Abstract
Deviation from a balanced genome by either gain or loss of entire chromosomes is generally poorly tolerated in all eukaryotic systems studied to date. Errors in mitotic or meiotic cell division lead to aneuploidy, which places a burden of additional or insufficient gene products from the mis-segregated chromosomes on the daughter cells. The burden of aneuploidy often manifests itself as impaired fitness of individual cells and whole organisms, where abnormal development is also characteristic. Yet, the vast majority of human cancers, noted for their rapid growth, also display various levels of aneuploidy. Here we discuss the detrimental, potentially beneficial, and sometimes puzzling effects of aneuploidy on cellular and organismal fitness, tissue function, and its role in diseases such as cancer and neurodegeneration.
Keywords: Aneuploidy, chromosome mis-segregation, proteotoxic stress, cancer, neurodegenerative disease
I INTRODUCTION
Maintenance of a balanced euploid genome is a key requisite for the success of all multicellular organisms. While almost all mammals are diploid (the exception being some species of viscacha rat reported to be tetraploid (Gallardo et al. 1999)), there exist species of fungi, insects, reptiles, amphibians, and plants with chromosomal copy numbers ranging from one up to at least twelve (Comai 2005, Chen 2007). Regardless of ploidy, possession of an equal number of each chromosome ensures a balanced genome where genes on different chromosomes are present in equal numbers.
In contrast, aneuploidy, defined as a karyotype that is not a multiple of the haploid complement, results in an unbalanced genome with different copy numbers for genes on different chromosomes. Several studies have shown that gene expression levels are, for the most part, well correlated with genome copy number, meaning that aneuploid cells are burdened by either an excess or deficit of gene products located on the afflicted chromosomes. This is generally not well tolerated in nature, as evidenced by the impaired fitness of aneuploid cells and organisms. Notably, one type of aneuploidy where this is not typically the case is sex chromosome aneuploidy. In contrast to autosomes, sex chromosomes naturally vary in copy number between sexes. Dosage compensation mechanisms exist to equalize the dosages of sex chromosome-encoded genes between the two sexes. These mechanisms also seem able to partially compensate for abnormal numbers of sex chromosomes.
While dosage compensation may help to mitigate the effects of sex chromosome aneuploidy, this response is unavailable for autosomal aneuploidy in most species, meaning that the success of the organism depends on the accurate segregation of chromosomes. The importance of propagating equal numbers of chromosomes is underscored by the fact that surveillance mechanisms are in place that ensure that chromosome duplication and attachment to the mitotic spindle occur accurately before the cell cycle proceeds. Errors in DNA replication and checkpoint pathways that monitor the completion of DNA replication and DNA integrity result in compromised genome integrity. This rarely results in whole chromosomal mis-segregation, however, instead manifesting as chromosomal deletions, amplifications and non-reciprocal translocations. On the other hand, defects in mitosis, be it defects in chromosome attachment to the mitotic spindle, in the spindle structure or in the spindle-assembly checkpoint, the surveillance mechanism that monitors these events, lead to chromosome mis-segregation and aneuploidy.
In this review we will first briefly discuss how whole-chromosome aneuploidy arises. We will then focus on the consequences of aneuploidy, discussing its impact on organismal and cell physiology. We will end with a discussion of the potential contributions of aneuploidy to diseases such as cancer and neurodegeneration.
II ORIGINS OF WHOLE CHROMOSOME ANEUPLOIDY
To understand the origins of whole chromosome aneuploidies, we will first review how chromosomes are segregated. Cells begin to prepare for chromosome segregation during S phase, when replicated DNA strands, known as sister chromatids, are linked to each other via ring-like cohesin molecules (Nasmyth & Haering 2009). During prophase the pairs of sister chromatids attach to the mitotic spindle such that each sister kinetochore attaches to microtubules emanating from opposite spindle poles. In metaphase, sister chromatids are said to be bi-oriented, with sister kinetochores under tension created by the pulling forces of microtubules and cohesins that resist this pulling force. A checkpoint pathway known as the spindle-assembly checkpoint (SAC) monitors kinetochore attachment and tension, and halts cell cycle progression until every sister chromatid pair is correctly attached to the mitotic spindle (reviewed in Musacchio & Salmon, 2007). The target of the SAC is a ubiquitin ligase known as the Anaphase Promoting Complex or Cyclosome bound to its activating subunit Cdc20 (APC/C-Cdc20). Once all sister chromatids are accurately attached, SAC inhibition is relieved and APC/C-Cdc20 degrades the inhibitory subunit (called Securin) of a protease known as Separase. Active Separase then cleaves cohesins, thereby initiating chromosome segregation.
In all organisms analyzed to date, cells with compromised spindle-assembly checkpoint function, or with hyperactive APC/C-Cdc20, give rise to aneuploid daughter cells because chromosome segregation occurs in the presence of unattached or incorrectly attached chromosomes (Figure 1a). Defects in sister chromatid cohesion or hyperactivation of Separase, either by overexpression of Separase or inactivation of Securin, can also cause aneuploidy. Under defective cohesion, individual sister chromatids can segregate as they attach to microtubules, leading to virtually random segregation of chromosomes (Figure 1b; Nasmyth & Haering 2009). Aberrant microtubule-kinetochore attachments represent a major source of aneuploidy. Merotelic attachment, where one kinetochore is attached to microtubules emanating from both spindle poles, is a frequent occurrence in early mitosis and, although usually corrected before the onset of anaphase, can result in lagging chromosomes (Cimini et al. 2001; Thompson & Compton 2008; Bakhoum et al. 2009; Figure 1c). Recent work indicates that the majority of lagging chromosomes do not mis-segregate, but instead segregate accurately to end up in micronuclei in daughter cells (Thompson & Compton 2011). Only merotelic attachments that are unequal, meaning that kinetochores attach to more microtubules emanating from one pole than the other are thought to mis-segregate causing aneuploidy (Figure 1c). It is thought that, in contrast to unattached kinetochores or sister kinetochores attached to microtubules emanating from the same pole, merotely is more difficult to detect. Subtle differences in tension must be detected and therefore can be “overlooked” by the SAC, leading to entry into anaphase with faulty attachment.
Figure 1.
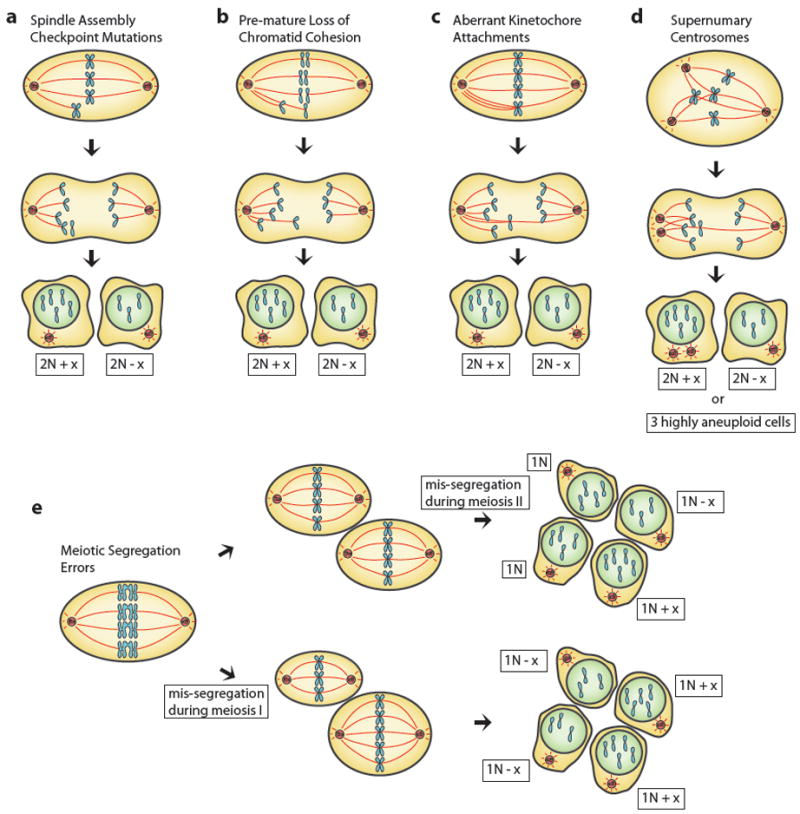
Cell division errors that result in aneuploidy. Mitotic cells mis-segregate one or multiple chromosomes by (a) mutations to the spindle assembly checkpoint pathway, where aberrantly attached chromosomes do not trigger a cell cycle arrest, (b) premature loss of chromatid cohesion, where single chromatids attach to microtubules and are randomly segregated, (c) merotelic attachment, where kinetochores attach to microtubules emanating from both centrosomes, or by (d) transition through a multi-polar spindle. Kinetochores are attached to several centrosomes, which eventually cluster, resulting in merotelic and syntelic attachments. (e) Aneuloidy can also result form mis-segregation of homologous chromosomes during meiosis I (bottom) or sister chromatids in meiosis II (top). “x” denotes any number of mis-segregated chromosomes.
Several conditions are known to greatly enhance merotely. The formation of multipolar spindles resulting from the cell possessing more than two centrosomes or fractured centrosomes gives rise to aberrant kinetochore attachment and lagging chromosomes (Ganem et al. 2009; Figure 1d). Multipolar spindles are generated through a variety of mechanisms including cell fusion, endoreduplication (duplication of the genome without a subsequent mitosis), failed cytokinesis and mitotic slippage (Brito & Rieder 2006), which describes a “giving up phenomenon”, where cells exit from mitosis after prolonged mitotic arrest creating tetraploid G1 cells (reviewed in Davoli & de Lange 2011). In all circumstances save when endoreduplication is not accompanied by centrosome duplication, the resultant tetraploid cell possesses extra centrosomes, which can then form multipolar spindles (Ganem et al. 2009). Recently, the creation of binucleate cells following entosis, the process of one cell being internalized by another cell, has also been shown to result in increased numbers of centrosomes (Krajcovic et al. 2011).
Chromosome mis-segregation can also arise during meiosis causing the creation of aneuploid gametes, which, when fertilized lead to entire organisms being aneuploid. Failure of several meiotic events has been shown to lead to aneuploidy, including premature separation of sister chromatids during meiosis I or II, failure to establish cross-overs between homologous chromosomes during meiosis I, and chromosome attachment defects in either meiosis I or II (Figure 1e). Additionally, highly aneuploid gametes are generated when the organism harbors a karyotype of odd ploidy.
Whole chromosomal aneuploidy can occur through many different mechanisms. Understanding how they arise and how they impact cellular and organismal physiology is critically important. In what follows, we will focus on what happens to cells and organisms in which the safe-guard mechanisms that prevent chromosome mis-segregation have failed and become aneuploid. We will see that the impact of the condition is severe and most frequently detrimental to cells and organisms.
III IMPAIRED FITNESS AND ABNORMAL DEVELOPMENT IN ANEUPLOID MULTICELLULAR ORGANISMS
Aneuploidy that arises during gamete formation or during the early embryonic divisions results in entire organisms with an aberrant karyotype. Studies of organismal aneuploidy across several species have shown that aneuploidy is frequently lethal early in development and that those karyotypes that are not lethal usually have substantial developmental defects.
The perhaps most famous early studies on aneuploidy were conducted by Marcella O’Grady and Theodor Boveri, who would later propose aneuploidy as a potential cause of cancer (Boveri 1902). They examined the effects of two sea urchin sperm fertilizing the same egg and noted that the vast majority of embryos derived from such fertilizations underwent a tripolar first mitosis, which generates extensive aneuploidy and leads to severe developmental defects and death.
Plants, likewise, have a long tradition in the study of organismal aneuploidy due to viability of polyploidy in many plant species and the frequent viability of offspring resulting from cross breeding of plants with different ploidies. Matings between diploid and triploid jimson weed (Datura stramonium) plants resulted in aneuploid plants that displayed differing phenotypes depending on the identity of the trisomic chromosome (Blakeslee 1921; Blakeslee et al. 1922). Despite phenotypic differences, it was, however, clear that all aneuploid plants grew more poorly than euploid plants and the severity of the growth defect scaled with the size of the trisomic chromosome, a phenomenon also later observed in other organisms. Similarly, growth defects resulting from aneuploidy have been observed in maize, rice, and Arabidosis thaliana. (McClintock 1929; Singh et al. 1996; Henry et al. 2010; Figure 2a). It is noteworthy that plants appear to be remarkably resistant to the adverse effects of aneuploidy. As discussed in more detail below, some type of dosage compensation may occur on autosomes in plants, allowing them to better tolerate gene copy imbalances.
Figure 2.

Examples of the effects of aneuploidy on organisms. (a) Aneuploid Arabidopsis thaliana rosettes (right, trisomic for chromosomes 3 and 5) show thinner, distorted leaves in comparison with diploid rosettes (left), from Henry et al. 2010. Tetrasomy IV Drosophila melanogaster wings (b, right) are smaller and more pointed than wild type (left), from Grell 1961. Trisomy 16 (Ts16) mouse embryos (c, right) show a smaller size compared to diploid littermates (left), as well as a variety of developmental abnormalities including nuchal edema, from Williams et al. 2008. “Ts” indicates trisomy and “Tet” indicates tetrasomy.
In the fruit fly Drosophila melanogaster, all whole chromosome aneuploidies are lethal save for aneuploidies of chromosome IV, the smallest chromosome. Monosomy of chromosome IV is viable and results in a mutant known as Diminished, with phenotypes including smaller size and delayed hatching (Bridges 1921a). In addition, these flies are frequently sterile. Flies trisomic for chromosome IV are also viable, with the morphological differences between trisomic and diploid flies being subtle (Bridges 1921b; Li 1927). Finally, in rare cases, diploid flies tetrasomic for chromosome IV survive to adulthood. These flies have more obvious morphological defects (Grell 1961; Figure 2b). Substantial work has also been performed on segmental aneuploidies in Drosophila. Lindsley et al. described the identification of 57 loci throughout the fruit fly genome that result in phenotypic variation, the most common of which was a Minute (small) fly (Lindsley et al. 1972). Highlighting an additional trend conserved across species, monosomies (even segmental monosomies) produced more pronounced phenotypes than trisomies.
In the nematode Caenorhabditis elegans, which has five autosomes of roughly equal size and one sex chromosome, three aneuploidies are viable: trisomy of the X chromosome (Hodgkin et al. 1979), trisomy of chromosome IV (Sigurdson et al. 1984), and a triploid animal monosomic for the X chromosome (Hodgkin 1987). Both sex chromosome aneuploids are abnormal, but animals trisomic for chromosome IV exhibit few phenotypes beyond reduced fertility. While no other aneuploid worms have been described, C. elegans has been shown to tolerate large duplications and deletions throughout much of the genome (reviewed in Hodgkin 2005), although the tolerated duplications and deletions are still smaller than the smallest chromosome.
The study of aneuploidy in vertebrates has been extensive. In mouse, all autosomal aneuploidies are embryonic lethal with the exception of Trisomy 19, which is the smallest mouse autosome and even these mice die shortly after birth (Gropp et al. 1983; Dyban & Baranov 1987). Trisomic embryos exhibit various degrees of developmental retardation, as well as a variety of developmental defects including cranio-facial defects, hypoplasia and nuchal edema (Gropp et al. 1975; Krushinskii et al. 1986; Figure 2c).
By far the best-known and most-studied organismal aneuploidy in vertebrates is Trisomy 21 in humans, the genetic condition responsible for Down syndrome. Similar to viable aneuploids in other species, individuals with Down syndrome frequently display stunted growth. Other characteristics associated with Down syndrome are mental retardation, decreased fertility, and increased rates of congenital heart disease, hypothyroidism, epilepsy, Alzheimer’s disease, and acute leukemias. Down syndrome individuals do, however, exhibit a decreased risk of solid tumors (Hasle et al. 2000; Satge et al. 2003). Two other trisomies survive to birth in humans; these are Trisomy 18 (Edwards syndrome) and Trisomy 13 (Patau syndrome). Only approximately 10% of children born with these syndromes live to one year of age (Rasmussen et al. 2003). Edwards and Patau syndrome fetuses display severe developmental abnormalities including microcephaly, cleft palate, and cardiac abnormalities. Autosomal monosomies are inviable in humans. Several segmental monosomies, however, have been described to cause a variety of developmental abnormalities (Shaffer & Lupski 2000). Finally, a rare condition exists in humans, called mosaic variegated aneuploidy (MVA), in which a systemic predisposition to mitotic non-disjunction (via mutations of the spindle assembly checkpoint component BUBR1 (Hanks et al. 2004) or the centrosomal protein CEP57 (Snape et al. 2011)) leads to individuals with more than 25% aneuploid cells. MVA is associated with growth deficiencies, microcephaly, developmental delay, and an increased risk for childhood cancers.
As observed in Drosphila, C. elegans and mice, aneuploidies of the sex chromosomes in humans are viable. Turner syndrome (monosomy of the X chromosome) is the only whole chromosomal monosomy viable in humans, in which common symptoms include short stature, infertility, and cardiac or kidney abnormalities. Klinefelter’s (XXY) and Triple X (XXX) syndromes usually result in mild physical or developmental symptoms and reduced fertility. Rare instances of individuals with four or five sex chromosomes have also been observed. This clear difference in severity of phenotypes between autosomal aneuploidies and sex chromosome aneuploidies highlights the contribution of copy number imbalances to the phenotypes of aneuploidy. For chromosomes where dosage compensation occurs, i.e. the X chromosome (reviewed in Berletch et al. 2011; Prestel et al. 2010), the phenotypes are mild, for autosomes where dosage compensation is less common and gene expression generally correlates well with copy number, the consequences of aneuploidy are severe.
In summary, it is clear that in all species analyzed to date, as described for multicellular organisms here and for single celled yeast below, aneuploidy is detrimental, causing growth retardation and developmental abnormalities. Understanding the basis of these phenotypes has been the focus of much investigation in recent years and current progress is highlighted below.
IV MODEL SYSTEMS TO STUDY ANEUPLOIDY IN CELLS
The cellular effects of aneuploidy have been the subject of an intense investigation that hopes to connect aneuploid cellular physiology to observed characteristics of whole-organism aneuploidy and to diseases such as cancer. Recent efforts have established a variety of yeast strains and mammalian cell lines with defined aneuploidies or with random, varying aneuploidies.
Several methods have been used to derive aneuploid yeast strains and mammalian cell lines to study the effects of aneuploidy at the cellular level. These can be broadly divided into two categories: (1) random aneuploidies generated through mutations that cause increased chromosome mis-segregation or through meiosis of cells with an odd ploidy and (2) defined aneuploidies created through chromosome transfers or meiotic non-disjunction. We will briefly explain the predominant techniques for generating aneuploid strains and mammalian cell lines below.
In yeast, random aneuploidies can be generated by two means: through employing strains with mutations that cause increased chromosome mis-segregation and by sporulating strains harboring an odd number of genetic complements (i.e. triploid strains; Figure 3a). This method has been employed in fission (Niwa & Yanagida 1985; Niwa et al. 2006) and budding yeast (Parry & Cox 1970; St. Charles et al. 2010; Pavelka et al. 2010a) to create highly aneuploid strains. These strains are, however, karyotypically unstable (Niwa & Yanagida 1985; Niwa et al. 2006; St. Charles et al. 2010; Sheltzer et al. 2011a), complicating their use in examining the effects of aneuploidy on cell physiology. Strains with defined aneuploidies have been created in budding yeast by transferring single chromosomes from a donor strain into a recipient strain (Conde & Fink 1976; Torres et al. 2007; Figure 3b). In this manner, near-haploid strains have been created that harbor two copies of one chromosome, referred to as disomes, which carry different selectable markers at the same locus on each homologous chromosome to allow stable propagation through selection. Only a limited number of mostly single chromosomal aneuploidies can be created using this approach, however, precluding the study of effects of complex aneuploidies.
Figure 3.

Laboratory models of aneuploidy. Several methods have been used to create aneuploid yeast strains and mammalian cells. (a) Yeast strains with uneven ploidy produce highly aneuploid meiotic products (spores). (b) Karyogamy defective yeast strains (kar1Δ15) can be used to produce rare chromosome transfers between nuclei during abortive matings by simultaneously selecting for two different markers present on homologous chromosomes. (c) Aneuploid mouse embryos can be generated by relying on meiotic non-disjunctions during gamete formation in male mice with two heterozygous Robertsonian fusion chromosomes (for example the fusions of 11/13 and 13/16 can create sperm with two copies of chromosome 13). When such males are crossed to wild-type females, trisomic embryos are generated. (d) Chromosome transfer by fusion of micro-cell encapsulated chromosomes to recipient cells (Saxon & Stanbridge 1987) can be used to create aneuploid cell lines. “x” denotes any number of mis-segregated chromosomes.
Karyotypically complex but unstable aneuploidies and stable defined single chromosomal abnormalities have also been developed in the mouse. Mice harboring mutations in spindle assembly checkpoint components (Michel et al. 2001; Dobles et al. 2000; Kalitsis et al. 2000; Babu et al. 2003; Perera et al. 2007; Putkey et al. 2002; Jeganathan et al. 2005; Jeganathan et al. 2007; Weaver et al. 2003; Baker et al. 2004; Diaz-Rodríguez et al. 2008; Schliekelman et al. 2011) or checkpoint insensitive APC/C-Cdc20 (Rao et al. 2005; Li et al. 2009) have been generated. Cell lines derived from such mice are populations of karyotypically diverse cells that are ideally suited for the study of chromosomal instability (CIN), but have, in the instances where this was examined, also shed light on the effect of aneuploidy on cell physiology (i.e. Baker et al. 2004; Li et al. 2010).
Stable single chromosomal aneuploidies have been created by two different methods. Mouse embryonic fibroblasts (MEFs) harboring a defined trisomy have been created from crosses of male mice heterozygous for two separate Robertsonian fusion chromosomes (which cause meiotic non-disjunction) with wild-type female mice (Gropp et al. 1975; Williams et al., 2008; Figure 3c). Specific aneuploidies have also been introduced into diploid immortalized and cancer cells via microcell-mediated chromosome transfer (Upender et al. 2004; Figure 3d).
The study of these different types of aneuploidy has greatly enhanced our understanding of the effects of aneuploidy on cells. Chromosome-specific effects have been revealed, as well as phenotypes shared between many different aneuploidies indicating that a response to the aneuploid state exists in cells. In what follows we will summarize these efforts, highlighting commonalities between discoveries made in yeast and mammalian tissue culture cells.
V IMPACT OF ANEUPLOIDY ON GENE EXPRESSION
Before discussing the effects aneuploidy on cell physiology we must consider the important question of whether the observed phenotypes are due to the mere presence of additional DNA in the form of chromosomes or lack of chromosomes, or whether they are due to changes in gene expression levels. It is thus essential to know whether aneuploid chromosomes are expressed according to gene copy number or whether compensatory mechanisms exist that attempt to “balance out” gene copy number imbalances caused by aneuploidy, as they do for sex chromosome aneuploidies. Studies of cellular systems across several species indicate that, with few exceptions, aneuploid autosomes are both actively transcribed and translated in proportion to their gene copy number.
In 1979, David Kurnit showed that total mRNA from chromosome 21 in patient derived fibroblasts with monosomy 21, disomy 21 (normal) and trisomy 21 (Down syndrome) was present in proportion to the genomic copy number of the chromosome (Kurnit). Microarray technology provided direct evidence for a genome proportional up-regulation of all genes across the aneuploid chromosome 21 (Mao et al. 2003). A similar genome proportional expression of aneuploid chromosomes was observed in aneuploid fission yeast, budding yeast, MEFs, and adult partially trisomic mouse tissues (Chikashige et al. 2007; Torres et al. 2007; Williams et al. 2008; Kahlem et al. 2004; Lyle et al. 2004; Vacík et al. 2005). The correlation between chromosome copy number and relative gene expression levels is so strong that gene expression arrays can be used to accurately karyotype aneuploidy, as shown in S. cerevisae (Hughes et al. 2000) and the pathogenic fungus Candida albicans (Bouchonville et al. 2009). Expression profiling has also been shown to reflect karyotypes in several types of cancers, including glioblastoma (Gao et al. 2007), lung adenocarcinoma (Lu et al. 2011), oral squamous cell carcinoma (Xu et al. 2010), colorectal cancer (Tsafrir et al. 2006), and breast cancer (Pollack et al. 2002).
In yeast and mammals, gene expression appears to correlate well with gene copy number, however this appears not to be the case in Drosophila and plants. In Drosophila, a specific mechanism exists to normalize the expression of genes on the fourth chromosome. The Painting of fourth protein binds to and normalizes the expression level of chromosome four to diploid levels (Larsson et al. 2004). A general mechanism for dosage compensation also appears to exist for segmental aneuploids (Zhang et al. 2010). In addition to this general buffering mechanism and the chromosome four-specific mechanism, Drosophila also has a sex chromosome specific dosage compensation mechanism, meaning that fruit flies contain not one, but three distinct mechanisms to ensure balanced gene expression across even aneuploid genomes (reviewed in Stenberg & Larsson 2011).
Due to the high viability of aneuploidy in plants, dosage compensation has long been studied. In maize, compensation occurs on most genes on aneuploid chromosomes at the RNA level (Guo & Birchler 1994). A study of gene expression in leaves of developing aneuploid Arabidopsis plants has shown that aneuploidy alters the expression levels of several genes dispersed across the genome (Huettel et al. 2008). A similar effect has also been shown to occur in aneuploid maize (Makarevitch & Harris 2010). Thus, mechanisms exist in some organisms that dampen the gene dosage imbalances caused by aneuploidy. Why these mechanisms appear more prevalent in plants is an important question that deserves further investigation.
Clearly, aneuploid autosomes are at least active to some extent in all organisms where this has been investigated, but there remains ambiguity as to whether these increases in gene expression translate into increased levels of protein. Quantitative proteomic analysis showed that at least in budding yeast the majority of genes are not only transcribed according to gene copy number but that protein levels also largely reflect gene copy number (Torres et al. 2010a; Pavelka et al. 2010a; Figure 4a). There is disagreement, however, as to what extent this is the case. Pavelka et al. reported that virtually all genes are expressed according to gene copy number also at the protein levels. Torres et al. reported that approximately 20% of proteins are not. The proteins that were not found to be expressed in proportion to gene copy number were predominantly found to be components of large protein complexes which is in agreement with an earlier report from the same group that specifically looked at a small number of proteins that function in large protein complexes (Torres et al. 2007). Differences in methodology, SILAC (Ong et al. 2002) used by Torres et al. versus MudPIT (Ghaemmaghami et al. 2003) by Pavelka et al., are most likely the reason for the apparent discrepancies.
Figure 4.

Cellular systems of aneuploidy display growth defects and proteotoxic stress phenotypes. (a) Micro-array DNA content analysis, micro-array gene expression data, and SILAC protein profiling of a haploid budding yeast strain disomic for chromosome V shows transcript and protein levels of chromosome V-encoded genes to be increased by approximately 1.8 fold over wild-type (data from Torres et al. 2007 & Torres et al. 2010a). (b) Growth of aneuploid budding yeast shows a proliferation defect in most aneuploid strains (data from Torres et al. 2007). (c) Increased basal expression of the inducible protein chaperone Hsp72 in trisomic (Ts) MEFs (data from Tang et al. 2011).
Whether or not protein expression broadly correlates with gene copy number in other organisms is not clear. Some individual proteins in aneuploid plants and Drosophila are expressed at levels corresponding to their gene copy number, whereas others are not. As indicated by enzyme level analysis in partially triploid Drosophila pupae, the aforementioned general buffering mechanism in Drosophila normalizes autosomal expression levels for some, but not all proteins (Devlin et al. 1982). Total protein analysis of aneuploid maize showed a similar effect (Birchler & Newton 1981). A study of total protein levels in trisomic mouse embryos as analyzed by two-dimensional gel electrophoresis, however, suggested that protein levels did not correlate well with gene copy number in this system (Klose & Putz 1983). Clearly, quantitative proteomic approaches, as have been applied to budding yeast, will be needed to clarify the extent to which protein levels correlate with gene copy numbers in higher eukaryotes.
In summary, it appears that species-specific differences exist in the ability to attenuate gene expression from aneuploid chromosomes, both at the RNA and protein levels. Better understanding these differences will likely provide key insights into the effects of aneuploidy on cell and organismal physiology.
VI EFFECTS OF ANEUPLOIDY OF CELL PHYSIOLOGY
Cellular studies have revealed that, in addition to chromosome specific effects, aneuploidy causes several general effects. These are largely detrimental and appear to be conserved across species. In rare circumstances, however, beneficial effects of aneuploidy have also been observed. These rare beneficial aneuploidies may help to explain why aneuploid cells are observed in some normally functioning, healthy tissues in addition to diseased cells.
Aneuploidy causes chromosome specific and general effects and is largely detrimental to cells
It is clear that there are phenotypes that are unique to specific chromosomal aneuploidies. These cases can be traced to imbalances in one or a few gene products on a particular chromosome and will not be discussed here. Instead we will focus on general aspects of aneuploidy to better understand the consequences of carrying an unbalanced karyotype.
Among the key phenotypes shared by aneuploid cells is their slower proliferation compared to euploid cells. This effect was first observed in fibroblasts derived from individuals with Down syndrome (Segal & McCoy 1974). The generality of this phenomenon, however, was not appreciated until a systematic analysis began of cells harboring a variety of different aneuploidies. Studies of S. pombe aneuploid strains derived from triploid meioses (Niwa et al. 2006) and S. cerevisiae strains carrying one or two additional chromosomes (“disomes”; Torres et al. 2007) or derived from triploid meioses (Pavelka et al. 2010a) showed that aneuploidy impairs proliferation (Figure 4b). Similar slowed growth was observed in mouse cells. Aneuploid mouse embryonic fibroblasts (MEFs) trisomic for either chromosome 1, 13, 16, or 19 (Williams et al. 2008) exhibit proliferation defects, as do cells harboring random aneuploidies caused by impaired SAC function. MEFs derived from mice homozygous for a hypomorphic allele of the SAC component BubR1 showed slower proliferation than wild-type MEFs once aneuploidies had accumulated in the culture (passage 7), but not at the earlier passage 3 when the degree of aneuploidy was less severe (Baker et al. 2004). Aneuploidies obtained through chemically-induced mitotic non-disjunction or MEFs harboring mutations in the SAC component Bub1 or mutations that render the checkpoint effector Cdc20 unresponsive to the checkpoint signal also exhibit proliferation defects and are outcompeted by diploid cells in cultures (Thompson & Compton 2008; Li et al. 2009). Impaired proliferation of aneuploid cells has not been described in aneuploidy models that spawn low-grade aneuploidies at a lower frequency such as cells heterozygous for deletions in CENP-E (Weaver et al. 2007), Bub3, or Rae1 (Babu et al. 2003), or cells that overexpress the ubiquitin conjugating enzyme UbcH10 (van Ree et al. 2010). We suspect that aneuploidy of a small fraction of cells could go unnoticed in population doubling time measurements.
Beyond proliferation rates, aneuploid cells also exhibit a number of other phenotypes that can be broadly summarized as an “aneuploidy stress response” (Figure 5). The metabolism of aneuploid cells is altered. Biomass production is decreased in aneuploid yeast strains (Torres et al. 2007) and metabolic changes are also seen in aneuploid mammalian cells. MEF cultures containing the aneuploidy-inducing Cdc20AAA mutation exhibit increased glucose uptake and increased production of lactate and reactive oxygen species (Li et al. 2010). The increased use of glucose was not observed in the majority of trisomic MEF lines, but these cell lines did show an increased uptake of glutamine, as well as increased production of lactate and ammonium (Williams et al. 2008). Trisomic MEFs were also shown to be under energy stress, and this stress could be magnified to the point of aneuploid-specific cell death by treatment with the AMP kinase activator AICAR (Tang et al. 2011).
Figure 5.

Observed characteristics of aneuploidy in (a) yeast and (b) mammalian cells. Purple boxes indicate conditional changes resulting from aneuploidy, while blue boxes indicate observed physiological stresses. See text for details.
In addition to energy stress, proteotoxic stress, i.e. physiological strain accrued from an abundance of misfolded proteins, is present in aneuploid cells (Figure 5). By and large, aneuploid S. cerevisiae grew slower under conditions of proteotoxic stress: elevated temperature or treatment with the protein synthesis inhibitors cycloheximide and hygromycin, and the Hsp90 inhibitor geldanamycin (Torres et al. 2007; Pavelka et al. 2010a). Furthermore, aneuploid yeast cells form protein aggregates and appear challenged in their ability to fold proteins (A.B. Oromendia, S. Dodgson, G. Brar, C. Gonzalez, J Weissman & A. Amon, unpublished data). Some aneuploids also have been shown to up-regulate genes involved in regulating proteasome activity (Jung et al. 2011). In addition, evolution of disomic yeast strains identified several components of the proteasome pathway that were mutated in aneuploid strains evolved to exhibit increased proliferative abilities. Specifically, loss of function mutations in UBP6, a gene encoding a deubiquitinase that antagonizes proteasome function, improved the fitness of several different disomic strains and partially attenuated the increased protein expression on the aneuploid chromosomes, presumably by increasing protein turnover rates (Torres et al. 2010a).
The proteotoxic stress observed in aneuploid yeast strains seems to be due to protein stoichiometry imbalances brought about by gene copy number imbalances, because increasing the basic ploidy of cells mitigates the proteotoxic effects of aneuploidy. Furthermore, chromosome sized mouse or human DNA fragments do not cause proteotoxicity in yeast (Torres et al. 2007; A.B. Oromendia, S. Dodgson, G. Brar, C. Gonzalez, J Weissman & A. Amon, unpublished data). Evidence for proteotoxic stress also exists in aneuploid mammalian cells. Trisomic MEFs harbor increased rates of autophagy and increased basal levels of the inducible chaperone Hsp72, as well as exhibit increased sensitivity to the Hsp90 chaperone inhibitor 17-AAG (Tang et al. 2011; Figure 4c).
The observed stresses and phenotypic changes in aneuploid cells beg the question of whether cells launch a system wide response to aneuploidy. While the expression levels of genes on aneuploid chromosomes scale with genomic copy number, additional changes to the transcriptome have been observed to result from aneuploidy and the slow-growth that accompanies the condition. Disomic yeast display an expression profile indicative of the environmental stress response in yeast (Gasch et al. 2000; Torres et al. 2007; Jung et al. 2011). A recent investigation suggests that this effect is conserved across species (J.M. Sheltzer, E.M. Torres, M.J. Dunham & A. Amon, unpublished data).
Aneuploidy has also been shown to increase genomic instability. All disomic budding yeast strains investigated were found, through a variety of assays, to have some combination of increased rates of chromosome mis-segregation, mitotic recombination, mutation, increased DNA damage, or increased sensitivity to genotoxins (Sheltzer et al. 2011a). In the same report, fission yeast disomic for chromosome III were shown to harbor increased sensitivity to DNA damaging agents. Two recent studies in mammalian cells demonstrate that genomic instability, especially DNA damage, is the result of chromosome mis-segregation. Merotelically attached chromosomes, induced by compounds that interfere with mitotic spindle formation, remain in the center of the cell during anaphase and are broken during cytokinesis (Janssen et al. 2011). Lagging chromosomes can also form micronuclei, which then experience substantial DNA damage during subsequent replication (Crasta et al. 2012).
In mammalian cells aneuploidy appears to induce a p53 response (Figure 5b), although the reported mechanisms differ. Janssen et al. found that lagging chromosomes suffer DNA damage during cytokinesis, which activates p53 through the ATM pathway (Janssen et al. 2011; Figure 5b). A second study using the same treatment found no evidence for DNA damage in lagging chromosomes after treatment, however p53 was still activated and halted cell cycle progression, but through a p38 kinase dependent stress response, presumably triggered by aneuploidy-induced stresses such as metabolic alterations and protein imbalances (Thompson & Compton 2010; Figure 5b). A third report, studying spindle assembly checkpoint mutant MEFs, found that p53 was activated by ATM, but in response to aneuploidy itself rather than DNA damage. In Cdc20AAA and BubR1 mutant MEFs, loss of the spindle assembly checkpoint pathway led to p53 activation that persisted even after the checkpoint was restored. These cells showed alterations in metabolism, in particular, an increase in reactive oxygen species (ROS) led to the activation of p53 through activation of ATM (Li et al. 2010). Interestingly, this study also found increased levels of p53 activity correlating with the severity of the aneuploidy, which may explain why p53 activity was not found to be increased in trisomic MEFs (Tang et al. 2011). These results may also help explain the high coincidence of aneuploidy and loss of p53 function in cancer (Tomasini et al. 2008).
How aneuploidy leads to the observed phenotypes is, perhaps, the key open question in the field (reviewed in Sheltzer & Amon 2011b, Torres et al. 2010b; Pavelka et al. 2010b). Several reports suggest that copy number changes of individual genes can cause severe phenotypes and might drive aneuploidies of specific chromosomes. For example, amplifications of the oncogene Myc are thought to be the main driving factor for the high frequency of Trisomy 8 in human acute myeloid leukemia (Jones et al. 2010). Likewise, phenotypes of aneuploid yeast evolved from a yeast strain defective in cytokinesis were found to be phenocopied by duplications of a specific transcription factor, RLM1, and its activator, MKK2, which are both located on the aneuploid chromosome (Rancati et al. 2008). Another, not mutually exclusive, possibility is that the cumulative effect of a large number of small effects resulting from changes in copy number of a large number of genes might drive certain phenotypes. One example is the proteotoxic stress observed in many aneuploid cells. Another example is the activation of p53 by reactive oxygen species. Proteins on aneuploid chromosomes are translated, which increases ROS levels in proportion to the severity of the aneuploidy. This increased ROS is sensed by ATM, which phosphorylates p53, again, in proportion to the severity of the aneuploidy (Li et al. 2010). Additional evidence for the cumulative effect of several small effects is that the severity of phenotypes observed in disomic budding yeast largely scales with the size of the aneuploid chromosome (Torres et al. 2007; Torres et al. 2008). Mouse models of Down syndrome suggest that both cumulative effects and individual genes are responsible for the complex phenotypes of the disease (Reeves et al. 1995; Sago et al. 1998; O’Doherty et al. 2005; Reeves et al. 2006; Dierssen et al. 2009). Individual genes appear to be responsible for some phenotypes, i.e. duplication of the gene APP, which encodes a protein that when cleaved forms the main component of amyloid-β plaques, is thought to cause the Alzheimer’s Disease phenotype (reviewed in Kingsbury et al. 2006), however neither duplication of one gene nor the originally defined “Down Syndrome Critical Region” is sufficient to recapitulate all phenotypes. The ~82% of human chromosome 21 present in the Tc1 mouse is likewise insufficient (see Lana-Elola et al. 2011 for a review). Most likely, some phenotypes resulting from aneuploidy are the result of cumulative effects of multiple gene duplications whereas others are caused by changes in the dosage of single genes.
(Potentially) beneficial effects to cellular fitness
Despite the clearly detrimental effects of aneuploidy on cellular fitness, the condition can, in rare cases of strong selective pressures, give cells a competitive edge. How this potential of aneuploidy to provide adaptive power could contribute to the development of new traits will be discussed next.
The pathogenic fungus C. albicans can develop resistance to the antifungal fluconazole through the acquisition of a segmental aneuploidy of the left half of chromosome 5 (Selmecki et al. 2006; Figure 6a). Potential beneficial effects of aneuploidy under extreme selective pressure have also been observed in several other systems. For example, in budding yeast, specific aneuploidies have been shown to provide resistance to toxic agents (Figure 6b; Torres et al. 2007; Pavelka et al. 2010a; Sheltzer et al. 2011a; Chen et al. 2012), facilitated increased survival under nutrient limiting conditions (Dunham et al. 2002; Gresham et al. 2008) and arose in the evolution of new traits (Rancati et al. 2008). In human fibroblasts, the introduction of an additional copy of chromosome 8 caused loss of contact inhibition, but cells still proliferated more slowly than euploid cells (Nawata et al. 2011).
Figure 6.

Aneuploidy can be beneficial under certain circumstances and could be a useful therapeutic target in the treatment of cancer. (a) An additional copy of the left arm of chromosome 5 confers increased resistance to fluconazole in C. albicans, (data from Selmecki et al. 2009). (b) Some aneuploid budding yeast strains show increased resistance to the microtubule poison benomyl, while others show increased sensitivity (data from Torres et al. 2007). (c) Xenografts of highly aneuploid human colon cancer cells (Aneuploid, SW620 cells) were reduced in size compared to xenografts of near diploid colon cancer cells (Near Diploid, HCT15 cells) after treatment with AICAR and 17-AAG, drugs that preferentially target aneuploid cells (data from Tang et al. 2011). (d) Spectral Karyotype Analysis of the pancreatic cell line Capan-2, (left) (data from Sirivatanauksorn et al. 2001) and aligned chromosomes (right) of same cell (data from http://www.path.cam.ac.uk/~pawefish/index.html).
Recent studies suggest that aneuploidy may not be the optimal mechanism to respond to an extreme selective pressure, but instead provides a quick means of adaptation that is replaced over time by more subtle genetic changes that achieve the same goal. Analysis of C. albicans fluconazole resistance over time from in vivo samples during continuous drug treatment suggests that acquisition of the segmental aneuploidy conferring drug resistance does not necessarily correlate with changes in drug resistance and can be a transient occurrence in some cases (J. Funt & A. Regev, personal communication). Acquired aneuploidy likewise appears to be a quick way for mammalian cells to adapt to stressful conditions before context-specific genomic aberrations take hold. Murine epithelial cells were found to progress through specific aneuploidies before obtaining specific focal duplications when fully transformed. Before the cells had immortalized, extra copies of chromosomes 1, 10, 15, and 19 were found along with monosomies for chromosomes 4, 9, 12, 13, 16, and loss of the Y chromosome. Once transformed, focal duplications of the oncogenes MYC and MDM2 were observed, suggesting again that gain or loss of chromosomes is a faster way for cells to adapt to new conditions but over time more subtle genetic changes are selected for to obtain improved fitness (Padilla-Nash et al. 2011).
Together these results indicate that aneuploidy can provide an effective means of quickly adapting to a selective pressure. This selective advantage of a specific aneuploidy comes at a price, however. Changes in gene copy number of an entire chromosome induced by aneuploidy disrupt protein and energy homeostasis and cause proliferation defects in addition to chromosome specific detrimental effects. It is easy to envision that in the presence of mutations that mitigate the adverse effects of aneuploidy, the full adaptive and genome-instability inducing potential of aneuploidy comes into play. Aneuploidy-tolerating mutations have been found in yeast (Torres et al. 2010a). Loss of p53 also increases the proliferative abilities of aneuploid mammalian cells (Li et al. 2010; Thompson & Compton 2010; Janssen et al. 2011). Identifying genetic alterations that ameliorate the adverse effects of aneuploidy could yield dramatic insight into tumorigenesis.
When aneuploidy is normal
Perhaps the best example for potential beneficial effects of aneuploidy is the observation that the condition is prevalent in certain tissues in mammals (Iourov et al. 2010). Cytotrophoblasts in the developing placenta are frequently aneuploid (Weier et al. 2005), as well as hepatocytes of the liver (Duncan et al. 2010), and both neuronal and glial cells of the brain (Kingsbury et al. 2006). Aneuploidy in these cells is thought to be a normal aspect of development and function.
Polyploidy has long been known to exist in liver cells, however the significance of this phenomenon is still unknown. Aneuploidy in liver hepatocytes was first described in mice in 2010 as a result of error prone multipolar mitoses of polyploid cells. Approximately one quarter of hepatocytes were found to be aneuploid in three week old mice and this frequency jumped to between sixty and seventy percent in mice aged between 5 and 12 months (Duncan et al. 2010). Subsequent studies of healthy livers from adult humans showed a rate of aneuploidy between twenty-five and fifty percent. Loss of chromosomes was more frequently observed than chromosome gain and all chromosomes were found to be lost at approximately the same frequency (1-7% of cells), with the exception of Chromosome 12, which was never found to be lost (Duncan et al. 2012). Aneuploidy in hepatocytes may promote adaptation to changes in nutrition or detoxification needs, and may therefore outweigh the detrimental effects of aneuploidy.
The high incidence of aneuploidy in brain cells was first reported in mice in 2001. Spectral karyotyping of cells in developing embryonic mouse brains showed that approximately one third of developing neuroblasts were aneuploid. As observed in liver hepatocytes, loss of chromosomes was more common than gain of chromosomes (Rehen et al. 2001). The percentage of aneuploid neurons declines during development, but aneuploid neurons were found to be functional and integrated into the circuitry of the adult mouse brain (Kingsbury et al. 2005). Aneuploid neuronal and glial cells also exist in the developing and adult brain of humans (Rehen et al. 2005; Yurov et al. 2005; Yurov et al. 2007). The reason for this aneuploidy is not known, however it is tempting to speculate that loss of heterozygosity for entire chromosomes could play an important role during learning and memory.
Aneuploidy in the brain and the liver seems to allow cells to specify and modify their functional capacities. Perhaps the reduction of detrimental effects of aneuploidy in these tissues is due to the functions of these cell types. Neurons (although not glial cells) are no longer dividing cells. In this post-mitotic state, the anti-proliferative effects of aneuploidy likely have a limited impact. Aneuploidy may still have dire consequences, however, as aneuploidy-induced proteotoxic stress, which if present chronically and without dilution by cell division, could alter neuronal activity with age. As discussed in more detail below, most aging-associated neurodegenerative diseases are characterized by the presence of protein aggregates and the aneuploid state of neurons may contribute to disease formation and progression.
Unlike neurons, liver cells retain the ability to proliferate even after they become aneuploid. Liver cells may be naturally more tolerant of aneuploidy than other cell types or it could be that the functional benefits of genomic variability in the liver outweigh the detrimental effects. Determining whether different cell types exhibit different sensitivities to the aneuploid state and understanding why in naturally aneuploid tissues aneuploidy is not associated with transformation will provide critical insights into whether and how aneuploidy contributes to diseases such as cancer.
VII ANEUPLOIDY AND DISEASE
Aneuploidy has been implicated in several diseases, with the most striking correlation between aneuploidy and disease being found in cancer. While Down syndrome and other constitutional aneuploid conditions are, obviously, directly caused by aneuploid chromosomes, evidence is also mounting for a role of aneuploidy in Alzheimer’s disease and other neurodegenerative diseases. In this section we will discuss studies of chronic and acute aneuploidies in both humans and mice that shed light on the role of aneuploidy in cancer and neurodegeneration.
Aneuploidy in cancer
The occurrence of aneuploidy in cancer has long been known. David van Hansemann first noted that tumors have unbalanced mitoses over 120 years ago (Hansemann 1890). This work influenced Theodor Boveri to expand upon his earlier characterization of aneuploidies in sea urchins to suggest that a single aneuploid cell might cause cancer (Boveri 1914; reviewed in Hardy & Zacharias 2005). It is now known that greater than ninety percent of solid tumors and seventy-five percent of blood cancers show some degree of aneuploidy (Weaver & Cleveland 2006; Figure 6d). While aneuploidy is highly prevalent in cancer, many different types of chromosomal abnormalities are observed in cancer. This complexity and variability of cancers is perhaps best illustrated by the Mitelman database that currently harbors karyotype information of over 60 000 human cancers (Mitelman et al. 2012).
The evolution of cancer cells from benign tumor to invasive metastasis appears to correlate with increased aneuploidy and karyotypic complexity (reviewed in Albertson et al. 2003). Neoplasms are thought to be heterogeneous with a variety of karyotypes (Fidler 2003), become more complex as the cancer progresses, but eventually the karyotype stabilizes in advanced cancers (Albertson et al. 2003). Kuukasjärvi et al. found that seventy percent of breast cancer metastases were clonal and similar to the primary tumor, whereas the rest of the metastases showed distinct changes from the primary tumors suggesting that karyotypic stabilization occurred after metastasis (Kuukasjärvi et al. 1997). Single cell analysis of cells from paired primary breast tumors and associated metastases suggests that cancers also evolve in a punctuated fashion, where a karyotype eventually evolves that proliferates much faster than it evolves (Navin et al. 2011). This may help to explain the lack of conserved karyotypes within individual types of cancers even though specific chromosome aberrations appear frequently in certain cancers, for instance, the high occurrence of loss of the q arm of chromosome 16 in estrogen receptor positive breast cancers (Hungermann et al. 2011). Specific chromosomal gains and losses have also been identified as useful indicators of clinical outcome in colorectal cancer (Bomme et al. 2001; Bardi et al. 2004; Lozynska 2009), and a recent study of cancer karyotypes identified many chromosome pairs that are frequently either lost or gained together across all cancers (Ozery-Flato et al. 2009).
The tremendous variability and complexities of aneuploidy in cancer, makes determining whether aneuploidy is a causal driver of cancers difficult. Several characteristics of cancer cells are, however, observed in aneuploid cells. The frequently observed changes in metabolism caused by aneuploidy (increased glucose uptake and increased lactate production) are similar to the Warburg Effect of increased aerobic glycolysis observed in cancer cells (reviewed in Vander Heiden et al. 2009). Likewise, the elevated proteotoxic stress and genomic instability observed in aneuploid cells are also key characteristics of cancer cells (reviewed in Luo et al. 2009; Hanahan & Weinberg 2011). This correlation is intriguing, but proof that aneuploidy indeed contributes to tumorigenesis comes from the study of mouse models of chromosomal instability. The situation is, however, not as simple as “aneuploidy causes cancer”, as Boveri suggested almost 100 years ago. It appears that aneuploidy sometimes promotes tumorigenesis, sometimes seems inconsequential, and sometimes inhibits disease initiation and progression. The subject of the effects of chromosome instability on mouse models of human cancers has recently been reviewed by others and us in depth (Holland & Cleveland 2009; Schvartzman et al. 2010; Pfau & Amon submitted). Here, we will only briefly summarize the key findings.
Several studies have shown that chromosomal instability increases tumorigenesis in mouse models, indicating that aneuploidy can act as a tumor promoter (Iwanaga et al. 2007; Michel et al. 2001; Sotillo et al. 2007; Sotillo et al. 2010; Weaver et al. 2007; Baker et al. 2009; Li et al. 2010; Zhang et al. 2008, Dai et al. 2004; Ricke et al. 2011). In humans, mosaic variegated aneuploidy (MVA), predisposes one to cancer, and a recent study has identified inactivating mutations in the cohesin subunit gene STAG2 as a cause of aneuploidy in a wide variety of different cancers (Solomon et al. 2011). Aneuploidy can also act as a tumor suppressor, however. Aneuploidy induced by the loss of one copy of CENP-E inhibited tumorigenesis in some tissues (Weaver et al. 2007). Furthermore, introducing additional aneuploidy can prevent tumorigenesis, presumably by inducing sufficient levels of aneuploidy to cause cell death (Weaver et al. 2007). Finally, individuals with trisomy 21 have a lower likelihood of developing solid tumors (Hasle et al. 2000; Satge et al. 2003), as do mouse models of the disease.
In addition to aneuploidy, loss of p53 function frequently occurs in cancer and evidence is mounting that the two may be related. As discussed above, loss of p53 allows highly aneuploid cells to proliferate in vitro (Li et al. 2010; Thompson and Compton 2010; Janssen et al. 2011), but does not directly cause euploid cells to become aneuploid (Bunz et al. 2002). This observation together with the data described above suggests a model for the complex effects of aneuploidy on tumorigenesis. High levels of aneuploidy leads to cell death. Minor to moderate aneuploidy on its own has only a modest positive impact on tumorigenesis and, in case of constitutional aneuploidies (i.e. Down syndrome), has a tumor protective function. This is unsurprising given that aneuploidy generally interferes with cell proliferation. When the anti-proliferative effects of aneuploidy are mitigated through the inactivation of p53 or the acquisition of other aneuploidy tolerating mutations, the tumorigenesis-promoting effects of the condition dominate. The genome instability-inducing effects of aneuploidy recently described in yeast and mammals are likely to have a major impact on disease progression (Sheltzer et al. 2011a; Janssen et al. 2011; Crasta et al. 2012). Increased double strand breaks, either as a result of aneuploidy per se, DNA breaks during cytokinesis or of inadequate replication in micronuclei, could be a significant source of oncogenic mutations. A critical role of DNA damage in tumor evolution is well documented (Mitelman et al. 2007), and the DNA damage-inducing features of whole-chromosome mis-segregation could be a critical aspect of the tumor-promoting effects of aneuploidy. Lastly, aneuploidy is a way to generate phenotypic diversity. This feature of aneuploidy could play an important role in the stages of tumorigenesis where a cancer cell must adapt to a new environment, such as during metastasis. Owing to these potential positive effects of aneuploidy on tumorigenesis, mutations that mitigate the adverse effects of aneuploidy may be critical factors in disease progression.
Neurodegenerative diseases are linked to increased aneuploidy
Beyond its pervasiveness in cancer, increased aneuploidy has also recently been associated with the neurodegenerative disorder Alzheimer’s disease. Early onset of Alzheimer’s disease is caused by the formation of amyloid ß plaques, the main component of which is a cleaved form of the protein encoded by the APP gene found on chromosome 21 (reviewed in Kinsgbury et al. 2006). Indeed, Alzheimer’s disease is observed in nearly all Down syndrome individuals by the age of 40 (Gardiner et al. 2010; Lana-Elola et al. 2011). Normal patients with Alzheimer’s disease were found to have a 1.5 fold increase in trisomy 21 cells in the brain and a 1.2 fold increase in trisomy 17 compared to non-diseased individuals (Thomas & Fenech 2008) and genes associated with Alzheimer’s disease were recently mapped to chromosome 17 in addition to chromosome 21 (Faggioli et al. 2011). Furthermore, as discussed above, the brain is a naturally aneuploid organ and neurons do not turn over throughout the life-time of the organism. Since aneuploid cells experience proteotoxic stress and have defects in misfolded protein turnover in culture, it is tempting to speculate that aneuploidy sensitizes neurons to plaque formation. In addition to Alzheimer’s disease patients, aneuploidy has also been observed to be increased in patients with Ataxia-telangietesia (Aguilar et al. 1968), a neurodegenerative disorder caused by loss of function of the ATM protein (Kingsbury et al. 2006). There are even reports to suggest that brain cell aneuploidy is increased in patients with Schizophrenia (Yurov et al. 2008). It is thus possible that aneuploidy may contribute to disease progression in a wide range of neurological and psychiatric diseases.
VIII FUTURE PERSECTIVES
In this review, we have discussed the detrimental effects of aneuploidy on organisms and cells, the cases where aneuploidy may benefit cells, and the role of aneuploidy in disease. Many questions remain to be answered, such as why some organisms and tissues are more apt to tolerate aneuploidy than others. How the ATM-p53 and/or p38-p53 pathways antagonize the growth of highly aneuploid cells is a critical question that we must understand to get a full picture of the impacts of the condition on normal mammalian cells. The full impact of aneuploidy in cancer must also be addressed. Identifying and understanding the combinations of aberrant chromosome numbers in various cancers and mutations that allow cells to tolerate aneuploidy and take advantage of the tumorigenesis-promoting aspects of the condition will be critical. The possibility of exploiting traits common to most if not all aneuploidies or to specific aneuploidies in cancer therapy should be addressed. A proof-of principle screen in trisomic MEFs showed that such compounds exist and that they are effective against aneuploid colon cancer and lung cancer cells (Tang et al. 2011; Figure 6c). Aneuploidy has been observed to induce senescence within a tumor, leading to tumor dormancy, which may later be reactivated (Kusumbe et al. 2009). Targeting aneuploidy may thus not only serve to kill growing cancers; it may also prevent recurrence. Perhaps the troubles of aneuploidy may in the end help in the treatment of a wide variety of cancers.
Acknowledgments
We thank Sarah Pfau, Jason Sheltzer, Eduardo Torres, and other members of the Amon lab for comments on this review. We also thank Eduardo Torres for help with Figure 4a. Work in the Amon lab was supported by NIH grant GM56800. A.A. is also an Investigator of the Howard Hughes Medical Institute.
Key Terms
- Aneuploidy
a chromosome number that is not a multiple of the haploid complement. Also referred to as “whole chromosome aneuploidy.”
- Euploidy
the species-defining karyotype.
- Polyploidy
a multiple of the haploid chromosome complement.
- Spindle-Assembly Checkpoint (SAC)
a surveillance mechanisms that prevents anaphase entry when chromosomes are not accurately attached to the mitotic or meiotic spindle.
- Organismal Aneuploidy
an organism in which all cells possess the same aneuploidy, for example Down syndrome (Trisomy 21) in humans.
- Chromosomal instability (CIN)
condition where genetic alterations, such as chromosome gains/losses or chromosomal rearrangements, occur at an increased frequency.
- Segmental aneuploidy
deletion or amplification of a region of a chromosome.
- Autosome
non-sex chromosome, all autosomes are present in equal copy numbers in euploid cells.
- Dosage Compensation
mechanisms that equalize the dosages of sex chromosome-encoded genes between the two sexes. Similar mechanisms may also exist to adjust the expression levels of genes on aneuploid chromosomes in order to dampen the effects of gene copy number imbalances.
LITERATURE CITED
- Aguilar MJ, Kamoshita S, Landing BH, Boder E, Sedgwick RP. Pathological observations in ataxia-telangiectasia. A report of five cases. J Neuropathol Exp Neurol. 1968;27:659–76. [PubMed] [Google Scholar]
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–76. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–53. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, et al. BubR1 insifficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–9. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drive tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubules dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–42. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardi G, Fenger C, Johansson B, Mitelman F, Heim S. Tumor karyotype predicts clinical outcome in colorectal cancer patients. J Clin Oncol. 2004;22:2623–34. doi: 10.1200/JCO.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130:237–45. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Newton KJ. Modulation of protein levels in chromosomal dosage series of maize: the biochemical basis of aneuploid syndromes. Genetics. 1981;99:247–66. doi: 10.1093/genetics/99.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF. The globe, a simple trisomic mutant in Datura. Proc Natl Acad Sci USA. 1921;7:148–52. doi: 10.1073/pnas.7.5.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF, Belling J, Farnham ME, Bergner AD. A haploid mutant in the jimson weed, “Datura stramonium”. Science. 1922;16:646–7. doi: 10.1126/science.55.1433.646. [DOI] [PubMed] [Google Scholar]
- Bomme L, Lothe RA, Bardi G, Fenger C, Kronorg O, et al. Assessments of clonal composition of colorectal adenomas by FISH analysis of chromosomes 1, 7, 13 and 20. Int J Cancer. 2001;92:816–23. doi: 10.1002/ijc.1275. [DOI] [PubMed] [Google Scholar]
- Bouchonville K, Forche A, Tang KES, Selmecki A, Berman J. Aneuploid chromosomes are highly unstable during DNA transformation of Candida albicans. 2009;8:1554–66. doi: 10.1128/EC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Ueber mehrpolige mitosen als mittel zur analyse des Zellkerns. Verh Phys-med Ges Würzburg NF. 1902;35:67–90. [Google Scholar]
- Boveri T. Zurfrage der entstehung maligner tumoren. Jena, Germany: Fischer; 1914. [Google Scholar]
- Bridges CB. Genetical and cytological proof of non-disjunction of the fourth chromosome of Drosophila melanogaster. Proc Nati Acad Sci USA. 1921a;7:186–92. doi: 10.1073/pnas.7.7.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB. Triploid intersexes in Drosophila melanogaster. Science. 1921b;44:252–4. doi: 10.1126/science.54.1394.252. [DOI] [PubMed] [Google Scholar]
- Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Fauth C, Speicher MR, Dutriaux A, Sedivy JM, et al. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 2002;62:1129–33. [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyloids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–50. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Okamasa K, Yamane M, Nakayama J, et al. Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct Funct. 2007;32:149–61. doi: 10.1247/csf.07036. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–46. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–5. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Wang Q, Liu T, Swamy M, Fang Y, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–5. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Holm DG, Grigliatti TA. Autosomal dosage compensation in Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc Natl Acad Sci USA. 1982;79:1200–4. doi: 10.1073/pnas.79.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rodríguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA. 2008;105:16719–24. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierssen M, Herault Y, Estivill X. Aneuploidy: from physiological mechanism of variance to Down syndrome. Physiol Rev. 2009;89:887–920. doi: 10.1152/physrev.00032.2007. [DOI] [PubMed] [Google Scholar]
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–45. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Newell AEH, Smith L, Wilson EM, Olson SB, et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–8. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Newell AEH, Lenzi ML, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–10. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:16144–9. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyban AP, Baranov VS. Cytogenetics of mammalian embryonic development. Oxford; New York: Clarendon Press; Oxford University Press; 1987. [Google Scholar]
- Faggioli F, Vijg J, Montagna C. Chromosomal aneuploidy in the aging brain. Mech Aging Dev. 2011;132:429–36. doi: 10.1016/j.mad.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisted. Nat Rev Cancer. 2003;3:1–6. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Gallardo MH, Bickham JW, Honeycutt RL, Ojeda RA, Köhler N. Discovery of tetraploidy in a mammal. Nature. 1999;401:341. doi: 10.1038/43815. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Purge K, Koeman J, Dykema K, Su Y, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci USA. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner K, Herault Y, Lott IT, Antonarakis SE, Reeves RH, et al. Down syndrome: from understanding the neurobiology to therapy. J Neurosci. 2010;30:14943–5. doi: 10.1523/JNEUROSCI.3728-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–57. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Grell EH. The tetrasomic for chromosome 4 in Drosophila melanogaster. Genetics. 1961;46:1177–83. doi: 10.1093/genetics/46.9.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp A. Proceedings: hypoplasia and malformation in trisomy syndromes of the mouse foetus. Mutat Res. 1975;29:216. doi: 10.1016/0027-5107(75)90137-2. [DOI] [PubMed] [Google Scholar]
- Gropp A, Winking H, Herbst EW, Claussen CP. Murine trisomy: developmental profiles of the embryo, and isolation of trisomic cellular systems. J Exp Zool. 1983;228:253–69. doi: 10.1002/jez.1402280210. [DOI] [PubMed] [Google Scholar]
- Guo M, Birchler JA. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266:1999–2002. doi: 10.1126/science.266.5193.1999. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- Hansemann D. Ueber asymmertriche zelltheilung in epithelkrebsen und biologische bedeutung. Arch Pathol Anat Physiol Klin Medicin. 1890;119:299–326. [Google Scholar]
- Hardy PA, Zacharias H. Reappraisal of the Hansemann-Boveri hypothesis on the origin of tumors. Cell Biol Int. 2005;29:983–92. doi: 10.1016/j.cellbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukemia and solid tumours in individuals with Down’s syndrome. Lancet. 2000;355:165–9. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L. Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics. 2010;186:1231–45. doi: 10.1534/genetics.110.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. Primary sex determination in the nematode C. elegans. Development. 1987;101(Suppl):5–16. doi: 10.1242/dev.101.Supplement.5. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, editor. WormBook. The C. elegans Research Community, WormBook; 2005. Karyotype, ploidy and gene dosage. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisted: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kreil DP, Matzke M, Matzke AJM. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000226. doi: 10.1371/journal.pgen.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–7. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Hungermann D, Schmidt H, Natrajan R, Tidow N, Poos K, et al. Influence of whole arm loss of chromosome 16q on gene expression patterns in oestrogen receptor-positive, invasive breast cancer. J Pathol. 2011;224:517–28. doi: 10.1002/path.2938. [DOI] [PubMed] [Google Scholar]
- Iourov IY, Vorsanova SG, Yurov YB. Somatic genome variations in health and disease. Curr Genomics. 2010;11:387–96. doi: 10.2174/138920210793176065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increase the incidence of tumors in mice. Cacner Res. 2007;67:160–6. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–8. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to support spontaneous tumorigenesis. J Cell Biol. 2007;179:255–67. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan KB, Malureanu L, van Deursen JM. The Rael-Nup98 complex prevent aneuploidy by inhibiting securing degradation. Nature. 2005;438:1036–9. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- Jones L, Wei G, Sevcikova S, Phan V, Jain S, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute pormyelocytic leukemia. J Exp Med. 2010;207:2581–94. doi: 10.1084/jem.20091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung PP, Fritsch ES, Blugeon C, Souciet JL, Potier S, et al. Ploidy influences cellular responses to gross chromosomal rearrangements in Saccharomyces cerevisiae. BMC Genomics. 2011;12:331. doi: 10.1186/1471-2164-12-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlem P, Sultan M, Herwig R, Steinfath M, Balzereit D, et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 2004;14:1258–67. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Earle E, Fowler KJ, Choo KHA. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:22777–82. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, et al. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci USA. 2005;102:6143–7. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury MA, Yung YC, Peterson SE, Westra JW, Chun J. Aneuploidy in the normal and diseased brain. Cell Mol Life Sci. 2006;63:2626–41. doi: 10.1007/s00018-006-6169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose J, Putz B. Analysis of two-dimensional protein patterns from mouse embryos with different trisomies. Proc Natl Acad Sci USA. 1983;80:3753–7. doi: 10.1073/pnas.80.12.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovic M, Johnson NB, Sun Q, Normand G, Hoover N, et al. A non-genetic route to aneuploidy in human cancers. Nat Cell Biol. 2011;13:324–30. doi: 10.1038/ncb2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krushinskii LV, Dyban AP, Baranov VS, Poletaeva II, Romanova LG. Behavior of mice with robertsonian translocations of chromosomes. Genetika. 1986;22:434–41. [PubMed] [Google Scholar]
- Kurnit DM. Down syndrome: gene dosage at the transcriptional level in skin fibroblasts. Proc Natl Acad Sci USA. 1979;76:2372–5. doi: 10.1073/pnas.76.5.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe AP, Bapat SA. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res. 2009;69:9245–53. doi: 10.1158/0008-5472.CAN-09-2802. [DOI] [PubMed] [Google Scholar]
- Kuukasjärvi T, Karhu R, Tanner M, Kähkönen M, Schäffer A, et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–1604. [PubMed] [Google Scholar]
- Lana-Elola E, Watson-Scales SD, Fisher EMC, Tybulewicz VLJ. Down syndrome: search for the genetic culprits. Dis Model Mech. 2011;4:586–95. doi: 10.1242/dmm.008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Svensson MJ, Stenberg P, Mäkitalo M. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc Natl Acad Sci USA. 2004;101:9728–33. doi: 10.1073/pnas.0400978101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JC. The effect of chromosome aberrations on development in Drosophila melanogaster. Genetics. 1927;12:1–58. doi: 10.1093/genetics/12.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Wei Z, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci USA. 2010;107:14188–93. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–94. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Sandler L, Baker BS, Carpenter ATC, Denell RE, et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–84. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozynska M. The prognostic value of cytogenetic markers for early diagnosis of colorectal cancer. Exp Oncol. 2009;31:237–241. [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogee and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TP, Lai LC, Tsai MH, Chen PC, Hsu CP, et al. Integrated analyses of copy number variations and gene expression in lung adenocarcinoma. PLoS One. 2011;9:e24829. doi: 10.1371/journal.pone.0024829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S, Antonarakis SE. Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res. 2004;14:1268–74. doi: 10.1101/gr.2090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Harris C. Aneuploidy causes tissue-specific qualitative changes in global gene expression patterns in maize. Plant Physiol. 2010;152:927–38. doi: 10.1104/pp.109.150466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Zielke CL, Zielke HR, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–67. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- McClintock B. A cytological and genetical study of triploid maize. Genetics. 1929;14:180–222. doi: 10.1093/genetics/14.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F, editors. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2012 http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechansisms. Annu Rev Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawata H, Kashino G, Tano K, Daino K, Shimada Y, et al. Dysregulation of gene expression in the artificial human trisomy cells of chromosome 8 associated with transformed cell phenotypes. PLos One. 2011;6:e25319. doi: 10.1371/journal.pone.0025319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Tange Y, Kurabayashi A. Growth arrest and chromosome instability in aneuploid yeast. Yeast. 2006;23:937–50. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- Niwa O, Yanagida M. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr Genet. 1985;53:165–73. [Google Scholar]
- O’Doherty A, Ruf S, Mulligan C, Hildreth V, Errington ML, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrom phenotypes. Science. 2005;309:2033–7. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ozery-Flato M, Linhart C, Trakhtenbrot L, Izraeli S, Shamir R. Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to cancer aneuploidy. Genome Biol. 2011;12:R61. doi: 10.1186/gb-2011-12-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Nash HM, Hathcock K, McNeil NE, Mack D, Hoeppner D, et al. Spontaneous transformation of murine epithelial cells requires the early acquisition of specific chromosomal aneuploidies and genomic imbalances. Genes Chromosomes Cancer. 2011;51:353–74. doi: 10.1002/gcc.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry EM, Cox BS. The tolerance of aneuploidy in yeast. Genet Res. 1970;16:333–40. doi: 10.1017/s0016672300002597. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010a;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Li R. Dr. Jekyll and Mr. Hyde: role of aneuploidy in cellular adaptation and cancer. Curr Opin Cell Biol. 2010b;22:809–15. doi: 10.1016/j.ceb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D, Tilston V, Hopwood JA, Barchi M, Boot-Handford RP, et al. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell. 2007;13:566–79. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sørlie T, Perou CM, Rees CA, Jeffrey SS, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11:216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, et al. Unstable kinetochore-mircotubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–65. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–93. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, et al. Colonic tumorigenesis in BubR1+/- ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci USA. 2005;102:4365–70. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Wong LY, Yang Q, May KM, Friedman JM. Population-based analyses of mortality in trisomy 13 and trisomy 18. Pediatrics. 2003;111:777–84. doi: 10.1542/peds.111.4.777. [DOI] [PubMed] [Google Scholar]
- Reeves RH. Down syndrome mouse models are looking up. Trends Mol Med. 2006;12:237–40. doi: 10.1016/j.molmed.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, et al. A mouse model for Down syndrome exhibits learning and behavior deficits. Nat Genet. 1995;11:177–84. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci USA. 2001;98:13361–6. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–80. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–64. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, Kilbridge J, Rubin EM, et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci USA. 1998;26:6256–61. doi: 10.1073/pnas.95.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satge D, Sasco AJ, Lacour B. Are solid tumours different in children with Down’s syndrome? Int J Cancer. 2003;106:297–8. doi: 10.1002/ijc.11212. [DOI] [PubMed] [Google Scholar]
- Saxon PJ, Stanbridge EJ. Transfer and selective retention of single specific human chromosomes via microcell-mediated chromosome transfer. Methods Enzymol. 1987;151:313–25. doi: 10.1016/s0076-6879(87)51026-6. [DOI] [PubMed] [Google Scholar]
- Schliekelman M, Cowley DO, O’Quinn R, Oliver TG, Lu L, et al. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 2009;69:45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modeling of the human disease. Nat Rev Cancer. 2010;10:102–15. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, McCoy EE. Studies on Down’s syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. J Cell Physiol. 1974;83:85–90. doi: 10.1002/jcp.1040830112. [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during evolution of antifungal drug resistance. PLoS Genet. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–70. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotypes. Trends Genet. 2011b;27:446–53. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011a;333:1026–30. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson DC, Spanier GJ, Herman RK. Caenorhabditis elegans deficiency mapping. Genetics. 1984;108:331–45. doi: 10.1093/genetics/108.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Multani DS, Khush GS. Secondary trisomics and telotrisomics of rice: origins, characterization, and use in determining the orientation of chromosome map. Genetics. 1996;143:517–29. doi: 10.1093/genetics/143.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirivatanauksorn V, Sirivatanauksorn Y, Gorman PA, Davidson JM, Sheer D, et al. Non-random chromosomal rearrangements in pancreatic cancer cell lines identified by spectral karyotyping. Int J Cancer. 2001;91:350–8. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1049>3.3.co;2-3. [DOI] [PubMed] [Google Scholar]
- Snape K, Hanks S, Ruark E, Barros-Núñez P, Elliot A, et al. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet. 2010;43:527–9. doi: 10.1038/ng.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 333:1039–43. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordón-Cardo C, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–40. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Charles J, Hamilton ML, Petes TD. Meiotic chromosome segregation in triploid strains of Saccharomyces cerevisiae. Genetics. 2010;186:537–50. doi: 10.1534/genetics.110.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P, Larsson J. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma. 2011;120:213–25. doi: 10.1007/s00412-011-0319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Fenech M. Chromosome 17 and 21 aneuploidy in buccal cells is increased with ageing and in Alzheimer’s disease. Mutagenesis. 2008;23:57–65. doi: 10.1093/mutage/gem044. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci USA. 2011;108:17974–8. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–52. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010a;143:1–13. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–46. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Tang YC, Amon A. Thoughts on aneuploidy. Cold Spring Harb Symp Quant Biol. 2010b;75:445–51. doi: 10.1101/sqb.2010.75.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–37. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, et al. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–49. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacík T, Ort M, Gregorová S, Strnad P, Blatny R, et al. Segmental trisomy of chromosome 17: a mouse model of human aneuploidy syndromes. Proc Natl Acad Sci USA. 2005;102:4500–5. doi: 10.1073/pnas.0500802102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Bonday ZQ, Putkey FR, Kops GJPL, Silk AD, et al. Centrosome-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–63. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Weier JF, Weier HUG, Jung CJ, Gormley M, Zhou Y, et al. Human cytotrophoblasts acquire aneuploidies as they differentiate to an invasive phenotype. Dev Biol. 2005;279:420–32. doi: 10.1016/j.ydbio.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–9. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liu Y, Wang P, Fan W, Rue TC, et al. Integrative analysis of DNA copy number and gene expression in metastatic oral squamous cell carcinoma idetifies genes associated with poor survival. Mol Cancer. 2010;9:143. doi: 10.1186/1476-4598-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Monakhov VV, Soloviev IV, Vostrikov VM, et al. The variation of aneuploidy frequency in the developing and adult human brain revealed by interphase FISH study. J Histochem Cytochem. 2005;53:385–90. doi: 10.1369/jhc.4A6430.2005. [DOI] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Vorsanova SG, Demidova IA, Kravetz VS, et al. The schizophrenia brain exhibits low-level aneuploidy involving chromosome 1. Schizophr Res. 2008;98:139–47. doi: 10.1016/j.schres.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;27:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Ge G, Meyer R, Sethi S, Basu D, et al. Overexpression of separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci USA. 2008;105:13033–38. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]


