Abstract
Exploiting the power of the RNAi pathway through the use of therapeutic siRNA drugs has remarkable potential for treating a vast array of human disease conditions. However, difficulties in delivery of these and similar nucleic acid-based pharmacological agents to appropriate organs or tissues, remains a major impediment to their broad clinical application. Synthetic nucleic acid ligands (aptamers) have emerged as effective delivery vehicles for therapeutic oligonucleotides, including siRNAs. In this review, we summarize recent attractive developments in creatively employing cell-internalizing aptamers to deliver therapeutic oligonucleotides (e.g., siRNAs, miRNAs, anti-miRs and antisense oligos) to target cells. We also discuss advancements in aptamer-siRNA chimera technology, as well as, aptamer-functionalized nanoparticles for siRNA delivery. In addition, the challenges and future prospects of aptamer-targeted oligonucleotide drugs for clinical translation are further highlighted.
When RNAi was first described in mammals about a decade ago [1], there was a great deal of enthusiasm that siRNAs, the effector molecules of the RNAi pathway [2], could be the next new class of drugs. The broad therapeutic potential of siRNAs stems from their ability to deplete the expression of virtually any gene in the genome, thus providing a means of inhibiting established target genes that have proven `undruggable' with other approaches. However, enthusiasm waned when it was realized that getting siRNAs to work as drugs was not as straightforward as originally thought. Fortunately, after substantial efforts to address problems with cellular uptake, considered a major obstacle to the clinical translation of siRNAs [3,4], there is now renewed and well-deserved optimism about RNAi-based drugs. Over the past year, Phase I and II studies have shown promising gene silencing and/or clinical benefit for several liver pathologies caused by aberrant gene expression including: hypercholesterolemia, transthyretin-related amyloidosis, hepatitis C, and liver metastasis [5–8]. Although promising, the application of siRNAs for treating other nonhepatic diseases still remains elusive. In these other clinical settings, human trials have highlighted the need for robust delivery techniques that will enable the application of RNAi therapeutics to increasingly complex disease and organ systems. In recent years, several approaches to enhance delivery of siRNAs to target tissues (besides the liver) in vivo have been investigated [9,10]. Among targeted approaches, aptamers (synthetic DNA/RNA ligands) are emerging as one potential solution to the problem of siRNA delivery to specific cell-types [11–14].
Since the first description of aptamer technology over 20 years ago [15–17] significant progress has been made in the development of aptamers for therapeutic or diagnostic applications. Over the years, methodological improvements, such as automated and microfluidic-based selections, have enabled researchers to select and characterize aptamers to multiple targets within a few weeks to days [18,19]. However, traditional selection techniques are still more commonly used and usually require several months before candidate aptamers can be identified. Recently, multiple research groups have used living cells to identify cell- and receptor-specific aptamers [20–29]. Because these aptamers bind to cell-surface proteins in their native state, they can be exploited for target validation, target inhibition and delivery of a variety of therapeutic agents into the cell. To date, aptamers for hundreds of targets have been discovered and investigated. For a complete list of these aptamers we refer you to the reviews by Keefe et al. [30] and Sundaram et al. [31] and to the aptamer database maintained by the Ellington laboratory [201]. The use of aptamers as therapeutics was first demonstrated in 1990 by Sullenger et al. [32]. Since then, one aptamer drug, Macugen® (pegaptanib) has been approved by the US FDA for the treatment of age-related macular degeneration. Importantly, several other aptamers are being evaluated clinically [30,31,33]. As more therapeutic aptamers make their way into the clinic, the prospect of aptamer-based therapeutics is increasingly more tangible.
While aptamer-based delivery approaches have yet to be thoroughly evaluated in a clinical setting, many outstanding preclinical studies have demonstrated the potential of exploiting aptamers as delivery agents for RNAi-based therapeutics [34–40]. This review will provide an overview of these studies with a focus on recent advances in aptamer-based siRNA delivery technology (also referred to as aptamer-siRNA chimera [AsiC] technology) (Table 1), as well as, novel developments in aptamer-targeted nanoparticles for siRNA delivery.
Table 1.
Aptamers for cell-specific delivery of RNA therapeutics.
| DNA/RNA | Aptamer target | siRNA target/miRNA | Chimera/nanoparticle | Ref. |
|---|---|---|---|---|
| RNA | PSMA | Plk1, Bcl-2, EEF2, DNAPK, Smg1, Upf2, Bcl-xL, miR-15a, miR-16–1, EGFP | Chimera and nanoparticle | [34,35,40,58,64,108,111,114] |
| RNA | αVβ3 | EEF2 | Chimera | [60] |
| RNA | BAFF-R | STAT3 | Chimera | [59] |
| RNA | MUC1 | miR-29b | Chimera | [61] |
| RNA | Axl | let-7g | Chimera | † |
| RNA | HER2 | Bcl-2 | Chimera | [62] |
| RNA | gp120 | HIV tat/rev, CD4, TNPO3 | Chimera and nanoparticle | [38,39,57,78,119] |
| RNA | CD4 | CCR5, HIV gag/vif, survivin | Chimera and nanoparticle | [36,37,116] |
| DNA | TLR9 | STAT3 | Chimera | [80] |
| DNA | Nucleolin | SSO, miR-221 MB | Chimera and nanoparticle | [88,115] |
| DNA | CD4 | HIV protease | Chimera | [95] |
| DNA | CD8 | granulysin | Chimera | [75] |
| RNA | CD30 | ALK | Nanoparticle | [110] |
| RNA | Transferrin (CD71) | EGFP | Nanoparticle | [112] |
| RNA | Malachite green dye | Survivin | Nanoparticle | [128] |
Unless specified, the therapeutic oligonucleotide refers to an siRNA/shRNA.
[Esposito CL, Cerchia L, Catuogno S et al. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy (2013), Submitted].
MB: Molecular beacon; PSMA: Prostate specific membrane antigen; SSO: Splice-switching oligonucleotides.
RNA-based therapeutics: advantages, challenges & progress
AsiC technology has important advantages over other approaches (e.g., protein-based or other multicomponent conjugated methods) including its simplicity (single-component system), ease of manufacturing, promising safety profile, robust serum stability and tissue penetrability. Multiple in vivo studies involving low dose (<1 mg/kg), systemic administration of AsiCs have reported efficacy, highlighting the promise of this approach [34,35,39]. Despite the demonstration that aptamers are effective drugs, several challenges common to most oligonucleotide drugs, must be overcome before aptamer-targeted siRNA reagents can be broadly translated [11,12,14]. These include:
■ Susceptibility of unmodified RNAs to nuclease-mediated degradation;
■ Potential toxicity due to non-specific immune stimulation and/or unintended off-target/on-target effects of aptamers and siRNAs;
■ Current state of synthesis/conjugation technologies and large-scale production of long, modified RNAs;
■ Rapid renal clearance of small molecular weight RNAs;
■ Inefficient cellular uptake and intracellular processing of endosome-targeted RNAs (e.g., endosomal escape and processing by the RNAi machinery).
Here we will briefly discuss these challenges and highlight solutions that have been successful at overcoming several of these hurdles.
In its native, unmodified form, RNA is rapidly degraded by serum nucleases. To increase its stability in human serum, RNA can be modified at several positions [41–43]. The most commonly used chemical modifications are:
■ Substituting S for O in the phosphate backbone (to produce RNAse-resistant phosphorothioate [PS] linkages);
■ Substituting 2′-fluoro (2′-F), 2′-O-methyl (2′-OMe), or 2-′O-methyoxyethyl (2′MOE) for the 2′-OH in the ribose;
■ A 2′-O, 4′-C methylene bridge modification known as a locked nucleic acid [30,42,44–46].
PS, 2′-F and 2′-OMe modifications have been used extensively in clinically tested and FDA-approved oligonucleotide drugs [31,43]. The 2′-OMe substitution occurs naturally in rRNA and tRNA and is therefore safe, and the 2′-F substitution is also well-tolerated [42]. Heavily PS-modified nucleotides (nt) are sticky and, while this modification promotes binding to serum proteins, leading to longer serum retention times, it is often associated with unwanted side effects. In contrast, lightly modified PS-RNAs are not toxic while still retaining the benefits imparted by the PS modification [42,44]. Chemical modifications have also been shown to decrease nonspecific immune stimulation and off-target effects, thereby decreasing the overall toxicity of modified RNA drugs [43,47,48]. For a more comprehensive discussion of these chemical modifications and their application we direct you to the reviews by Keefe and Cload [49], and by Behlke [42,43]. To date, the 2′-F modification is the chemical modification that has been more extensively applied to AsiC technology.
The success of any therapeutic will ultimately depend on the ease with which it can successfully be produced on a large scale. Currently, both DNA and RNA oligonucleotides are manufactured via phophoramidite solid-phase chemical synthesis. Although highly effective, the fidelity of this process is inversely correlated with the length of the oligonucleotide being synthesized [50]. Therefore, any aptamer-based RNAi therapeutic must use the shortest sequence possible (generally, under 60 nt in length) if it is going to be evaluated clinically. Therapeutic aptamers have typically been truncated post-selection using a trial-and-error process, which can often be time consuming and inefficient. However, recent studies have described computer-guided approaches that promise to streamline the process of aptamer truncations and expedite the design of functional AsiCs [21,51]. Fortunately, continued advances in nucleic acid synthesis and conjugation technologies are expected to make the large-scale GMP synthesis of AsiCs greater than 60 nt in length, both feasible and economical in the not so distant future [12].
While shorter aptamers and AsiCs are currently more amenable to large-scale chemical synthesis, their smaller size decreases serum circulating times by promoting rapid renal clearance [30,52]. Most clinically relevant aptamers are approximately 10–20 kDa in size, resulting in excretion via the kidneys. Thus, in order to optimize the bioavailability/circulating half-life, the overall size of the aptamer therapeutic must be increased without modifying the length of the aptamer sequence itself. To accomplish this, aptamers have been conjugated to various macromolecules including PEG and cholesterol [53–55]. Similar approaches to improve the pharmacokinetics of AsiCs have been described [34]. While these initial studies are encouraging and suggest that the PEG modification can be used to stabilize AsiC drugs in vivo, the optimal size PEG or macromolecule will have to be evaluated empirically for any given application.
A final consideration for any RNAi-based therapeutic is that of its intracellular processing [3,11,13,14,43]. Important considerations, with regards to efficient intracellular processing, are the subcellular localization and the ability of the RNA drug to be recognized and processed by the RNAi machinery. Most siRNA-delivery approaches direct/concentrate the siRNAs in endosomes. This is also true for the aptamer-targeted siRNA-delivery approach. Thus optimizing this step by facilitating or enhancing endosomal escape will undoubtedly increase the efficacy of the RNAi drug. Although different conjugates exist for this purpose (e.g., proton sponges) [56], to date, little work has been done to investigate the benefits of these approaches for enhancing endosomal escape in the context of AsiC technology. In addition, several studies have demonstrated that RNAi processing can be improved through slight alterations in the design of their siRNAs [34,57]. In one study, the modifications to improve the silencing potency of a siRNA in the context of an AsiC, translated into a 100-fold increase in silencing of the siRNA target gene [34]. Continued progress towards optimizing the in vivo disposition (serum stability, biodistribution and pharmacokinetics) and efficacy (targeting and silencing efficiency) of aptamer-siRNA drugs is likely to expedite the development of a clinically viable aptamer-targeted RNAi therapeutic.
Aptamer-siRNA cancer therapies
The first aptamer-targeted siRNA drug was described in 2006 [58]. This AsiC was designed to target a cytotoxic, cancer-specific siRNA to prostate cancer cells. Since then, many groups have harnessed and expanded on this approach to develop AsiCs with the ability to target different cancers (by varying the aptamer) [59–61], deliver siRNA drugs as neoadjuvant therapy (by varying the siRNA) [40,62] or deliver tumor-immunity-enhancing siRNAs (by varying the siRNA) [35]. These studies highlight the flexibility and broad clinical potential of the AsiC platform. By changing the aptamer, the AsiC platform challenges the delivery roadblock that has thwarted the application of RNAi-based therapy to most diseases, including cancer. In contrast, by interchanging the siRNA, the choice of genes to be silenced can be adjusted depending on the therapeutic need or the molecular characteristics of a diseased cell, rendering this approach ideal for personalized therapy (e.g., cancer therapy) or for treating diseases with a high mutational rate (e.g., cancer, HIV). In this section, we elaborate on the various studies that have employed AsiC technology as either a cytotoxic, neoadjuvant or immune-therapy for the treatment of various cancers.
Cytotoxic AsiC drugs
The initial study to describe AsiC technology designed their chimeric RNA drug to impart a cytotoxic insult to the target cancer cells. In this study, McNamara et al. synthesized AsiCs by covalently linking a previously described RNA aptamer (A10) [63], which binds to prostate specific membrane antigen (PSMA) expressed on the surface of prostate cancer cells, to siRNAs directed against prosurvival genes Plk1 and B-cell lymphoma 2 (Bcl2), known to promote the growth and survival of prostate cancer cells (blunt in Figure 1) [58]. The authors showed that when added to prostate cancer cells that express PSMA, the Plk1 and Bcl2 AsiCs bind to the cell surface target (PSMA), undergo receptor-mediated endocytosis and deliver the cytotoxic siRNA cargo to the RNAi machinery. The authors confirm processing of the Plk1 and Bcl2 AsiCs by the RNAi machinery by showing that the reduction of the Plk1 and Bcl2 transcripts is through an Ago2-mediated event. Importantly, the authors went on to show that silencing of the Plk1 prosurvival gene in these cells resulted in death of the cancer cells in culture and in a xenograft mouse model of PSMA+ prostate cancer. Although the RNA drug was delivered intratumoraly in this study, the therapeutic potential of this chimeric RNA drug stems from its exquisite selectivity: aptamer targets PSMA on cancer cells while delivering a cancer specific siRNA cargo. This dual level of specificity is imperative for developing effective cancer therapies with low-to-no toxicity to normal tissues. Thus, this work was an important first step in developing a one-component (all RNA), highly specific therapeutic approach for RNAi.
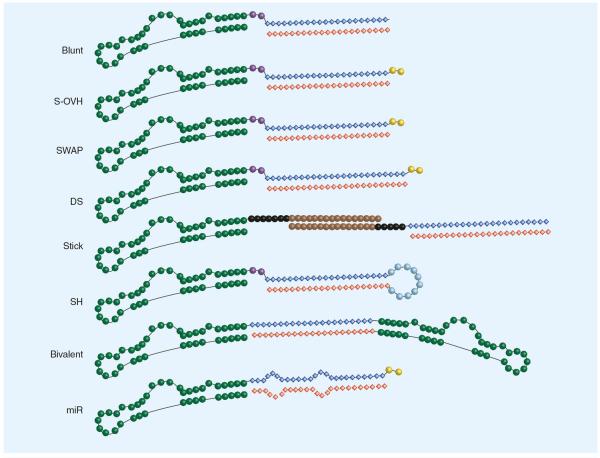
Figure 1. Aptamer-siRNA chimera design strategies.
Various aptamer-siRNA chimera (AsiCs) have been described that employ different design elements within their siRNA moiety. Each AsiC uses a unique aptamer, depicted here in green as a representative structure. Some designs use a linker region (purple) generally made up of 2–4 nucleotides (nt) in order to enable the proper folding of the aptamer structure. The siRNA moiety is made up of two strands. The antisense/guide strand (red) directs the RNAi machinery to degrade its target gene, while the sense/passenger strand (blue) is rapidly degraded. (A) The first generation AsiC contained a blunt [58]. (B) The S-OVH AsiC employed a 2 nt 3′ overhang (yellow) on the sense strand for increased Dicer recognition [35,36,62]. (C) The SWAP AsiC was designed by exchanging the position of the sense and antisense strands [34,80]. This favored processing of the guide strand by the RNAi machinery and allowed for terminal modifications (e.g., PEG conjugation) to the siRNA that would not interfere with target gene silencing.(D) The DS AsiC was designed with a longer 27mer dsiRNA, which was shown to increase Dicer processing [39,57]. (E) The stick AsiC was synthesized with a rigid seven three-carbon linker (black) to prevent steric interference followed by a 16 nt, G–C rich 2′OMe/2′F modified `stick' sequence (brown) to allow the facile exchange of various therapeutic siRNAs [78]. The second-generation stick design (depicted here), also included a five three-carbon linker adjacent to the therapeutic siRNA to further prevent steric hindrance [59]. (F) The SH AsiC is synthesized from a single RNA strand and therefore does not need to be combined and annealed with other RNAs (e.g., antisense strand) to form a functional AsiC [40,60,61]. Here, the siRNA moiety folds into a short hairpin structure, which closely resembles endogenous miRNA. This has been demonstrated to be more readily processed by the RNAi machinery. (G) This AsiC consists of two aptamers connected by a linker region consisting of a therapeutic siRNA. The bivalent AsiC was demonstrated to increase internalization of the reagent, resulting in a more effective therapeutic response [64]. (H) The miR was designed to mimic the natural structure of endogenous miRNAs. Unlike the perfect complementarity of the short hairpin AsiC, this design contained regions of mismatched base pairing within the miRNA sequence resulting in the two bulges [128]. Blunt: Blunted siRNA terminus; DS: Dicer substrate; miR: miRNA AsiC; SH: Short hairpin; S-OVH: Sense-overhang; SWAP: Second-generation AsiC.
In a follow up to this study, Dassie et al. made several modifications to the original A10-Plk1 AsiC in order to achieve therapeutic efficacy following systemic administration [34]. First, the aptamer portion of the first-generation AsiC was truncated from 71 to 39 nt in order to enable its large-scale chemical synthesis. Next, the siRNA was modified to increase its silencing potency. First, a 2 nt overhang on the 3′-end of the guide strand was included to increase Dicer recognition and overall RNAi processing (S-OVH in Figure 1). Second, the position of the guide and passenger strands relative to the original design were swapped (SWAP in Figure 1). This positioned the 2 nt overhang at the 3′-end of the guide strand, thereby increasing strand bias, thus favoring loading of the desired guide strand into the RNA induced silencing complex (RISC). These modifications resulted in a second-generation AsiC (SWAP) that was 100-fold more potent at silencing its siRNA target in cells in culture. Importantly, the SWAP AsiC demonstrated therapeutic efficacy following systemic administration (intraperitoneal injections) in a xenograft mouse model of prostate cancer. To further optimize the in vivo disposition of the chimeric RNA drug, the authors conjugated a 20-kDa PEG to the 5′-end of the siRNA passenger strand. This modification was intended to increase the serum circulating half-life of the chimeric RNA drug and provide stability from terminal nucleases. The authors performed extensive in vitro studies to confirm that the addition of the terminal PEG did not affect the ability of the chimeric RNA drug to undergo cell-internalization or to be processed by the RNAi machinery. As anticipated, conjugation of the 20 kDa PEG to the SWAP AsiC prolonged its circulating half-life (from minutes to several hours). Importantly, this modification significantly increased the in vivo efficacy of the AsiC, allowing the authors to decrease the therapeutic dose from ~1.4 to ~0.4 mg/kg and perform fewer administrations of the drug (five injections instead of ten). This study was the first demonstration that AsiC therapeutic technology has the potential to be translated clinically for treating diseases, which require intravenous delivery of the drug.
While the Giangrande group sought to improve efficacy of their AsiC primarily through modifications of the siRNA component, Wullner and colleagues demonstrated that altering the design of the aptamer/targeting component could also lead to a more effective AsiC [64]. To this end, the authors designed bivalent AsiCs by linking two PSMA aptamer (A10-3) monomers with a double-stranded RNA linker sequence [63]. The authors also compared different bivalent AsiC designs. One design incorporated the EEF2 siRNA sequence as part of the double-stranded RNA linker (bivalent in Figure 1). The second design placed an EEF2 shRNA sequence at the 3′-end of one of the monomers. The latter design is of interest as it can be adapted to deliver multiple therapeutic siRNAs/shRNAs at once. Both AsiC designs were shown to accelerate PSMA recycling and thus, enhance uptake of the EEF2 siRNA/shRNA into the target cells. Importantly, when incubated with PSMA+ cells these reagents were found to be more effective at silencing the siRNA target gene, than a monomeric A10-3/EEF2 AsiC. While encouraging, future studies addressing the in vivo efficacy of bivalent AsiCs are warranted.
The PSMA AsiC proof-of-concept studies are important for demonstrating the feasibility of the aptamer-mediated siRNA delivery approach; however, evidence that this technology can be broadly applied to treating various cancers came with the demonstration that aptamers directed against other cancer antigens can also function in the context of AsiCs. In one of these studies Hussain et al. designed a cytotoxic AsiC that used a previously described aptamer [65] targeting the αVβ3 integrin [60]. Elevated surface expression of this integrin has been reported in several cancers including cancers of the cervix [66], brain [67] and prostate [68]. The authors covalently linked the αVβ3 integrin aptamer to a siRNA directed against the EEF2 gene transcript. When administered to cells in culture, the αVβ3-EEF2 AsiC resulted in selective death of αVβ3+ cancer cells. Importantly, the authors demonstrated that the αVβ3-EEF2 AsiC was effective at inhibiting growth of several αVβ3+ cancer cell lines including: prostate cancer (PC3), brain cancer/glioblastoma (U-87 MG), and cervical cancer (SiHa) cells. While promising, future studies are necessary to evaluate the efficacy and safety of this approach in preclinical, animal models of cancer.
In a different study, Zhou et al. described the use of AsiC technology to treat non-solid cancers [59]. In this study, the authors identified a 2′-F-modified RNA aptamer (R-1APT) that binds to the BAFF receptor (BAFF-R) with high affinity and specificity. High expression of BAFF-R has been demonstrated in various B-cell malignancies, such as non-Hodgkin's lymphoma (NHL) [69]. The authors demonstrated that R-1APT behaved as an antagonist by interfering with the binding of the endogenous BAFF-R ligand to its receptor. Importantly, this resulted in inhibition of tumor growth in the absence of any added therapeutic cargo. The authors went on to develop a dual-function AsiC by covalently linking the inhibitory aptamer to a cytotoxic siRNA directed against STAT3 transcript. The AsiC was found to inhibit BAFF mediated proliferation to the same degree as the R-1APT aptamer alone, demonstrating that siRNA conjugation does not interfere with the function of the aptamer. Further studies demonstrated that this reagent could effectively deliver its siRNA cargo and decrease expression of STAT3. However, decreased STAT3 expression had no additional effect on cellular proliferation or apoptosis. While inhibition of STAT3 has been described to promote cell death in some settings, the precise mechanism and effect of STAT3 inhibition in other settings (e.g., NHL cells) is not well understood. The authors suggest that inhibition of STAT3 in NHL cells may increase sensitivity to chemotherapeutics. However, this was not directly assessed. Importantly, this study provides additional evidence of the broad potential of AsiC technology.
More recent examples of cytotoxic AsiCs for cancer therapy have been developed by conjugating cancer-targeting aptamers to miRNAs. A single miRNA has the advantage of targeting multiple transcripts, thus it can alter the expression profile of numerous genes simultaneously [70]. miRNAs have been extensively described in cancer and several studies have proposed that delivering miRNA mimics or anti-miRs to cancer cells can have a dramatic effect on cancer growth and survival [71]. Esposito et al. were the first to describe an RNA aptamer-targeted delivery of a miRNA. In a presentation given at the 4th RNAi Research and Therapeutics Conference in San Francisco, deFranciscis described a dual-function aptamer-miRNA chimera (AmiC) for treating cancers of the lung. The dual-function AmiC consists of an inhibitory RNA aptamer, directed against the RTK Axl, which is covalently linked to the miRNA, let-7g (miR in Figure 1). The aptamer had been previously selected for its ability to inhibit the kinase activity of Axl, leading to a reduction in the growth and motility of cancer cells [72]. Similarly, re-introduction of the let-7g miRNA into cancer cells was shown to decrease the expression of several known prosurvival gene targets (N-Ras and Bcl-XL) leading to cancer cell death. DeFranciscis demonstrated the combinatorial effect of the dual-function AmiC by confirming that when delivered simultaneously the aptamer and the miRNA were more effective at inducing cell-specific apoptosis than either treatment alone. Importantly, the work by deFranciscis and colleagues is the first demonstration of in vivo efficacy of an AmiC following systemic administration in a xenograft mouse model of human lung cancer. Another important contribution of this work is the demonstration of design flexibility and broad clinical potential of the aptamer-targeted RNAi platform. By swapping the Axl aptamer with the PSMA-specific A10 aptamer (PSMA-let-7g AmiC), deFranciscis and colleagues were able to demonstrate efficient delivery of the let-7g miRNA to PSMA+ prostate cancer cells. Similarly, conjugation of a different therapeutic miRNA (miR-16) to the Axl aptamer resulted in a functional Axl-miR-16 AmiC capable of efficient cell-targeted miRNA delivery. The contribution from this study is significant because it highlights the flexibility of the aptamer-delivery platform for developing novel and effective cancer therapies and thus constitutes critical progress towards addressing an unmet need in the field. Taken together, these studies highlight the utility of the aptamer-based delivery technology as a cytotoxic therapeutic modality for the treatment of various neoplasms.
Neoadjuvant AsiC therapy
While monotherapy can be a powerful approach for eradicating cancer, many cancer patients are currently given multiple treatments (combination therapies), such as radiation in combination with chemotherapy, to increase the success of eradicating their cancers. Towards this end, Ni et al. adapted the aptamer-delivery technology to target radiosensitizing siRNAs to prostate cancer cells prior to treatment with the established oncologic treatment of ionizing radiation (IR) [40]. It is known that IR induces DNA double strand breaks that, if not repaired via cellular DNA repair pathways, elicit cell death. Thus, the authors performed a high-throughput siRNA screen in order to identify siRNAs that could inhibit DNA repair and induce radio-sensitivity. From this screen, they identified a siRNA that targets the DNAPK gene. They then engineered an AsiC by covalently linking the DNAPK siRNA to a truncated version of the PSMA A10 aptamer (A10–3) [63]. This study employed a different AsiC design, from the studies detailed above. Similar to the design first reported by Wullner and colleagues [64], the authors conjugated a shRNA, and not a 21–27-mer double-stranded duplex, to the aptamer (Figure 1). This allowed the AsiC to be synthesized from a single RNA strand, rather than having to anneal the short complementary siRNA strand to the AsiC, as is done with conventional AsiC designs. The PSMA-DNAPK AsiC was capable of delivering the shRNA in a cell-specific manner both in vitro and in vivo. Importantly, when this AsiC was administered (via intratumoral injections in a xenograft mouse model of prostate cancer) as a neoadjuvant to IR treatment, it significantly increased the radiosensitivity of all tumors assessed. Consequently, AsiC treated tumors showed an impressive tenfold reduction in growth rate compared with control treated tumors. Given that IR treatment is the standard-of-care for recurrent late-stage prostate cancer, radiosensitization via neoadjuvant treatment with PSMA-DNAPK AsiCs has the potential to significantly improve clinical outcomes.
In a further attempt to demonstrate the use of AsiC drugs as neoadjuvants for cancer therapy, Thiel et al. described the use of cell-specific RNA aptamers to selectively deliver chemo-sensitizing siRNAs to HER2+ breast cancer cells [62]. In the study by Giangrande and colleagues, addition of HER2-specific AsiCs to HER2+ breast cancer cells in culture increased the sensitivity of these cells to low-dose cisplatin. The AsiCs were engineered by coupling various HER2-specific aptamers to siRNAs directed against Bcl2. When applied to cells in culture, the HER2-Bcl2 AsiCs internalized via a HER2-dependent, receptor-mediated endocytic mechanism and silenced Bcl2 expression as measured by RT-qPCR and Ago2 cleavage of the siRNA target gene. This resulted in moderate cell death. In contrast, when given as a neoadjuvant with the chemo-therapeutic cisplatin, the HER2-Bcl2 AsiCs increased sensitivity to low-dose cisplatin (albeit to varying degrees) resulting in an increase in cell death. Importantly, this study described the first application of the aptamer selection process termed, cell-internalization systematic evolution of ligands by exponential enrichment (SELEX). This modified SELEX approach was developed to favor the identification of RNA aptamers with the ability to internalize into specific cell types. Because, the cell-internalization SELEX approach can be performed on any cell-type of interest, in principle, this could result is a repertoire of aptamers for delivering a therapeutic cargo (e.g., siRNAs/miRNAs) to any diseased cell. Thus, the approach described by Giangrande and colleagues, which couples cell-based methodologies with novel bioinformatics analyses, is significant because it provides a platform for identifying cell-targeted aptamers for the broad application of targeted-RNAi therapeutics. Indeed, the application of this selection platform to cancer is paramount given the cell-type multiformity of this disease.
The studies by Lupold, Giangrande and colleagues expand the scope of AsiC technology as a cancer therapeutic modality. Reduced sensitivity and acquired resistance to therapy are common complications of many cancers that warrant the development of novel more effective therapies. The use of cell-targeted AsiCs as neoadjuvants for cancer therapy has the potential to greatly improve clinical outcomes for patients with unresponsive or refractory cancers.
AsiC-based immunotherapy
In a groundbreaking study by Gilboa and colleagues, the aptamer-targeted delivery technology was adapted to develop AsiCs for cancer immunotherapy [35]. In order to successfully mount an immune response against tumors, unique antigens must be expressed by tumor cells to distinguish them from normal cells. As a way to avoid an immune attack, cancer cells intercede with the nonsense-mediated mRNA decay (NMD) pathway to prevent expression of aberrant mRNAs containing premature termination codons, commonly expressed in these cells [73]. Gilboa and colleagues used AsiC technology to show that cell-targeted inhibition of the NMD pathway promotes the generation of unique antigens expressed on the surface of cancer cells that can be recognized by the immune system as foreign. This recognition results in the specific destruction of the cancer cell. The authors conjugated the PSMA A10 aptamer [63] to siRNA directed against genes implicated in the NMD pathway (Upf2 and Smg1). Silencing of both of these genes resulted in the expression of aberrant epitopes on the surface of the target cells in vitro. Importantly, the authors demonstrated therapeutic efficacy following systemic administration of the AsiCs in an immune-competent mouse model of prostate cancer. Of note, therapeutic efficacy was sustained following tumor challenge, implicating an AsiC-specific, immune-mediated response against the tumors and was more effective than vaccination with GM-CSF-expressing irradiate syngeneic tumor cells (GVAX®) [74], an established immunomodulatory approach. The authors went on to confirm that the mechanism of tumor inhibition was immune-mediated as silencing of Smg1 had no effect on tumor burden in an immune-deficient mouse model. This study, was also the first to demonstrate the effectiveness of RNA AsiCs in treating the underlying cause of cancer mortality, metastasis. To date, this is the only study to apply the AsiC technology to cancer immunotherapy.
RNA aptamer-siRNA therapies for HIV
Since the first demonstration that aptamers can be used to target siRNAs to specific cell-types (e.g., cancer cells) in vivo, efforts from several groups have focused on expanding the aptamer-delivery technology to develop RNAi-drugs for the eradication of diseases other than cancer [36–39,75]. Due to the availability of aptamers against HIV and HIV host factors, it became possible to evaluate RNAi drugs for the treatment and prevention of HIV infections. In this section, we will discuss the various studies that have used aptamer-delivery technology to develop RNAi-based drugs for the treatment or prevention of HIV. We will also highlight design optimizations and delivery formulations to streamline the development and enhance the efficacy of AsiCs in vivo.
HIV AsiC therapy
In 2008, Rossi and colleagues described an AsiC for the treatment of established HIV infections [57]. This AsiC was engineered by covalently linking a neutralizing aptamer directed against the HIV glycoprotein (gp120) [76], to a siRNA targeting tat/rev, two viral genes which drive viral replication. Treatment of HIV-infected cells with the HIV AsiC was shown to decrease HIV-1 replication, as well as, overall viral infectivity in vitro. Importantly, this was the first study to develop a dual-function AsiC, where both the aptamer and the siRNA have therapeutic properties. Furthermore, binding of the aptamer to gp120 expressed on infected T-cells results in the uptake of the aptamer and its therapeutic siRNA cargo into the cells. Silencing of tat/rev viral transcripts in the infected T-cells was shown to abrogate viral replication. A significant contribution of this work was the development of a one-component reagent with the ability to reduce HIV infectivity while simultaneously inhibiting viral replication in pre-infected T cells. An additional contribution of this work was the use of Dicer substrate (DS) siRNAs (DsiRNAs) linked to the 3′-end of the aptamer (DS in Figure 1). DsiRNAs differ from standard siRNAs in the length of the duplex. Previous studies by the same group demonstrated that a longer duplex (27-mer) is a better substrate for Dicer compared with the standard 21-mer siRNA duplex [77]. Enhanced Dicer recognition translates into more efficient loading of the siRNA duplex into RNA-induced silencing complex and improved silencing efficiency. Based on these findings, the authors concluded that AsiCs composed of DsiRNAs are more efficiently processed by the RNAi machinery compared with aptamer-siRNA (23-mer) AsiCs.
A subsequent study by the same group identified novel neutralizing aptamers to gp120 and also described a `sticky-bridge' approach for appending siRNAs to the aptamer [78]. The `sticky-bridge' approach consists of a poly-carbon linker positioned at the 3′-end of the aptamer, followed by a 16-nucleotide GC-rich (stick) sequence (stick in Figure 1). The carbon linker adds rigidity to the aptamer, decreasing the likelihood that the inclusion of additional sequences would interfere with proper aptamer folding. The 16-nt GC-rich sequence was designed to allow facile interchange of any siRNA sequence for another, by simply adding the complementary 16-nt sequence to the desired siRNA. This strategy is compelling given the high mutagenic rate of HIV. In principle, the `sticky-bridge' approach can accommodate the inclusion of next-generation siRNA drugs into AsiCs, in order to rapidly target those mutant virions that developed insensitivity to the first-line AsiC treatment.
In a 2011 follow up study, the same group demonstrated initial proof of in vivo efficacy of an HIV AsiC. Here, Neff et al. evaluated the ability of the first-generation gp120 AsiC (non-stick form) [57] to reduce viral load in a humanized Rag2−/− γc−/− (Rag-hu) mouse model of HIV [39]. The gp120 AsiC was delivered by systemic administration to mice with established HIV infections. Of significance, viral loads in treated mice were decreased to below detectable levels after 5 weekly intravenous AsiC injections of 0.38 mg/kg. In addition, AsiC treatment also protected mice from a decline in CD4+ helper T cells, the contributing factor to morbidity and mortality in humans. This was specific for the gp120 AsiC and was not observed with a control AsiC composed of a nonbinding, mutant gp120 aptamer conjugated to the tat/rev DsiRNA. While treatment with the gp120 aptamer alone was capable of significantly decreasing viral loads, conjugation to the anti-tat/rev DsiRNA increased the duration of HIV suppression. The AsiC was therefore, more effective overall than the aptamer alone. Importantly, this was the first in vivo demonstration of an effective dual-function AsiC.
In a subsequent study, Rossi and colleagues evaluated the in vivo efficacy of their second-generation `sticky-bridge' AsiCs (stick; Figure 1) [38]. Using the aptamer-stick sequence previously described by them, the authors were able to readily engineer three distinct therapeutic AsiCs. Each AsiC differed only by the DsiRNA that was covalently linked to the gp120 aptamer via the stick sequence. One DsiRNA was designed to silence the expression of HIV tat/rev (HIV stick AsiC previously described by the authors). The other two DsiRNAs were designed to silence the expression of essential host-derived proteins, CD4 and transportin-3, integral players in promoting HIV infectivity. What was unique about this in vivo study was the use of chemically synthesized rather than in vitro transcribed reagents. This is of importance since preclinical studies performed with chemically synthesized RNA oligonucleotides promise to expedite the future clinical development of these RNA drugs. Using the Rag-hu mice as a model for HIV infection, a reduction in the viral load and a subsequent increase in CD4+ helper T-cells was observed, particularly when the three AsiCs were used in combination. The studies by Rossi and colleagues highlight the power of AsiC technology as a therapeutic modality for the treatment of established HIV infections. In addition, these studies significantly advanced the field as a whole by describing optimizations for streamlining the design of high-potency, therapeutic AsiCs.
Prophylactics for HIV
To date, considerable effort has been placed on employing AsiC technology to develop therapeutics for eradicating established disease (e.g., HIV and cancer). However, for those diseases that can be easily transmitted from person to person, such as HIV, another effective strategy is prevention, by means of a prophylactic approach. To this end, Wheeler et al. were the first to describe AsiCs as prophylactic agents against HIV [36]. In this study, an RNA aptamer to CD4 [79] was used to deliver anti-HIV siRNAs to CD4+ T cells (HIV target cells). The authors described two AsiCs, differing only in the siRNA sequence that was conjugated to the CD4 aptamer. One AsiC used an siRNA designed to decrease expression of C-C chemokine receptor type 5 (CCR5), a protein on the surface of white blood cells that is critical for enabling HIV entry into cells. The other AsiC was designed to deliver an siRNA to silence the expression of the HIV gag and vif genes, required for viral replication. Both AsiCs were effective at cell-specific targeting and gene knock down in vitro and in vivo. Importantly, intravaginal application of these AsiCs in a humanized mouse model prevented transmission of HIV.
In a follow up study, Wheeler et al. described the durability of gene silencing by the HIV AsiCs [37]. The authors observed that two AsiC treatments, administered 24 h apart, were sufficient to achieve silencing and protection from HIV infection. Importantly, silencing persisted for several weeks following treatment. The implications of this study are significant, as durable treatments (i.e., ones that do not have to be administered frequently), like the one describe by Lieberman and colleagues, increase the likelihood of patient compliance and consequently the efficacy of the prophylactic agent. In the same study, the authors employ an FDA approved hydrogel (hydroxyethyl cellulose gel) for administering the AsiC prophylactic agent. The use of the hydrogel has several advantages: it stabilizes the AsiCs in vaginal fluid, it enhances cell-uptake and it facilitates the vaginal application of the prophylactic agent. Importantly, combining the HIV AsiCs with the hydrogel did not affect overall targeting or silencing efficiency. More importantly, the authors report an impressive outcome where 100% of treated mice are protected from infection. Interestingly, although silencing was demonstrated to last several weeks following the last AsiC-gel treatment, complete protection from HIV infection was only possible if the treatment was administered several days before HIV challenge. While the reasons for this are not clear, the authors offer several explanations for this observation including: the migration of new CD4+ cells to the vagina after treatment that can serve as viral hosts, dilution of siRNAs due to activated T-cell division and the potential for more rapid siRNA degradation in activated versus resting T cells. In future studies, Lieberman and colleagues propose to evaluate several dosing schedules/regimens, as well as, evaluating the efficacy and safety of the AsiC prophylactic agents in larger preclinical models of HIV (e.g., Rhesus macaques model of HIV infection). Together, these studies highlight the possibility of exploiting AsiC technology for developing prophylactics for various sexually transmitted diseases including HIV. Furthermore, given the side-effects and hurdles with HIV therapy, prevention of the initial infection is an attractive and powerful alternative for eradicating this disease.
DNA aptamer-RNA conjugates
Much like RNA aptamers, DNA aptamers can be used to deliver siRNAs or other therapeutic oligonucleotides to cells. DNA-RNA AsiCs are typically chemically synthesized as it is not possible to in vitro transcribe a single hybrid molecule composed of DNA and RNA. One downside of this is that the cost of synthesis and the oligonucleotide-length constraints limit the number of DNA-RNA AsiCs that can be thoroughly evaluated in preclinical studies. Even so, several groups have been able to take advantage of existing, short DNA aptamers for conjugating to therapeutic oligonucleotides for various therapeutic applications. This section will center on recent DNA-aptamer delivery studies and comment on the potential of this novel class of AsiCs as a therapeutic modality in the setting of cancer, HIV infection and several immune disorders.
Kortylewski and colleagues were the first to describe the development and in vivo characterization of a DNA-RNA AsiC [80]. Here, the authors employed the known toll like receptor 9 (TLR9)-specific agonist (CpG 1668) as a DNA aptamer [81,82]. CpG 1668 is known to internalize into various TLR9 positive immune cells, including those that promote oncogenesis in the tumor microenvironment such as myeloid cells and B-cells. Upon internalization and TLR9 binding, CpG 1668 activates TLR9 signaling cascades resulting in immune activation [83–85]. The authors sought to further modulate immune function by appending to the CpG DNA aptamer, an siRNA against the immune-suppressive gene, STAT3. The CpG-STAT3 AsiC was shown to enhance immune function in vivo resulting in antitumor immune activation. This ultimately led to an impressive reduction in tumor burden. Importantly, the authors validated the efficacy of the CpG-STAT3 AsiC in several mouse models of cancer, including melanoma and colon carcinoma. Of note, the studies by Kortylewski [80], Gilboa [35] and colleagues provide strong evidence for the use of DNA-RNA or all RNA AsiCs as future cancer immunotherapy drugs.
A different application of the DNA-RNA AsiC approach was recently described by Kotula et al. In this study, the authors used an existing G-quartet DNA aptamer to nucleolin (AS1411) [86,87] to deliver antisense oligonucleotides to cancer cells [88]. Nucleolin is overexpressed on the surface of many carcinomas. In cancer cells, nucleolin rapidly cycles from the cell surface to the nucleus where it is thought to play a transport role, shuttling extracellular ligands to the nuclear compartment [89]. Sullenger and colleagues exploited this property of nucleolin to deliver splice-switching oligonucleotides (SSOs) (a form of antisense technology) to the nucleus of cancer cells. SSOs are single stranded RNA oligonucleotides that are designed to bind their complementary sequence (splice-site or splice enhancer) within a target pre-mRNA. Binding of the SSO to its pre-mRNA prevents the endogenous splicing machinery from being recruited to that splice-site, giving rise to a different splice-transcript [90–92]. In this study, the authors validated the proof-of-concept for their approach by using the nucleolin aptamer [86,87] to deliver an SSO to the nucleus of target cells and correct an aberrant stop codon within the Luciferase reporter gene. Following nucleolin-mediated cellular uptake, the nucleolin-SSO AsiC redirected splicing of the target gene in the nuclei of treated cells and restored Luciferase expression. Importantly, the combination of the SSO with the aptamer was more effective than the SSO alone, suggesting that proper targeting of an SSO can significantly enhance its efficiency. While promising, future studies addressing: the ability of the nucleolin DNA aptamer to target SSOs directed against endogenous transcripts; the splice-switching efficiency of the nucleolin-SSO in vivo and the in vivo efficacy of a therapeutic nucleolin-SSO are warranted. Of note, this study further highlights the flexibility of the AsiC platform for targeting a range of therapeutic oligonucleotides (e.g., siRNAs, miRNAs and antisense oligonucleotides) to cells.
In a separate study, Dai et al. [61] employed a previously characterized DNA aptamer to Mucin (MUC1) [93], a transmembrane glycoprotein that is highly expressed on the cell surface of adenocarcinomas of the breast, colon and lung [94], to deliver a therapeutic miR. Specifically, the authors engineered an AmiC by covalently linking the MUC1 aptamer to miR-29b [61]. Of note, this was the first description of AmiC technology. When applied to OVCAR-3 cells in culture, the DNA AmiC internalized into the target cells leading to silencing of known miR-29b target genes (e.g., DNA MT 3a and 3b). This, in turn, resulted in decreased methylation of the tensin homolog deleted on chromosome ten (PTEN) promoter, subsequent re-expression of the PTEN gene product and an increase in cellular apoptosis. Given the expression profile of MUC1 in many adenocarcinomas, this reagent has the potential to be evaluated as a therapeutic for various solid cancers. Thus, future studies addressing the in vivo efficacy of this reagent in the setting of ovarian cancer as well as other MUC+ cancers are warranted.
To date, most studies have focused on evaluating DNA-RNA AsiCs as potential cancer agents. Recently, Zhu et al. described the synthesis of a DNA-RNA AsiC composed of a DNA aptamer specific to CD4 and a siRNA against the HIV protease gene for the treatment of HIV [95]. What was unique about this study is that the authors chose to convert a known RNA aptamer against CD4 into a DNA aptamer. They then conjugated the DNA and RNA aptamers to the therapeutic siRNA and compared the efficiency of each AsiC in HIV infected cells in culture. While both AsiCs resulted in efficient gene silencing, the DNA AsiC appeared to be more effective than the RNA AsiC at silencing its target gene. Future studies will hopefully provide some insight as to why the DNA-RNA AsiC seems to work better than the all-RNA AsiC and evaluate the efficacy of the DNA-RNA AsiC in vivo.
A more recent application of the DNA-aptamer delivery technology has been the use of DNA-RNA AsiCs for treating various immune disorders [75]. In immune disorders such as Stevens–Johnson syndrome, toxic epidermal necrolysis and graft-versus-host disease, activation of cytotoxic T lymphocytes by drugs or allogeneic antigens, is thought to play a significant role in the pathogenesis of disease [96,97]. Specifically, production of granulysin by cytotoxic T lymphocytes has been implicated as a major cause of these disorders [98]. Wang et al. sought to exploit DNA AsiC technology as a potential therapeutic intervention in these disorders. Initially, the authors describe the identification of a DNA aptamer specific to the CTL marker, CD8. The CD8 aptamer was conjugated (via a `sticky-bridge') to a DsiRNA targeting the granulysin gene (stick in Figure 1). Importantly, the CD8-granulysin AsiC was shown to inhibit pathologic T-cell responses in several in vitro models of Stevens–Johnson syndrome/toxic epidermal necrolysis and graft-versus-host disease. This work has significant implications, as currently there are no safe and effective therapeutic strategies for any of these immune disorders, therefore, future in vivo studies are warranted.
Together these studies highlight the broad potential of the aptamer-delivery technology as many more therapeutic DNA aptamers have been described and can now be evaluated in the contexts of AsiCs. For a complete list of these DNA aptamers we direct you to the reviews by Zhou and Rossi [11], Thiel and Giangrande [14], or Ni et al. [99].
Aptamer-targeted siRNA/shRNA nanoparticle conjugates
Nanoparticles have been proposed as a potential means to deliver siRNA to cells [100–104]. The benefits of nanoparticles include protection of siRNA from serum nucleases, good tissue penetration, relatively good cell uptake, and stability [105–107]. Different types of reagents that can be collectively categorized as nanoparticles or nanoplexes have been tested for their ability to deliver nucleic acids to cells. While these reagents show promise at enhancing cellular uptake of siRNAs in vitro and in vivo, one major problem with nanoparticle delivery is lack of specificity for target cells. In this section we will highlight recent studies that have taken advantage of the targeting properties of aptamers for delivering nanoparticles to specific cell types.
One of the first studies to use aptamer-targeted nanoparticle technology to deliver therapeutic RNAs was performed by Kim et al. [108]. Here, the authors used the PSMA aptamer (A10–3) [63] to specifically target prostate cancer cells. Initially, the aptamer was conjugated to a polyethyleneimine (PEI)-PEG scaffold. The aptamer-functionalized nanoparticle was loaded with shRNAs directed against the pro-survival gene Bcl-xL to generate the final therapeutic particle (Figure 2a). Interestingly, the authors also sought to include the chemotherapeutic drug, doxorubicin (DOX), which, they showed, intercalated into the PSMA aptamer itself. Importantly, addition of DOX did not affect aptamer function (target binding and cell-internalization). Using this approach, the authors demonstrated delivery of two therapeutic moieties (chemotherapy and shRNA) to PSMA+ target cells in culture, leading to the death of the cancer cells. Interestingly, delivery of either the shRNA or DOX alone had no effect on cell viability, demonstrating the effectiveness of the combination therapy. Although, the efficacy of this multi-component drug was only evaluated in vitro, the encouraging results from these studies warrant future in vivo testing.
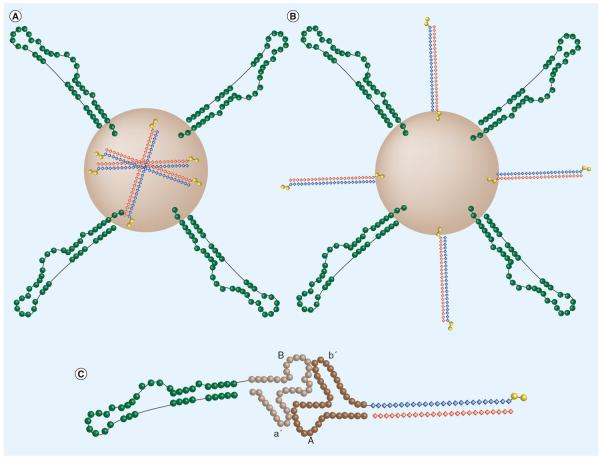
Figure 2. Aptamer functionalized nanoparticle design.
Representative aptamer functionalized nanoparticle structure where the therapeutic siRNA is (A) within the nanoparticle itself [108,112] or (B) conjugated to the nanoparticle surface [110,114,115]. (C) Representative RNA scaffold nanoparticle structure. Exploiting the loop–loop binding interaction of pRNA, this scaffold can be harnessed to conjugate an aptamer to a therapeutic siRNA [116,118,119].
Since the first description of an aptamerfunctionalized therapeutic nanoparticle in 2004 [109], several studies have used aptamers to target nanoparticles to specific cell types. While these studies share a common approach to targeting, the composition of the nano particle (e.g., scaffold, aptamer, therapeutic cargo) varies between the studies. Zhao et al. used a CD30 aptamer to deliver siRNAs against the ALK gene to human anaplastic large cell lymphoma cells [110]. The aptamer and siRNAs were coupled to a PEI-citrate carrier, to generate the therapeutic nanoparticle (Figure 2b). The authors confirmed binding and uptake of the therapeutic nanoparticle into target cells in culture. In a similar study, Wu et al. conjugated the PSMA aptamer (A10–3.2) [34] to polyamidoamine and PEG [111]. The PSMA aptamer-functionalized nanoparticle was used to target miRNAs, miR-15a and miR-16–1, to PSMA+ prostate cancer cells. Proof-of-efficacy was reported when the therapeutic nanoparticle was added to PSMA+ prostate cancer cells in culture. A similar approach was also described by Wilner et al. in 2012 [112]. In this study, the authors coupled the transferrin receptor aptamer (c2.min) to stable nucleic acid lipid particles (SNALPs). SNALPs have been shown to efficiently bind, stabilize and deliver siRNAs to the cytoplasm of target cells in vitro and in vivo [113].Addition of the aptamer-SNALP nanoparticle to cells resulted in silencing of the siRNA target gene. Given the promise of SNALPs for RNAi therapeutics, future studies to address the in vivo efficacy and potential advantages of aptamer-targeted SNALPs are needed.
Recently, efforts have been focused on adapting the aptamer-targeted nanoparticle technology for developing targeted image-guided approaches for RNAi. To this end, Baglkot and Gao describe the coupling of PSMA AsiCs onto polymer coated quantum dots (QDs) [114]. When applied to PSMA+ prostate cancer cells, the AsiCs underwent cell-specific uptake leading to silencing of the siRNA target gene. Given the intrinsic fluorescent properties of QDs, QD-functionalized AsiC nanoparticles enabled the authors to visualize internalization of their nanoparticles into PSMA+ prostate cancer cells. This approach is significant given the paucity of information on endosomal uptake and cellular trafficking of AsiCs. In a similar study, Kim et al. coupled the nucleolin DNA aptamer [86,87] to a magnetic fluorescence nanoparticle [115]. This was used to deliver a miR-221 molecular beacon (MB), which contains a fluorophore on one strand of the miR-221 dsRNA and a fluorescent quencher on the opposing strand. Upon binding, the guide strand is released from its fluorescent quencher, eliciting Cy5 fluorescence. Interestingly, the miRNA MB was designed not for the delivery of a functional miRNA but rather, to inhibit the expression of an endogenous miRNA via complementary binding to the miRNA MB. In this regard, the miRNA MB functioned as an anti-miR (or antago miR). Importantly, the authors demonstrated the efficacy of their approach both in vitro and in vivo. This study is significant since it is the first demonstration of a targeted anti-miR therapy and it combined aptamer-delivery technology with molecular beacon technology to enable biogenesis tracking of miR-221 in targeted cells.
Recently, several groups have described a unique method, based on an RNA nano particle scaffold, for coupling targeting-aptamers to therapeutic-siRNAs. These authors use what is known as a packaging RNA (pRNA) as the scaffold for the nanoparticle. pRNAs are derived from bacteriophage phi29 small RNAs that normally package DNA into procapsids and contain a 5′/3′ helical domain and an intermolecular interaction domain, which serves to form dimeric pRNA molecules [116]. By substituting the 5′/3′ helical domains with an aptamer on one pRNA molecule and an siRNA on another pRNA molecule, an aptamer-siRNA conjugate can be generated through dimerization of the intermolecular interaction domains of the two pRNAs (Figure 2C).
In proof-of-concept studies by Guo and colleagues, an aptamer for CD4 [117], a receptor predominantly expressed on T cells, was used to specifically deliver a 29-mer DsiRNA against survivin, a prosurvival gene whose expression is elevated in many cancers [116,118]. This delivery mechanism resulted in reduced viability of a model thymocyte cell line engineered to express CD4, but had no effect on CD4-thymocytes in culture. In addition, when these CD4+ cells were incubated with this reagent prior to injection into the flanks of mice, a significant decrease in tumor burden was observed. Using a similar approach, Zhou et al. designed a pRNA dimer where one monomer was conjugated to an anti-HIV gp120 aptamer and the other monomer was coupled to a therapeutic siRNA against the HIV tat/rev gene [119]. These monomers were combined at a 1:1 ratio. The aptamer-siRNA-pRNA nanoparticle was shown to bind to gp120 on the surface of infected T cells, undergo cellular uptake and silence target gene (tat/rev) expression. Future work will focus on optimizing the dimerization efficiency and stability of these chimeric constructs. These pRNA studies are significant given the modular nature of the technology. Theoretically, these methods can be applied to any cell internalizing aptamer for the targeted, cell-specific delivery of a therapeutic siRNA. Future studies assessing the interchangeability of diverse aptamers and siRNAs in addition to thorough in vivo studies are warranted [120].
Conclusion
Aptamer-delivery technology promises to facilitate economical and versatile RNAi-based treatments for a variety of disorders. In this review we have discussed preclinical studies centered on aptamer-delivery technology, for the development of targeted oligonucleotide-based drugs (e.g., siRNAs, miRNAs, shRNAs, SSOs) for treating medical conditions including: cancer, HIV, and diseases of the immune system. We highlight the therapeutic potential of each aptamer-targeted drug and discuss advances in selection strategies and drug design for optimizing the uptake and processing of these oligonucleotide-drugs in cells. We have also considered major challenges facing manufacturing and utilization of chemically modified long oligonucleotides and multicomponent aptamer-targeted nanoparticle drugs. Fortunately, increased availability and lower costs of raw materials have significantly reduced costs of manufacturing chemically modified oligonucleotides. Likewise, advances in chemical strategies for incorporating modified nucleotides have dramatically improved resistance of aptamers and other therapeutic oligonucleotides (siRNAs and SSOs) to nuclease-mediated degradation while, conjugation of aptamer-targeted drugs to delivery vehicles (e.g., high-molecular-weight PEG and nanoparticles) effectively mitigates rapid renal clearance. Although aptamer-siRNA drugs seem to display minimal toxicity, new chemistries and conjugation strategies used for production of these chimeric oligonucleotides warrant further toxicity studies. As investment in therapeutic oligonucleotides (e.g., aptamers, siRNAs and antisense technology) is constantly increasing, it is projected that clinical testing of aptamer-targeted drugs will materialize over the next 2–5 years.
Future perspective
Before aptamer-delivery technology can be broadly translated, continued efforts in several key areas will need to be considered. As such, one limiting factor to the broad clinical application of this delivery technology is the relative paucity of aptamers capable of undergoing efficient, cell-specific uptake. Traditionally, aptamers have been identified using in vitro selection methodologies that rely on the availability of purified, recombinant proteins. Although effective in several contexts, these in vitro selection strategies are not optimal for identifying aptamers that: recognize cell-surface receptors in their native milieu (cell membrane), undergo cellular uptake and are released in the cytoplasm of target cells. Current progress in cell-based selection methodologies has made it possible to overcome several of these hurdles. Aptamers with the ability to bind to cell-surface receptors on cells have been described. Furthermore, by slightly modifying the conditions of the cell-selection protocol Giangrande, McNamara and colleagues have been able to enrich for aptamers capable of undergoing cell-specific uptake (Figure 3) [21,62,121]. In one of these studies, the authors used the cell-internalizing aptamers to demonstrate delivery of therapeutic siRNAs to HER2+ breast cancer cells in culture [62]. While still in their infancy, these cell-internalization approaches are certainly poised to increase the repertoire of aptamers capable of cell-specific uptake. Modifications to the cell-internalization approach pioneered by Giangrande and colleagues, such as the inclusion of cell-fractionation steps to enrich for cytoplasmic material, may favor the isolation of cytoplasmic-targeting aptamers for RNAi-delivery applications.
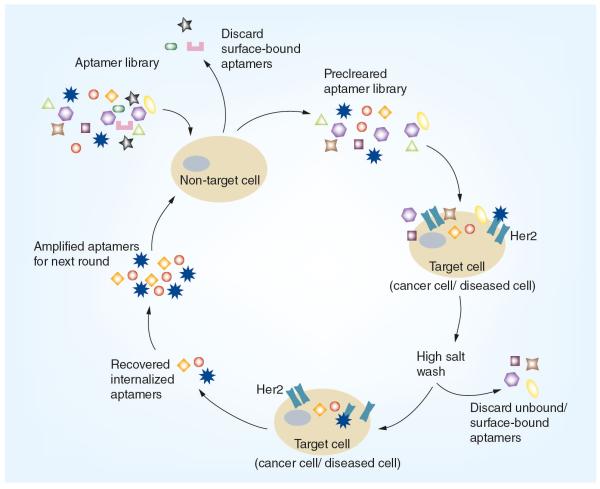
Figure 3. Cell-internalization systematic evolution of ligands by exponential methodology.
This cell-based systematic evolution of ligands by exponential methodology enriches for aptamers that bind a target cell in a more physiologically relevant setting and internalize into cells [62]. These two properties allow any identified aptamers to more readily be used as delivery vehicles for therapeutic reagents such as siRNAs, miRNAs or antisense technology. The process starts with a complex (generally greater than 1012 individual sequences) aptamer library (depicted as polygons) that is incubated with a non-target cell to remove sequences that bind or internalize into cells non-specifically. Those aptamers that do not bind the non-target cell make up the precleared aptamer library. This pool of aptamers is then incubated on a target cell where the aptamer target is known (e.g., HER2). This can also be performed on cells where the target receptor is unknown [21]. Following a high salt wash to remove unbound and surface bound aptamers, internalized aptamers are extracted from whole-cell lysates. The recovered aptamers are enzymaticaly amplified and then used as the aptamer library in another round of selection. This iterative process is carried out for several rounds in order to enrich for sequences that bind and internalize specifically into the selected target cell.
Alternatively, conjugation of aptamers with cationic amphipathic peptides [121–123] or nanoparticles [124] may facilitate the release of cell-internalizing aptamers from endocytic compartments into the cytoplasm of target cells. Nanoparticles have been extensively investigated for their ability to deliver siRNAs to the cytoplasm of cells. Two recent studies reported on the endosomal escape mechanisms of siRNAs delivered via lipid nanoparticles [125,126]. While these studies did not use aptamer-functionalized nanoparticles, they address ways to quantify the amount of siRNA that is efficiently released into the cytoplasm. While the efficiency and mechanism of release is likely to vary based on the nature of the nanoparticle and targeting approach (aptamer-mediated), the methodologies described in these studies are of importance. Similar studies performed on aptamer-targeted drugs are predicted to advance the understanding of the subcellular trafficking of these reagents and expedite their clinical application.
Because efficient cytoplasmic delivery is a major challenge for using aptamers to deliver siRNAs to cells, understanding the mechanism by which aptamers release their siRNA therapeutic cargo in the cytoplasm of target cells is critical. In a recent study, Nechaev et al. reported that endosomal release of a DNA-RNA AsiC was dependent on TLR9 [127]. The authors showed that the DNA-RNA AsiC was taken up into cells by a non-specific, scavenger receptor-mediated endocytic mechanism. Rapidly after internalization, the siRNA was uncoupled from the CpG DNA aptamer within Dicer and TLR9 containing early endosomes. Here, the siRNA was bound by Dicer and shuffled to the endoplasmic reticulum (ER). In the absence of TLR9, the siRNA was trapped in endosomal compartments resulting in loss of RNAi. It is known that TLR9 can regulate the transport of endosomes toward the ER. Previous studies have demonstrated the localization of RNAi machinery at the ER. Thus, the authors suggest that TLR9 may be required for transportation of the therapeutic siRNA from endosomes to the ER where it can be processed by the RNAi machinery.
In the case of aptamer-targeted drugs, where the aptamer is directly conjugated to the therapeutic siRNA moiety, optimizing drug design to enhance processing by the RNAi machinery is imperative. As discussed in previous sections, several designs have been explored (Figure 1). These include the use of a `sticky bridge' to facilitate the exchange of the therapeutic siRNA cargo without affecting aptamer function or a DsiRNA to enhance Dicer processing. Systematic studies, which focus on the direct comparison of current AsiC design strategies in order to identify the optimal design, are likely to accelerate the development of novel AsiC therapeutics.
Recent advancements in the field of aptamer-delivery technology promise to bring significant changes to the field of medicine by facilitating the development of targeted therapeutic agents for the treatment of inherently challenging diseases (cancer and HIV) and reducing side-effects inherent to traditional (non-targeted) therapies. With the 10 year anniversary of the aptamer-delivery technology approaching, we look forward to the eventual clinical evaluation of the first aptamer-targeted drug and to the successful application of this technology in the clinic.
Executive summary.
Progress and challenges of aptamer-delivery technology
■ Aptamers have emerged as effective reagents for the delivery of therapeutic oligonucleotides (siRNAs, shRNAs, antisense, miRNAs and anti-miRs) to specific diseased tissue/cells.
■ Aptamer therapeutics can be augmented to overcome various challenges such as susceptibility to nuclease-mediated degradation, potential toxicity due to off-target effects, efficient large-scale production via chemical synthesis, rapid renal clearance of small molecules, and inefficient cellular uptake and intracellular therapeutic processing.
Application of aptamer-delivery technology
■ Therapeutic aptamer-siRNA chimeras have been developed as cytotoxic, neo-adjuvant, or immune-modulatory drugs for the treatment of various malignancies.
■ Therapeutic aptamer-siRNA chimeras have been developed as anti-viral drugs for the treatment of established HIV infections as well as the prevention of HIV infection through prophylactic use.
Aptamer-nanoparticle delivery platforms
■ Aptamers have been successfully functionalized onto various nanoparticle platforms complexed with therapeutic oligonucleotides as another means to specifically deliver these therapeutic molecules.
Optimization of systematic evolution of ligands by exponential technology
■ Novel cell-based selection strategies coupled with a more in depth understanding of aptamer therapeutic processing is expected to lead to the clinical evaluation of the first aptamer-targeted drug within the next 5 years.
Acknowledgements
The authors acknowledge T Ruggle at the University of Iowa Design Center, IA, USA for assistance with figure design.
JP Dassie is supported by a Postdoctoral Fellowship from the NIH (T32 HL07734). PH Giangrande acknowledges support from the NIH (1RO1CA138503), the Mary Kay Ash Foundation (MKACF 033–12) and the Elsa U Pardee Foundation. PH Giangrande is a co-inventor on patent applications: PCT/US09/53023_ NUCLEIC ACIDS APTAMERS and PCT/US2011/034169_ HER2 NUCLEIC ACID APTAMERS.
Glossary
- Aptamer-siRNA chimera
An aptamer-siRNA chimera is a two-component delivery vehicle composed of a targeting moiety (aptamer) and a therapeutic moiety (siRNA). Aptamer-siRNA chimeras bearing aptamers with both targeting and inhibitory functions have also been described.
Footnotes
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Kole R, Krainer AR, Altman S. RNA therapeutics. beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug. Discov. 2012;11(2):125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juliano RL, Ming X, Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug. Chem. 2012;23(2):147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36(12):4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coelho T, Adams D, Silva A, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369(9):819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 6.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 7.Barros SA, Gollob JA. Safety profile of RNAi nanomedicines. Adv. Drug Deliv. Rev. 2012;64(15):1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Sehgal A, Vaishnaw A, Fitzgerald K. Liver as a target for oligonucleotide therapeutics. J. Hepatol. 2013 doi: 10.1016/j.jhep.2013.05.045. doi:10.1016/j.jhep.2013.05.045. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Gavrilov K, Saltzman WM. Therapeutic siRNA. principles, challenges, and strategies. Yale J. Biol. Med. 2012;85(2):187–200. [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey JM, Hibbitts A, Barlow J, Kelly C, Sivadas N, Cryan SA. `Smart' non-viral delivery systems for targeted delivery of RNAi to the lungs. Ther. Deliv. 2013;4(1):59–76. doi: 10.4155/tde.12.133. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Rossi JJ. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides. 2011;21(1):1–10. doi: 10.1089/oli.2010.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Bobbin ML, Burnett JC, Rossi JJ. Current progress of RNA aptamer-based therapeutics. Front. Genet. 2012;3:234. doi: 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Rossi JJ. Aptamer-targeted cell-specific RNA interference. Silence. 2010;1(1):4. doi: 10.1186/1758-907X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiel KW, Giangrande PH. Intracellular delivery of RNA-based therapeutics using aptamers. Ther. Deliv. 2010;1(6):849–861. doi: 10.4155/tde.10.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344(6265):467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 16.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment. RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]; ■■ First study to coin the term systematic evolution of ligands by exponential.
- 17.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; ■ This study and the one by Tuerk and Gold (1990) provided the first demonstration of an in vitro selection methodology (systematic evolution of ligands by exponential) for identifying RNA aptamers with affinity and specificity for a given target.
- 18.Stoltenburg R, Reinemann C, Strehlitz B. SELEX – a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Mosing RK, Bowser MT. Microfluidic selection and applications of aptamers. J. Sep. Sci. 2007;30(10):1420–1426. doi: 10.1002/jssc.200600483. [DOI] [PubMed] [Google Scholar]
- 20.Thiel KW, Hernandez LI, Dassie JP, et al. Delivery of chemo-sensitizing siRNAs to HER2+−breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40(13):6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiel WH, Bair T, Peek AS, et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS ONE. 2012;7(9):e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerchia L, Duconge F, Pestourie C, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3(4):e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001;276(19):16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 24.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX. Systematic evolution of ligands by exponential enrichment. Proc. Natl Acad. Sci. USA. 2003;100(26):15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shamah SM, Healy JM, Cload ST. Complex Target SELEX. Acc. Chem Res. 2008;41(1):130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 26.Sefah K, Tang ZW, Shangguan DH, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23(2):235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shangguan D, Meng L, Cao ZC, et al. Identification of liver cancer-specific aptamers using whole live cells. Anal. Chem. 2008;80(3):721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 28.Guo K-T, Paul A, Schichor C, Ziemer G, Wendel HP. CELL-SELEX. Novel perspectives of aptamer-based therapeutics. Int. J. Mol. Sci. 2008;9(4):668–678. doi: 10.3390/ijms9040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris KN, Jensen KB, Julin CM, Weil M, Gold L. High affinity ligands from in vitro selection: complex targets. Proc. Natl Acad. Sci. USA. 1998;95(6):2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat. Rev. Drug. Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013;48(1–2):259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990;63(3):601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Yigit MV, Mazumdar D, Lu Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv. Drug Deliv. Rev. 2010;62(6):592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dassie JP, Liu XY, Thomas GS, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27(9):839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■ Optimization of the first-generation prostate-specific membrane antigen aptamer-siRNA chimera achieved systemic delivery in an in vivo model of prostate cancer.
- 35.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465(7295):227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■ This groundbreaking tumor immunotherapy study used aptamers to deliver siRNAs against components of the nonsense-mediated mRNA decay pathway, which generated novel antigens on the surface of PSMA+ and tumor cells and resulted in an immune response to the tumor.
- 36.Wheeler LA, Trifonova R, Vrbanac V, et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Invest. 2011;121(6):2401–2412. doi: 10.1172/JCI45876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler LA, Vrbanac V, Trifonova R, et al. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol. Ther. 2013;21(7):1378–1389. doi: 10.1038/mt.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Neff CP, Swiderski P, et al. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 2013;21(1):192–200. doi: 10.1038/mt.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neff CP, Zhou J, Remling L, et al. An aptamersiRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci. Transl. Med. 2011;3(66):66ra6. doi: 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni X, Zhang Y, Ribas J, et al. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J. Clin. Invest. 2011;121(6):2383–2390. doi: 10.1172/JCI45109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burnett JC, Rossi JJ. RNA-based therapeutics. current progress and future prospects. Chem. Biol. 2012;19(1):60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18(4):305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 43.Rettig GR, Behlke MA. Progress toward in vivo use of siRNAs-II. Mol. Ther. 2012;20(3):483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge Q, Dallas A, Ilves H, Shorenstein J, Behlke MA, Johnston BH. Effects of chemical modification on the potency, serum stability, and immunostimulatory properties of short shRNAs. RNA. 2010;16(1):118–130. doi: 10.1261/rna.1901810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolkenbeck K, Zundel G. The significance of the 2′ OH group and the influence of cations on the secondary structure of the RNA backbone. Biophys. Struct. Mech. 1975;1(3):203–219. doi: 10.1007/BF00535757. [DOI] [PubMed] [Google Scholar]
- 46.Campbell MA, Wengel J. Locked vs. unlocked nucleic acids (LNA vs. UNA). contrasting structures work towards common therapeutic goals. Chem. Soc. Rev. 2011;40(12):5680–5689. doi: 10.1039/c1cs15048k. [DOI] [PubMed] [Google Scholar]
- 47.Henry S, Stecker K, Brooks D, Monteith D, Conklin B, Bennett CF. Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J. Pharmacol. Exp. Ther. 2000;292(2):468–479. [PubMed] [Google Scholar]
- 48.Peacock H, Fucini RV, Jayalath P, et al. Nucleobase and ribose modifications control immunostimulation by a microRNA-122-mimetic RNA. J. Am. Chem. Soc. 2011;133(24):9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keefe AD, Cload ST. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008;12(4):448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 50.Reese CB. Oligo- and poly-nucleotides. 50 years of chemical synthesis. Org. Biomol. Chem. 2005;3(21):3851–3868. doi: 10.1039/b510458k. [DOI] [PubMed] [Google Scholar]
- 51.Rockey WM, Hernandez FJ, Huang SY, et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011;21(5):299–314. doi: 10.1089/nat.2011.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19(3):209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boomer RM, Lewis SD, Healy JM, Kurz M, Wilson C, Mccauley TG. Conjugation to polyethylene glycol polymer promotes aptamer biodistribution to healthy and inflamed tissues. Oligonucleotides. 2005;15(3):183–195. doi: 10.1089/oli.2005.15.183. [DOI] [PubMed] [Google Scholar]
- 54.Healy JM, Lewis SD, Kurz M, et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004;21(12):2234–2246. doi: 10.1007/s11095-004-7676-4. [DOI] [PubMed] [Google Scholar]
- 55.Bozza M, Sheardy RD, Dilone E, Scypinski S, Galazka M. Characterization of the secondary structure and stability of an RNA aptamer that binds vascular endothelial growth factor. Biochemistry. 2006;45(24):7639–7643. doi: 10.1021/bi0521442. [DOI] [PubMed] [Google Scholar]
- 56.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151(3):220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008;16(8):1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ An inhibitory aptamer was combined with a silencing siRNA to deliver a two-hit punch to HIV-infected T cells.
- 58.Mcnamara JO, 2nd, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24(8):1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]; ■■ Established aptamer-siRNA chimeras as a platform technology to specifically deliver siRNAs against prosurvival genes to prostate specific membrane antigen+ human prostate cancer tumors.
- 59.Zhou J, Tiemann K, Chomchan P, et al. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res. 2013;41(7):4266–4283. doi: 10.1093/nar/gkt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hussain AF, Tur MK, Barth S. An aptamer-siRNA chimera silences the eukaryotic elongation factor 2 gene and induces apoptosis in cancers expressing avb3 integrin. Nucleic Acid Ther. 2013;23(3):203–212. doi: 10.1089/nat.2012.0408. [DOI] [PubMed] [Google Scholar]
- 61.Dai F, Zhang Y, Zhu X, Shan N, Chen Y. Anticancer role of MUC1 aptamer-miR-29b chimera in epithelial ovarian carcinoma cells through regulation of PTEN methylation. Target. Oncol. 2012;7(4):217–225. doi: 10.1007/s11523-012-0236-7. [DOI] [PubMed] [Google Scholar]
- 62.Thiel KW, Hernandez LI, Dassie JP, et al. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acid Res. 2012;40(13):6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62(14):4029–4033. [PubMed] [Google Scholar]; ■ Prostate-specific membrane antigen A9 and A10 aptamers characterized in this study were the initial aptamers used, in subsequent studies, to deliver siRNAs to target cells.
- 64.Wullner U, Neef I, Eller A, Kleines M, Tur MK, Barth S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr. Cancer Drug Targets. 2008;8(7):554–565. doi: 10.2174/156800908786241078. [DOI] [PubMed] [Google Scholar]
- 65.Mi J, Zhang X, Giangrande PH, et al. Targeted inhibition of alphavbeta3 integrin with an RNA aptamer impairs endothelial cell growth and survival. Biochem. Biophys. Res. Commun. 2005;338(2):956–963. doi: 10.1016/j.bbrc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 66.Chattopadhyay N, Chatterjee A. Studies on the expression of alpha(v)beta3 integrin receptors in non-malignant and malignant human cervical tumor tissues. J. Exp. Clin. Cancer Res. 2001;20(2):269–275. [PubMed] [Google Scholar]
- 67.Takano S, Tsuboi K, Tomono Y, Mitsui Y, Nose T. Tissue factor, osteopontin, alphavbeta3 integrin expression in microvasculature of gliomas associated with vascular endothelial growth factor expression. Br. J. Cancer. 2000;82(12):1967–1973. doi: 10.1054/bjoc.2000.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goel HL, Li J, Kogan S, Languino LR. Integrins in prostate cancer progression. Endocr. Relat. Cancer. 2008;15(3):657–664. doi: 10.1677/ERC-08-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Novak AJ, Grote DM, Stenson M, et al. Expression of BLyS and its receptors in B-cell non-Hodgkin lymphoma. Correlation with disease activity and patient outcome. Blood. 2004;104(8):2247–2253. doi: 10.1182/blood-2004-02-0762. [DOI] [PubMed] [Google Scholar]
- 70.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 71.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer. rationale, strategies and challenges. Nat. Rev. Drug. Discov. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerchia L, Esposito CL, Camorani S, et al. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 2012;20(12):2291–2303. doi: 10.1038/mt.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. implications for physiology and disease. Biochim. Biophys. Acta. 2013;1829(6–7):624–633. doi: 10.1016/j.bbagrm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nemunaitis J. Vaccines in cancer. GVAX, a GM-CSF gene vaccine. Expert Rev. Vaccines. 2005;4(3):259–274. doi: 10.1586/14760584.4.3.259. [DOI] [PubMed] [Google Scholar]
- 75.Wang CW, Chung WH, Cheng YF, et al. A new nucleic acid-based agent inhibits cytotoxic T lymphocyte-mediated immune disorders. J. Allergy Clin. Immunol. 2013;132(3):713–722. doi: 10.1016/j.jaci.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 76.Khati M, Schuman M, Ibrahim J, Sattentau Q, Gordon S, James W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2′F-RNA aptamers. J. Virol. 2003;77(23):12692–12698. doi: 10.1128/JVI.77.23.12692-12698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23(2):222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 78.Zhou J, Swiderski P, Li H, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37(9):3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■ New anti-gp120 inhibitory aptamers were identified and a unique `sticky bridge' strategy was introduced to non-covalently conjugate siRNAs to the gp120 aptamer.
- 79.Davis KA, Lin Y, Abrams B, Jayasena SD. Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acid Res. 1998;26(17):3915–3924. doi: 10.1093/nar/26.17.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kortylewski M, Swiderski P, Herrmann A, et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat. Biotechnol. 2009;27(10):925–932. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klinman DM, Yi AK, Beaucage SL, Conover J, Krieg AM. CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc. Natl Acad. Sci. USA. 1996;93(7):2879–2883. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krieg AM, Yi AK, Matson S, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374(6522):546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 83.Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27(2):161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 84.Barchet W, Wimmenauer V, Schlee M, Hartmann G. Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr Opin. Immunol. 2008;20(4):389–395. doi: 10.1016/j.coi.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 85.Verthelyi D, Wang VW, Lifson JD, Klinman DM. CpG oligodeoxynucleotides improve the response to hepatitis B immunization in healthy and SIV-infected rhesus macaques. AIDS. 2004;18(7):1003–1008. doi: 10.1097/00002030-200404300-00007. [DOI] [PubMed] [Google Scholar]
- 86.Girvan AC, Teng Y, Casson LK, et al. AGRO100 inhibits activation of nuclear factor-κB (NF-κB) by forming a complex with NF-κB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;5(7):1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 87.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5(12):2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 88.Kotula JW, Pratico ED, Ming X, Nakagawa O, Juliano RL, Sullenger BA. Aptamer-mediated delivery of splice-switching oligonucleotides to the nuclei of cancer cells. Nucleic Acid Ther. 2012;22(3):187–195. doi: 10.1089/nat.2012.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hovanessian AG, Soundaramourty C, El Khoury D, Nondier I, Svab J, Krust B. Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS ONE. 2010;5(12):e15787. doi: 10.1371/journal.pone.0015787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang SH, Cho MJ, Kole R. Up-regulation of luciferase gene expression with antisense oligonucleotides. implications and applications in functional assay development. Biochemistry. 1998;37(18):6235–6239. doi: 10.1021/bi980300h. [DOI] [PubMed] [Google Scholar]
- 91.Kole R, Vacek M, Williams T. Modification of alternative splicing by antisense therapeutics. Oligonucleotides. 2004;14(1):65–74. doi: 10.1089/154545704322988067. [DOI] [PubMed] [Google Scholar]
- 92.Kole R, Williams T, Cohen L. RNA modulation, repair and remodeling by splice switching oligonucleotides. Acta Biochim. Pol. 2004;51(2):373–378. [PubMed] [Google Scholar]
- 93.Ferreira CS, Matthews CS, Missailidis S. DNA aptamers that bind to MUC1 tumour marker. design and characterization of MUC1-binding single-stranded DNA aptamers. Tumour Biol. 2006;27(6):289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- 94.Constantinou PE, Danysh BP, Dharmaraj N, Carson DD. Transmembrane mucins as novel therapeutic targets. Expert Rev. Endocrinol. Metab. 2011;6(6):835–848. doi: 10.1586/eem.11.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu Q, Shibata T, Kabashima T, Kai M. Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA. Eur. J. Med. Chem. 2012;56:396–399. doi: 10.1016/j.ejmech.2012.07.045. [DOI] [PubMed] [Google Scholar]
- 96.Mockenhaupt M. The current understanding of Stevens-Johnson syndrome and toxic epidermal necrolysis. Expert Rev. Clin Immunol. 2011;7(6):803–813. doi: 10.1586/eci.11.66. quiz 814-815. [DOI] [PubMed] [Google Scholar]
- 97.Shlomchik WD. Graft-versus-host disease. Nat. Rev. Immunol. 2007;7(5):340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 98.Chung WH, Hung SI, Yang JY, et al. Granulysin is a key mediator for disseminated keratinocyte death in Stevens-Johnson syndrome and toxic epidermal necrolysis. Nat. Med. 2008;14(12):1343–1350. doi: 10.1038/nm.1884. [DOI] [PubMed] [Google Scholar]
- 99.Ni X, Castanares M, Mukherjee A, Lupold SE. Nucleic acid aptamers: clinical applications and promising new horizons. Curr. Med. Chem. 2011;18(27):4206–4214. doi: 10.2174/092986711797189600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shi B, Abrams M. Technologies for investigating the physiological barriers to efficient lipid nanoparticle-siRNA delivery. J. Histochem. Cytochem. 2013;61(6):407–420. doi: 10.1369/0022155413484152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Musacchio T, Torchilin VP. siRNA delivery: from basics to therapeutic applications. Front. Biosci. 2013;18:58–79. doi: 10.2741/4087. [DOI] [PubMed] [Google Scholar]
- 102.Kesharwani P, Gajbhiye V, Jain NK. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials. 2012;33(29):7138–7150. doi: 10.1016/j.biomaterials.2012.06.068. [DOI] [PubMed] [Google Scholar]
- 103.Ye X, Hemida M, Zhang HM, Hanson P, Ye Q, Yang D. Current advances in Phi29 pRNA biology and its application in drug delivery. Wiley Interdiscip. Rev. RNA. 2012;3(4):469–481. doi: 10.1002/wrna.1111. [DOI] [PubMed] [Google Scholar]
- 104.Nimesh S. Recent patents in siRNA delivery employing nanoparticles as delivery vectors. Recent Pat. DNA Gene Seq. 2012;6(2):91–97. doi: 10.2174/187221512801327406. [DOI] [PubMed] [Google Scholar]
- 105.Higuchi Y, Kawakami S, Hashida M. Strategies for in vivo delivery of siRNAs: recent progress. BioDrugs. 2010;24(3):195–205. doi: 10.2165/11534450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 106.Leucuta SE. Systemic and biophase bioavailability and pharmacokinetics of nanoparticulate drug delivery systems. Curr. Drug Deliv. 2013;10(2):208–240. doi: 10.2174/1567201811310020007. [DOI] [PubMed] [Google Scholar]
- 107.Marcus ME, Leonard JN. FedExosomes: Engineering therapeutic biological nanoparticles that truly deliver. Pharmaceuticals (Basel) 2013;6(5):659–680. doi: 10.3390/ph6050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim E, Jung Y, Choi H, et al. Prostate cancer cell death produced by the co-delivery of Bcl-xL shRNA and DOXorubicin using an aptamer-conjugated polyplex. Biomaterials. 2010;31(16):4592–4599. doi: 10.1016/j.biomaterials.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 109.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64(21):7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 110.Zhao N, Bagaria HG, Wong MS, Zu Y. A nanocomplex that is both tumor cell-selective and cancer gene-specific for anaplastic large cell lymphoma. J. Nanobiotechnology. 2011;9:2. doi: 10.1186/1477-3155-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu X, Ding B, Gao J, et al. Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. Int. J. Nanomedicine. 2011;6:1747–1756. doi: 10.2147/IJN.S23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wilner SE, Wengerter B, Maier K, et al. An RNA alternative to human transferrin. a new tool for targeting human cells. Mol. Ther. Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bagalkot V, Gao X. siRNA-aptamer chimeras on nanoparticles: preserving targeting functionality for effective gene silencing. ACS Nano. 2011;5(10):8131–8139. doi: 10.1021/nn202772p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim JK, Choi KJ, Lee M, Jo MH, Kim S. Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials. 2012;33(1):207–217. doi: 10.1016/j.biomaterials.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 116.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum. Gene. Ther. 2005;16(9):1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kraus E, James W, Barclay AN. Cutting edge: novel RNA ligands able to bind CD4 antigen and inhibit CD4+ T lymphocyte function. J. Immunol. 1998;160(11):5209–5212. [PubMed] [Google Scholar]
- 118.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5(9):1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou J, Shu Y, Guo P, Smith DD, Rossi JJ. Dual functional RNA nanoparticles containing phi29 motor pRNA and anti-gp120 aptamer for cell-type specific delivery and HIV-1 inhibition. Methods. 2011;54(2):284–294. doi: 10.1016/j.ymeth.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv. Drug Deliv. Rev. 2007;59(2–3):134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 121.Huang YZ, Hernandez FJ, Gu B, et al. RNA aptamer-based functional ligands of the neurotrophin receptor, TrkB. Mol Pharmacol. 2012;82(4):623–635. doi: 10.1124/mol.112.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang S, Zhao Y, Zhi D. Non-viral vectors for the mediation of RNAi. Bioorg. Chem. 2012;40(1):10–18. doi: 10.1016/j.bioorg.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 123.Meade BR, Dowdy SF. Enhancing the cellular uptake of siRNA duplexes following noncovalent packaging with protein transduction domain peptides. Adv. Drug Deliv. Rev. 2008;60(4–5):530–536. doi: 10.1016/j.addr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tamura A, Nagasaki Y. Smart siRNA delivery systems based on polymeric nanoassemblies and nanoparticles. Nanomedicine (Lond.) 2010;5(7):1089–1102. doi: 10.2217/nnm.10.76. [DOI] [PubMed] [Google Scholar]
- 125.Gilleron J, Querbes W, Zeigerer A, et al. Image-based ana lysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31(7):638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 126.Sahay G, Querbes W, Alabi C, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31(7):653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nechaev S, Gao C, Moreira D, et al. Intracellular processing of immunostimulatory CpG-siRNA. Toll-like receptor 9 facilitates siRNA dicing and endosomal escape. J. Control. Release. 2013;170(3):307–315. doi: 10.1016/j.jconrel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011;6(10):658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 201.Ellington A. The Ellington Lab Aptamer Database. http://aptamer.icmb.utexas.edu.