Abstract
Bacterial contamination of food products presents a challenge for the food industry and poses a high risk for the consumer. Despite increasing awareness and improved hygiene measures, foodborne pathogens remain a threat for public health, and novel methods for detection of these organisms are needed. Bacteriophages represent ideal tools for diagnostic assays because of their high target cell specificity, inherent signal-amplifying properties, easy and inexpensive production, and robustness. Every stage of the phage lytic multiplication cycle, from the initial recognition of the host cell to the final lysis event, may be harnessed in several ways for the purpose of bacterial detection. Besides intact phage particles, phage-derived affinity molecules such as cell wall binding domains and receptor binding proteins can serve for this purpose. This review provides an overview of existing phage-based technologies for detection of foodborne pathogens, and highlights the most recent developments in this field, with particular emphasis on phage-based biosensors.
Keywords: foodborne pathogens, bacterial detection, diagnostics, reporter phage, biosensor, phage amplification, cell wall binding domain, receptor binding protein
Introduction
Foodborne bacterial pathogens such as E. coli, Salmonella, Campylobacter, and Listeria monocytogenes are major causes of disease and mortality worldwide, and generate high costs for both the food industry and health care systems. Despite increasing awareness and improved hygiene conditions in most western countries, numbers of foodborne disease outbreaks have remained constant or are even increasing.1 The rising demand particularly in the western world for convenience food and ready-to-eat products poses additional challenges for food production and processing environments, and higher risks for the consumers. Therefore, there is a need for the development of novel procedures for detection of these pathogens in food that are fast and reliable and enable rapid implementation of suitable control strategies.
Conventional culture-based diagnostic protocols still represent the gold standard for detection of foodborne bacteria2 since they are most sensitive and, as an added benefit, yield colonies that can be further subjected to additional tests and used for source tracking. Some major drawbacks of these traditional methods are, however, that they are time-consuming (i.e., often require 48 to 72 h for preliminary results1), and labor-intensive. For any newly developed diagnostic test, rapidity, sensitivity, and specificity are key issues. In food analysis, legal requirements often make it necessary to demonstrate the absence of an organism from a food product (i.e., the method must be able to reliably detect single cells in 25 g samples), which poses an additional challenge for such culture-independent methods. Furthermore, such tests should be cost-effective (i.e., they should not require expensive reagents or equipment) and should ideally be simple to perform, under various different conditions, and with minimal pre-processing of sample material.
There are a number of culture-independent methods that have been used in diagnostics, such as polymerase chain reaction (PCR)-based, immunological (e.g., enzyme-linked immunosorbent assay, ELISA), and mass spectrometry (MS) techniques. However, these rapid methods often require lengthy pre-enrichment steps; are hampered by the requirement of expensive machinery and difficult handling and interpretation of results (MS); or lack the ability to distinguish between living and dead cells, as is the case for PCR, which detects the mere presence of DNA. Since many food products undergo processing in order to inactivate bacteria, it is of particular importance for detection methods used in food analysis to be able to identify viable cells. This problem may be solved by a combined approach of reverse transcription and PCR (RT-PCR) detecting mRNA instead of DNA, but technical challenges and costs prevent RT-PCR-based detection methods from being routinely used.2 An alternative approach is the use of propidium monoazide in combination with PCR to differentiate between viable and dead bacteria.3 Examples for commercial molecular detection systems that do not rely on PCR include the 3MTM Molecular Detection Assay (3M) and the ANSR Pathogen Detection System (Neogen Corporation).
Bacteriophages present ideal tools which can be used for bacterial detection. These viruses have co-evolved with their bacterial hosts to recognize and infect their target cells with an extraordinary specificity that can be harnessed for various rapid detection formats. The complete infection cycle of a virulent phage usually takes only 1–2 h and, by multiplication inside the host cell, offers an inherent amplification step,4 which in many detection assays makes it possible to shorten or completely dispense with lengthy pre-enrichment procedures. In addition, phages are easy and inexpensive to produce, robust (e.g., they show low susceptibility to variations of temperature and pH, organic solvents,5 and proteases6), and are able to distinguish between live and dead cells (i.e., only multiply in viable cells). There is a multitude of reports in the literature on different phage-based detection techniques, exploiting every step of the phage infection cycle, from host cell recognition to lysis, and these methods have been reviewed extensively.1,2,4,7-14 However, despite the various bacteriophage-based diagnostic protocols developed to date, and the obvious advantages of harnessing phage for bacterial detection, only a few of these tests have been developed into commercial products.10 This review gives an overview of the current state of bacteriophage-based detection of food-borne pathogens, with a particular focus on recent developments in the field of biosensor technologies.
Detection on the Basis of Phage-Induced Lysis
Lysis of the host cell “from within” constitutes the last step in the lytic multiplication cycle of most phages, and is commonly mediated by two components of the lytic cassette of the phage: the holin, which creates pores in the cytoplasmic membrane, and the endolysin, which accesses the peptidoglycan through these pores and degrades its substrate, resulting in destabilization of the murein sacculus and rapid lysis.15 Besides liberation of the progeny phage particle, host cell lysis also results in the release of other intracellular components, some of which may be used as markers for measuring the lysis event. This can be exploited in a variety of bacterial detection methods, in which the bacteriophage provides the necessary specificity.
Detection of ATP release
Adenosine triphosphate (ATP) is the intracellular marker most widely used for determination of bacterial numbers in a sample through cell lysis. Since the amount of ATP in an average living bacterium is approximately 10−15 g and quite consistent between different species, quantification of released ATP via a bioluminescent assay allows determination of the number of viable cells present in a sample.2,16,17 In this assay, ATP drives a reaction catalyzed by the firefly luciferase, which converts its substrate luciferin in presence of oxygen into oxyluciferin, adenosine monophosphate (AMP), pyrophosphate, and carbon dioxide, emitting detectable light at femtomolar concentrations of ATP.18 Traditionally, unspecific membrane-disrupting agents (detergents) have been used for the assay to determine the total bacterial number in a sample.19 Employing bacteriophage as lytic agents can turn this general assay into a specific method for detection of target cells. A drawback of this assay format is the high background concentration of ATP present in many food samples, which results in insufficient detection limits (104 to 105 CFU/ml).4,19 One strategy addressing this problem employs a biosorbent consisting of bacteriophage T4 particles immobilized on a nano-aluminum fiber-based DisruptorTM filter, coupled with the bioluminescent ATP assay for detection of E. coli.20 This phage-based biosorbent allowed capture and concentration of target pathogens on the filter surface and, as a consequence, detection of a significantly lower number of bacteria (6 × 103 CFU/ml) within 2 h. The method was shown to be highly accurate and robust, with sample background microflora at concentrations 60-fold higher than that of the target pathogen not affecting the results. An alternative strategy to increase sensitivity of the bioluminescent ATP assay is signal amplification. This can be achieved by measuring the activity of adenylate kinase (AK), an intracellular enzyme released by phage-mediated cell lysis, which equilibrates concentrations of adenine nucleotides, i.e., it produces ATP in the presence of excess adenosine diphosphate (ADP).21 Addition of ADP to the sample increases sensitivity by one to two orders of magnitude compared with the classical bioluminescent ATP assay, and detection limits reported for E. coli and Salmonella were lower than 103 CFU/ml under optimal conditions.19,22 The method was further improved by including an immunomagnetic separation (IMS) step prior to the AK/ATP assay, in which target cells are captured by antibody-coated magnetic beads, concentrated, and partially purified.18,23 This combined protocol was once commercialized for detection of Salmonella, E. coli O157, Listeria, and other foodborne pathogens. However, this so-called FastrAK assay has not been a commercial success, since the time advantage offered compared with traditional methods is not significant, and it does not provide viable bacteria required for further studies.
Detection of other bacterial cytoplasmic markers
Besides ATP and AK, other cytoplasmic markers have been harnessed for bacterial detection by phage-mediated cell lysis. Neufeld et al. described detection of E. coli at concentrations as low as 1 CFU/100ml within 6 to 8 h, by amperometric measurement of the activity of bacterial β-D-galactosidase released after phage lysis of target cells.24 The substrate of the enzyme, p-aminophenyl-β-D-galactopyranoside, has to be added externally and is converted into p-aminophenol, whose subsequent oxidation can be detected by change of the electric current in a potentiostat device. The high sensitivity of the assay was achieved by filtration and preincubation of the sample prior to phage infection. Similarly, Yemini et al. reported detection of Bacillus cereus and Mycobacterium smegmatis after phage lysis (detection limit of 10 CFU/ml), using α- and β-glucosidase as cytoplasmic markers, respectively, within 8 h.25 The potentiostat used in this study allowed the simultaneous measurement of 8 samples.
Measurement of impedance or conductivity
The growth of microorganisms causes changes in conductivity of the growth medium, mostly through transformation of large uncharged metabolites (such as carbohydrates) to smaller charged molecules (e.g., acids). The term impedance is inversely related to conductivity and can be defined as resistance to the flow of an alternating current through conducting material such as bacterial growth media.26 Bacteriophage are suitable tools for specific impedimetric detection of bacteria, since addition of phage to a sample results in retardation of changes in impedance if the target organism is present. One prominent example for employing a phage for impedimetric detection of foodborne pathogens is a study by Chang et al.,27 who were able to detect E. coli O157:H7 cells through the absence of changes in conductivity of a MacConkey-sorbitol medium in presence of the phage AR1, which is specific for this pathogen. The method proved highly sensitive (with 41 of 41 O157:H7 strains giving a positive signal) and specific (99.4% of non-O157:H7 E. coli strains tested yielded a negative result). The drawbacks of direct impedance-based detection methods are the requirement of suitable growth media optimized for conductivity measurement, whose development is often time-consuming, the potential of contamination by microorganisms of the background flora, and the fact that not all target organisms produce highly charged metabolites resulting in measurable changes in conductivity.13,26,28 Some of these limitations may be circumvented by indirect impedimetric methods, in which carbon dioxide produced during growth of the target organisms is absorbed by a potassium hydroxide solution separated from the growth medium, and the resulting change in conductivity is detected.26 This principle has been applied for detection of a variety of foodborne pathogens, including Staphylococcus aureus, Listeria monocytogenes, Enterococcus faecalis, Escherichia coli, Salmonella, and Campylobacter species.29,30 While many reports of classical impedance-based methods for bacterial detection date back to the 1990s, there has been considerable progress in recent years on the development of impedimetric biosensors,31 which will be discussed in more detail below.
Detection by the Phage Amplification Assay
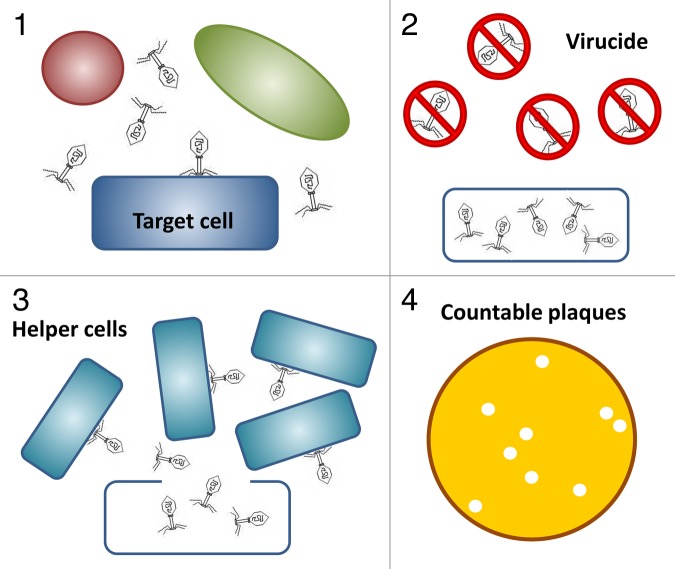
The use of unmodified phage particles for generation and enumeration of plaques within a bacterial lawn is certainly the most obvious and direct way of utilizing these viruses for bacterial detection. In the so-called Phage Amplification Assay (Fig. 1),32-34 samples to be tested for the presence of certain target pathogens are mixed with bacteriophages specific for these bacteria, which are then allowed sufficient time to infect their host cells. The subsequent addition of a virucide to the sample ensures survival only of those phages currently engaged in the infection process, whereas all extracellular phage particles are inactivated. Since the number of target cells present in a sample is rarely sufficient to produce a bacterial lawn when plated directly, the sample is then mixed with an ample amount of helper cells (of a propagating strain for the respective phage) following neutralization of the virucide. Plating of the mixture in a soft agar overlay results in formation of plaques as a consequence of phage liberated from lysed target cells infecting surrounding helper cells. The number of plaques directly corresponds to the number of target cells initially present in the sample, and the nature of the assay ensures that only living cells are detected. Other benefits of the assay include low costs, high specificity, speed, and the use of unmodified phage, which avoids genetic engineering and legal issues associated with the application of GMOs.2,9 A critical point is the efficacy of the virucide chosen for inactivation of phage particles, since any residual free phage will yield false-positive results, by infecting the added helper cells. One agent commonly employed for this purpose is ferrous ammonium sulfate.34,35 Difficulties may also arise when the concentration of competing microflora in a sample is high enough to overgrow the helper cells or when the matrix itself inhibits phage infection, which can often be circumvented by dilution or decontamination steps prior to performing the assay.8 In certain cases, the assay may provide an important time advantage when rapid signal amplification is achieved by using fast-growing helper cells for detection of a slow-growing target pathogen. A prime example is the previously reported detection of Mycobacterium tuberculosis in human sputum samples, by using fast-growing Mycobacterium smegmatis as helper cells, which constitutes the basic principle of the FastPlaqueTB and FastPlaque-Response (for detection of rifampicin-resistant Mycobacteria) tests.36 Unfortunately, these tests have not been commercially successful, which is partly due to the disadvantage of the culture-based method as opposed to more user-friendly solutions for clinical labs. For food analysis laboratories, however, plate-based protocols do not pose an obstacle. In fact, the phage amplification assay has been developed for a broad spectrum of bacteria, including food-borne pathogens such as Salmonella, E. coli, Staphylococcus aureus, Listeria, and Mycobacterium avium.32,37-41 M. avium ssp. paratuberculosis (MAP) is an obligate pathogen that occurs in milk, and is the causative agent of Johne’s disease in ruminants.42 There is also a possible link to Crohn’s disease in humans.43,44 The FastPlaqueTB assay has been reported to be applicable for the detection of MAP and Mycobacterium bovis in milk samples. In this case, the assay is coupled with plaque PCR testing for the presence of certain MAP and M. bovis-specific signature elements, which provides the specificity necessary to differentiate between the two species.38 Compared with the use of PCR alone, the combined method offers the advantage of detecting viable cells only.45 Another recent development based on the Phage Amplification Assay for MAP detection employs MAP-specific affinity peptides for peptide-mediated magnetic separation (PMS) of target cells from the sample prior to the assay,46 and replaces the classical plaque formation step by ELISA detection of released progeny phage using phage-specific polyclonal antibodies.47 In this setup, the PMS ensures high specificity, at the same time circumventing the aforementioned problems related to background microflora or unfavorable matrix properties of the sample, while the ELISA-based endpoint detection offers advantages in speed and sensitivity compared with the classical method. Magnetic separation in combination with phage amplification was also utilized for detection of Salmonella cells, in this case using Salmonella-specific antibodies as affinity molecules coated on magnetic beads.37 When applied to a variety of food samples, this assay was able to detect an average of 3 CFU of Salmonella in 25 g food within 20 h, and was also adapted for detection of E. coli O157:H7.48 Another report describes the simultaneous detection of two different pathogens (Salmonella and E. coli) in one sample by coupling phage amplification with matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF). Here, structural proteins of the amplified phage serve as secondary biomarkers for the respective target bacteria.39,49 Phage amplification protocols using MALDI-TOF analysis as endpoint detection method are, however, limited by the low inoculation phage titers that have to be used to ensure that initial phage concentrations are below the detection limit of the MS instrument, which results in longer incubation times. Pierce at al. attempted to circumvent this limitation by designing an assay for detection of S. aureus in which 15N-isotopically labeled phages are used for inoculation. These can be distinguished by mass from the wild-type 14N progeny viruses.50,51 One further attempt to shorten the time-consuming last step (i.e., the actual detection step) of the classical phage amplification assay with plaque enumeration was reported by Jassim et al.,52 who applied a Live/Dead staining protocol to the helper cells 1–2 h after addition to the sample. This way, the authors were able to correlate the fluorescence signal indicating the proportion of dead helper cells to the initial number of target cells present in the original sample, allowing detection of 10 CFU/ml within 4 h without prior enrichment. Further endpoint detection methods that have been used to replace the traditional plaque formation and enumeration include quantitative real-time PCR (qPCR)53 and competitive ELISA (phage replication-competitive enzyme-linked immunosorbent assay, PR-cELISA).54
Figure 1. Phage Amplification Assay using unmodified phage for detection of target cells (modified from ref. 9). 1) Target cells in a mixed bacterial culture are specifically recognized and infected by the phage; 2) extracellular phage particles are inactivated by addition of a virucide; 3) phage progeny liberated from target cells infect added helper cells, resulting in signal amplification; 4) countable plaques on an agar plate correspond to initial number of target cells in the sample.
Genetically Modified Reporter Phage
Besides using native, unaltered phage particles for bacterial detection, molecular cloning techniques makes it possible to engineer phage to carry a specific reporter gene. Upon infection of a target cell, the gene is expressed and allows detection of its product, e.g., by measuring bioluminescence, fluorescence, or enzymatic conversion of a chromogenic substrate. Table 1 summarizes reporter genes that have been used for construction of reporter phages, with their respective advantages and disadvantages. Reporter phage assays are fast and sensitive and, like the phage amplification assay using native phage, only detect active and viable cells. Another advantage is that the robustness of the assay and the high specificity of the phage eliminate the need for lengthy sample preparation or purification. There is a large body of literature on the many different reporter phages constructed and evaluated, and recent reviews of this topic are available.1,4,9,11 However, certain disadvantages associated with the construction and application of reporter phages should also be mentioned. First, their construction is labor-intensive and requires detailed genetic knowledge of the phage. Furthermore, the volume of the phage capsid sets a natural limit to the amount of genetic material that can be introduced into the phage genome. Common ways of introducing reporter genes include direct cloning (feasible only with a limited number of phages), transposition, and homologous recombination.66 Finally, the fact that reporter phages create genetically modified organisms (GMOs) by infecting their host cells poses potential hurdles with regard to consumer acceptance and regulatory approval of reporter phage products. As a consequence, despite the large amount of effort that has gone into developing reporter phages, there is, as yet, no commercially available product based on this technology. However, the US-based company Sample6 is currently implementing this technology in their Sample6 Detect portfolio for rapid diagnostics of bacteria in food industry environments (sample6.com).67 This approach uses synthetic biology tools such as yeast-based recombineering allowing rapid high-throughput engineering of environmentally isolated phages, thereby generating large synthetic phage libraries; optimization of reporter molecules by genetic engineering; and close integration of these reporters with suitable detectors. The Sample6 Bioillumination PlatformTM enables highly sensitive (single cell) in-plant detection of foodborne pathogens without enrichment and generates results within few hours.
Table 1. Reporter genes used for detection of foodborne pathogens by engineered reporter phages.
| Gene | Encoded product | Advantages | Disadvantages | Refs. |
|---|---|---|---|---|
| luxCDABE | complete bacterial luciferase operon (encodes luciferase holoenzyme LuxAB and proteins providing its aldehyde substrate) | • high sensitivity • high signal-to-noise ratio (low luminescence background in food samples) • no substrate addition required |
• packaging constraints (large DNA fragment) • relatively weak luminescence signal |
55 |
| luxAB | bacterial luciferase genes encoding the two subunits of the holoenzyme LuxAB | • high sensitivity • high signal-to-noise ratio (low luminescence background in food samples) • smaller than the luxCDABE operon |
• addition of substrate required • transient luminescence signal |
56, 57 |
| luxI | Quorum sensing; LuxI generates the autoinducer AHL, which stimulates transcription of luxCDABE | • high sensitivity • no substrate addition required • smaller compared with luxCDABE • longer signal emission due to presence of bioreporter cells |
• bioreporter strain carrying luxCDABE required • false-positive results possible (bioreporter luminescence can be induced by compatible autoinducers present in the environment) |
58 |
| luc | firefly luciferase Luc | • high sensitivity • high signal-to-noise ratio (low luminescence background in food samples |
• addition of substrate required • substrate expensive • short luminescence signal |
59 |
| inaW | INP | • very high sensitivity • high signal-to-noise ratio • no specialized equipment required |
• addition of phase-sensitive fluorescent dye required • target cell must present INP on its surface |
60 |
| lacZ | β-galactosidase enzyme | • No specialized equipment needed • works with various substrates (colorimetric, fluorescent, luminescent) |
• addition of substrate required • high background of β-galactosidase activity in the environment |
61 |
| gfp | GFP | • no substrate addition required • long-lasting signal increasing over time • multiplexed detection possible when different fluorescent markers are used • high stability and low toxicity |
• posttranslational chromophore formation required • low signal-to-noise ratio due to background fluorescence • detection by fluorescence microscopy or FACS requires expensive equipment and trained personnel |
62 |
| celB | Pyrococcus furiosus thermostable β-glycosidase CelB | • thermostable enzyme: elimination of background enzyme activity possible by heating, resulting in high signal-to-noise ratio • works with various substrates (colorimetric, fluorescent, luminescent) • long-lasting signal increasing over time |
• addition of substrate required | 63 |
| biotinylation tag coding sequence (used in combination with QDs) | 25 amino acid biotinylation peptide | • small reporter gene (75 bp) • when used with QDs: very high sensitivity due to superior optical properties • multiplexed assays possible |
• addition of functionalized QDs required • detection by fluorescence microscopy or FACS requires expensive equipment and trained personnel |
64 |
| tetracysteine tag coding sequence | 12 amino acid tetracysteine peptide | • very small reporter gene (36 bp) | • addition of dye required • detection by fluorescence microscopy or FACS requires expensive equipment and trained personnel |
65 |
Luciferase reporter phage
Among all reporter phage described to date, those carrying a luciferase gene for bacterial detection account for the largest share. Major advantages of using luciferase genes as reporters are the highly sensitive detection of the bioluminescent signal they generate, and the fact that bioluminescence is practically absent from food samples, resulting in a high signal-to-noise ratio.4 The first luciferase reporter phage (LRP) was constructed by Ulitzur and Kuhn in 1987, who inserted the lux operon from Vibrio fischeri (i.e., bacterial luciferase genes) into a lambda-based cloning vector and demonstrated detection of as few as 10 E. coli cells in milk within 1 h.55 Nowadays, LRPs are available for a multitude of foodborne pathogens, including E. coli, Salmonella, Listeria, Staphylococcus aureus, and Mycobacterium avium.55-57,59,63,66,68-74 One of the most prominent examples with relevance for food safety is the A511::luxAB, which is based on the broad host range A511 phage infecting the foodborne pathogen Listeria monocytogenes.56 This reporter phage features a luxAB fusion gene from Vibrio harveyi immediately downstream of the gene coding for the major capsid protein of A511, resulting in emission of light during the expression of late phage genes inside infected host cells, and allowing detection of very low numbers of Listeria cells in food samples within less than 24 h.72 This significantly reduced time requirement compared with the standard plating method (4 d) constitutes the major advantage of this assay, while at the same time the high specificity of the phage ensures reliable results even in samples with high levels of background flora. More recently, a similar approach was used to create luxAB reporter phages for detection of the potential bioterrorism agents Yersinia pestis and Bacillus anthracis, offering advantages with regard to ease of use, speed, and the possibility of testing multiple samples simultaneously, compared with the respective traditional detection protocols.75-77 Kuhn et al. designed a LRP for detection of Salmonella based on the virulent broad host range phage Felix-O1, which is able to infect > 95% of clinically relevant strains.70,78 The distinctive feature of this phage is the replacement of native genes, one of which is essential for propagation of the phage on wild type strains, by the luxAB genes. As a consequence, the phage is genetically locked, meaning that infection of wild type strains (and consequently generation of the luminescent signal) is possible, but multiplication can only be achieved in an engineered propagation strain which provides the missing genes in trans.70 This characteristic is interesting since it may circumvent some GMO-related concerns. Another interesting concept is the use of luxI, one regulatory gene of the bioluminescence cassette of Vibrio fischeri, as a reporter gene.58,79-81 The luxI gene encodes a synthase of acyl-homoserine lactone (AHL), which functions as diffusible quorum sensing molecule that induces transcription of the lux operon resulting in bioluminescence of neighboring cells carrying the essential genes. By using an engineered luxI incorporated lambda phage and an E. coli lux bioreporter strain, the authors were able to detect as few as 1 E. coli cell per ml in pure culture within slightly more than 10 h and 130 CFU/ml in less than 24 h in artificially contaminated lettuce leaf washings.58 A similar reporter system was also designed for E. coli O157:H7, with similarly low detection limits.80,81
In summary, detection limits for the reported LRPs range from 102 to 106 CFU/ml, and short pre-enrichment steps allowed detection of even fewer cells. The time requirement is between one and several hours.4
Other reporter phage
Several other reporter genes have been used to construct reporter phage. Genes encoding fluorescent proteins such as the green fluorescent protein (GFP) have been found particularly suitable for this purpose. The GFP combines many desirable properties such as high stability, low toxicity, and the fact that fluorescence is triggered by excitation light, abolishing the requirement for adding a substrate as required for luciferases.11,82 Furthermore, the availability of a wide range of fluorescent proteins differing in their excitation and emission properties opens new possibilities for multiplexed detection of various different target pathogens in one sample.11,83 The first GFP reporter phage was constructed by Funatsu et al., who modified the lambda phage to express the gfp gene, allowing specific detection of E. coli by fluorescence within 4–6 h.62 The T-even type phage PP01 served as a specific reporter for E. coli O157:H7 in a similar setup. Here, the gfp gene was fused to the gene coding for the small outer capsid protein of the phage.84 Detection of target cells by fluorescent microscopy was possible 10 min after mixing the sample with the phage, and was not influenced by presence of non-O157:H7 E. coli cells. The same strategy was used to create a T4-based reporter phage. In addition to carrying the GFP reporter gene, this phage was genetically locked, i.e., capable of infection but not of lysing the infected host cell, through inactivation of its endolysin gene.85 It should be noted that with these reporter phages, detection of target pathogens relies both on adsorption of previously produced fluorescently tagged phage particles to the cell surface and subsequent additional production of the reporter protein within the infected target cell. This makes it possible to distinguish between live cells, in which the fluorescent signal increases over time, and inactivated cells, where fluorescence is only due to the surface decoration and does not increase during the course of the assay.
One further possibility is the utilization of the lacZ gene encoding β-galactosidase as a reporter, which can be used with a variety of different substrates, allowing detection of colorimetric, fluorescent, or luminescent signals.7 A very practical device termed PhastSwab was developed by Willford and Goodrige, and contains a lacZ reporter phage specific for E. coli O157:H7, as well as a suitable substrate (colorimetric or luminescent) within the cap of a SnapValve tube.61 The bottom of the tube contains a swab for sample collection, suitable enrichment broth, and immunomagnetic beads for separation of target cells. Following sample collection, the swab is returned to the enrichment broth compartment and incubated for 8 h before concentration of target cells by IMS. The reporter phage is then added to the immobilized cells, and after 1 h, the cap is snapped to release the substrate, and the resulting signal can be measured. When used with a luminescent substrate, the PhastSwab was able to detect 10 CFU/100 cm2 within 10 h.
Even lower detection limits were achieved with a Salmonella-specific reporter phage carrying the inaW gene, which encodes a Pseudomonas-derived ice nucleation protein (INP).60 When Salmonella target cells are infected with this phage, the INP is produced and presented on the surface of the bacterium by integration into its outer membrane. As a consequence, samples containing Salmonella cells freeze at slightly higher temperatures, which can be detected by adding a phase-sensitive indicator dye that turns orange in case of elevated temperature freezing and remains green otherwise. This so-called BIND (bacterial ice nucleation detection) assay was shown to detect as few as 2 CFU/ml of Salmonella within 3 h in buffer and raw eggs, and 10 CFU/ml in samples with high background flora.86 Sensitivity of the assay was further improved by combining it with IMS, achieving detection limits as low as 5 CFU/ml.87 The BIND assay has been commercialized as diagnostic tool for Salmonella, but it is no longer available.11
Another useful reporter gene is the sequence encoding the highly thermostable β-glycosidase CelB from the hyperthermophilic archaeon Pyrococcus furiosus, which can be used with a variety of different substrates to generate a measurable signal.63 An important advantage of this system compared with other reporter phages is the possibility to eliminate background activity of other enzymes present in the sample, simply by heating. When used with chromogenic substrates, the assay offers an additional advantage with regard to ease-of-use, by generating a long-lasting signal that increases over time, in contrast to the transient signals produced by luminescence-based assays [even though there are new luciferase variants that produce a strong and stable light signal, such as the Gaussia luciferase GLuc88].
Edgar et al. reported the construction of reporter phages carrying a small (approximately 75 base pairs) gene encoding a biotinylation peptide, which allowed tagging of the phage by streptavidin-coated quantum dots (QDs).64 QDs are fluorescent semiconductor nanocrystals that feature broad excitation and narrow emission spectra, and are superior in photostability to organic fluorescent molecules such as GFP. This strategy involves i) infection of E. coli target cells present in a sample by the engineered phage; ii) biotinylation of the progeny phage (presenting biotinylation peptides on their surface) inside the cells by the bacterial biotin-ligase (a process highly conserved throughout evolution); iii) fluorescent tagging of the progeny phage by streptavidin-functionalized QDs binding to the biotinylated capsids with high affinity; and iv) detection of the QD-tagged phage by flow cytometry. This method allows specific detection of 10 CFU/ml and a 100-fold amplification of the signal over background within 1 h.
An even smaller reporter gene (36 nucleotides) was used to create a filamentous phage that displays tetracysteine tags fused to its major coat protein.65 Upon infection of a host cell, tetracysteine-tagged progeny phages are produced, which can then be stained by adding fluorescein arsenical helix binder (FlAsH-EDT2), a membrane-permeable biarsenical dye, and detected by flow cytometry or fluorescence microscopy.
Methods Using Phage or Phage Components as Affinity Molecules
The previous sections focused on detection techniques that rely on native or genetically engineered phages infecting and (for some methods) lysing their host cells. Another possibility is to use complete phage particles or phage components such as receptor binding proteins or cell wall binding domains from phage endolysins as affinity molecules to tag and subsequently detect target pathogens without the necessity of infection.
Labeled phage particles
The first report of directly labeled phage particles for bacterial detection dates back to the 1960s, when Watson and Eveland used fluorescently labeled antibodies directed against a phage coat protein. This “phage-fluorescent antiphage staining system” was successfully used to specifically identify Listeria monocytogenes cells.89 More than 30 y later, Goodridge et al. reported the fluorescent labeling of an E. coli O157:H7 specific phage. In this case, the phage DNA was tagged with the fluorescent nucleic acid dye YOYO-1.90 When used in combination with IMS for concentration of target cells and flow cytomentry for measuring the fluorescent signal, this fluorescent-bacteriophage assay (FBA) had a detection limit of 104 CFU/ml. When attempting E. coli O157:H7 detection in food samples, the sensitivity of the assay could be further increased by including short enrichment steps. After 6 h enrichment, 2.2 cells per gram of artificially contaminated ground beef were successfully detected, and a detection limit between 10 and 100 CFU/ml was achieved in raw milk following a 10 h enrichment step.91 A similar approach was also reported for detection of Salmonella, this time using the fluorescent dye SYBR gold, which was found to be superior to other dyes in terms of fluorescence intensity, quantum yield and difference between excitation and emission wavelengths.92 In this study, the fluorescent DNA was found to be injected into the host cells, which were then detected by epifluorescence microscopy. A recent report describes the construction of a SnapValve-based device similar to the aforementioned PhastSwab, for detection of Shiga toxin producing E. coli (STEC). Instead of a lacZ carrying reporter phage, this device involves so-called Phazymes, i.e., phage particles specific for the target pathogen that are chemically labeled with horse radish peroxidase (HRP), and are contained within the cap of the SnapValve tube.93 As for the PhastSwab, the bottom of the tube contains a swab for sample collection, suitable enrichment broth, and beads for IMS of STEC cells. Following IMS, the Phazymes are added to the immobilized cells by snapping the cap, and after several washing steps, STEC can be detected by addition of either a colorimetric or luminescent HRP substrate. The Phazyme assay was able to detect E.coli O157:H7 at levels of 1 CFU/g in spinach and 1 CFU/100 cm2 on swabbed meat samples.
Cell wall binding proteins
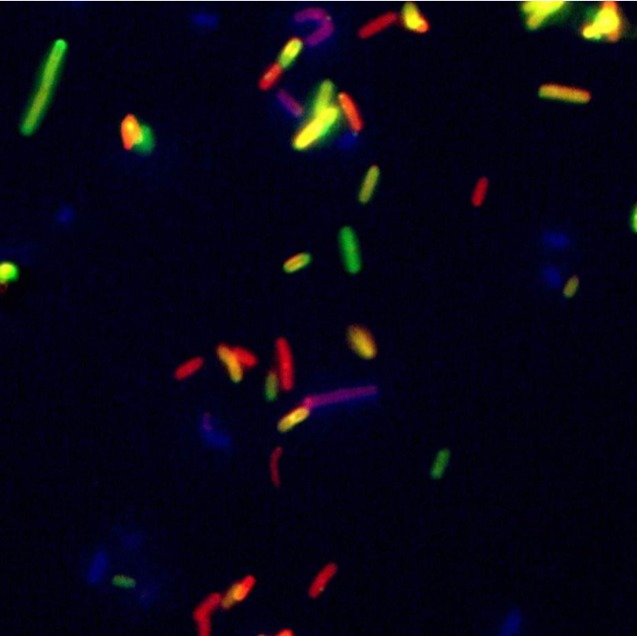
Besides entire phage particles, individual components of the phage or proteins produced during its multiplication cycle can also be harnessed as affinity molecules for specific immobilization and detection of pathogens. In the case of Gram-positive bacteria, where the cell wall is not shielded from the outside by an outer membrane, cell wall binding domains (CBDs) of phage endolysins have emerged as promising tools for this purpose. Endolysins from a Gram-positive background show a modular architecture in which catalytic activity and cell wall recognition are embodied by two or more distinct functional domains.94-96 CBDs, which are C-terminally located in most cases, recognize and noncovalently bind to certain ligands within the bacterial cell wall, thereby bringing the catalytic domains in proximity of their substrate. The binding spectra of CBDs are usually broader than the host ranges of the corresponding phage, and can encompass entire bacterial genera.97 In other cases, these binding modules can exhibit specificity down to the serovar or even strain level, as has been reported for a set of CBDs from Listeria phage endolysins.83,98 This high specificity and the exceptionally high affinity of the CBDs to the bacterial cell wall (Kd in the nano- to picomolar range)83,98,99 make them ideal tools for immobilization, detection, and differentiation of Listeria cells. In an effort to reduce the time required for detection of Listeria in food samples following standard plating protocols, Kretzer et al. developed a CBD-based magnetic separation (CBD-MS) procedure, in which two different Listeria phage CBDs featuring complementary binding spectra (together encompassing all serovar groups) have been immobilized on paramagnetic beads via chemical coupling. These CBD-coated beads can then be used for high-affinity binding of target cells from food samples, and recovery via a magnet.100 This method was shown to be superior to IMS in terms of specificity and recovery rates, with more than 90% of Listeria cells recovered from suspensions within 20–40 min. When applied to food samples, detection limits between 1 and 100 CFU/g were achieved after 6 h enrichment, and extension of the enrichment period to 24 h made it possible to detect less than 1 cell per gram food. In the original protocol, detection of Listeria was accomplished by plating the beads after magnetic separation and enumeration of colonies. This CBD-MS procedure was demonstrated to outperform the standard plating protocol in terms of time requirement (maximum of 48 h vs. 96 h) and sensitivity, and is also applicable for detection of other foodborne pathogens, such as Bacillus cereus and Clostridium perfringens.100 As an alternative to direct plating of the beads, Walcher et al. combined CBD-MS with real-time PCR-based quantification (qPCR) of Listeria in raw milk.101 The detection limit of this combined approach ranged from 102 to 103 CFU/ml, which is close to the practical limit of this method. In a further modification of the CBD-MS assay, the high specificity of some CBDs for certain serovar groups was exploited for differentiation of Listeria cells of different strains following recovery from artificially contaminated food samples.83 To this end, CBDs of different specificity were fused to differently-colored fluorescent reporter proteins such as GFP, CFP, and dsRed. After CBD-MS using beads coated with a CBD featuring a broad binding spectrum and plating on selective agar, emerging colonies were collectively resuspended in buffer. Incubation of the cell suspension with mixtures of differently-tagged CBDs of higher specificity for 15 min allowed subsequent differentiation of strains by fluorescent microscopy (Fig. 2). Another advantage of using domains from bacteriophage endolysins is given by the modular structure of these enzymes, which makes it possible to optimize their lytic or binding properties for certain applications by molecular engineering and module shuffling. Combining CBDs from various origins yielded chimeric fusion proteins with altered binding spectra or increased affinities which hold promise for application in CBD-based detection methods.99
Figure 2. Mixed culture of Listeria cells of different serovar groups labeled with differently-colored fluorescent reporter-CBD fusion proteins and visualized by fluorescence microscopy.
For detection of Gram-negative bacteria (which are not directly accessible by CBDs from the outside because of the presence of an outer membrane), receptor binding proteins (RBPs) from phage tail fibers or spikes can assume the role of the CBDs.102 These proteins mediate attachment of the phage to its receptor on the bacterial surface. A fluorescently labeled phage tail fiber specific for E. coli O157:H7 has been harnessed in the VIDAS UP technology (BioMerieux SA). This sandwich assay setup utilizes antibodies for immobilization of the Gram-negative pathogens followed by detection using the fluorescent tail fiber. Similar assays are available for Salmonella and Listeria detection (http://www.biomerieux-industry.com/vidasup/). RBPs have also found application in various biosensors for detection of Gram-negative foodborne pathogens (see below).102-104
Biosensor Technology
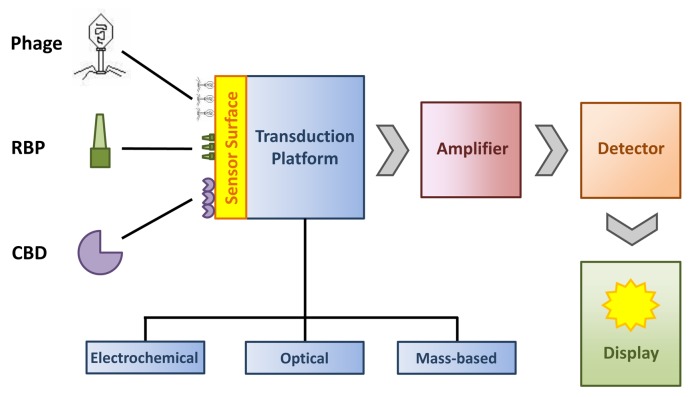
Considerable progress has been made in the past 5–10 y in the development of biosensor devices for detection of bacterial pathogens that use phage particles or phage-derived affinity molecules as recognition elements. A classical biosensor consists of the following components (Fig. 3):12 i) a surface to which the recognition elements are attached; ii) a transduction platform which can be electrochemical (e.g., impedimetric), optical (e.g surface plasmon resonance-based), or mass-based (e.g., magnetoelastic); iii) a signal amplifier; iv) a signal detector; and v) a signal display. A crucial point for the performance of a phage-based biosensor is the effective immobilization of the phage or phage-derived recognition elements to the sensor surface. Strategies include physical absorption, covalent but undirected immobilization by chemical functionalization, and oriented immobilization by genetic modification of the recognition element.12 While all three possibilities have been used, immobilization by physical absorption is often hampered by weak binding and inconsistent density of recognition elements attached to the sensor surface, which may be significantly improved by chemical coupling.105 Oriented immobilization offers the additional advantage that a larger subset of immobilized molecules on the surface are fully functional (i.e., the binding sites/domains are exposed and therefore available for capturing target cells). As an example, genetically modified phages featuring biotinylated capsids can be efficiently immobilized on surfaces coated with streptavidin.106,107
Figure 3. Composition of a bacteriophage-based biosensor for detection of bacterial pathogens (modified from ref. 12). The various recognition elements (phage, RBP, CBD) can be attached to the sensor surface by physical adsorption, covalent binding, or affinity tags.
One subgroup of biosensors is based on measurement of impedance, which can be classified in two different types:31 i) biosensors that measure changes in impedance of growth media as a result of the production of metabolites during bacterial growth, and are therefore based on the same principle as the classical impedimetric assays described above; and ii) biosensors measuring the change in impedance caused by target cells binding to certain receptors immobilized on the surface of an electrode. The receptors endow the sensor with the required specificity and can be constituted by various types of recognition elements, such as antibodies, lectins, complete phage particles, or phage-derived affinity molecules. One example for utilizing complete phages as recognition elements is provided by Shabani et al., who covalently immobilized T4 phages on the surface of functionalized, screen-printed carbon electrode microarrays.108 Binding of E. coli cells to the phages was measured by shifts in impedance, whereas no significant change was observed in the presence of Salmonella. With this biosensor, the authors were able to reach a detection limit of 104 CFU/ml when using 50 µl samples. In an effort to improve the sensitivity of this system, the detection procedure was modified by involving paramagnetic beads coated with the same phage as the sensor surface, and the possibility of applying a magnetic field to the carbon electrode microarray.109 Through these modifications, it became feasible to selectively capture E. coli cells from a sample with magnetic beads, and bring them in proximity to the electrode surface via magnetic forces, thereby increasing the chance of interaction with the surface-immobilized phage. The detection limit of this modified biosensor was 103 CFU/ml, i.e., one order of magnitude lower than that of the original device. In addition, the modified sensor proved superior in terms of specificity and enabled detection of target cells in more complex samples such as milk. Another interesting approach is the use of impedance biosensors based on CBDs from bacteriophage endolysins instead of complete phage particles. In a recent study, Tolba et al. reported immobilization of high-affinity binding domains from Listeria phage endolysins on a gold screen-printed electrode, allowing bacterial detection by electrochemical impedance spectroscopy. This setup enabled rapid and direct detection of Listeria cells from pure culture or 2% artificially contaminated milk with detection limits of 1.1 × 104 and 1 × 105, respectively.110 The use of phage-derived affinity molecules for coating of biosensor surfaces avoids some of the problems associated with the use of whole phage particles. These include the large size of phages, which can be disadvantageous for integration into certain biosensor platforms,12 and the observation that infection and lysis of immobilized target cells may occur after certain periods of time, resulting in decreasing signal intensities.102
There are several studies in which surface plasmon resonance (SPR) was employed as a transduction platform in bacteriophage-based biosensors. Nanduri et al. demonstrated the detection of Listeria monocytogenes using a biosensor that features filamentous phage displaying antibodies against the listerial actin polymerization protein (ActA), fused to phage surface proteins.111 The phages were immobilized on the sensor surface by physical adsorption, and binding of target cells to the displayed antibodies was measured in real-time by SPR. The detection limit of the biosensor, however, was high (2 × 106 CFU/ml). While in this approach, phages only play an indirect role for bacterial detection by displaying antibody-based recognition elements, a biosensor developed in the same year for detection of S. aureus exploits the actual specific interaction between phage and host cell.112 In this study, a staphylococcal phage was physically absorbed to the gold surface of a SPR-based SPREETATM sensor, and a detection limit of 104 CFU/ml was achieved. More recently, Arya et al. used a self-assembled monolayer of dithiobis(succimidylpropionate) (DTSP) for covalent immobilization of T4 phage on the gold surface of an SPR sensor.113 This method ensured uniform and strong binding of the phages to the surface, and allowed specific detection of E. coli cells at concentrations as low as 7 × 102 CFU/ml. Furthermore, the sensor surfaces could be regenerated and repeatedly used. Besides complete phages, RBPs have also been utilized for SPR-based pathogen detection. A genetically engineered tailspike protein (lacking its native endorhamnosidase activity) from Salmonella phage P22 was anchored to a gold surface via an N-terminal cysteine tag, using thiol chemistry.102 The resulting biosensor was able to detect Salmonella cells through measurement of real-time interaction at a concentration of 103 CFU/ml, whereas E. coli was not recognized. In a similar approach, an RBP from Campylobacter phage NCTC 12673 was fused to glutathione S transferase (GST), and attached to an SPR surface using glutathione self-assembled monolayers (GSH-SAM), resulting in oriented immobilization of the recognition elements.103 This strategy improved capturing of target cells by 2–3 times compared with an undirected DTSP-mediated chemical coupling approach tested in parallel, which is reflected by a low detection limit of 102 CFU/ml.
A number of reports describe the design and construction of phage-based magnetoelastic (ME) biosensors, mostly for detection of Salmonella. This type of transduction platform, in which signals can be detected by either magnetic, acoustic, or optical techniques, offers several advantages, including wireless interrogation (through magnetic fields), possible miniaturization, and the capability of detecting multiple different targets simultaneously in a multiplexed fashion.114 In one study, filamentous phage specific for S. Typhimurium were physically adsorbed to the sensor surface, and binding of target cells to the phage could be detected by changes in resonance frequency of the sensor as a consequence of increasing mass, with a detection limit of 103 CFU/ml.115 Using multiple ME sensors that are simultaneously interrogated allowed detection of two different pathogens, S. Typhimurium and spores of Bacillus anthracis.116 This setup comprises a reference sensor as control, a Salmonella-specific sensor as described above, and a sensor coated with B. anthracis spore-specific filamentous phages. When sequentially exposed to the different pathogenic agents, only the sensors coated with phages specific for the respective pathogens generated measurable signals. The Salmonella-specific biosensor was further used to detect S. Typhimurium directly on the surface of fresh tomatoes.117 The tomato surfaces were spiked with the pathogens at different concentrations, air-dried, and the phage-coated sensor (plus a control sensor without phages) was placed directly on the surfaces in a humid environment for 30 min. Surfaces that had been spiked with Salmonella suspensions at concentrations as low as 5 × 102 CFU/ml generated responses of the phage-coated sensor significantly different from those of the control sensor. The same principle was applied for detection of Salmonella on eggshells, and a similarly low detection limit (1.4 × 102 CFU/cm2) was achieved, with a total assay time of 30 min.118
Various other detection concepts using phage-based biosensor systems have been reported, and some selected examples will be briefly described here. Olsen et al. constructed acoustic wave biodetectors by immobilizing filamentous phage that display target cell-specific affinity molecules to piezoelectric transducers.119 Binding of bacteria to the phage resulted in resonance frequency changes, and allowed detection of 102 CFU/ml. Another recent study by Tay et al. reports the conjugation of Salmonella tailspike proteins to silica-encapsulated Raman-reporter-embedded nanoprobes, using surface enhanced Raman scattering (SERS) for highly specific and sensitive detection of Salmonella down to a single cell.104 A biosensor concept reported by Smietana utilizes optical fiber long-period gratings, which allow detection of resonance wavelength shifts upon binding of bacteria to physically adsorbed bacteriophage.120 Finally, Pacheco-Gomez et al. utilized the principle of linear dichroism (LD) for measuring the interaction of bacterial pathogens with (non-immobilized) filamentous M13 phage functionalized with affinity molecules specific for these target cells.121 In this setup, the LD signal is dependent on the degree of alignment of molecules within the sample. While phages carrying the affinity molecule are perfectly aligned within a shear flow (yielding a high LD signal), binding of (several) phage particles to a target pathogen abolishes this alignment, resulting in a reduced LD signal. This principle can also work in a multiplexed assay by using differentially labeled phage particles.
Conclusions
Bacteriophages combine several properties that are desirable for the purpose of detecting bacterial pathogens from food samples: i) they are highly specific for their target cells and therefore largely unaffected by organisms of the background flora; ii) they can “distinguish” between living and dead cells; iii) they work under many different, including harsh, environmental conditions, eliminating the need for laborious sample pre-processing; iv) they act as signal-amplifiers when running through an infection cycle; and v) they are inexpensive and easy to produce. Furthermore, it should be considered that bacteriophages are the most abundant self-replicating entities on earth, with an estimated 1031 particles,122 and therefore offer a virtually unlimited selection of tools (including complete phage particles and phage-derived affinity molecules such as CBDs and RBPs) we can harness for different detection strategies. Any newly developed detection method must provide a significant advantage in at least one of the key points (rapidity, sensitivity, specificity) over existing techniques, and at the same time, offer sufficient economic benefit to justify the costs. Some of the phage-based detection formats described in this review meet these requirements and may have the potential to pass the hurdle toward commercialization. This particularly appears to be true for biosensor technologies, which have developed exponentially during the past decade.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Catherine Rees (The University of Nottingham, UK) for sharing her knowledge on bacteriophage-based detection methods.
Glossary
Abbreviations:
- PCR
polymerase chain reaction
- ELISA
enzyme-linked immunosorbent assay
- MS
mass spectrometry
- RT-PCR
reverse transcription and PCR
- ADP
adenosine diphosphate
- IMS
immunomagnetic separation
- MAP
M. avium ssp. paratuberculosis
- PMS
peptide-mediated magnetic separation
- MALDI-TOF
matrix-assisted laser desorption-ionization time-of-flight mass spectrometry
- qPCR
quantitative real-time PCR
- PR-cELISA
phage replication-competitive enzyme-linked immunosorbent assay
- GMO
genetically modified organism
- LRP
luciferase reporter phage
- AHL
acyl-homoserine lactone
- ATP
adenosine triphosphate
- AMP
adenosine monophosphate
- AK
adenylate kinase
- GFP
green fluorescent protein
- INP
ice nucleation protein
- QD
quantum dot
- FlAsH-EDT2
fluorescein arsenical helix binder
- FBA
fluorescent-bacteriophage assay
Citation: Schmelcher M, Loessner MJ. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 2014; 4:e28137; 10.4161/bact.28137
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/28137
References
- 1.Hagens S, Loessner MJ. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol. 2007;76:513–9. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- 2.Rees CE, Dodd CE. Phage for rapid detection and control of bacterial pathogens in food. Adv Appl Microbiol. 2006;59:159–86. doi: 10.1016/S0065-2164(06)59006-9. [DOI] [PubMed] [Google Scholar]
- 3.Nogva HK, Drømtorp SM, Nissen H, Rudi K. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques. 2003;34:804–8, 810, 812-3. doi: 10.2144/03344rr02. [DOI] [PubMed] [Google Scholar]
- 4.Brovko LY, Anany H, Griffiths MW. Bacteriophages for detection and control of bacterial pathogens in food and food-processing environment. Adv Food Nutr Res. 2012;67:241–88. doi: 10.1016/B978-0-12-394598-3.00006-X. [DOI] [PubMed] [Google Scholar]
- 5.Olofsson L, Ankarloo J, Nicholls IA. Phage viability in organic media: insights into phage stability. J Mol Recognit. 1998;11:91–3. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<91::AID-JMR397>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 6.Schwind P, Kramer H, Kremser A, Ramsberger U, Rasched I. Subtilisin removes the surface layer of the phage fd coat. Eur J Biochem. 1992;210:431–6. doi: 10.1111/j.1432-1033.1992.tb17438.x. [DOI] [PubMed] [Google Scholar]
- 7.Goodridge L, Griffiths M. Reporter bacteriophage assays as a means to detect foodborne pathogenic bacteria. Food Res Int. 2002;35:863–70. doi: 10.1016/S0963-9969(02)00094-7. [DOI] [Google Scholar]
- 8.Rees CED, Loessner MJ. Phage for the Detection of Pathogenic Bacteria. In: Kutter E, Sulakvelidze A, eds. Bacteriophages: Biology and Applications. Boca Raton: CRC Press, 2005:267-84 [Google Scholar]
- 9.Schmelcher M, Loessner M. Bacteriophage: Powerful Tools for the Detection of Bacterial Pathogens. In: Zourob M, Elwary S, Turner A, eds. Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems. New York, USA: Springer, 2008:731-54 [Google Scholar]
- 10.Henry M, Debarbieux L. Tools from viruses: bacteriophage successes and beyond. Virology. 2012;434:151–61. doi: 10.1016/j.virol.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Smartt AE, Xu T, Jegier P, Carswell JJ, Blount SA, Sayler GS, Ripp S. Pathogen detection using engineered bacteriophages. Anal Bioanal Chem. 2012;402:3127–46. doi: 10.1007/s00216-011-5555-5. [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Arutyunov D, Szymanski CM, Evoy S. Bacteriophage based probes for pathogen detection. Analyst. 2012;137:3405–21. doi: 10.1039/c2an35371g. [DOI] [PubMed] [Google Scholar]
- 13.Mandeville R, Griffiths M, Goodridge L, McIntyre L, Ilenchuk TT. Diagnostic and therapeutic applications of lytic phages. Anal Lett. 2003;36:3241–59. doi: 10.1081/AL-120026569. [DOI] [Google Scholar]
- 14.Petty NK, Evans TJ, Fineran PC, Salmond GP. Biotechnological exploitation of bacteriophage research. Trends Biotechnol. 2007;25:7–15. doi: 10.1016/j.tibtech.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–81. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley PE. A review of bioluminescent ATP techniques in rapid microbiology. J Biolumin Chemilumin. 1989;4:375–80. doi: 10.1002/bio.1170040151. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths MW. The role of ATP bioluminescence in the food industry: New light on old problems. Food Technol. 1996;50:62. [Google Scholar]
- 18.Squirrel DJ, Price RL, Murphy MJ. Rapid and specific detection of bacteria using bioluminescence. Anal Chim Acta. 2002;457:109–14. doi: 10.1016/S0003-2670(01)01495-7. [DOI] [Google Scholar]
- 19.Blasco R, Murphy MJ, Sanders MF, Squirrell DJ. Specific assays for bacteria using phage mediated release of adenylate kinase. J Appl Microbiol. 1998;84:661–6. doi: 10.1046/j.1365-2672.1998.00393.x. [DOI] [PubMed] [Google Scholar]
- 20.Minikh O, Tolba M, Brovko LY, Griffiths MW. Bacteriophage-based biosorbents coupled with bioluminescent ATP assay for rapid concentration and detection of Escherichia coli. J Microbiol Methods. 2010;82:177–83. doi: 10.1016/j.mimet.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Corbitt AJ, Bennion N, Forsythe SJ. Adenylate kinase amplification of ATP bioluminescence for hygiene monitoring in the food and beverage industry. Lett Appl Microbiol. 2000;30:443–7. doi: 10.1046/j.1472-765x.2000.00744.x. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Brovko L, Griffiths MW. Influence of phage population on the phage-mediated bioluminescent adenylate kinase (AK) assay for detection of bacteria. Lett Appl Microbiol. 2001;33:311–5. doi: 10.1046/j.1472-765X.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- 23.Kannan P, Yong HY, Reiman L, Cleaver C, Patel P, Bhagwat AA. Bacteriophage-based rapid and sensitive detection of Escherichia coli O157:H7 isolates from ground beef. Foodborne Pathog Dis. 2010;7:1551–8. doi: 10.1089/fpd.2010.0634. [DOI] [PubMed] [Google Scholar]
- 24.Neufeld T, Schwartz-Mittelmann A, Biran D, Ron EZ, Rishpon J. Combined phage typing and amperometric detection of released enzymatic activity for the specific identification and quantification of bacteria. Anal Chem. 2003;75:580–5. doi: 10.1021/ac026083e. [DOI] [PubMed] [Google Scholar]
- 25.Yemini M, Levi Y, Yagil E, Rishpon J. Specific electrochemical phage sensing for Bacillus cereus and Mycobacterium smegmatis. Bioelectrochemistry. 2007;70:180–4. doi: 10.1016/j.bioelechem.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Silley P, Forsythe S. Impedance microbiology--a rapid change for microbiologists. J Appl Bacteriol. 1996;80:233–43. doi: 10.1111/j.1365-2672.1996.tb03215.x. [DOI] [PubMed] [Google Scholar]
- 27.Chang TC, Ding HC, Chen S. A conductance method for the identification of Escherichia coli O157:H7 using bacteriophage AR1. J Food Prot. 2002;65:12–7. doi: 10.4315/0362-028x-65.1.12. [DOI] [PubMed] [Google Scholar]
- 28.Wawerla M, Stolle A, Schalch B, Eisgruber H. Impedance microbiology: applications in food hygiene. J Food Prot. 1999;62:1488–96. doi: 10.4315/0362-028x-62.12.1488. [DOI] [PubMed] [Google Scholar]
- 29.Bolton FJ. An investigation of indirect conductimetry for detection of some food-borne bacteria. J Appl Bacteriol. 1990;69:655–61. doi: 10.1111/j.1365-2672.1990.tb01559.x. [DOI] [PubMed] [Google Scholar]
- 30.Falahee MB, Park SF, Adams MR. Detection and enumeration of Campylobacter jejuni and Campylobacter coli by indirect impedimetry with an oxygen scavenging system. J Food Prot. 2003;66:1724–6. doi: 10.4315/0362-028x-66.9.1724. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Ye Z, Ying Y. New trends in impedimetric biosensors for the detection of foodborne pathogenic bacteria. Sensors (Basel) 2012;12:3449–71. doi: 10.3390/s120303449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart GS, Jassim SA, Denyer SP, Newby P, Linley K, Dhir VK. The specific and sensitive detection of bacterial pathogens within 4 h using bacteriophage amplification. J Appl Microbiol. 1998;84:777–83. doi: 10.1046/j.1365-2672.1998.00408.x. [DOI] [PubMed] [Google Scholar]
- 33.Wilson SM, al-Suwaidi Z, McNerney R, Porter J, Drobniewski F. Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat Med. 1997;3:465–8. doi: 10.1038/nm0497-465. [DOI] [PubMed] [Google Scholar]
- 34.McNerney R, Wilson SM, Sidhu AM, Harley VS, al Suwaidi Z, Nye PM, Parish T, Stoker NG. Inactivation of mycobacteriophage D29 using ferrous ammonium sulphate as a tool for the detection of viable Mycobacterium smegmatis and M. tuberculosis. Res Microbiol. 1998;149:487–95. doi: 10.1016/S0923-2508(98)80003-X. [DOI] [PubMed] [Google Scholar]
- 35.Park DJ, Drobniewski FA, Meyer A, Wilson SM. Use of a phage-based assay for phenotypic detection of mycobacteria directly from sputum. J Clin Microbiol. 2003;41:680–8. doi: 10.1128/JCM.41.2.680-688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mole RJ, Maskell TWO. Phage as a diagnostic - the use of phage in TB diagnosis. J Chem Technol Biotechnol. 2001;76:683–8. doi: 10.1002/jctb.439. [DOI] [Google Scholar]
- 37.Favrin SJ, Jassim SA, Griffiths MW. Development and optimization of a novel immunomagnetic separation-bacteriophage assay for detection of Salmonella enterica serovar enteritidis in broth. Appl Environ Microbiol. 2001;67:217–24. doi: 10.1128/AEM.67.1.217-224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanley EC, Mole RJ, Smith RJ, Glenn SM, Barer MR, McGowan M, Rees CE. Development of a new, combined rapid method using phage and PCR for detection and identification of viable Mycobacterium paratuberculosis bacteria within 48 hours. Appl Environ Microbiol. 2007;73:1851–7. doi: 10.1128/AEM.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rees JC, Voorhees KJ. Simultaneous detection of two bacterial pathogens using bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:2757–61. doi: 10.1002/rcm.2107. [DOI] [PubMed] [Google Scholar]
- 40.Almeida PE, Almeida RCC, Barbalho TCF, Melo CG, Almeida AO, Magalhaes ER, Oliveira IC, Hofer E. Development of protocols for the bacteriophage amplification assay for rapid, quantitative and sensitive detection of viable Listeria cells. Abstr Gen Meet Am Soc Microbiol. 2003;103:P–052. [Google Scholar]
- 41.de Siqueira RS, Dodd CE, Rees CE. Evaluation of the natural virucidal activity of teas for use in the phage amplification assay. Int J Food Microbiol. 2006;111:259–62. doi: 10.1016/j.ijfoodmicro.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 42.Ott SL, Wells SJ, Wagner BA. Herd-level economic losses associated with Johne’s disease on US dairy operations. Prev Vet Med. 1999;40:179–92. doi: 10.1016/S0167-5877(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 43.Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn’s disease. Lancet. 2004;364:1039–44. doi: 10.1016/S0140-6736(04)17058-X. [DOI] [PubMed] [Google Scholar]
- 44.Bull TJ, McMinn EJ, Sidi-Boumedine K, Skull A, Durkin D, Neild P, Rhodes G, Pickup R, Hermon-Taylor J. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn’s disease. J Clin Microbiol. 2003;41:2915–23. doi: 10.1128/JCM.41.7.2915-2923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Botsaris G, Slana I, Liapi M, Dodd C, Economides C, Rees C, Pavlik I. Rapid detection methods for viable Mycobacterium avium subspecies paratuberculosis in milk and cheese. Int J Food Microbiol. 2010;141(Suppl 1):S87–90. doi: 10.1016/j.ijfoodmicro.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 46.Foddai A, Elliott CT, Grant IR. Maximizing capture efficiency and specificity of magnetic separation for Mycobacterium avium subsp. paratuberculosis cells. Appl Environ Microbiol. 2010;76:7550–8. doi: 10.1128/AEM.01432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart LD, Foddai A, Elliott CT, Grant IR. Development of a novel phage-mediated immunoassay for the rapid detection of viable Mycobacterium avium subsp. paratuberculosis. J Appl Microbiol. 2013;115:808–17. doi: 10.1111/jam.12275. [DOI] [PubMed] [Google Scholar]
- 48.Favrin SJ, Jassim SA, Griffiths MW. Application of a novel immunomagnetic separation-bacteriophage assay for the detection of Salmonella enteritidis and Escherichia coli O157:H7 in food. Int J Food Microbiol. 2003;85:63–71. doi: 10.1016/S0168-1605(02)00483-X. [DOI] [PubMed] [Google Scholar]
- 49.Madonna AJ, Van Cuyk S, Voorhees KJ. Detection of Escherichia coli using immunomagnetic separation and bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:257–63. doi: 10.1002/rcm.900. [DOI] [PubMed] [Google Scholar]
- 50.Pierce CL, Rees JC, Fernández FM, Barr JR. Viable Staphylococcus aureus quantitation using ¹⁵N metabolically labeled bacteriophage amplification coupled with a multiple reaction monitoring proteomic workflow. Mol Cell Proteomics. 2012;11:012849. doi: 10.1074/mcp.M111.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pierce CL, Rees JC, Fernández FM, Barr JR. Detection of Staphylococcus aureus using 15N-labeled bacteriophage amplification coupled with matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal Chem. 2011;83:2286–93. doi: 10.1021/ac103024m. [DOI] [PubMed] [Google Scholar]
- 52.Jassim SA, Griffiths MW. Evaluation of a rapid microbial detection method via phage lytic amplification assay coupled with Live/Dead fluorochromic stains. Lett Appl Microbiol. 2007;44:673–8. doi: 10.1111/j.1472-765X.2007.02115.x. [DOI] [PubMed] [Google Scholar]
- 53.Sergueev KV, He Y, Borschel RH, Nikolich MP, Filippov AA. Rapid and sensitive detection of Yersinia pestis using amplification of plague diagnostic bacteriophages monitored by real-time PCR. PLoS One. 2010;5:e11337. doi: 10.1371/journal.pone.0011337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan J, Chan M, Allain B, Mandeville R, Brooks BW. Detection of multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 by phage replication-competitive enzyme-linked immunosorbent assay. J Food Prot. 2006;69:739–42. doi: 10.4315/0362-028x-69.4.739. [DOI] [PubMed] [Google Scholar]
- 55.Ulitzur S, Kuhn J. Introduction of lux genes into bacteria, a new approach for specific determination of bacteria and their antibiotic susceptibility. In: Schlomerich J, Andreesen R, Kapp A, Ernst M, Wood WG, eds. Bioluminescence and chemiluminescence, new perspectives. New York, NY: John Wiley & Sons, 1987:463-72 [Google Scholar]
- 56.Loessner MJ, Rees CE, Stewart GS, Scherer S. Construction of luciferase reporter bacteriophage A511:luxAB for rapid and sensitive detection of viable Listeria cells. Appl Environ Microbiol. 1996;62:1133–40. doi: 10.1128/aem.62.4.1133-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thouand G, Vachon P, Liu S, Dayre M, Griffiths MW. Optimization and validation of a simple method using P22:luxAB bacteriophage for rapid detection of Salmonella enterica serotypes A, B, and D in poultry samples. J Food Prot. 2008;71:380–5. doi: 10.4315/0362-028x-71.2.380. [DOI] [PubMed] [Google Scholar]
- 58.Ripp S, Jegier P, Birmele M, Johnson CM, Daumer KA, Garland JL, Sayler GS. Linking bacteriophage infection to quorum sensing signalling and bioluminescent bioreporter monitoring for direct detection of bacterial agents. J Appl Microbiol. 2006;100:488–99. doi: 10.1111/j.1365-2672.2005.02828.x. [DOI] [PubMed] [Google Scholar]
- 59.Sasahara KC, Gray MJ, Shin SJ, Boor KJ. Detection of viable Mycobacterium avium subsp. paratuberculosis using luciferase reporter systems. Foodborne Pathog Dis. 2004;1:258–66. doi: 10.1089/fpd.2004.1.258. [DOI] [PubMed] [Google Scholar]
- 60.Wolber PK, Green RL. Detection of bacteria by transduction of ice nucleation genes. Trends Biotechnol. 1990;8:276–9. doi: 10.1016/0167-7799(90)90195-4. [DOI] [PubMed] [Google Scholar]
- 61.Willford J, Goodridge LD. An integrated assay for rapid detection of Escherichia coli O157:H7 on beef samples. Food Prot Trends. 2008;28:468–72. [Google Scholar]
- 62.Funatsu T, Taniyama T, Tajima T, Tadakuma H, Namiki H. Rapid and sensitive detection method of a bacterium by using a GFP reporter phage. Microbiol Immunol. 2002;46:365–9. doi: 10.1111/j.1348-0421.2002.tb02708.x. [DOI] [PubMed] [Google Scholar]
- 63.Hagens S, de Wouters T, Vollenweider P, Loessner MJ. Reporter bacteriophage A511:celB transduces a hyperthermostable glycosidase from Pyrococcus furiosus for rapid and simple detection of viable Listeria cells. Bacteriophage. 2011;1:143–51. doi: 10.4161/bact.1.3.16710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar R, McKinstry M, Hwang J, Oppenheim AB, Fekete RA, Giulian G, Merril C, Nagashima K, Adhya S. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc Natl Acad Sci U S A. 2006;103:4841–5. doi: 10.1073/pnas.0601211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu L, Huang T, Yang L, Pan J, Zhu S, Yan X. Sensitive and selective bacterial detection using tetracysteine-tagged phages in conjunction with biarsenical dye. Angew Chem Int Ed Engl. 2011;50:5873–7. doi: 10.1002/anie.201100334. [DOI] [PubMed] [Google Scholar]
- 66.Ulitzur S, Kuhn J. Construction of lux bacteriophages and the determination of specific bacteria and their antibiotic sensitivities. Methods Enzymol. 2000;305:543–57. doi: 10.1016/S0076-6879(00)05512-9. [DOI] [PubMed] [Google Scholar]
- 67.Lu TK, Bowers J, Koeris MS. Advancing bacteriophage-based microbial diagnostics with synthetic biology. Trends Biotechnol. 2013;31:325–7. doi: 10.1016/j.tibtech.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 68.Waddell TE, Poppe C. Construction of mini-Tn10luxABcam/Ptac-ATS and its use for developing a bacteriophage that transduces bioluminescence to Escherichia coli O157:H7. FEMS Microbiol Lett. 2000;182:285–9. doi: 10.1111/j.1574-6968.2000.tb08909.x. [DOI] [PubMed] [Google Scholar]
- 69.Stewart GSAB, Smith T, Denyer S. Genetic engineering for bioluminescent bacteria. Food Sci Technol Today. 1989;3:19–22. [Google Scholar]
- 70.Kuhn J, Suissa M, Wyse J, Cohen I, Weiser I, Reznick S, Lubinsky-Mink S, Stewart G, Ulitzur S. Detection of bacteria using foreign DNA: the development of a bacteriophage reagent for Salmonella. Int J Food Microbiol. 2002;74:229–38. doi: 10.1016/S0168-1605(01)00683-3. [DOI] [PubMed] [Google Scholar]
- 71.Kodikara CP, Crew HH, Stewart GS. Near on-line detection of enteric bacteria using lux recombinant bacteriophage. FEMS Microbiol Lett. 1991;67:261–5. doi: 10.1111/j.1574-6968.1991.tb04474.x. [DOI] [PubMed] [Google Scholar]
- 72.Loessner MJ, Rudolf M, Scherer S. Evaluation of luciferase reporter bacteriophage A511:luxAB for detection of Listeria monocytogenes in contaminated foods. Appl Environ Microbiol. 1997;63:2961–5. doi: 10.1128/aem.63.8.2961-2965.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pagotto F, Brovko L, Griffiths MW. Phage-mediated detection of Staphylococcus aureus and Escherichia coli O157:H7 using bioluminescence. Symposium on Bacteriological Quality of Raw Milk, Proceedings 1996:152-6 [Google Scholar]
- 74.Chen J, Griffiths MW. Salmonella detection in eggs using Lux(+) bacteriophages. J Food Prot. 1996;59:908–14. doi: 10.4315/0362-028X-59.9.908. [DOI] [PubMed] [Google Scholar]
- 75.Schofield DA, Molineux IJ, Westwater C. ‘Bioluminescent’ reporter phage for the detection of Category A bacterial pathogens. J Vis Exp. 2011;53:e23740. doi: 10.3791/2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schofield DA, Molineux IJ, Westwater C. Diagnostic bioluminescent phage for detection of Yersinia pestis. J Clin Microbiol. 2009;47:3887–94. doi: 10.1128/JCM.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schofield DA, Westwater C. Phage-mediated bioluminescent detection of Bacillus anthracis. J Appl Microbiol. 2009;107:1468–78. doi: 10.1111/j.1365-2672.2009.04332.x. [DOI] [PubMed] [Google Scholar]
- 78.Fey H, Bürgi E, Margadant A, Boller E. An economic and rapid diagnostic procedure for the detection of salmonella/shigella using the polyvalent salmonella phage O-1. Zentralbl Bakteriol Orig A. 1978;240:7–15. [PubMed] [Google Scholar]
- 79.Birmele M, Ripp S, Jegier P, Roberts MS, Sayler G, Garland J. Characterization and validation of a bioluminescent bioreporter for the direct detection of Escherichia coli. J Microbiol Methods. 2008;75:354–6. doi: 10.1016/j.mimet.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 80.Brigati JR, Ripp SA, Johnson CM, Iakova PA, Jegier P, Sayler GS. Bacteriophage-based bioluminescent bioreporter for the detection of Escherichia coli 0157:H7. J Food Prot. 2007;70:1386–92. doi: 10.4315/0362-028x-70.6.1386. [DOI] [PubMed] [Google Scholar]
- 81.Ripp S, Jegier P, Johnson CM, Brigati JR, Sayler GS. Bacteriophage-amplified bioluminescent sensing of Escherichia coli O157:H7. Anal Bioanal Chem. 2008;391:507–14. doi: 10.1007/s00216-007-1812-z. [DOI] [PubMed] [Google Scholar]
- 82.Zimmer M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chem Rev. 2002;102:759–81. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 83.Schmelcher M, Shabarova T, Eugster MR, Eichenseher F, Tchang VS, Banz M, Loessner MJ. Rapid multiplex detection and differentiation of Listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl Environ Microbiol. 2010;76:5745–56. doi: 10.1128/AEM.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oda M, Morita M, Unno H, Tanji Y. Rapid detection of Escherichia coli O157:H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl Environ Microbiol. 2004;70:527–34. doi: 10.1128/AEM.70.1.527-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanji Y, Furukawa C, Na SH, Hijikata T, Miyanaga K, Unno H. Escherichia coli detection by GFP-labeled lysozyme-inactivated T4 bacteriophage. J Biotechnol. 2004;114:11–20. doi: 10.1016/j.jbiotec.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 86.Wolber PK. Bacterial ice nucleation. Adv Microb Physiol. 1993;34:203–37. doi: 10.1016/S0065-2911(08)60030-2. [DOI] [PubMed] [Google Scholar]
- 87.Irwin P, Gehring A, Tu SI, Brewster J, Fanelli J, Ehrenfeld E. Minimum detectable level of Salmonellae using a binomial-based bacterial ice nucleation detection assay (BIND) J AOAC Int. 2000;83:1087–95. [PubMed] [Google Scholar]
- 88.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–43. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 89.Watson BB, Eveland WC. The application of the phage-fluorescent antiphage staining system in the specific identification of Listeria monocytogenes. I. Species specificity and immunofluorescent sensitivity of Listeria monocytogenes phage observed in smear preparations. J Infect Dis. 1965;115:363–9. doi: 10.1093/infdis/115.4.363. [DOI] [PubMed] [Google Scholar]
- 90.Goodridge L, Chen J, Griffiths M. Development and characterization of a fluorescent-bacteriophage assay for detection of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:1397–404. doi: 10.1128/aem.65.4.1397-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goodridge L, Chen J, Griffiths M. The use of a fluorescent bacteriophage assay for detection of Escherichia coli O157:H7 in inoculated ground beef and raw milk. Int J Food Microbiol. 1999;47:43–50. doi: 10.1016/S0168-1605(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 92.Mosier-Boss PA, Lieberman SH, Andrews JM, Rohwer FL, Wegley LE, Breitbart M. Use of fluorescently labeled phage in the detection and identification of bacterial species. Appl Spectrosc. 2003;57:1138–44. doi: 10.1366/00037020360696008. [DOI] [PubMed] [Google Scholar]
- 93.Willford JD, Bisha B, Bolenbaugh KE, Goodridge LD. Luminescence based enzyme-labeled phage (Phazyme) assays for rapid detection of Shiga toxin producing Escherichia coli serogroups. Bacteriophage. 2011;1:101–10. doi: 10.4161/bact.1.2.15666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7:1147–71. doi: 10.2217/fmb.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loessner MJ, Wendlinger G, Scherer S. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol Microbiol. 1995;16:1231–41. doi: 10.1111/j.1365-2958.1995.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 96.Fischetti VA. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 2005;13:491–6. doi: 10.1016/j.tim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 97.Gu J, Lu R, Liu X, Han W, Lei L, Gao Y, Zhao H, Li Y, Diao Y. LysGH15B, the SH3b domain of staphylococcal phage endolysin LysGH15, retains high affinity to staphylococci. Curr Microbiol. 2011;63:538–42. doi: 10.1007/s00284-011-0018-y. [DOI] [PubMed] [Google Scholar]
- 98.Loessner MJ, Kramer K, Ebel F, Scherer S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol. 2002;44:335–49. doi: 10.1046/j.1365-2958.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- 99.Schmelcher M, Tchang VS, Loessner MJ. Domain shuffling and module engineering of Listeria phage endolysins for enhanced lytic activity and binding affinity. Microb Biotechnol. 2011;4:651–62. doi: 10.1111/j.1751-7915.2011.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kretzer JW, Lehmann R, Schmelcher M, Banz M, Kim KP, Korn C, Loessner MJ. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl Environ Microbiol. 2007;73:1992–2000. doi: 10.1128/AEM.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walcher G, Stessl B, Wagner M, Eichenseher F, Loessner MJ, Hein I. Evaluation of paramagnetic beads coated with recombinant Listeria phage endolysin-derived cell-wall-binding domain proteins for separation of Listeria monocytogenes from raw milk in combination with culture-based and real-time polymerase chain reaction-based quantification. Foodborne Pathog Dis. 2010;7:1019–24. doi: 10.1089/fpd.2009.0475. [DOI] [PubMed] [Google Scholar]
- 102.Singh A, Arya SK, Glass N, Hanifi-Moghaddam P, Naidoo R, Szymanski CM, Tanha J, Evoy S. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens Bioelectron. 2010;26:131–8. doi: 10.1016/j.bios.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 103.Singh A, Arutyunov D, McDermott MT, Szymanski CM, Evoy S. Specific detection of Campylobacter jejuni using the bacteriophage NCTC 12673 receptor binding protein as a probe. Analyst. 2011;136:4780–6. doi: 10.1039/c1an15547d. [DOI] [PubMed] [Google Scholar]
- 104.Tay LL, Huang PJ, Tanha J, Ryan S, Wu X, Hulse J, Chau LK. Silica encapsulated SERS nanoprobe conjugated to the bacteriophage tailspike protein for targeted detection of Salmonella. Chem Commun (Camb) 2012;48:1024–6. doi: 10.1039/c1cc16325f. [DOI] [PubMed] [Google Scholar]
- 105.Singh A, Glass N, Tolba M, Brovko L, Griffiths M, Evoy S. Immobilization of bacteriophages on gold surfaces for the specific capture of pathogens. Biosens Bioelectron. 2009;24:3645–51. doi: 10.1016/j.bios.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 106.Gervals L, Gel M, Allain B, Tolba M, Brovko L, Zourob M, Mandeville R, Griffiths M, Evoy S. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens Actuators B Chem. 2007;125:615–21. doi: 10.1016/j.snb.2007.03.007. [DOI] [Google Scholar]
- 107.Tolba M, Minikh O, Brovko LY, Evoy S, Griffiths MW. Oriented immobilization of bacteriophages for biosensor applications. Appl Environ Microbiol. 2010;76:528–35. doi: 10.1128/AEM.02294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shabani A, Zourob M, Allain B, Marquette CA, Lawrence MF, Mandeville R. Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria. Anal Chem. 2008;80:9475–82. doi: 10.1021/ac801607w. [DOI] [PubMed] [Google Scholar]
- 109.Shabani A, Marquette CA, Mandeville R, Lawrence MF. Magnetically-assisted impedimetric detection of bacteria using phage-modified carbon microarrays. Talanta. 2013;116:1047–53. doi: 10.1016/j.talanta.2013.07.078. [DOI] [PubMed] [Google Scholar]
- 110.Tolba M, Ahmed MU, Tlili C, Eichenseher F, Loessner MJ, Zourob M. A bacteriophage endolysin-based electrochemical impedance biosensor for the rapid detection of Listeria cells. Analyst. 2012;137:5749–56. doi: 10.1039/c2an35988j. [DOI] [PubMed] [Google Scholar]
- 111.Nanduri V, Bhunia AK, Tu SI, Paoli GC, Brewster JD. SPR biosensor for the detection of L. monocytogenes using phage-displayed antibody. Biosens Bioelectron. 2007;23:248–52. doi: 10.1016/j.bios.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 112.Balasubramanian S, Sorokulova IB, Vodyanoy VJ, Simonian AL. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus--A surface plasmon resonance spectroscopic study. Biosens Bioelectron. 2007;22:948–55. doi: 10.1016/j.bios.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 113.Arya SK, Singh A, Naidoo R, Wu P, McDermott MT, Evoy S. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst. 2011;136:486–92. doi: 10.1039/c0an00697a. [DOI] [PubMed] [Google Scholar]
- 114.Grimes CA, Roy SC, Rani S, Cai Q. Theory, instrumentation and applications of magnetoelastic resonance sensors: a review. Sensors (Basel) 2011;11:2809–44. doi: 10.3390/s110302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lakshmanan RS, Guntupalli R, Hu J, Kim DJ, Petrenko VA, Barbaree JM, Chin BA. Phage immobilized magnetoelastic sensor for the detection of Salmonella typhimurium. J Microbiol Methods. 2007;71:55–60. doi: 10.1016/j.mimet.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 116.Huang S, Yang H, Lakshmanan RS, Johnson ML, Wan J, Chen IH, Wikle HC, 3rd, Petrenko VA, Barbaree JM, Chin BA. Sequential detection of Salmonella typhimurium and Bacillus anthracis spores using magnetoelastic biosensors. Biosens Bioelectron. 2009;24:1730–6. doi: 10.1016/j.bios.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 117.Li S, Li Y, Chen H, Horikawa S, Shen W, Simonian A, Chin BA. Direct detection of Salmonella typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens Bioelectron. 2010;26:1313–9. doi: 10.1016/j.bios.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 118.Chai Y, Li S, Horikawa S, Park MK, Vodyanoy V, Chin BA. Rapid and sensitive detection of Salmonella Typhimurium on eggshells by using wireless biosensors. J Food Prot. 2012;75:631–6. doi: 10.4315/0362-028X.JFP-11-339. [DOI] [PubMed] [Google Scholar]
- 119.Olsen EV, Sorokulova IB, Petrenko VA, Chen IH, Barbaree JM, Vodyanoy VJ. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens Bioelectron. 2006;21:1434–42. doi: 10.1016/j.bios.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 120.Smietana M, Bock WJ, Mikulic P, Ng A, Chinnappan R, Zourob M. Detection of bacteria using bacteriophages as recognition elements immobilized on long-period fiber gratings. Opt Express. 2011;19:7971–8. doi: 10.1364/OE.19.007971. [DOI] [PubMed] [Google Scholar]
- 121.Pacheco-Gómez R, Kraemer J, Stokoe S, England HJ, Penn CW, Stanley E, Rodger A, Ward J, Hicks MR, Dafforn TR. Detection of pathogenic bacteria using a homogeneous immunoassay based on shear alignment of virus particles and linear dichroism. Anal Chem. 2012;84:91–7. doi: 10.1021/ac201544h. [DOI] [PubMed] [Google Scholar]
- 122.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A. 1999;96:2192–7. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]