Abstract
Cancer can take hundreds of different forms depending on the location, cell of origin and spectrum of genomic alterations that promote oncogenesis and affect therapeutic response. Although many genomic events with direct phenotypic impact have been identified, much of the complex molecular landscape remains incompletely charted for most cancer lineages. For that reason, The Cancer Genome Atlas (TCGA) Research Network has profiled and analyzed large numbers of human tumours to discover molecular aberrations at the DNA, RNA, protein, and epigenetic levels. The resulting rich data provide a major opportunity to develop an integrated picture of commonalities, differences, and emergent themes across tumour lineages. The Pan-Cancer initiative compares the first twelve tumour types profiled by TCGA. Analysis of the molecular aberrations and their functional roles across tumour types will teach us how to extend therapies effective in one cancer type to others with a similar genomic profile.
Molecular Profiling of Single Tumour Types
That cancer is fundamentally a genomic disease is now well established. Early on, large numbers of oncogenes were identified using functional assays on genetic material from tumours in positive selection systems1-3, and a subset of tumour suppressor genes were identified by analyzing loss of heterozygosity4. More recently, systematic cancer genomics projects have applied emerging technologies to the analysis of specific tumour types including the Cancer Genome Atlas Project (TCGA; Box 1). That disease-specific focus has identified novel oncogenic drivers, those genes contributing to functional change5-7, established molecular subtypes8-13 and identified new biomarkers based on genomic, transcriptomic and proteomic alterations. Some of those biomarkers have clinical implications14,15. For example, we now view ductal breast cancer as a collection of distinct diseases whose major subtypes (e.g. luminal A, luminal B, HER2, basal-like) are managed differently in the clinic; the outcomes for metastatic melanoma have changed as a result of therapeutic targeting of BRAFV600 mutations16; and the fraction of lung cancers treated with targeted agents is increasing with the discovery of likely driver aberrations in most lung tumours17,18. Large-scale processes that shape cancer genomes have similarly been identified. Chromothripsis19 and chromoplexy20, which involve breakage and rearrangement of chromosomes at multiple loci, kataegis21, which describes hypermutational processes associated with genomic rearrangements, are providing insight into tumour evolution (see Garraway and Lander (2013)22 for a review).
Analysis Across Tumour Types
Increases in the number of tumour sample data sets enhance our ability to detect and analyze molecular defects in cancers. For example, driver genes can be pinpointed more precisely by narrowing amplifications and deletions to smaller regions of the chromosome using recurrent events across tumour types. Large cohorts have enabled DNA sequencing to uncover a list of recurrent genomic aberrations (mutations, amplifications, deletions, translocations, fusions and other structural variants), both known and novel, as common themes across tumour types23. However, “long tails” in the distributions of aberrations among samples have also been uncovered24. Indeed, a majority of the TCGA samples have distinct alterations not shared with others in their cohort. Despite the apparent uniqueness of each individual tumour in this regard, the set of molecular aberrations often integrate into known biological pathways that are shared by sets of tumour samples. In other cases, rare somatic mutations can be implicated as drivers by aggregating events across tumour types to improve detection of patterns, for example hotspot mutations in protein domains, leading to identification of potential new drug targets.
Determining whether the rare aberrations are drivers (oncogenic contributors) or just passengers (clonally propagated with neutral effect), and whether they are clinically actionable, will require further functional evaluation as well as analysis of additional tumours to increase power. The identification of more driver aberrations and acquired vulnerabilities for each individual tumour will undoubtedly boost personalized care. Developing treatments that target the ~140 drivers23 validated to date, however daunting, appears possible; devising one-off therapies for the thousands of aberrations in the “long tail” will be much more challenging.
Although important general principles have emerged from decades of study25,26, until recently most research on the molecular, pathological and clinical nature of cancers has been “silo-ed” by tumour type27. One has only to glance at the directory of oncology departments in any major cancer center to realize that medical and surgical cancer care are, for the most part, also divided by disease as defined by organ-of-origin. That framework has made sense for generations, but molecular analysis is now calling this view into question; cancers of disparate organs reveal many shared features, and, conversely, cancers from the same organ are often quite distinct.
Important similarities among tumour subtypes from different organs have already been identified. For example, TP53 mutations drive high-grade serous ovarian, serous endometrial and basal-like breast carcinomas, all of which share a global transcriptional signature of activation of similar oncogenic pathways10,28. Similarly, ERBB2/HER2 is mutated and/or amplified in subsets of glioblastoma, gastric, serous endometrial, bladder and lung cancers. The result, at least in some cases, is responsiveness to HER2-targeted therapy analogous to that previously observed for HER2-amplified breast cancer. Other commonalities across tumour types include inherited and somatic inactivation of the BRCA1/2 pathway in both serous ovarian and basal-like breast cancer, microsatellite instability in colorectal and endometrial tumours, and the recently identified POLE-mediated ultramutator phenotype characterized by extremely high mutations rates, common to both colon and endometrial cancers12,28,29. Conversely, there are important cases in which the same genetic aberrations have very different effects depending on the organ within which they arise. A prime example is NOTCH, which is inactivated in some squamous cell cancers of the lung, head and neck30, skin31 and cervix32 but activated by mutation in liquid tumours33.
Such examples illustrate the importance of developing a comprehensive perspective across tumours, independent of histopathologic diagnosis; shared molecular patterns will enable etiologic and therapeutic discoveries in one disease that can be applied to another. Importantly, integrative interpretation of the data will help identify how the consequences of mutations vary across tissues, with important therapeutic implications. Relatively rare cancers, such as the childhood malignancies, particularly stand to benefit from such an approach.
We know much more about the molecular details of major cancers than we did just a few years ago, but once a cancer is metastatic it remains incurable, with few exceptions. Only time will tell whether the integration of molecular characteristics with histology, organ site and metastatic location will contribute to an improvement in patient outcomes. But the balance is shifting in that direction. Hence, the goal of the Pan-Cancer Project is to identify and analyze aberrations in the tumour genome and phenotype that define cancer lineages and those that transcend them. This report outlines the scope of the project and introduces the first coordinated set of manuscripts to be published from the enterprise.
The Pan-Cancer Project
To gain analytical breadth – defining commonalities, differences and emergent themes across cancer types and organs of origin – TCGA launched the Pan-Cancer analysis project at a meeting held on October 26-27, 2012 in Santa Cruz, California. Pan-Cancer is a coordinated initiative whose goals are to assemble coherent, consistent TCGA data sets across tumour types, as well as across platforms, and then to analyze and interpret those data (Box 2). Within two months of the launch a data “freeze” was declared, based on the first twelve TCGA tumour types, each profiled using six different genomic, epigenomic, transcriptional and proteomic platforms (Figure 1). Since that time, the aggregated data sets have been quality-controlled, analyzed statistically and interpreted by a consortium of researchers, principally members of the TCGA Research Network.
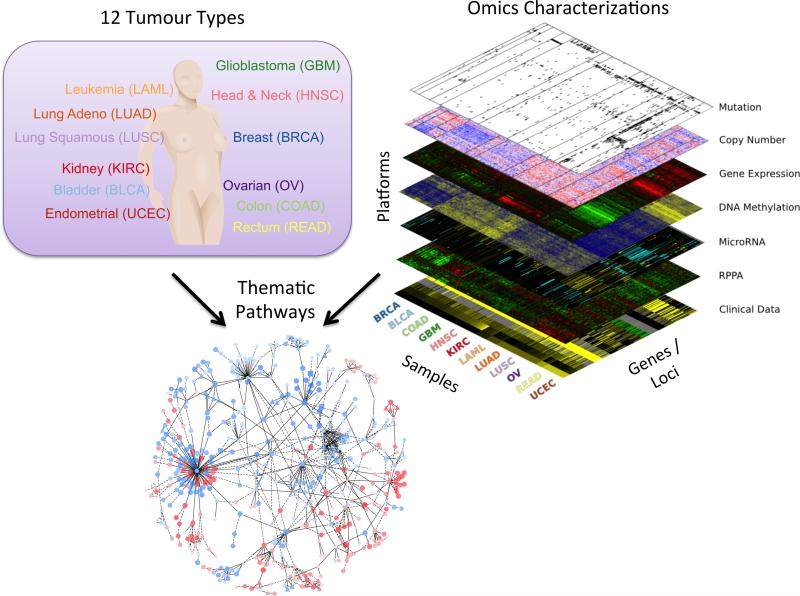
Figure 1. Integrated data set for the comparison and contrast of multiple tumour types.
The Pan-Cancer project assembled data from thousands of patients with primary tumours occurring in different sites of the body covering twelve tumour types (upper left panel) including glioblastoma multiform (GBM), lymphoblastic acute myeloid leukemia (LAML), head and neck squamous carcinoma (HNSC), lung adenocarcinoma (LUAD), lung squamous carcinoma (LUSC), breast carcinoma (BRCA), kidney renal clear cell carcinoma (KIRC), ovarian carcinoma (OV), bladder carcinoma (BLCA), colon adenocarcinoma (COAD), uterine cervical and endometrial carcinoma (UCEC), and rectal adenocarcinoma (READ). Six platforms of omics characterizations were performed creating a “data stack” (upper right panel) in which data elements across the platforms are linked by the fact that tissue material from the same samples were assayed, thus maximizing the potential of integrative analysis. Use of the data enables the identification of general trends including common pathways (lower panel) revealing master regulatory hubs activated (red) or deactivated (blue) across different tissue types.
The Pan-Cancer project lays the framework for an analytic process that, in the future, will include integration of new tumour types and data from TCGA and other such enterprises. There are currently major consortial efforts in pediatric cancers (TARGET; Therapeutically Applicable Research to Generate Effective Treatments) and adult cancers (ICGC; the International Cancer Genomics Consortium), as well as smaller projects by research teams around the world. A critical component will be the functional validation of aberrations in individual genes in team science efforts such as the CTD2 (Cancer Target Discovery and Development) and elucidation of pathway and network relationships in programs like the ICBP (Integrative Cancer Biology Program).
A number of major questions in cancer biology that go beyond the single-tumour perspective are being addressed in the collection of Pan-Cancer manuscripts. For example:
Can increases in statistical power help new driver mutations be distinguished from the background of passenger mutations as the sample size is increased by aggregating the 12 tumour types together? The assembled Pan-Cancer data have, in fact, enabled the identification of new patterns of genomic drivers. New computational approaches that leverage cross-tumour principles of replication timing and gene expression correlates with background mutation rates now enable the identification of frequently mutated genes while eliminating many false positive and negative calls made in several single tumour-type projects34. Further, the power to identify recurrent mutations and mutual exclusivity has strengthened the ability to distinguish “driver” from “passenger” aberrations (Lopez-Bigas, Scientific Reports 2013, personal communication).
What tissue associations underlie the major genomic structural changes in cancer? Improved methods for the analysis of structural variation of large chromosome segments have refined the ability to identify genomic and epigenetic regulators in multiple peak regions seen only by collating data across different cancer types. Tissue-associated patterns have now been established for the rate and timing of whole-genome duplication events35.
What pathways emerge as critical and potentially actionable when all mutational events across many tissues are considered together? New classes of mutations such as those in chromatin remodeling are emerging as pan-cancer drivers revealed only by: 1) collecting less-frequent events across tumour-types, 2) integrating event types such as mutations, copy number changes, and epigenetic silencing, 3) combining multiple algorithms to identify predicted drivers36, and 4) aggregating genes using gene networks and pathways (Ideker, Nature Methods 2013, personal communication).
Can the increase in numbers of samples enhance the analysis of co-occurrence and mutual exclusivity of gene aberrations and improve our ability to distinguish drivers from passengers? A bird's-eye view of genomic and epigenomic events reveals a “fate map” of the alternative routes to carcinogenesis in a decision tree that spans tissue boundaries (Ciriello, Nature Genetics 2013, personal communication).
Can molecular subtypes be delineated to disentangle tissue-specific from tissue-independent components of disease? Analysis of the epigenome, transcriptome, and proteome reveals a strong influence of tissue on the state of the altered pathways in tumour cells. For instance, the gene expression landscape reinforces the dominant tissue-dependence of altered pathways, including a view at the level of over a hundred proteins of high cancer import37 . Using all of the tumour types together allows for any tumour-specific signals to be subtracted from the datasets. Intriguingly, subtracting the tissue signal from DNA microarray gene expression datasets reveals signatures of immune stromal influence that transcend tumour-type boundaries38. Further, events that are common across lineages become apparent in a Pan-Cancer analysis37. Examples are the hormonal dependencies of breast, ovarian and endometrial cancers and a common “squamous” signature across head and neck, lung, cervical and bladder cancers.
Which events actionable in one tumour lineage are also actionable in another tumour lineage, potentially increasing the range of indications for specific targeted therapeutics? A systematic evaluation of machine-learning approaches reveals methodological principles for predicting patient outcomes using integrated information across tissues39.
Limitations of Pan-Cancer Analysis
Several data integration challenges place unavoidable limitations on the Pan-Cancer analysis at the current time. A key challenge is the integration of data that have been generated on different platforms, or updates of the same platform, as the technologies improve. In the Pan-Cancer studies for example, there have been transitions to much higher density DNA methylation arrays, use of different exome capture technologies, addition of RNA-Seq to microarray-based RNA characterization and increases in the quality and number of antibodies available for reverse-phase proteomic arrays (RPPA). A series of batch effects analyses have been carried out to assess systematic platform-specific biases. However, more work is needed to establish best practices for minimizing unwanted batch effects while preserving biological signals.
The kind and quality of clinical data available for the cancer types varies widely. The differences limit the ability to establish one-size-fits-all norms for demographic information, histopathologic characterization, behavioral context, and clinical outcomes. For example, our survival data are relatively robust for serous ovarian cancer because of its poor prognosis, but still immature for breast and endometrial cancers because (thankfully) most of the patients do better for longer. Certain data elements are routinely collected only when they are anticipated to be relevant (for example, the smoking history of lung, bladder and head-and-neck cancer patients). Clear viral etiologies have been identified in several solid tumours types, including head and neck cancer, cervical cancer, Kaposi's sarcoma and hepatocellular carcinoma. However, a Pan-Cancer analysis of the infectious etiologies of other cancers could not be conducted at present because infection status was recorded for only some tumours and tumour types (as an optional data element). Finally, tumour stage and grade are not easily comparable across different tumour types because, for good reason, each has its own system. This set of challenges to Pan-Cancer analysis highlights the fact that current clinical practice is largely conducted according to tissue or organ.
Statistically speaking, care must be taken to ensure that the increased sample size achieved by cross cancer comparison does not lead to increased false negative rates for discovery (e.g. by ‘diluting out’ an important mutation specific to one disease) or false-positive rates (e.g. by compounding on false-positives known to result from current single-tumour investigations34.
Rare events must not be obscured by disease-associated events. Tumour lineage plays an important role in the observed patterns of co-aberrations and gene expression profiles that indicate different consequences of seemingly similar events, for example involving the same gene(s) or amplicon(s). Likewise, new methods for accurately probing cross-tumour trends will need to account explicitly for the differences across tissues in mutation rates, copy number changes at the focal and arm-level scales, and the prevalence of other co-occurring events in the genetic and epigenetic background.
Despite those challenges, this collection of Pan-Cancer publications represents a landmark in the continuing effort to understand the common and contrasting biology of cancers from a molecular perspective. Still, major questions amenable to further Pan-Cancer investigations remain (Box 3), and the techniques used to compare different tumours will undoubtedly improve with use, time and further collaborative efforts.
Future Directions
The Pan-Cancer project represents one of the first of what will surely be many efforts to coordinate analysis across the molecular landscape of cancer, especially as additional tumour types are investigated in large numbers. Further increasing the number of samples per tumour type and the variety of these tumour types will improve our ability to detect rare driver events in heterogeneous tumour samples. But the true power will come from a detailed analysis across types -- with links to high quality clinical outcomes and eventual experimental validation and clinical trials to test the hypotheses that emerge. Technologies such as laser capture microdissection and cell sorting will improve our ability to distinguish whether omic signals arise from malignant or stromal cells. Histone profiling, protein analysis based on mass spectrometry and de-convolution of tumour heterogeneity through single-cell sequencing are examples expected to add important new dimensions of information. Continued efforts to identify the progenitor cells of tumours will enable distinguishing parochial from universal properties. Clone-level and single-cell cross-tumour comparisons may reveal even further connections among tumour types. Longitudinal genomic studies on primary resected tumours paired with their local recurrences and/or metastases will be undertaken by large consortial efforts, which have heretofore been restricted to primary disease and have lacked information about response to treatment. The characteristics of primary tumours may change markedly when tumours metastasize to distant sites, particularly bone and brain. Pan-Cancer analyses of metastases will therefore be highly informative for mapping out the relationships of metastatic tumours to primaries and to normal tissues, establishing potential rules for invasion and homing.
The power of pan-cancer analysis will increase as technologies for monitoring individual tumour cells at high resolution come into play. Now that the price of genome sequencing has fallen, the next pan-cancer enterprise will be able to analyze large numbers of whole-genome sequences across tumour types. Whole-genome analysis will complement the current studies by shedding light on mutational processes in the non-coding parts of the genome, which are largely unexplored to date. That expanded analysis will bring focus to disruptions in promoter and enhancer sites and aberrations in non-coding RNAs, as well as genomic integration processes at work in tumour evolution that result from mobile endogenous and exogenous DNA elements such as retrotransposons and viruses. Whole-genome sequencing will create a backdrop against which genome-wide association studies can relate inherited predispositions to particular forms of cancer. Systems-oriented approaches, based on relevant pathways and networks, will add to the therapeutic opportunities that arise from the wealth of data. Experimental follow-up will be critical to assess the functional consequences and therapeutic liabilities of these new findings.
From Many Tumours to the Individual Patient
The hope is that cross-tumour investigations such as the Pan-Cancer project will ultimately inform clinical decision-making. We hope they will enable discovery of novel therapeutic agents that can be tested clinically -- perhaps in novel adaptive, biomarker-based clinical trials that cross tumour boundaries. Toward those ends, Pan-Cancer TCGA data sets have been made available publicly in one location. Although coordination remains a challenge, the data sets comprise an unequalled resource for integrative analysis of cancer in its many forms.
A key challenge is the development of clinical trial strategies for connecting subsets of tumours from different tissues in terms of molecular signatures. Recent analyses of pharmacological profiling experiments across a diverse panel of cancer cell lines has suggested that common genetic alterations predict response to therapy across multiple cell lineages40-43. Biomarker-based design of clinical trials can increase statistical power, greatly decreasing the size, expense, and duration of clinical trials.
The number and size of omic datasets on cancer available to the research community for mining and exploring continue to expand rapidly, and computational tools to derive insights into the fundamental causes of cancer are becoming more powerful. It is important to note that the full potential of the enterprise will be realized only over time and with broader efforts. Still, the collection of TCGA Pan-Cancer publications represents a significant contribution to a new period of discovery in cancer research.
Box 1: TCGA: Mission and Strategy.
Important information about the biological relevance of the molecular changes in cancer can be obtained through combined analysis of multiple different types of data at the DNA, RNA, and protein levels.
For that reason, TCGA's principal aims are to generate, quality control, merge, analyze, and interpret molecular profiles at the DNA, RNA, protein and epigenetic levels for hundreds of clinical tumours from various tumour types and their subtypes. Cases that meet quality assurance specifications are characterized using technologies that assess the sequence of the exome, copy number variation (measured by single-nucleotide polymorphism arrays), DNA methylation, mRNA expression and sequence, miRNA expression, and transcript splice variation. Additional platforms applied to a subset of the tumours, including whole genome sequencing and reverse phase protein arrays, provide additional layers of data to complement the core genomic datasets and clinical/pathological data. By the end of 2015, the TCGA Network plans to have achieved the ambitious goal of analyzing the genomic, epigenomic, and gene expression profiles of more than 10,000 specimens from 25 different tumour types.
TCGA's has other, complementary purposes as well: to promote the development and application of new technologies, to detect cancer-specific molecular alterations, to make the data and results freely available to the scientific community, to develop tools and standard operating procedures that can serve other large-scale profiling projects, and to build cadres of individuals (including experimentalists, computational biologists, statistical analysts, computer scientists, and administrative staff) with the expertise to carry out such large scale team science projects. As of July 24, 2013, TCGA has mapped molecular patterns across 7,992 total cases representing 27 tumour types. The data, along with tools for exploring them, are publicly available at cancergenome.nih.gov. Eight ‘marker papers’ (i.e., comprehensive initial publications on each of the tumour types) have been published to date8-13,15,28.
Box 2: Coordination of Data and Results.
The first goal of the Pan-Cancer Analysis Working Group was to assemble data from the separate disease projects to build a well-coordinated joint data set spanning multiple tumour types. A data “freeze” (Dec 21, 2012) based on six different genomic and epigenomic characterization platforms was made available as the “pancan12” data set to all analysis groups. Twelve tumour types (GBM, OV, BRCA, LUSC, LUAD, COAD, READ, KIRC, UCEC, BLCA, HNSC and LAML) were selected based on: data maturity, adequate sample size, and publication or submission for publication of the primary analyses. The pancan12 data set includes a total of 5,074 tumour samples, for which at least one platform from each of genomic, epigenomic, and gene expression data had been assessed for 93% (i.e., 4,705, listed in Table 1 by measurement platform). The essential purpose of such a joint data set is twofold: to increase the statistical power to detect functional genomic determinants of disease and to reveal both tissue-specific aspects of cancer and intrinsic molecular commonalities across tumour types.
The Pan-Cancer analysis project started as an informal collaboration among members of the TCGA Network but then quickly expanded to include many other interested researchers. Ensuring standardization and consistency of the data and annotations across multiple platforms and clinical data elements was a necessity for the project. To coordinate analyses across this large group of researchers, formal pipelines were created to establish a coherent working base of data and results.
The process of TCGA data generation and Pan-Cancer analysis is as follows (Figure 2). First, tumour and germline samples are obtained from a large number of tissue source sites and processed by the Biospecimen Core Resource (with sample selection according to criteria established for each tumour type and with extensive quality controls) to generate purified DNA, RNA and protein preparations. The preparations are sent to Genome Characterization Centers (GCCs) and Genome Sequencing Centers (GSCs) for molecular profiling, and the resulting data are deposited in the TCGA Data Coordinating Center (DCC) to provide a primary source of data, at four levels of data processing. Seven Genome Data Analysis Centers (GDACs), along with analysts in the GCCs, GSCs, and in the external research community, share analysis and interpretation of the data, coordinating activities through face-to-face meetings and regular (usually weekly) teleconferences.
A “data freeze” was created by pulling higher levels of interpreted data (“Level 3”) from the DCC into a coordinating repository called Synapse created by Sage Bionetworks. To create a coherent dataset, a sample “white list” was created by synchronizing flagged samples with the DCC, based on annotations and criteria from the individual disease working groups. The Pan-Cancer project leverages the TCGA infrastructure for sample acquisition, sample processing and data generation on individual tumour types, as well as the production of derived data sets and a variety of analysis results assembled in the Broad Institute's Firehose system (citation). Assembled robust, self-consistent data sets across all 12 Pan-Cancer tumour types were deposited into Synapse. The Synapse system implements mechanisms for tracking provenance and metadata, stable digital object identifiers (DOIs) for data referencing, and flexible methods for data access, either through a wiki-like web-based environment or programmatically through application programming interfaces (APIs). The pancan12 datasets and selected results are available at https://www.synapse.org/#!Synapse:syn300013 (doi:10.7303/syn300013).
Box 3. Examples of additional major questions amenable to further Pan-Cancer analyses.
● What is the spectrum of nucleotide- and dinucleotide-level changes associated with different carcinogenic etiologies (e.g. tobacco, pathogens, or inflammation) operating in different parts of the body?
● Will integration of additional data sources including additional tumour types from TCGA and other projects increase the power of analysis?
● How can molecular changes complement pathological analysis for classification into tumour lineages with potentially different management?
● Can molecular profiles effectively categorize cancers for therapeutic decision-making?
● Are there predictive expression-based signatures for genomic events that transcend tissues, reflecting pathways disrupted by the alterations?
● Will comprehensive protein analysis through emerging mass spectrometry approaches in the CPTAC and other efforts extend the power of the genomic, transcriptomic, and proteomic analyses in TCGA.
● Will emerging technologies such as mass spectrometric metabolomics improve our ability to identify actionable processes?
● How are changes in protein families distributed across different tumour types?
● Are aberrations in specific protein domains or pathways distributed differentially across tumour lineages?
● Beyond the known examples including in cervical, head-and-neck, esophageal and hepatocellular cancers, can we identify other cancer types that show virally-mediated initiation?
● Are bacteria associated with different cancer lineages (as arefusebacteria in colorectal cancer)?
● Can the answers to any of these questions help us design novel therapies and clinical trials, with the ultimate goal of improving patient outcomes?
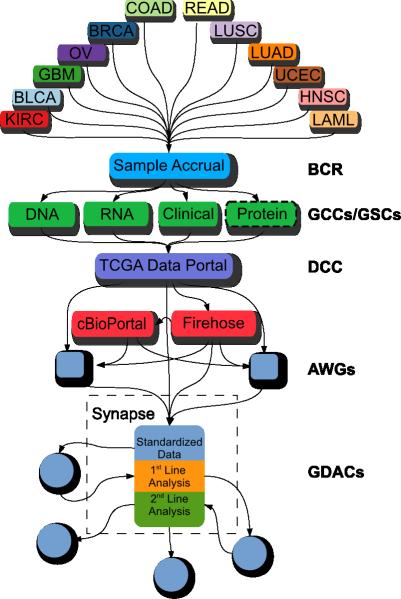
Figure 2. Data coordination for the Pan-Cancer TCGA project.
Data were collected by the biospecimen collection resource (BCR) from 12 different tumour types, characterized on six major platforms by the genome characterization and sequencing centers (GCC/GSC). Datasets are deposited into the TCGA data coordination center (DCC) from which it is then distributed to the Broad Institute's Firehose and Memorial Sloan Kettering Cancer Center's cBioPortal for various automated processing pipelines. Analysis working groups (AWG) conduct focused analyses on individual tumour types. Results from the DCC, Firehose, and AWGs were collected and stored in Sage Bionetworks’ Synapse system to create a “data freeze.” Genome data analysis centers (GDACs) accessed and deposited both data and results through Synapse to coordinate distributed analyses.
Table 1. The data “freeze” used by the Pan-Cancer project defined on December 21, 2012.
Tabulated are the numbers of unique tumour samples available for each tumour type (rows) and each measurement platform (columns).
| RPPAa | DNA Methylationb | Copy Numberc | Mutationd | miRNAe | Expressionf | |
|---|---|---|---|---|---|---|
| LUSC | 195 | 358 | 345 | 178 | 332 | 227 |
| READ | 130 | 162 | 164 | 69 | 143 | 71 |
| GBM | 214 | 405 | 578 | 290 | 501 | 495 |
| LAML | 194 | 198 | 197 | 187 | 179 | |
| HNSC | 212 | 310 | 310 | 277 | 309 | 303 |
| BLCA | 54 | 126 | 126 | 99 | 121 | 96 |
| KIRC | 423 | 457 | 457 | 417 | 442 | 431 |
| UCEC | 200 | 512 | 511 | 248 | 497 | 333 |
| LUAD | 237 | 431 | 357 | 229 | 365 | 355 |
| OV | 332 | 592 | 577 | 316 | 454 | 581 |
| BRCA | 408 | 888 | 887 | 772 | 870 | 817 |
| COAD | 269 | 420 | 422 | 155 | 407 | 192 |
| Total | 2674 | 4855 | 4932 | 3247 | 4628 | 4080 |
RPPA: Reverse-phase protein arrays measuring protein and phosphoprotein abundance.
DNA Methylation, DNA methylation at CpG islands.
Copy Number. Microarray-based measurement of copy number.
Mutation. Samples subjected to whole-exome sequencing to determine single nucleotide and structural variants.
miRNA. Sequencing of microRNA.
Expression. RNA Sequencing and microarray gene expression.
ACKNOWLEDGEMENTS
We thank J. Zhang for administrative coordination of TCGA Pan-Cancer Analysis Working Group activities, C. Perou and K. Hoadley for contributions to Figure 1, and D. Wheeler, M. Meyerson, and L. Ding for comments on early drafts of the manuscript. The study was funded by the National Cancer Institute and the National Human Genome Research Institute.
REFERENCES
- 1.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 3.Payne GS, Bishop JM, Varmus HE. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982;295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 4.Baker SJ, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. doi:10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. doi:10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. doi:10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TCGA Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.TCGA Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. doi:10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.TCGA Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. doi:10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.TCGA Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. doi:10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.TCGA Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. doi:10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.TCGA Network Comprehensive molecular characterization of urothelial carcinoma of the bladder. Nature. 2013 submitted. [Google Scholar]
- 14.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. doi:10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 15.TCGA Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. doi:10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. doi:10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. doi:10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi K, et al. RET, ROS1 and ALK fusions in lung cancer. Nature medicine. 2012;18:378–381. doi: 10.1038/nm.2658. doi:10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 19.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. doi:10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. doi:10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013 doi: 10.1038/nature12477. doi:10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. doi:10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. doi:10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler DA, Wang L. From human genome to cancer genome: The first decade. Genome research. 2013;23:1054–1062. doi: 10.1101/gr.157602.113. doi:10.1101/gr.157602.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. doi:10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 27.McDermott U, Settleman J. Personalized cancer therapy with selective kinase inhibitors: an emerging paradigm in medical oncology. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:5650–5659. doi: 10.1200/JCO.2009.22.9054. doi:10.1200/JCO.2009.22.9054. [DOI] [PubMed] [Google Scholar]
- 28.Kandoth C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. doi:10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nature genetics. 2013;45:136–144. doi: 10.1038/ng.2503. doi:10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. doi:10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. doi:10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. doi:10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. doi:10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zack T, et al. Pan-cancer patterns of somatic copy number alteration. Nature In review. 2013 doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez A, Ding L, Bader GD, Bigas-Lopes N. Identification of cancer driver genes using complementary criteria. Nature Scientific Reports Under review. 2013 [Google Scholar]
- 37.Li J, et al. A Proteomic View of The Cancer Genome Atlas. Nature methods. 2013 [Google Scholar]
- 38.Verhaak RG. Estimating the presence of tumor-associated normal cells relates the tumor microenvironment to genomic correlates. Nature Communications Under review. 2013 [Google Scholar]
- 39.Yuan Y, et al. How much can we gain in predicting patient prognosis by integrating cancer genomics data? Nature biotechnology Under review. 2013 [Google Scholar]
- 40.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. doi:10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. doi:10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinstein J. Cell lines battle cancer. Nature. 2012;483:2. doi: 10.1038/483544a. [DOI] [PubMed] [Google Scholar]
- 43.Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1018854108. doi:10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]