Abstract
NDG-4 is a predicted transmembrane acyltransferase protein that acts in the distribution of lipophilic factors. Consequently, ndg-4 mutants lay eggs with a pale appearance due to lack of yolk, and they are resistant to sterility caused by dietary supplementation with the long-chain omega-6 polyunsaturated fatty acid dihommogamma-linolenic acid (DGLA). Two other proteins, NRF-5 and NRF-6, a homolog of a mammalian secreted lipid binding protein and a NDG-4 homolog, respectively, have previously been shown to function in the same lipid transport pathway. Here, we report that mutation of the NDG-4 protein results in increased organismal stress resistance and lifespan. When NDG-4 function and insulin/IGF-1 signaling are reduced simultaneously, maximum lifespan is increased almost fivefold. Thus, longevity conferred by mutation of ndg-4 is partially overlapping with insulin signaling. The nuclear hormone receptor NHR-80 (HNF4 homolog) is required for longevity in germline less animals. We find that NHR-80 is also required for longevity of ndg-4 mutants. Moreover, we find that nrf-5 and nrf-6 mutants also have extended lifespan and increased stress resistance, suggesting that altered lipid transport and metabolism play key roles in determining lifespan.
Keywords: aging, C. elegans, lipid transport, NDG-4, insulin signaling
Introduction
A growing number of genes and signaling pathways are being identified that determine lifespan in Caenorhabditis elegans (Lapierre & Hansen, 2012). With the detailed characterization of these genes, it is becoming increasingly clear that many of them function either in common signaling pathways or in overlapping molecular pathways that have extensive cross-talk. Several pathways have also been shown to function synergistically and additively in extending lifespan. Here, we report that the gene ndg-4, encoding a predicted transmembrane protein, is a novel gene acting additively with reduced insulin signaling to extend lifespan when inactivated.
The insulin/IGF-1-signaling (IIS) pathway is a central phosphatidylinositol-3-kinase (PI3-Kinase) signaling cascade determining C. elegans longevity, but it also influences many other biological processes such as development, dauer formation (an alternative hibernating larval stage), stress responses and metabolism (Kenyon, 2010). daf-2 encodes the only insulin/IGF-1 receptor in C. elegans (Kenyon et al., 1993). Upon binding of insulin-like ligands DAF-2 phosphorylates the PI3-Kinase homolog AGE-1 (Morris et al., 1996). AGE-1 activates several other kinases, including AKT-1, SGK-1 and PDK-1, ultimately controlling the forkhead-family (FoxO) transcription factor, DAF-16. In its phosphorylated form, DAF-16 is sequestered in the cytoplasm and prevented from activating transcription or repression of target genes. Translocation of DAF-16 to the nucleus and subsequent activation is required for increased lifespan due to reduced IIS, and many DAF-16 target genes have been identified as modulators of lifespan (Lin et al., 1997; Ogg et al., 1997). In addition to IIS, DAF-16 also responds to signals from stress (Oh et al., 2005) and nutrient deprivation sensing cascades (Pan et al., 2007). Due to the importance of DAF-16 in such essential biological processes, a complex regulation of its activity has evolved involving expression of different isoforms (Kwon et al., 2010) as well as various DAF-16-binding proteins capable of regulating its activity (Cahill et al., 2001; Wang et al., 2006; Wolff et al., 2006; Li et al., 2008; Alam et al., 2010). Other proteins contribute to longevity of IIS mutants, such as the AMP-activated protein kinase α subunit AAK-2 (Apfeld et al., 2004), heat-shock factor HSF-1 (Hsu et al., 2003), the xenobiotic-response factor SKN-1/NRF (Tullet et al., 2008), and the ER unfolded-protein-response regulator XBP-1 (Henis-Korenblit et al., 2010). Interestingly, many of the genes regulated by these proteins are involved with responses to various types of stress, such as molecular chaperones, other heat-shock proteins, antioxidants, and detoxifying enzymes.
While there is growing evidence that both altered lipid metabolism and lipid signaling also play important roles in longevity, these are complex processes and the underlying molecular mechanisms are only beginning to be understood. In C. elegans, fat is stored in lipid droplets primarily in the intestine and in the hypodermal cells (Ashrafi, 2007). The long-lived daf-2 mutants have increased fat content, and this is also a hallmark of the extremely long-lived dauers (Ogg et al., 1997). However, there is no direct correlation between increased fat accumulation and longevity, as not all mutants with increased fat accumulation are long lived, and likewise not all long-lived mutants have increased fat levels. In fact, the long-lived eat-2 mutants have reduced fat stores (Brooks et al., 2009). Lipophilic hormonal signals from the C. elegans germ cells have an inhibitory effect on longevity and germline ablation increases lifespan (Hsin & Kenyon, 1999). The exact nature of this signal and how it is mediated is still largely unknown, but longevity due to reduced germline signaling requires a number of proteins including the nuclear hormone receptor DAF-12 (Hsin & Kenyon, 1999), DAF-16 and several others (McCormick et al., 2011). Genes involved in cellular checkpoint control are also involved in stress resistance and longevity (Bauer et al., 2005; Olsen et al., 2006; Arum & Johnson, 2007). As some checkpoint genes influence germ cell division, it is possible that they influence longevity via an altered germline signaling, but the underlying mechanisms still remain to be solved at the molecular level.
To identify novel genes determining longevity and stress resistance by mechanisms similar to checkpoint proteins, we performed a whole-genome RNAi screen for resistance to stalled replication forks using the chemotherapeutic drug hydroxyurea (HU; data not shown). One of the genes conferring resistance to HU after RNAi knock down was ndg-4, a gene encoding a protein containing an acyl transferease domain and 12 predicted transmembrane domains (Choy & Thomas, 1999). Prior to our study, ndg-4 mutants were isolated in two different genetic screens. The first screen for resistance to nor dihydroguaiaretic acid (NDG) isolated the ndg-4(lb108) allele (Shreffler et al., 1995). NDG is a nonspecific lipoxygenase inhibitor that prevents synthesis of prostaglandins and leukotrienes. An additional allele ndg-4(sa529) was later isolated in a screen for fluoxetine resistance (Choy & Thomas, 1999). Fluoxetine is a serotonin reuptake inhibitor commonly used as an antidepressant (Prozac). Wild-type C. elegans worms exposed to fluoxetine contract their noses whereas a group of nose resistance to fluoxetine (Nrf) mutants, including ndg-4, do not exhibit this response (Choy & Thomas, 1999). Genetic analysis suggests that at least two independent pathways can cause resistance to fluoxetine (Choy et al., 2006). ndg-4 may function in a pathway together with the Nrf mutants nrf-5 and nrf-6 (Choy & Thomas, 1999). nrf-6 encodes an ndg-4 homolog and nrf-5 have homology to a mammalian secreted lipid binding protein. While ndg-4 and nrf-6 are expressed both in the intestine and in hypodermis, only their expression in the intestine is responsible for fluoxetine resistance (Choy & Thomas, 1999; Choy et al., 2006). nrf-5 is also expressed in the intestine, but the NRF-5 protein is thought to be secreted into the pseudocoelomic fluid (Choy et al., 2006).
An independent study supports that ndg-4, nrf-5 and nrf-6 function in a common pathway in fat metabolism (Watts & Browse, 2006). Addition of the long-chain omega-6 polyunsaturated fatty acid dihommogamma-linolenic acid (DGLA, 20:3n-6) to the diet of the worms causes germ cell depletion and sterility in C. elegans (Watts & Browse, 2006). By contrast, DGLA does not interfere with the development and survival of the somatic gonadal cells. Interestingly, ndg-4, nrf-5, and nrf-6 mutants are resistant to dietary addition of DGLA consistent with their gene products functioning in a common pathway transporting dietary lipids into the reproductive tract (Watts & Browse, 2006).
In this study, we report that ndg-4, nrf-5, and nrf-6 are novel genes determining lifespan in C. elegans. Moreover, when NDG-4 function and insulin/IGF-1 signaling are reduced simultaneously, maximum lifespan is increased almost fivefold.
Results
ndg-4 mutants are stress resistant and long lived
As RNAi against ndg-4 caused resistance to replication stress (Fig. S1A), and ndg-4 mutants had previously been shown to be resistant to NDG (Shreffler et al., 1995) and fluoxetine (Choy & Thomas, 1999), we hypothesized that ndg-4 mutants had generally improved stress defense systems. To test whether ndg-4 was involved with other types of stress resistance, we performed longitudinal thermotolerance assays. We found that ndg-4 knockdown by RNAi caused significant resistance to heat stress at 35 °C compared with control worms fed empty vector (Fig. 1A; Table S1). In a parallel set of experiments, we examined the ndg-4(lb108) mutant strain originally isolated in a screen for resistance to NDG (Shreffler et al., 1995) and found that these mutants were also significantly thermotolerant compared with wild-type N2 worms (Fig. 1B; Table S1). We also observed that the ndg-4(lb108) mutants required an extra day to reach egg laying adulthood compared to wild-type N2 worms and that they laid pale eggs and had reduced brood size (data not shown), confirming previous observations (Choy & Thomas, 1999).
Figure 1.
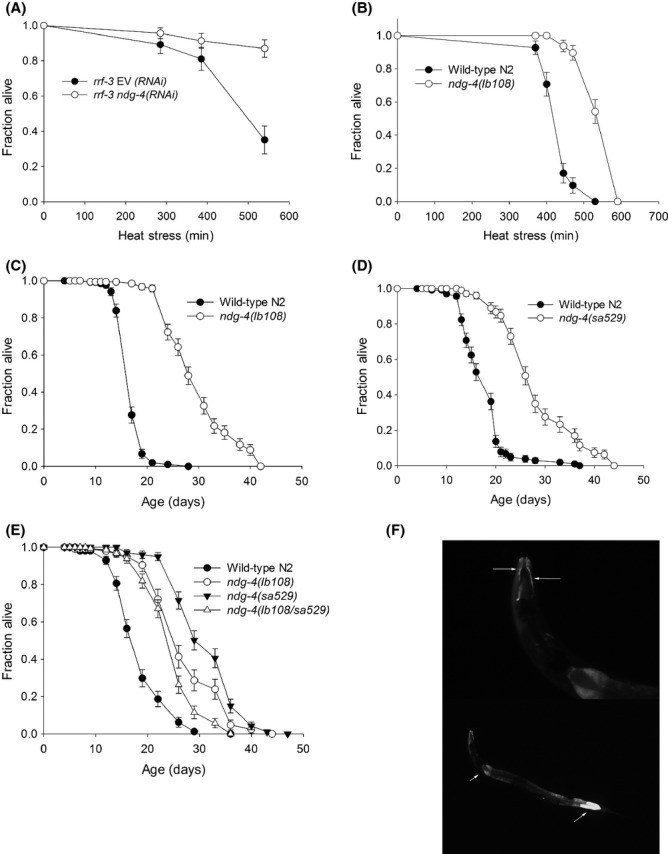
Loss of NDG-4 causes increased stress resistance and lifespan. (A) RNAi against ndg-4 causes a significant increase in thermotolerance of rrf-3(pk1426) mutants. (B) ndg-4(lb108) mutants have significantly increased thermotolerance compared with wild-type N2 worms. (C) ndg-4(lb108) mutants have significantly increased lifespan compared with wild-type N2 worms. (D) ndg-4(sa529) mutants have significantly increased lifespan compared with wild-type N2 worms. (E) Lifespan of transheterozygous ndg-4(sa529/lb108) mutants, ndg-4(lb108) mutants, ndg-4(sa529) mutants and wild-type N2 worms. (F) Expression pattern of ndg-4 shown by transgenic expression of the transcriptional reporter construct Pndg-4::gfp. Bottom: Strong ndg-4 expression is seen in the intestine. Top: Enlargement showing Pndg-4::gfp expression in hypodermal cells in the nose region.
Because of the general relationship between longevity and stress resistance (Benedetti et al., 2008) the significant thermotolerance caused by reduced NDG-4 function prompted us to examine the lifespan of the ndg-4(lb108) mutants. We found that the ndg-4(lb108) mutants lived significantly longer than the wild-type N2 animals, with a mean lifespan of 29 ± 2 days compared with 17 ± 1 days for the wild-type N2 at 20 °C (Fig. 1C; Table S2).
Whereas RNAi against ndg-4 resulted in thermotolerance (Fig. 1A; Table S1), no increase in lifespan was seen for worms treated from eggs with RNAi against ndg-4 (the first generation; Table S2). Worms subjected to RNAi against ndg-4 for two generations showed a small but significant increase in lifespan suggesting some maternal rescue (Fig. S1B; Table S2). It is not uncommon that reducing gene expression by RNAi does not phenocopy a mutation. However, to rule out the possibility of other unknown mutations in the genome being responsible for the observed phenotypes, we examined development and lifespan of mutants harboring another ndg-4 allele, sa529. The ndg-4(sa529) mutant was originally isolated in a screen for resistance to fluoxetine (Choy & Thomas, 1999). We found that the ndg-4(sa529) mutation also conferred thermotolerance (Fig. S1C; Table S1) and longevity (Fig. 1D; Table S2) with lifespan increases similar to those seen for the ndg-4(lb108) allele (27 ± 2.0 days compared with 18 ± 0.5 for the wild-type N2). The ndg-4(sa529) mutants also developed slower than wild-type N2, had reduced brood sizes and laid pale eggs (data not shown). To confirm that the longevity was indeed conferred by mutation of ndg-4 in both mutant strains, we examined the lifespan of trans-heterozygous ndg-4(lb108/sa529) mutants carrying one copy of each of the mutant ndg-4 alleles. We confirmed that both alleles are recessive and found that these trans-heterozygous mutants also had significantly increased lifespan (25 ± 5 days compared with18 ± 5 for the wild-type N2; Fig. 1E).
NDG-4 functions in the intestine and in hypodermal cells
To determine the temporal expression pattern of ndg-4, we generated a transcriptional reporter strain expressing green fluorescent protein (GFP) under the promoter of ndg-4. Strong GFP expression was seen in the intestine (Fig. 1F, lower panel) and in hypodermal cells, especially in the nose region (Fig. 1F, top panel), confirming previous observations (Choy & Thomas, 1999). This suggests that increased lifespan and stress resistance result from lack of ndg-4 in these tissues. However, as our transgenic strain was made by microinjection, we cannot rule out possible germline expression of endogenous ndg-4, as expression of transgenes in the germline is often silenced.
NDG-4 functions in a pathway partially overlapping with insulin signaling
In terms of magnitude, the increases in lifespan conferred by the ndg-4 alleles are comparable with those seen for the commonly used long-lived mutants, age-1(hx546) and daf-2(e1370), from the IIS pathway. Inactivation of the IIS pathway causes lifespan increase in a DAF-16-dependent manner (Kenyon et al., 1993). To establish whether stress resistance and lifespan extension by ndg-4 mutation occur via the IIS pathway, we constructed daf-16(mu86);ndg-4(lb108) double mutants and examined their lifespan and thermotolerance. The daf-16(mu86) allele is thought to be null (Lin et al., 1997), and we found that lack of DAF-16 activity significantly reduced the thermotolerance and lifespan of ndg-4(lb108) mutants (Fig. 2A,B; Tables S1 and S2). However, the daf-16(mu86);ndg-4(lb108) double mutants were still significantly more stress resistant and longer lived than daf-16 (mu86) single mutants. The daf-16(mu86) mutation did not suppress the pale egg phenotype or the slow growth phenotype of the ndg-4 mutants (data not shown). Therefore, we conclude that NDG-4 is functioning in a manner partially overlapping with the IIS pathway.
Figure 2.
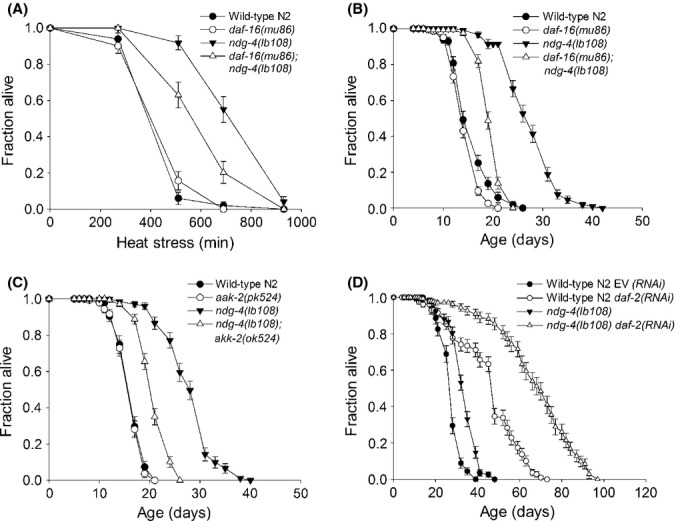
ndg-4 and insulin-like signaling extend stress resistance and lifespan additively. (A) Thermotolerance of ndg-4(lb108) mutants is partially daf-16-dependent. (B) Lifespan extension of ndg-4(lb018) mutants is partially daf-16-dependent. (C) The lifespan extension of ndg-4(lb108) mutants is partially dependent on aak-2. (D) daf-2 knockdown by RNAi increases the lifespan of ndg-4 mutants by nearly 100%.
To further study involvement of IIS signaling, we investigated dependence on the AMP-activated protein kinase α subunit aak-2, as AAK-2 is also necessary for lifespan extension of daf-2 mutants. AAK-2 responds to stressors, energy levels and insulin signaling. AAK-2 has been proposed to act in parallel to DAF-16 to extend lifespan in daf-2 mutants (Apfeld et al., 2004). Therefore, we predicted that AAK-2 would be necessary for the increased lifespan of ndg-4 mutants. To test this, we constructed an aak-2(ok524);ndg-4(lb108) double mutant and performed lifespan analysis. The aak-2(ok524) allele significantly, but not completely, reduced the lifespan of ndg-4 mutants (Fig. 2C), supporting the notion that the longevity of ndg-4 mutants is partially overlapping with IIS. The reduction in lifespan was similar to that seen by introduction of the daf-16(mu86) mutation. The aak-2(ok524);ndg-4(lb108) double mutants had a mean lifespan of 20.9 ± 2.9 days compared to 33.0 ± 6.8 days for ndg-4(lb108) mutants.
As a major part of the lifespan increase in ndg-4 mutants could be attributed to IIS dependent mechanisms, we predicted that RNAi against daf-2 would only result in a modest increase in the lifespan of ndg-4 mutants compared with RNAi against daf-2 in a wild-type N2 background. Intriguingly, we observed an additive effect on lifespan when ndg-4 mutants were treated with RNAi against daf-2 (Fig. 2D; Table S2). The longest lived ndg-4;daf-2(RNAi) worms had a maximum lifespan of close to 100 days. The lifespan increase in ndg-4 mutants due to RNAi against daf-2 was fully dependent on daf-16 (Table S2). Other additive effects with insulin signaling with regards to longevity have previously been reported for other genes and pathways, including for example reduced translation (Pan et al., 2007) and signals from the germline (Spanier et al., 2010).
Inactivation of chk-1 does not increase stress resistance of ndg-4 mutants
As ndg-4(RNAi) worms were able to grow to adulthood in the presence of the chemotherapeutic drug HU (Fig. S1A), we speculated that ndg-4 mutants might be checkpoint deficient and that this could be responsible for their stress resistance and longevity. Checkpoint proteins secure proper cell division and play key roles in regulating apoptosis. Using the CED-1::GFP marker (Zhou et al., 2001), we found that ndg-4(lb108) mutants had a slightly elevated number of apoptotic cells per gonad arm compared with the wild-type N2 controls (Mean 2.14, n = 51 compared with 0.18, n = 60, P < 0.05 Student’s t-test). Following exposure to DNA damaging ionizing radiation we observed a similar increase in the number of apoptotic cells per gonad arm in ndg-4(lb108) mutants (6.47, n = 30) and wild-type N2 controls (7.00, n = 27). Thus, ndg-4 is not required for mounting a normal apoptotic checkpoint response to DNA damage (Fig. S1C).
The serine threonine kinase CHK-1 is required for the DNA damage and S-M checkpoints and is necessary for germline development (Kalogeropoulos et al., 2004). chk-1 knockdown by RNAi confers increased thermotolerance and lifespan as does knockdown of the downstream phosphatase cdc-25 (Olsen et al., 2006; de Lencastre et al., 2010). To further test a possible involvement of checkpoint proteins in the longevity of the ndg-4 mutants, we examined the effect of inactivating chk-1. RNAi against chk-1 significantly increased the thermotolerance of wild-type worms but had no or limited effect on ndg-4(lb108) mutants (Fig. 3A and Table S1). The same phenomenon was observed by knockdown of cdc-25.3 (Fig. 3A; Table S1). These data are suggesting that stress resistance in ndg-4 mutants and checkpoint mutants may be conferred by the same mechanism. Although not completely resolved at the molecular level, it has been suggested that the increase in longevity following knockdown of checkpoint proteins is due to lack of germline signaling (de Lencastre et al., 2010).
Figure 3.

HNR-80 is required for longevity of ndg-4 mutants. (A) Increased thermotolerance is observed when the S-M checkpoint genes chk-1 and cdc-25.3 are RNAi inactivated in wild-type N2 worms. The thermotolerance of ndg-4 is not further increased following RNAi inactivation of chk-1 and cdc-25.3. (B) RNAi against nhr-80 completely abolishes longevity of ndg-4 mutants whereas RNAi against lipl-4 has no effect.
ndg-4 mutants are long lived due to altered germline signaling
As lifespan extension following germline removal acts synergistically with the IIS pathway (Hsin & Kenyon, 1999), and as RNAi against chk-1 (germline less) did not increase thermotolerance of ndg-4 mutant worms (Fig 3A; Table S1), we next investigated if ndg-4 mutants had increased lifespan due to altered germline signaling. The nuclear hormone receptor NHR-80 (HNF4 homolog) has recently been shown to be required specifically for longevity in germline less animals but not for other life extending pathways such as IIS and dietary restriction (Goudeau et al., 2011). NHR-80 regulates oleic acid synthesis via the desaturases fat-5, fat-6, and fat-7 and links fatty acid synthesis and longevity resulting from germline removal (Lapierre & Hansen, 2012). We found that RNAi inactivation of nhr-80 completely abolished the increased lifespan of the ndg-4 mutants (Fig. 3B; Table S1). This is consistent with ndg-4 mutants being long lived due to altered germline signaling. However, it should be noted that in our experiments RNAi against nhr-80 also significantly decreased the lifespan of wild-type N2 control animals, suggesting that it may not uniquely be required for longevity due to altered germline signaling as previously reported (Goudeau et al., 2011).
Several independent pathways have been described as regulators of longevity in germline less animals. The lipase lipl-4 (K04A8.4) is induced in germline less animals in a daf-16-dependent manner (Wang et al., 2008; Lapierre et al., 2012) in contrast to nhr-80, which acts independently of daf-16 (Goudeau et al., 2011). We did not observe any significant decrease in the lifespan of ndg-4 mutants following RNAi against lipl-4 (Fig. 3B; Table S2). Hence, our data support that nhr-80 and lipl-4 influence germline signals via separate pathways and that ndg-4 is part of the former pathway.
nrf-5 and nrf-6 also determine stress resistance and lifespan
In addition to ndg-4, mutations in six other genes named nrf-1 to -6 were reported to confer the Nrf phenotype (Choy & Thomas, 1999). Of these, nrf-5, nrf-6, and ndg-4 define a subclass of Nrf mutants sharing an additional second phenotype of producing pale eggs with fewer yolk granules. Members of this subclass also accumulate large globules of yolk in the pseudocoelomic space, develop slower than wild-type N2 worms and produce a large percentage of dead embryos. The genes in this subclass have been suggested to function in the same pathway or complex to confer fluoxetine-induced nose contraction (Choy & Thomas, 1999). None of the members of the other subclass comprised by nrf-1 to nrf-4 were reported to produce pale eggs (Choy & Thomas, 1999). To determine whether stress resistance and longevity might be correlated with the pale egg or Nrf phenotype, we examined lifespan and thermotolerance of these mutants. We found that nrf-5 and nrf-6 mutants (pale egg) were long-lived and that nrf-1, nrf-2, nrf-3, and nrf-4 mutants (Nrf but not pale egg) had wild-type or shortened lifespan (Table S2). In agreement with this, we found that nrf-1, nrf-5, and nrf-6 mutants were more thermotolerant than wild-type worms, whereas nrf-2, nrf-3, and nrf-4 mutants had wild-type thermotolerance (Table S1). Thus, with nrf-1 being the exception, it seems that longevity is associated with the pale egg phenotype. We cannot rule out that the lifespan of the Nrf mutants would change upon backcrossing, but our data show that the Nrf phenotype per se is likely not linked to longevity or stress resistance.
ndg-4 and nrf-5 might function in a common pathway to determine stress resistance and lifespan
We decided to investigate nrf-5 mutants in more detail because of reports placing ndg-4 and nrf-5 in the same genetic pathway or complex (Choy et al., 2006; Watts & Browse, 2006). In addition to the pale egg phenotype, we noticed that the ndg-4 and nrf-5 mutants also have slow development in common (data not shown). We found that the nrf-5 mutants had a significant increase in both thermotolerance (Fig. 4A) and lifespan (Fig. 4B) with a mean lifespan of 20.8 ± 3 days compared with 17.8 ± 3 days for the wild-type N2. The increase in lifespan observed for nrf-5 mutants was smaller than that observed for ndg-4 mutants, whereas their levels of thermotolerance were comparable (Figs 1B and 4A). Consistent with ndg-4 and nrf-5 functioning in a common pathway, we found that thermotolerance of the nrf-5 mutants were also dependent on the FoxO transcription factor DAF-16 (Fig. 4C). To further investigate whether ndg-4 and nrf-5 might function in a common pathway in terms of thermotolerance, we inactivated ndg-4 using RNAi in an nrf-5 mutant background. We found that RNAi against ndg-4 did not further increase thermotolerance of nrf-5 mutants (Fig. 4D). In terms of fluoxetine resistance and DGLA transport, ndg-4 and nrf-5 have been placed in a common pathway (Choy et al., 2006; Watts & Browse, 2006). Our data are consistent with ndg-4 and nrf-5 also functioning in a common pathway in terms of longevity and stress resistance. However, protein interaction studies need to be performed to establish if they actually function in a protein complex as previously suggested (Watts & Browse, 2006).
Figure 4.
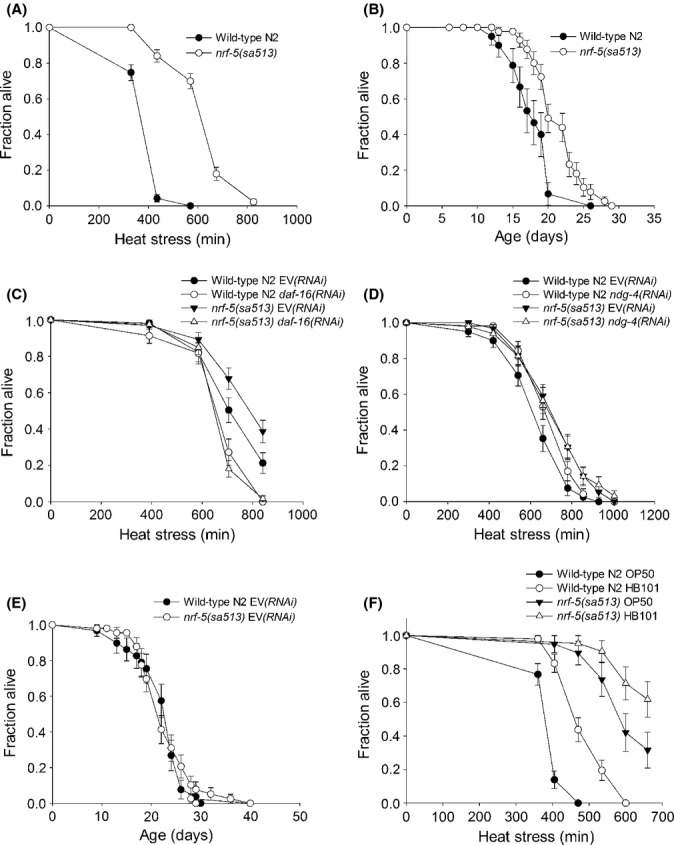
nrf-5 mutants are stress resistant and long-lived. (A) nrf-5(sa513) mutants are significantly thermotolerant compared with and wild-type N2 worms. (B) nrf-5(sa513) mutants are significantly longer lived than wild-type N2 worms at 20 °C. (C) nrf-5(sa513) mutants are thermotolerant when fed HT115 empty vector RNAi bacteria and this is dependent on daf-16. (D) Thermotolerance of nrf-5(sa513) mutants is not further increased by RNAi against ndg-4. (E) nrf-5(sa513) mutants are not long lived when fed HT115 empty vector RNAi bacteria. (F) Feeding a HB101 diet cause significant thermotolerance of wild-type N2 worms and nrf-5 mutants compared with feeding an OP50 diet.
The bacterial diet influences stress resistance
We also investigated the effect of RNAi against nrf-5 but found that it did not cause increased stress resistance or lifespan; despite a knockdown efficiency of nearly 98% (data not shown). The bacterial diet affects fat stores and lifespan in C. elegans. Specifically, feeding with HT115 or HB101 bacteria instead of OP50 leads to reduced fat stores and increased lifespan (Brooks et al., 2009). Intriguingly, we observed that the longevity of the nrf-5 mutant strain was either abolished or significantly reduced when the mutants were fed HT115 RNAi feeding bacteria rather than the normal food strain OP50 (Fig. 4E; Table S2). We speculated that perhaps nrf-5 mutants would only be long-lived when fed a diet not leading to reduced fat stores. To test this, we investigated feeding with HB101 bacteria. We found that the HB101 diet accelerated development of both wild-type N2 and nrf-5 mutants (data not shown), but that nrf-5 mutants were still long-lived when fed a HB101 diet (Table S2), showing that their longevity does not depend on a diet that leads to reduced fat stores. Supporting this, we found that nrf-5 mutants fed either HT115 or HB101 bacteria were significantly thermotolerant compared with wild-type N2 (Fig. 4A,C,D,F; Table S1). Moreover, feeding with HB101 bacteria significantly increased thermotolerance of wild-type N2 worms compared to feeding an OP50 diet (Fig. 4F). Thus, in agreement with previous studies showing that the bacterial diet can increase lifespan (Brooks et al., 2009), our data show that also stress resistance can be influenced by the bacterial diet.
Discussion
In this study, we describe how mutation of the gene ndg-4 leads to significant increases in lifespan and stress resistance. The increases in lifespan observed in ndg-4 mutants are more than 50% compared to wild-type controls. These phenotypes are partially dependent on daf-16, and simultaneous reduction of insulin signaling and ndg-4 leads to nearly a doubling of the already long lifespan with maximum lifespan reaching 100 days. The ndg-4 gene encodes a transmembrane protein with predicted acyltransferase activity that has not previously been described as having a role in lifespan determination in C. elegans. Earlier studies have shown that mutation of ndg-4 confers resistance to fluoxetine (nose resistant to fluoxetine Nrf phenotype) and NDG. We find that two other Nrf mutants, nrf-5, and nrf-6 encoding a putative lipid-binding protein and a ndg-4 homolog, respectively, also have increased stress resistance and lifespan, but that other Nrf mutants do not have this phenotype. It has previously been suggested that ndg-4 and nrf-5 might function in a common pathway or even in a complex in determining fluoxetine resistance, and our data support this notion also in terms of lifespan. We generally observe that the animals live longer when fed HT115 bacteria instead of OP50. These results are consistent with a previous study showing that feeding C. elegans HT115 or HB101 bacteria rather than OP50 increases mean lifespan and changes the fatty acid content (Brooks et al., 2009). Interestingly, the nrf-5 mutants fed HT115 bacteria did not have increased lifespan. Thus, the underlying mechanism for longevity in nrf-5 mutants could be the same mechanism causing longevity due to a HT115 diet. In contrast, feeding on a HB101 diet increased the lifespan of nrf-5 mutants. Stress resistance and increased lifespan are tightly linked in C. elegans. Supporting this notion, we find that feeding a HT115 or HB101 diet increases thermotolerance compared to feeding an OP50 diet. However, in contrast to lifespan, we find that feeding a HT115 or HB101 diet also increases thermotolerance in nrf-5 mutants. This suggests that a HT115 and a HB101 diet can confer thermotolerance and longevity through two distinct pathways.
There is growing evidence that fat metabolism and lipid-signaling might play important roles in longevity (Lapierre & Hansen, 2012). Given the previously described roles of ndg-4 and nrf-5 in fatty acid and yolk transport (Watts & Browse, 2006) and that ndg-4 longevity depends on nhr-80, we suggest that altered fat metabolism and/or germline signaling is responsible for the longevity and stress resistance of these mutants. This is consistent with the observation that only Nrf mutants with defective yolk transport (pale eggs) were found to be long-lived.
Hormonal signals from the germline are involved in lifespan determination and linked to lipid metabolism. One possibility is that in ndg-4 and nrf-5 mutants such hormonal signal is either not produced due to impaired delivery of a required substrate, or that the hormone does not correctly reach its intended target after it has been synthesized. Lifespan extension due to germline ablation requires several proteins including the FoxO transcription factor DAF-16 and the nuclear hormone receptor DAF-12 (Hsin & Kenyon, 1999). The increases in lifespan and stress resistance of the ndg-4 mutants are only partially dependent on DAF-16. Hence, a signal identical to that in germline ablated animals cannot completely explain their longevity because longevity due to germline ablation is completely suppressed by mutation of daf-16. However, it is possible that more than one signal from the germline can influence lifespan and that such signals will elicit different molecular responses. This view is supported by the recent finding that overexpression of NHR-80 can extend the lifespan of germline ablated animals in a DAF-16 independent manner, but it requires the nuclear hormone receptor DAF-12 (Goudeau et al., 2011).
The synergistic effect observed between ndg-4 and reduced insulin signaling is striking when considering the already impressive lifespan observed due to reduced insulin signaling. However, there are other examples of similar synergistic effects between reduced insulin signaling and other longevity pathways such as reduced TOR signaling/translation (Hansen et al., 2007; Pan et al., 2007; Robida-Stubbs et al., 2012) and altered germline signaling (Arantes-Oliveira et al., 2003). Germline ablation also significantly increases the lifespan of insulin-signaling mutants (Hsin & Kenyon, 1999) and altered signaling from the germline to the intestine is responsible for the long lifespan of double mutants between daf-2 and the intestinal di- and tripeptide transporter pept-1 (Spanier et al., 2010). Thus, the synergistic effect between an ndg-4 mutation and reduced insulin signaling is consistent with altered lipid signaling being responsible for the longevity. In C. elegans, fat is stored in lipid droplets primarily in the intestine and in the hypodermal cells (Ashrafi, 2007). Thus, a role of NDG-4 in lipid metabolism/signaling is supported by ndg-4 being expressed in the intestine, which has also been suggested in previous studies (Choy et al., 2006; Watts & Browse, 2006).
The NDG-4 protein contains an acyltransferase domain (Choy & Thomas, 1999). Based upon homology, there are several putative acyl transferases in C. elegans, and as these are not redundant to ndg-4 in terms of longevity, it is likely that they have different substrates. It is important to identify such substrates of NDG-4 and determine if they play conserved roles in longevity in other organisms. In this regard, it is interesting to note that in Drosophila melanogaster mutation of the NDG-4 homolog drop-dead (drd gene) causes accelerated aging and abrupt and early onset of death (Rogina et al., 1997). The drop-dead mutants have defects in movement of food in the gut (Peller et al., 2009), but they also suffer from severe neurodegeneration (Blumenthal, 2008). The neuronal damage is possibly associated with hypoxia in the brain (Kim et al., 2012). It is possible that these severe developmental defects mask an otherwise positive effect on longevity. These studies further stress the importance of identifying targets of NDG-4 because they could uncover novel mechanisms determining longevity.
Experimental procedures
Caenorhabditis elegans strains and culture conditions
Strains of C. elegans were cultured using standard protocols. The N2 Bristol strain was used as wild-type. For most experiments with RNAi, the RNAi hypersensitive strain NL2099 rrf-3(pk1426) II was used. The strain MP108 ndg-4 (lb108) III was backcrossed twice to our wild-type N2 strain to generate the strain OLS11 used in all ndg-4 experiments. We specifically ensured that a bus-1 mutation was eliminated using PCR. The strain JT513 nrf-5(sa513) V was backcrossed 2 and 3 times to our wild-type N2 strain following the slow development and pale egg phenotypes to generate the strains OLS53 and OLS51, respectively. We choose these phenotypes because they have previously been rescued by transgenic expression of the nrf-5 gene (Choy et al., 2006). Furthermore, to test that the slow development and low embryonic survival of the nrf-5 mutant was indeed due to the nrf-5(sa513) V mutation we took advantage of the mutant strain RB786 containing a large deletion in the gene stdh-1 located 0.09 cm away from nrf-5 on chromosome V. As the genes are located within such close proximity, the possibility of cross-over between them is minimal. We next analyzed the F2 progeny from a cross between nrf-5 hermaphrodites and stdh-1 males for slow development and the presence or absence of the stdh-1 deletion. All progeny that were homozygous for the nrf-5 mutation had slow development and low embryonic survival, as did the F3 and F4 progeny. All F2 progeny that was homozygous or heterozygous for the stdh-1 deletion had wild-type development and embryonic survival, as did the F3 and F4 progeny confirming that nrf-5 is indeed responsible for the phenotype. The ndg-4(sa529) III mutant was backcrossed once to our wild-type N2 to ensure the mutation was recessive before generating the transheterozygote ndg-4 mutants. JT524 nrf-1(sa524) III, JT366 nrf-2/vhp-1(sa366) II, JT363 nrf-3(sa363) IV, JT528 nrf-4(sa528) I, and JT525 nrf-6(sa525) II were not backcrossed because they showed no phenotypes relevant for this study. The slow growth and the pale egg phenotypes previously shown to be rescued by transgenic expression of ndg-4 (Choy & Thomas, 1999) were used to follow the ndg-4(lb108) mutation in all crosses. PCR was used to follow the daf-16(mu86) deletion. The transgenic strain OLS22 expressing GFP under control of the ndg-4 promotor (pndg-4::GFP) was generated by means of microinjection in DP38 unc-119 mutants. The pENTRY ndg-4 promotor clone was obtained from Open Biosystems and inserted in the pDEST DDO4 using a Gateway LR reaction. Several independent lines showed similar expression patterns.
RNAi treatment
RNAi feeding bacteria were obtained from Open Biosystems except bacteria expressing daf-2 dsRNA, which was a kind gift from Dr. Andrew Dillin, and bacteria expressing lipl-4 and nhr-80 dsRNA were kind gifts from Dr. Nils Færgeman. The bacteria expressing nrf-5 dsRNA were generated as described (Timmons, 2006). RNAi bacteria were maintained on Luria Broth (LB) plates with 100 μg mL−1 ampicillin and 12.5 μg mL−1 tetracycline. For experiments, RNAi bacteria were grown at 37 °C overnight in LB with 100 μg mL−1 ampicillin. The next day cultures were spotted onto RNAi plates consisting of NGM agar with 100 μg mL−1 ampicillin and 1 mM isopropyl 1-thio-β-D-galactopyranoside. After incubation at room temperature for at least 1 day, eggs were placed on spotted RNAi plates. Control animals were fed bacteria carrying an empty L4440 vector. For RNAi knockdown of chk-1, cdc-25.3, and ndg-4 (except where otherwise stated), experiments were performed in the second generation of growth on RNAi bacteria.
Lifespan analysis
Lifespan assays were performed as previously described (Olsen et al., 2006). Briefly, during the reproductive period, worms were transferred to fresh plates every day or every other day. After the reproductive period, worms were transferred every 3–5 days. Survival was scored as touch provoked movement. Bagged or exploded worms were scored as lost and censored. Survival data were analyzed using Graphpad Prism version 5 (GraphPad Software, Inc., La Jolla, CA, USA). Survival was plotted as surviving fraction ± SEM.
Thermotolerance assays
Longitudinal thermotolerance assays were performed as previously described (Olsen et al., 2006). Briefly, 3 cm NGM plates with each 25–30 synchronous four- or 5-day-old adults were shifted from their normal growth temperature to 35 °C and survival scored as indicated as touch provoked movement. Data were analyzed and plotted as described for lifespan assays.
Germline apoptosis and ionizing radiation
The ndg-4;ced-1::gfp strain was made by crossing MD701 ced-1::gfp with OLS11 ndg-4(lb108). Worms were grown at 25 °C on OP50 and scored for apoptotic cells 24 h post the L4 stage by mounting them i S-basal [0.1 m NaCl, 0.05 m H2PO4 (pH 6)] on slides with 2% agarose. Slides were sealed with nail polish, and the worms were immobilized by a brief exposure to heat. For ionizing radiation, the worms were irradiated at the L4 stage with 90 Gy and apoptotic cells in the death zone of the germline were scored 24 h later.
Acknowledgments
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Author contributions
JB, SN, LS, THM, HJ, and AO performed the experiments. JB, SN, LTJ, GJL, and AO designed the study, analyzed the data and wrote the manuscript. All authors contributed critical feedback to the manuscript.
Conflict of interest
None declared.
Funding
This study was supported by The Danish Research Council FNU (272-07-0162), Danish National Research Infrastructures Program (DAGMAR), The Lundbeck Foundation (R9-A968), The Carlsberg Foundation, The Novo Nordic Foundation, and the Interdisciplinary Research Consortium on Geroscience (UL1 RR024917).
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
Fig. S1. (A) RNAi against ndg-4 confers resistance to hydroxyurea at a concentration (10.16 mm) where control animals arrest their development. Animals were cultured for 4 days at 20 °C before their development was scored. (B) RNAi against ndg-4 causes a small but significant (P < 0.05) increase in lifespan of rrf-3(pk1426) mutants when kept on RNAi bacteria for two generations compared to controls. (C) ndg-4(sa529) mutants have significantly increased (P < 0.0007) thermotolerance compared to wild-type N2 worms. (D) Expression of the apoptosis marker CED-1:.GFP in wild-type N2 animals and ndg-4(lb108) mutants was used to quantify apoptosis in the death zone of the germline. ndg-4 mutants have significantly (P < 0.0001) higher levels of apoptotic cells (2.2 ± 0.1, n = 40) compared to controls (0.6 ± 0.5, n = 40). Error bars are standard error of mean.
Table S1 Summary of longitudinal thermotolerance assays.
Table S2 Summary of lifespan assays at 20 °C except experiment S1B#, which was at 25 °C.
References
- Alam H, Williams TW, Dumas KJ, Guo C, Yoshina S, Mitani S, Hu PJ. EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab. 2010;12:30–41. doi: 10.1016/j.cmet.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Berman JR, Kenyon C. Healthy animals with extreme longevity. Science. 2003;302:611. doi: 10.1126/science.1089169. [DOI] [PubMed] [Google Scholar]
- Arum O, Johnson TE. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:951–959. doi: 10.1093/gerona/62.9.951. [DOI] [PubMed] [Google Scholar]
- Ashrafi K. Obesity and the regulation of fat metabolism. Worm Book. 2007;1:20. doi: 10.1895/wormbook.1.130.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr. Biol. 2005;15:2063–2068. doi: 10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Benedetti MG, Foster AL, Vantipalli MC, White MP, Sampayo JN, Gill MS, Olsen A, Lithgow GJ. Compounds that confer thermal stress resistance and extended lifespan. Exp. Gerontol. 2008;43:882–891. doi: 10.1016/j.exger.2008.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal EM. Cloning of the neurodegeneration gene drop-dead and characterization of additional phenotypes of its mutation. Fly (Austin) 2008;2:180–188. doi: 10.4161/fly.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PLoS ONE. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J. Biol. Chem. 2001;276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- Choy RK, Thomas JH. Fluoxetine-resistant mutants in C. elegans define a novel family of transmembrane proteins. Mol. Cell. 1999;4:143–152. doi: 10.1016/s1097-2765(00)80362-7. [DOI] [PubMed] [Google Scholar]
- Choy RK, Kemner JM, Thomas JH. Fluoxetine-resistance genes in Caenorhabditis elegans function in the intestine and may act in drug transport. Genetics. 2006;172:885–892. doi: 10.1534/genetics.103.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, Aguilaniu H. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9:e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M, Kenyon C. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl Acad. Sci. USA. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Kalogeropoulos N, Christoforou C, Green AJ, Gill S, Ashcroft NR. chk-1 is an essential gene and is required for an S-M checkpoint during early embryogenesis. Cell Cycle. 2004;3:1196–1200. [PubMed] [Google Scholar]
- Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kim JY, Jang W, Lee HW, Park E, Kim C. Neurodegeneration of Drosophila drop-dead mutants is associated with hypoxia in the brain. Genes Brain Behav. 2012;11:177–184. doi: 10.1111/j.1601-183X.2011.00743.x. [DOI] [PubMed] [Google Scholar]
- Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 2012;23:637–644. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Melendez A, Hansen M. Autophagy links lipid metabolism to longevity in C. elegans. Autophagy. 2012;8:144–146. doi: 10.4161/auto.8.1.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs both promote and antagonize longevity in C. elegans. Curr. Biol. 2010;20:2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend C. elegans’ lifespan in response to reproductive signals. Aging Cell. 2011;11:192–202. doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl Acad. Sci. USA. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peller CR, Bacon EM, Bucheger JA, Blumenthal EM. Defective gut function in drop-dead mutant Drosophila. J. Insect Physiol. 2009;55:834–839. doi: 10.1016/j.jinsphys.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Benzer S, Helfand SL. Drosophila drop-dead mutations accelerate the time course of age-related markers. Proc. Natl Acad. Sci. USA. 1997;94:6303–6306. doi: 10.1073/pnas.94.12.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreffler W, Magardino T, Shekdar K, Wolinsky E. The unc-8 and sup-40 genes regulate ion channel function in Caenorhabditis elegans motorneurons. Genetics. 1995;139:1261–1272. doi: 10.1093/genetics/139.3.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier B, Rubio-Aliaga I, Hu H, Daniel H. Altered signalling from germline to intestine pushes daf-2;pept-1 Caenorhabditis elegans into extreme longevity. Aging Cell. 2010;9:636–646. doi: 10.1111/j.1474-9726.2010.00591.x. [DOI] [PubMed] [Google Scholar]
- Timmons L. Construction of plasmids for RNA interference and in vitro transcription of double-stranded RNA. Methods Mol. Biol. 2006;351:109–117. doi: 10.1385/1-59745-151-7:109. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Wang MC, O’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Browse J. Dietary manipulation implicates lipid signaling in the regulation of germ cell maintenance in C. elegans. Dev. Biol. 2006;292:381–392. doi: 10.1016/j.ydbio.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) RNAi against ndg-4 confers resistance to hydroxyurea at a concentration (10.16 mm) where control animals arrest their development. Animals were cultured for 4 days at 20 °C before their development was scored. (B) RNAi against ndg-4 causes a small but significant (P < 0.05) increase in lifespan of rrf-3(pk1426) mutants when kept on RNAi bacteria for two generations compared to controls. (C) ndg-4(sa529) mutants have significantly increased (P < 0.0007) thermotolerance compared to wild-type N2 worms. (D) Expression of the apoptosis marker CED-1:.GFP in wild-type N2 animals and ndg-4(lb108) mutants was used to quantify apoptosis in the death zone of the germline. ndg-4 mutants have significantly (P < 0.0001) higher levels of apoptotic cells (2.2 ± 0.1, n = 40) compared to controls (0.6 ± 0.5, n = 40). Error bars are standard error of mean.
Table S1 Summary of longitudinal thermotolerance assays.
Table S2 Summary of lifespan assays at 20 °C except experiment S1B#, which was at 25 °C.


