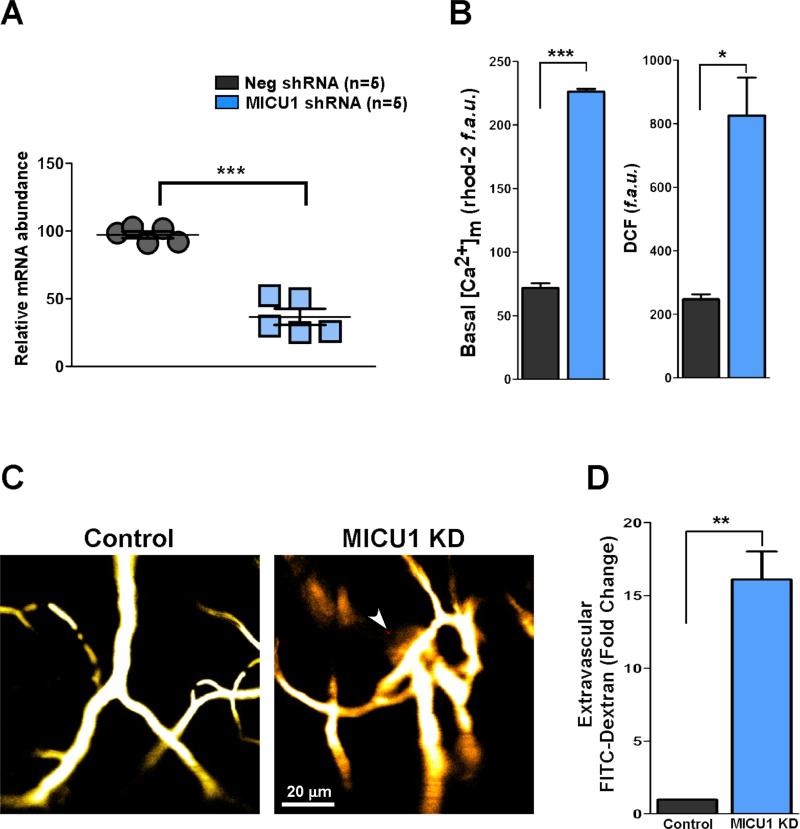
To explore the human MICU1 phenotype, we silenced MICU1 in mice and investigated the mitochondrial Ca2+ handling and vascular integrity. In vivo lentiviral delivery of MICU1 shRNA effectively knocked down MICU1 mRNA levels in ECs (Figure 7A). We also tested the MICU1 mRNA levels in cardiomyocytes, smooth muscle cells (SMCs) and splenocytes. Although SMCs and splenocytes, but not cardiomyocytes, showed partial knockdown of MICU1 following shRNA delivery, the MICU1 knockdown is more pronounced in ECs (Figure S5). Similar to cell based and CVD-derived studies, downregulation of MICU1 expression promoted higher basal [Ca2+]m and ROS production (Figure 7B) culminating as a loss of vascular integrity (Figure 7C and 7D). Collectively, these results show that loss of MICU1 leads to aberrant EC mitochondrial Ca2+ handling and vascular pathology.
Figure 7. In Vivo Knockdown of MICU1 Promotes Endothelial [Ca2+]m Accumulation, ROS Elevation and Vascular Integrity Loss.
(A) Knockdown of MICU1 mRNA levels in mouse endothelial cells after MICU1 shRNA lentiviral delivery. Total mRNA was isolated from ECs and qRT-PCR was performed. Mean ± SEM; ***p < 0.001, n = 5.
(B) Quantitation of basal [Ca2+]m accumulation in control and MICU1KD mouse ECs (left panel). Quantitation of basal ROS levels in control and MICU1KD mouse ECs (right panel). Mean ± SEM; *p < 0.05, ***p < 0.001; n = 5.
(C) 0.1- to 0.15-ml bolus of FITC-dextran (70 kDa, 5% w/v) was injected into the animals via facial vein. Anesthetized animals were placed under an intravital 2-photon imaging system (Zeiss 710 META NLO), and images were acquired. Images are representative of multiple independent experiments. n = 5.
(D) Quantitation of vascular leakage was assessed as extra-vascular FITC-dextran fluorescence intensity. Mean ± SEM; **p < 0.01; n = 5.
DISCUSSION
These findings establish that mitochondrial matrix resident, MICU1, forms regulatory oligomers that require the MICU1 polybasic region for MCU binding and suppression of IMCU. However, the polybasic region of MICU1 does not determine MICU1 oligomerization. Additionally, MICU1 EF hands which confer functional regulation of IMCU do not determine MICU1/MCU complex formation. Our experiments also establish that the coiled-coil domains of MCU are essential for MICU1 binding.
Identifying the physiological function of protein regions remains a challenge for advanced bioinformatics and requires broad-based biochemical techniques. One such new technique, the protein dynamic flux assay, provided a tool to discover MICU1 protein localization in the mitochondria while circumventing current conventional microscopy resolution limits. The major advantages of this method are that it is rapid, specific, real-time imaging, without RNAi silencing, subcellular fractionation, or antibodies, and it can resolve compartmentalization-determined protein functions such as residues of MCU accessible to MICU1. Taking the next step from localization, we show that the N-terminal region of the MICU1 polybasic motif determines MICU1/MCU binding. While the polybasic motif has been previously reported to enhance protein-protein interaction (Williams, 2003), to our knowledge this is the first polybasic motif identified in the mitochondria. The dominant negative MICU1-ΔK oligomer phenotype is likely a result of MICU1-ΔK interacting with wild-type MICU1, either sequestering wild-type MICU1 or causing a reduction of MICU1 oligomeric complex binding affinity for MCU. Together these data suggest that the polybasic region of MICU1 is not a MICU1/MICU1 interaction domain, but supports the polybasic region's role in MCU interaction. The loss of MICU1 regulation by polybasic mutation could also be explained by diminished interaction of the polybasic mutant with the inner leaflet of the IMM. In support of this, the conserved polybasic region contains a glycine residue that could undergo myristoylation for membrane anchoring (Mumby et al., 1990; Thelen et al., 1991). This anchor could provide a mechanism for MICU1 to come on and off at active and resting state. This loose association is supported by our FRAP studies which show that MICU1 as a population is more mobile than MCU (Figure 1R). These critically important and diverse roles of the polybasic region and its role in mitochondrial Ca2+ signaling will certainly be elucidated in future studies.
Despite physiological mitochondrial Ca2+ uptake participates in mitochondrial bioenergetics and cellular Ca2+ signaling, chronic Ca2+ accumulation is known to elicit mitochondrial dysfunction through mitochondrial permeability transition pore opening, mitochondrial ROS elevation, and cell death (Duchen et al., 2008; Hajnoczky et al., 2006). The mitochondrial Ca2+-dependent cellular damage pathways are shared in many pathophysiological models like cardiovascular disease, aging and neurodegeneration (Davidson and Duchen, 2007). In cardiovascular diseases, the endothelium suffers from Ca2+ dependent damage (Davidson and Duchen, 2007). Our results establish that endothelial cells derived from CVD subjects show low levels of MICU1 (Figures 6 and 7) and that the mitochondrial Ca2+ uptake component of the disease phenotype can be reversed by the reconstitution of MICU1, thus restoring endothelial cell migration.
Collectively, we established a unique resolution-independent confocal technique to characterize the localization of complex components in viable organelles. Most importantly, we revealed that MICU1 interacts with MCU and regulates MCU-dependent [Ca2+]m uptake with a polybasic region and Ca2+ sensing EF hand motifs. Additionally, we have shown that the coiled-coil domains of MCU interact with MICU1. Identification of these MICU1 and MCU domains provide the critical targets for mitochondrial Ca2+ uptake and potential therapeutic intervention in the pathogenesis of cardiovascular disease states.
EXPERIMENTAL PROCEDURES
Dynamic Protein Flux Assay
For all studies, MICU1, MCU and other plasmid constructs were generated from human sequences. HeLa cells were co-transfected with cyto c-GFP and COX8A-mRFP (mitochondrial targeting sequence; MTS), MICU1-YFP and COX8-mRFP or MCU-GFP and COX8-mRFP plasmid constructs. 48 hours post transfection, cell were permeabilized with digitonin (0.002% wt/vol) for seven minutes. Permeabilized cells were washed with digitonin free intracellular-like medium supplemented with 2 mM succinate. After 100 seconds of baseline recording, permeabilized cells were exposed to either mastoparan (20 μg/ml) or alamethicin (20 μg/ml) at the indicated time points. Confocal images were recorded every 10 s (510 Meta; Carl Zeiss, Inc.) at 488- and 561-nm excitation using a 40x oil objective. Experiments were performed under pinhole settings of approximately 2.3 μm to mitigate the impact of MICU1 rearrangement within the mitochondria. After completion of the time series, protein distribution maps were generated for before and after the mitochondrial membrane permeabilizing agents. Images were analyzed and flux rate was calculated. The colored histogram depiction represents the distribution of resolvable bit fluorescent intensities. Time is plotted from initial (top) to final (bottom) using the same individual experiment depicted in the representative individual trace. The intervals (bars) represent different time(s) during which the experiment was performed with the top bar indicating before the addition of either mastoparan or alamethicin. The protein flux images were analyzed using Zen 2010 software.
Fluorescence Recovery After Photobleaching (FRAP) Analysis
HeLa cells transfected with either MCU-GFP or MICU1-YFP were grown on glass coverslips for 48 hours before FRAP experiments. After baseline images were captured (one image/second), a small region (approximately 5% cytoplasmic area) was bleached with 10 iterations of 488 nm at 100% laser power using a Zeiss 510 confocal microscope. Recordings of fluorescence recovery were captured for the following 45 seconds at 488 nm excitation. Recovery in the bleached region was the sum of all diffusing molecules and normalized intensity was calculated as a percentage of the difference between the initial and bleached fluorescent arbitrary unit value.
Measurement of [Ca2+]m Uptake and Mitochondrial Membrane Potential (ΔΨm) in Permeabilized Cell System
Control, MICU1-ΔEF1ΔEF2 or MICU1-ΔK mutant HeLa cells were permeabilized using intracellular-like medium containing 2 μM thapsigargin and 40 μg/ml digitonin. The cell suspension supplemented with succinate (5 mM) was placed in a fluorimeter and permeabilized by gentle stirring. Fura-2FF (0.5 μM) was added at 0 s and JC-1 (800 nM) at 20 s to simultaneously measure extra-mitochondrial Ca2+ and ΔΨm, respectively. Fluorescence was monitored in a temperature-controlled (37°C) multi-wavelength-excitation dual wavelength-emission spectrofluorometer (Delta RAM, Photon Technology International) using 490-nm excitation/535-nm emission for the monomer, 570-nm excitation/595-nm emission for the J-aggregate of JC1and 340-nm/380-nm for Fura-2FF. The ratiometric dye, Fura-2FF was calibrated as previously described (Mallilankaraman et al., 2012b). At 450s, 1 μM Ca2+ pulse was added and ΔΨm and extra-mitochondrial Ca2+ were monitored simultaneously. CCCP was added at 750 s to determine mitochondrial Ca2+ content.
Mitoplast Patch-Clamp Recording
Mitoplast patch clamp recordings were performed at 30°C as detailed previously with modifications (Kirichok et al., 2004). Currents were recorded using a computer controlled Axon200B patch-clamp amplifier with a Digidata 1320A acquisition board (pClamp 10.0 software; Axon Instruments). The ionic composition of the pipette (2, 5 and 100 mM Ca2+) was chosen based on previous measurements. Mitoplasts with p-orbital morphology were used for patch-clamp recording. Mitoplasts were bathed with a solution containing sodium gluconate (150 mM), KCl (5.4 mM), CaCl2 (5 mM), Hepes (10 mM), pH 7.2. The pipette solution contained sodium gluconate (150 mM), NaCl (5 mM), sucrose (135 mM), HEPES (10 mM) and EGTA (1.5 mM), pH 7.2. After formation of GΩ seals (pipette resistance 20-35 MΩ), the mitoplasts were ruptured with 200-400 mV pulse varying from 2 to 6 ms duration. Mitoplast capacitance was measured (2.2-3.8 pF). After capacitance compensation, mitoplasts were held at 0 mV and IMCU were elicited with a voltage ramp (from −160 mV to 80 mV, 120 mV/s).
Isolation of Endothelial Cells From CVD And Control Subjects
Primary ECs were isolated from arteries and veins obtained from individuals undergoing surgical intervention for CAD/PAD at the Surgical Unit of Temple University Hospital after due ethical clearance. The tissues were collected in standard EC transport medium and transported to the laboratory through cold chain and processed immediately. The tissues were initially washed once with PBS containing 100 U/ml penicillin and 100 μg/ml streptomycin, then cleaned to remove the fibrous portion with sterile forceps. Using a sterile blade the veins were cut vertically and carefully flipped over. The tissues were cut into small squares of approximately 0.5 cm × 0.5 cm and placed in a 100 mm dish with medium just bathing the tissue pieces. The plates were incubated at 37°C, 5% CO 2. After a week, the tissue pieces were removed leaving the individual colonies to develop into a confluent monolayer. Endothelial cell phenotypes were characterized as previously described (Milovanova et al., 2008).
Endothelial Migration Assay
Control, CVD-derived ECs and CVD + MICU1 Rescue ECs were seeded at a density of 1×105 cells/well in 6-well plates overnight for confluent monolayer. A uniform 1.8 mm scratch running the entire length of the well was created using a sterile 200 μl tip. The wells were washed three times with PBS to remove the cell debris and then bathed in 2 ml complete endothelial medium. After 24 hours, cells were washed and fixed with CAMCO Quick Stain® II as per manufacturer instructions. The wells were photographed at multiple locations using phase contrast microscopy with a 4x objective. Migration was quantified using Image J software (NIH); results are expressed as percent gap closure (Craige et al., 2011).
In Vivo Knockdown of MICU1 and Vascular Integrity Measurement
To knockdown MICU1 in control mice we used lentivirus carrying MICU1-shRNA. Three daily doses of MICU1-shNRA (1×108 TU/ml as measured by p24 ELISA, and also confirmed by true infectious titer via puromycin resistance in EC culture; 100 μl/mice was delivered using saline as vehicle) lentivirus or control lentivirus negative shRNA (Neg shRNA) were administered intravenously and MICU1 expression in the endothelial cells, cardiomyocytes, smooth muscle cells (SMCs) and splenocytes were assessed by qRT-PCR after 5 days from the last injection (8 total days). Freshly isolated primary ECs were rapidly subjected to [Ca2+]m and ROS measurement. The vascular integrity studies were performed as described previously (Gandhirajan et al., 2013).
Statistical Analysis
Data were expressed as the means ± SE. Statistical significance was evaluated via one-way ANOVA and student's t test. A P value of < 0.05 was considered statistically significant. All experiments were conducted at least three times unless specified. Data were plotted either with GraphPad Prism version 5.0 or with Sigma Plot 11.0 software.
Supplementary Material
Highlights.
 Mitochondrial Ca2+ is maintained by MICU1 inhibition of mitochondrial uniporter
Mitochondrial Ca2+ is maintained by MICU1 inhibition of mitochondrial uniporter
 MICU1 localizes to the mitochondrial matrix and forms homo-oligomers
MICU1 localizes to the mitochondrial matrix and forms homo-oligomers
 MICU1/MCU binding is determined by a MICU1 N-terminal polybasic domain
MICU1/MCU binding is determined by a MICU1 N-terminal polybasic domain
 MICU1 EF-hands determine MCU activity but not binding
MICU1 EF-hands determine MCU activity but not binding
ACKNOWLEDGEMENTS
We thank Douglas R. Green for Cytochrome c - GFP plasmid construct. We thank Kevin J. Foskett, Donald L. Gill and Walter J. Koch for comments on the manuscript. This research was funded by the National Institutes of Health (HL086699, HL109920 and 1S10RR027327-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aichberger KJ, Mittermann I, Reininger R, Seiberler S, Swoboda I, Spitzauer S, Kopp T, Stingl G, Sperr WR, Valent P, et al. Hom s 4, an IgE-reactive autoantigen belonging to a new subfamily of calcium-binding proteins, can induce Th cell type 1-mediated autoreactivity. J Immunol. 2005;175:1286–1294. doi: 10.4049/jimmunol.175.2.1286. [DOI] [PubMed] [Google Scholar]
- Babcock DF, Herrington J, Goodwin PC, Park YB, Hille B. Mitochondrial participation in the intracellular Ca2+ network. J Cell Biol. 1997;136:833–844. doi: 10.1083/jcb.136.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- Balaban RS. The role of Ca(2+) signaling in the coordination of mitochondrial ATP production with cardiac work. Biochim Biophys Acta. 2009;1787:1334–1341. doi: 10.1016/j.bbabio.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi P. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev. 1999;79:1127–1155. doi: 10.1152/physrev.1999.79.4.1127. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, Vais H, Cheung KH, Yang J, Parker I, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri D, Sancak Y, Mootha VK, Clapham DE. MCU encodes the pore conducting mitochondrial calcium currents. eLife. 2013;2:e00704. doi: 10.7554/eLife.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Craige SM, Chen K, Pei Y, Li C, Huang X, Chen C, Shibata R, Sato K, Walsh K, Keaney JF., Jr. NADPH oxidase 4 promotes endothelial angiogenesis through endothelial nitric oxide synthase activation. Circulation. 2011;124:731–740. doi: 10.1161/CIRCULATIONAHA.111.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Fuente Perez S, Bogorad R, et al. MICU1 Controls Both the Threshold and Cooperative Activation of the Mitochondrial Ca(2+) Uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Duchen MR. Endothelial mitochondria: contributing to vascular function and disease. Circ Res. 2007;100:1128–1141. doi: 10.1161/01.RES.0000261970.18328.1d. [DOI] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton RM, McCormack JG. The role of calcium in the regulation of mitochondrial metabolism. Biochem Soc Trans. 1980;8:266–268. doi: 10.1042/bst0080266. [DOI] [PubMed] [Google Scholar]
- Drago I, Pizzo P, Pozzan T. After half a century mitochondrial calcium in- and efflux machineries reveal themselves. EMBO J. 2011;30:4119–4125. doi: 10.1038/emboj.2011.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR, Verkhratsky A, Muallem S. Mitochondria and calcium in health and disease. Cell Calcium. 2008;44:1–5. doi: 10.1016/j.ceca.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Fieni F, Lee SB, Jan YN, Kirichok Y. Activity of the mitochondrial calcium uniporter varies greatly between tissues. Nature communications. 2012;3:1317. doi: 10.1038/ncomms2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhirajan RK, Meng S, Chandramoorthy HC, Mallilankaraman K, Mancarella S, Gao H, Razmpour R, Yang XF, Houser SR, Chen J, et al. Blockade of NOX2 and STIM1 signaling limits lipopolysaccharide-induced vascular inflammation. J Clin Invest. 2013;123:887–902. doi: 10.1172/JCI65647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter TE, Gunter KK, Sheu SS, Gavin CE. Mitochondrial calcium transport: physiological and pathological relevance. Am J Physiol. 1994;267:C313–339. doi: 10.1152/ajpcell.1994.267.2.C313. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Csordas G, Das S, Garcia-Perez C, Saotome M, Sinha Roy S, Yi M. Mitochondrial calcium signalling and cell death: approaches for assessing the role of mitochondrial Ca2+ uptake in apoptosis. Cell Calcium. 2006;40:553–560. doi: 10.1016/j.ceca.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- Hansford RG. Physiological role of mitochondrial Ca2+ transport. J Bioenerg Biomembr. 1994;26:495–508. doi: 10.1007/BF00762734. [DOI] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner ML, Koval OM, Li J, He BJ, Allamargot C, Gao Z, Luczak ED, Hall DD, Fink BD, Chen B, et al. CaMKII determines mitochondrial stress responses in heart. Nature. 2012;491:269–273. doi: 10.1038/nature11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- Kluck RM, Esposti MD, Perkins G, Renken C, Kuwana T, Bossy-Wetzel E, Goldberg M, Allen T, Barber MJ, Green DR, et al. The pro-apoptotic proteins, Bid and Bax, cause a limited permeabilization of the mitochondrial outer membrane that is enhanced by cytosol. J Cell Biol. 1999;147:809–822. doi: 10.1083/jcb.147.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasters JJ, Theruvath TP, Zhong Z, Nieminen AL. Mitochondrial calcium and the permeability transition in cell death. Biochim Biophys Acta. 2009;1787:1395–1401. doi: 10.1016/j.bbabio.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P, Aikawa M, Jain MK. Vascular endothelium and atherosclerosis. Handbook of experimental pharmacology. 2006:285–306. doi: 10.1007/3-540-36028-x_9. [DOI] [PubMed] [Google Scholar]
- Mallilankaraman K, Cardenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenar T, Csordas G, Madireddi P, Yang J, Muller M, et al. MCUR1 is an essential component of mitochondrial Ca(2+) uptake that regulates cellular metabolism. Nat Cell Biol. 2012a;14:1336–1343. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, Hoffman NE, Gandhirajan RK, Molgo J, Birnbaum MJ, et al. MICU1 Is an Essential Gatekeeper for MCU-Mediated Mitochondrial Ca(2+) Uptake that Regulates Cell Survival. Cell. 2012b;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JG, Halestrap AP, Denton RM. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev. 1990;70:391–425. doi: 10.1152/physrev.1990.70.2.391. [DOI] [PubMed] [Google Scholar]
- Milovanova T, Chatterjee S, Hawkins BJ, Hong N, Sorokina EM, Debolt K, Moore JS, Madesh M, Fisher AB. Caveolae are an essential component of the pathway for endothelial cell signaling associated with abrupt reduction of shear stress. Biochim Biophys Acta. 2008;1783:1866–1875. doi: 10.1016/j.bbamcr.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby SM, Heukeroth RO, Gordon JI, Gilman AG. G-protein alpha- subunit expression, myristoylation, and membrane association in COS cells. Proc Natl Acad Sci U S A. 1990;87:728–732. doi: 10.1073/pnas.87.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG. The regulation of extramitochondrial free calcium ion concentration by rat liver mitochondria. Biochem J. 1978;176:463–474. doi: 10.1042/bj1760463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG. Mitochondria and calcium signaling. Cell Calcium. 2005;38:311–317. doi: 10.1016/j.ceca.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol. 2003;4:552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer DR, Gudz TI, Novgorodov SA, Erdahl WL. The peptide mastoparan is a potent facilitator of the mitochondrial permeability transition. J Biol Chem. 1995;270:4923–4932. doi: 10.1074/jbc.270.9.4923. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- Santo-Domingo J, Demaurex N. Calcium uptake mechanisms of mitochondria. Biochim Biophys Acta. 2010;1797:907–912. doi: 10.1016/j.bbabio.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai G, Krishnamurthy R, Hajnoczky G. Apoptosis driven by IP(3)- linked mitochondrial calcium signals. EMBO J. 1999;18:6349–6361. doi: 10.1093/emboj/18.22.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen M, Rosen A, Nairn AC, Aderem A. Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature. 1991;351:320–322. doi: 10.1038/351320a0. [DOI] [PubMed] [Google Scholar]
- Williams CL. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003;15:1071–1080. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- Williams GS, Boyman L, Chikando AC, Khairallah RJ, Lederer WJ. Mitochondrial calcium uptake. Proc Natl Acad Sci U S A. 2013;110:10479–10486. doi: 10.1073/pnas.1300410110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.